Abstract
The genetic background of extranodal marginal zone B-cell non-Hodgkin’s lymphoma (NHL) of mucosa-associated lymphoid tissue (MALT) type is poorly understood. In contrast to most entities of primary nodal lymphomas, few cytogenetic data are available, and gene rearrangements frequently encountered in and highly characteristic of certain entities of systemic NHL are absent in this type of lymphoma. Recently, it was suggested that MALT-type NHLs are associated with certain numerical chromosome aberrations and especially with trisomy 3. We performed an extensive study using a sensitive double (bicolor) fluorescence in situ hybridization technique for the analysis of trisomies for chromosomes 3, 7, 12, and 18 in 60 samples of low-grade and 45 high-grade MALT-type tumors. In the low-grade cases, trisomy 3 was found in a frequency of only 20%. High-grade lymphomas of MALT type were associated with trisomies 3, 7, 12, and 18 in 36, 20, 18, and 13% of the cases, respectively. Whereas no difference was encountered for trisomy 3 in primary and secondary/simultaneous high-grade lymphomas, +7 and +12 were associated with primary lymphomas, and a +18 was predominantly found in secondary/simultaneous high-grade NHL. These results challenge earlier reports describing a high frequency of +3 in low-grade MALT-type NHL and indicate a possibly different genetic evolution pattern of primary and secondary/simultaneous high-grade lymphomas of primary mucosal origin.
Extranodal marginal zone B-cell non-Hodgkin’s lymphoma of the mucosa-associated lymphoid tissue (MALT) type as defined in the Revised European-American Lymphoma classification system 1 is a malignant lymphoma arising in organs normally devoid of a regular lymphatic parenchyma. MALT is believed to be introduced in organs such as the stomach, the thyroid gland, or the salivary glands after chronic inflammatory processes triggered by autoimmune phenomena or chronic infections. 2,3 As a consequence of a clonal dysregulation of the cellular proliferation and/or survival ability, unlimited clonal cell growth occurs independent of outer regulatory influences. This clonal dysregulation is supposed to be acquired in several steps and results in the formation of, or may be the consequence of, specific alterations on the genetic level.
Unlike the situation in primary nodal lymphomas, few data are available concerning the chromosomal constitution of MALT-type lymphomas. A reciprocal chromosome translocation t(11;18)(q21;q21) has been reported as the most frequent structural aberration. 4-9 Other reports suggested trisomy 3 to be a crucial genetic alteration in MALT-type lymphomas, having been detected in up to 50% of cases studied in banding analysis. 10-13 Even higher frequencies were obtained by in situ hybridization (ISH) using chromosome-specific (peri-) centromeric DNA probes. 14-16
Recently, we reported on the largest series of both low- and high-grade lymphomas of MALT type studied so far by classical cytogenetics 8 and found that trisomy 3 occurs in only a minor number of cases. This discrepancy prompted us to carry out a study using a sensitive bicolor fluorescence ISH (FISH) technique on nuclei isolated from paraffin-embedded tissues. In this series, 105 samples of low- and high-grade MALT-type lymphomas arising at different sites were investigated to estimate the frequency and distribution of aneusomies for chromosomes 3, 7, 12, and 18.
Materials and Methods
Specimen Selection
One hundred five resection specimens of extranodal malignant non-Hodgkin’s lymphomas (NHLs) of MALT type were selected from the files of the Institute of Pathology at the University of Würzburg. The cases were independently reviewed by two of us (GO and HKMH) following the criteria of the Revised European-American Lymphoma classification. 1 The diagnosis of primary high-grade tumors arising in the stomach without a concomitant low-grade MALT-type component was based on histopathological features as well as on clinical data available as described earlier. 8 No Helicobacter pylori eradication therapy was carried out in the gastric cases. Table 1 ▶ gives an overview of histopathological diagnoses and sites of origin.
Table 1.
Localization and Grading of 105 MALT-Type Lymphomas Investigated by ISH for Chromosome Probes 3, 7, 12, and 18
| Site | Low-grade no. | Primary high-grade no. | Secondary/simultaneous high-grade no. |
|---|---|---|---|
| Stomach | 33 | 26 | 17 |
| Salivary gland | 13 | ||
| Orbit | 5 | ||
| Thyroid gland | 4 | ||
| Lung | 3 | ||
| Uterine corpus | 1 | ||
| Urinary bladder | 1 | ||
| Small intestine | 1 | ||
| Large intestine | 1 | ||
| Total | 60 | 26 | 19 |
Immunohistochemistry
Immunohistochemical investigations were performed for routine purposes on paraffin sections by a standard peroxidase anti-peroxidase method with preceding antigen retrieval by pressure cooking using antibodies against B- and T-cell markers (L26/CD20, CD3, MT1/CD43, and UCHL-1/CD45RO), immunoglobulin light and heavy chains, and macrophage markers, as well as pankeratin antibodies (CAM 5.2 and KL1) for the detection of lymphoepithelial lesions. Whenever fresh-frozen material was available, a panel of additional antibodies (CD2, CD3, CD4, CD5, CD7, CD8, CD10, CD11c, CD23, and others) was employed on cryostat sections.
ISH Studies
ISH on cells isolated from paraffin blocks was performed as previously described. 17 Briefly, cells were disaggregated from paraffin blocks according to the method of Visakorpi et al. 18 Up to 10 sections (10 μm each) were cut from a paraffin block containing sufficient lymphoma tissue. After dewaxing in xylene, the material was rehydrated and digested with 1 mg proteinase K in 2 ml Carlsberg solution. The tissue was then disaggregated mechanically and rinsed in phosphate-buffered saline, and the resulting nuclear pellet was resuspended in phosphate-buffered saline and dropped onto cleaned glass slides.
After washing twice in 2× standard saline citrate (SSC), a proteolytic treatment was performed with pepsin (4 mg/ml in 0.2 N HCl) with or without prior digestion using 1 mol/L sodium thiocyanate at 80°C according to the protocol of Hopman and associates. 19 Generally, digestion times had to be evaluated for each case ranging from 0 to 2 minutes. After digestion and rinsing in 2× SSC, dehydration followed.
Biotin- or digoxigenin-labeled centromeric repetitive satellite DNA probes specific for chromosomes 3, 7, 12, and 18 were purchased from Oncogene (Gaithersburg, MD). They were used in double-FISH experiments at concentrations of 1 to 2 ng DNA probe in approximately 10 μl hybridization mixture containing 60% formamide, 2× SSC, 10% dextrane sulfate, and 0.1% salmon sperm DNA. Simultaneous denaturation of the slides and the hybridization mixture containing the probe under a coverslip sealed with rubber cement was performed at 80°C for 5 minutes in a tin pan floating in a water bath. Hybridization was allowed to take place overnight at 37°C. After removing the coverslips, slides were washed three times for 10 minutes in 60% formamide in 2× SSC at 47°C and subsequently in 2× SSC at 42°C. Signal detection was accomplished using appropriate Cy3- and fluorescein isothiocyanate-conjugated antibodies. Visualization of the signals was performed with a Zeiss Axiophot fluorescence microscope. Illustrations were made using the ISIS imaging system (MetaSystems, Altlussheim, Germany). Disaggregated cells from 15 reactive lymph nodes or tonsil specimens served as negative controls. Nineteen of the cases investigated had also been subjected to classical banding analysis after short-term culture. 8
Signal Evaluation
Nuclear signals from at least 200 cells were analyzed per case. Generally, only slides with a good hybridization efficiency were evaluated, implying that nuclear signals had to be observed after hybridization in more than 80% of cells. Whenever more than 20% of nuclei were devoid of clearly discernible signals, the hybridization procedure was regarded as insufficient and was repeated. Because of this strategy, 32 more cases, in which ISH was also attempted, had to be discarded from the study due to poor signal quality. As a rule, only cells with clearly discernible, nonoverlapping nuclei were counted. Signals should have the same intensity, and split signals were counted as one signal.
Results
Reactive Lymphoid Tissues
To establish the cutoff level for the diagnosis of aneusomies, probes specific for centromeric repetitive sequences of chromosomes 3, 7, 12, and 18 were hybridized to disaggregated nuclei from 15 reactive lymphatic tissues. Up to 95% of nuclei showed two hybridization spots (mean, 74.7%; range, 56 to 95%). Cells with three signals were observed in 0 to 3% (mean 0.9% for chromosome 3) of at least 200 nuclei evaluated per case. A cutoff value for trisomies was established at a minimum of 5% nuclei (mean value + 3 SD) with three signals for avoiding false-positive results. Likewise, a tumor was regarded as tetraploid if more than 5% of nuclei showed four hybridization signals with at least three different chromosome probes.
Low-Grade Lymphomas
Of 60 low-grade MALT-type lymphomas, 16 (27%) demonstrated aneuploidy with significant (>4%) gains of hybridization signals for one or more of the chromosomes tested (Table 2) ▶ . Trisomy 3 was seen in 12 cases (20%), being the most frequent chromosome gain. Trisomies for chromosomes 7, 12, and 18 were seen in two, two, and four cases, respectively. Two lymphomas showed signal distributions indicative of trisomies for both chromosomes 3 and 18, and two cases were positive for a +3 and a +7. No tetraploid chromosome clones were detected.
Table 2.
Interphase Cytogenetic Results for 105 Cases of MALT-Type Lymphomas
| Type | Trisomy 3 | Trisomy 7 | Trisomy 12 | Trisomy 18 |
|---|---|---|---|---|
| Low grade | ||||
| Gastric (n = 33) | 4 (12%) | 1 (3%) | 1 (3%) | 2 (6%) |
| Extragastric (n = 27) | 8 (30%) | 1 (4%) | 1 (4%) | 2 (7%) |
| Total (n = 60) | 12 (20%) | 2 (3%) | 2 (3%) | 4 (7%) |
| Primary high grade (n = 26) | 8 (31%) | 7 (27%) | 6 (23%) | 2 (8%) |
| Simultaneous low and high grade (n = 19) | 8 (42%) | 2 (11%) | 2 (11%) | 4 (21%) |
High-Grade Lymphomas
Primary and secondary/simultaneous high-grade lymphomas of MALT type were characterized by chromosomal aneusomies in 27 cases (60%). Again, trisomy 3 was the most common chromosome gain (n = 16; 36%) followed by gains of chromosome 7 in 9 tumors (20%), of chromosome 12 in 8 cases (18%), and chromosome 18 in 6 cases (13%). More than one numerical aberration was detected in 12 tumors. One primary high-grade tumor showed a signal distribution indicative of a tetraploid chromosome clone. A certain difference was noted in the occurrence of trisomies 7, 12, and 18 in primary and simultaneous/secondary high-grade NHLs. Trisomy 7 was ascertained in 7 of 26 (27%) and trisomy 12 in 6 of 26 (23%) primary high-grade tumors as opposed to only 2 of 19 (11%) each in simultaneous/secondary high-grade lymphomas. Trisomy 18, on the other hand, was found more frequently in simultaneous/secondary high-grade lymphomas (4 of 19; 21%) than in primary high-grade tumors (2 of 26; 8%). These differences, however, are not statistically significant. No difference was found in the distribution of trisomy 3 among primary and simultaneous/secondary high-grade lymphomas (Table 2) ▶ .
In the tumors regarded as positive for trisomies in our series, the percentage of nuclei with three signals ranged from 5 to 63%. In cases without detectable trisomies, the mean number of cells showing two hybridization signals was 78% as compared with 76% in the trisomic cases, excluding the chromosome probe being aneuploid (and 75% in control lymph nodes). Therefore, a reduced sensitivity as a cause for the negative results in these cases could be ruled out.
Comparison to Banding Analyses
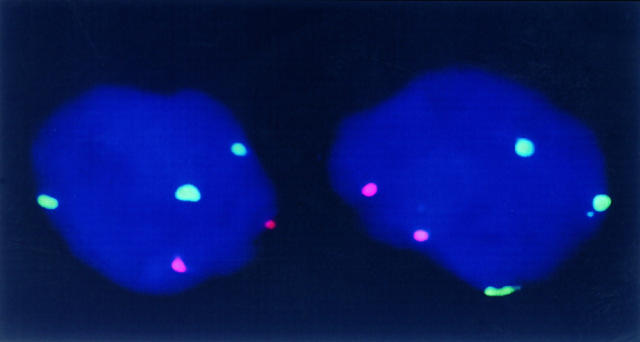
Of the tumors analyzed by FISH in the present series, 19 had also been studied in classical cytogenetics and were included in our previous report. 8 Comparison of banding analyses with the in situ data were congruent in 17 lymphomas. In two cases, FISH revealed aberrations that had been missed in classical cytogenetic analyses. In one case harboring a t(11;18) as the sole aberration in banding analysis, a +12 was detected by FISH in 34% of nuclei, and in the second case with a complex aberration, a trisomy 7 was present in 50% of cells that was not recognized in classical cytogenetics (Figure 1) ▶ .
Figure 1.
Bicolor FISH in a high-grade gastric MALT-type lymphoma. In two nuclei, three green signals for chromosome 7 and two red signals for chromosome 3 are discernible, indicating a trisomy 7.
Discussion
The concept of malignant NHLs as primary organ tumors arising from the MALT is new, and up to now few data are available concerning genomic alterations in MALT-type lymphomas. According to literature data, oncogene rearrangements similar to those found in nodal NHLs do not seem to play a particular role in the development of low-grade MALT-type lymphomas. Although single bcl-2-rearranged cases of MALT-type NHL have been reported, 20,21 the majority of investigators failed to find evidence for rearrangements of bcl-1, bcl-2, or bcl-3, 11,22-24 supporting the hypothesis that MALT-type NHLs have a genetic origin different from that of nodal lymphomas.
Compared with nodal malignant NHLs of all subtypes, few cytogenetic studies are available concerning the chromosomal constitution of MALT-type lymphomas. The recurring translocation t(11;18)(q21;q21.1) has been described as the most frequent structural chromosome aberration in 16 cases of low-grade MALT-type lymphoma or in small lymphocytic lymphoma with features of MALT. 4-9 According to conventional cytogenetic studies carried out in MALT-type lymphomas of low-grade malignancy, numerical aberrations of chromosomes 3, 12, and 18 seem to be common in this type of NHL. Especially, trisomy 3 has been reported in frequencies of about 50%. 10-13 This finding was corroborated by interphase cytogenetic studies using ISH with chromosome-specific α-satellite DNA probes, leading to the detection of trisomy 3 in even higher frequencies of 60 to 85%. 14-16
Two recent reports, however, failed to identify trisomy 3 as a frequent aberration in low-grade lymphomas of MALT type. In our own cytogenetic analyses of 13 cases with abnormal karyotypes, 8 trisomy 3 was found only twice (15%), and Auer and associates 9 recently found evidence for a +3 in 3 of their 9 cases (33%). In agreement with these cytogenetic studies, only 12 of 60 (20%) of low-grade NHLs of MALT type harbored a trisomy 3, which is in sharp contrast to previously published data. The fact that the lower frequency of trisomy 3 found in our cytogenetic series was now corroborated by FISH is even more intriguing, taking into account that conventional cytogenetics usually underestimates the rates of trisomies, because FISH can detect even small subclones of aberrant cell populations. 25
Because all hybridizations were carried out in bicolor FISH experiments, in contrast to previous ISH studies, an internal control was always available, assuring a high hybridization efficiency. Up to 94% of nuclei showed two signals when hybridized to probes specific for chromosomes 3, 7, 12, and 18, with a mean of 75% in controls and 77% in nonaneusomic cases. Therefore, an impaired sensitivity in our cases for the detection of trisomies can be ruled out. In addition, these results are consistent with a previous interphase cytogenetic study carried out on methanol/acetic glacial acid-fixed single cells with an even higher sensitivity. 8
Interestingly, the frequencies for trisomies 7, 12, and 18 in low-grade lymphomas in our hands (3%, 3% and 7%) are also lower than those reported by Wotherspoon and coworkers 14 and Brynes et al. 15 In both studies, however, no bicolor hybridization experiments have been carried out, avoiding the possibility of unspecific bindings for the probes tested in ISH. In addition, Brynes and coworkers 16 reported on high frequencies of trisomy 3 (up to 8.5%) in their control cases compared with 3% in our material.
Apart from technical differences in the studies carried out so far, the variation in results might also be due to different selection criteria for the cases investigated. In the recent cytogenetic and interphase cytogenetic studies by Dierlamm and associates, 13,15 reporting (partial) trisomies for chromosome 3 in a high number of MALT-type lymphomas, many complex chromosome alterations not regularly encountered in previous studies 4-9 were described. This may indicate higher tumor grades as compared with our cases. Alternatively, geographical variations might be the cause for the different results. In this context, the data obtained from classical or interphase cytogenetic studies in splenic marginal zone B-cell lymphoma are striking. Whereas Oscier et al 26 failed to find additional copies of chromosome 3 in 31 cases investigated by classical cytogenetics, other reports described the occurrence of a +3 in 18 to 50% of cases. 13-16 Significant geographic variations in the occurrence of a given chromosome abnormality have also been described in other hematological malignancies in Western populations. The detection of the t(4;11)(q21;q23) in acute lymphocytic leukemia by classical cytogenetics, for example, has been shown to vary from 2.5% 27 to 19%. 28 Using interphase FISH in cases of B-cell chronic lymphocytic leukemia, trisomy 12 was detected in frequencies of 12 to 54%. 29
Very few data are available concerning the chromosomal constitution of extranodal high-grade lymphomas of MALT type. In our series of 45 high-grade MALT-type lymphomas, chromosomal aneusomies were detected in a higher frequency than in the low-grade cases (60 versus 27%; P < 0.05). Trisomy 3 was the most common aberration (36%) followed by gains of chromosome 7 (20%) and +12 and +18 in 18% and 13%, respectively. These data, however, are not significantly different from those obtained in studies of large-cell lymphomas of nodal origin according to conventional cytogenetics or molecular investigations. In particular, gains of chromosome 3 have been reported in up to 33% of cases, of chromosome 7 in 22%, of chromosome 12 in 31%, and of chromosome 18 in 21%. 30-34 No difference was encountered in our series in the incidence of trisomy 3 between primary and secondary/simultaneous high-grade MALT-type lymphomas. Trisomies for chromosomes 7 and 12 were encountered in primary high-grade rather than in secondary/simultaneous high-grade lymphomas (27% and 23% versus 11%). These differences might indicate that additional copies of chromosomes 7 and 12 may contribute to tumorigenesis in primary high-grade MALT-type lymphomas in a manner similar to that in primary nodal large cell tumors. Trisomy 18, on the other hand, might rather be a progression factor in tumors with a low-grade component (21 versus 8%). Recently, Chan and associates, 35 using comparative genomic hybridization and FISH techniques, observed gains in chromosome 12 in all primary, but in only a minority of secondary/simultaneous high-grade gastric lymphomas investigated in their series from Hong Kong. A frequent overrepresentation of chromosome 12 in primary gastrointestinal large B-cell lymphomas was also found in the recent series of Barth et al. 36
It is not clear, however, by which underlying mechanism additional chromosomes could contribute to tumorigenesis and/or progression. One possible explanation would be an altered gene dosage effect, and in fact, several reports have provided evidence that the increase of chromosome copy numbers leads to enhanced gene expression. 37-39 Several candidate genes possibly being overexpressed by occurrence of a trisomy have been mapped to chromosome 18, among them bcl-2, ITF2, and YES. 40,41 Recently, the bcl-2 gene was shown to be amplified in cases of malignant lymphomas not characterized by a bcl-2/JH rearrangement by comparative genomic hybridization. It was suggested, therefore, that the bcl-2 protein overexpression might be the result of a gene amplification. 34 An alternative explanation would be that an additional chromosome or chromosomes could alter the spatial arrangement of the chromatin in the interphase nucleus 42,43 and thereby lead to an enhanced or decreased transcription of genes.
The results of our studies on a large number of low- and high-grade malignant lymphomas of MALT-type, all carried out using a sensitive double (bicolor) FISH technique, do question previous reports on the elevated occurrence of trisomy 3 in MALT-type lymphomas and indicate that a +3 is not a common feature at least in the majority of MALT tumors. Still, few cases have been studied for their chromosomal constitution so far, and resolving the problem, whether the differences reported in the literature rely on mere technical problems, geographical variations, or differing selection criteria, will have to await further studies.
Furthermore, our data indicate a possible different genetic background for the development of primary high-grade MALT-type lymphomas characterized by frequent gains of chromosomes 7 and 12 as opposed to secondary/simultaneous tumors, showing a +18.
Footnotes
Address reprint requests to Dr. German Ott, Pathologisches Institut der Universität, Josef-Schneider-Strasse 2, D-97080 Würzburg, Germany.
Supported by the Deutsche Forschungsgemeinschaft (DFG), Sonderforschungsbereich 172, Teilprojekt C-8 (to GO and HKM-H), and DFG grants Ot-168/1-1 and 1-2.
References
- 1.Harris NL, Jaffe ES, Stein H, Banks PM, Chan JKC, Cleary ML, Delsol G, De Wolf-Peeters C, Falini B, Gatter KC, Grogan TM, Isaacson PG, Knowles DM, Mason DY, Müller-Hermelink HK, Pileri SA, Piris MA, Ralfkiaer E, Warnke RA: A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood 1994, 84:1361-1392 [PubMed] [Google Scholar]
- 2.Hussell T, Isaacson PG, Crabtree JE, Spencer J: The response of cells from low-grade B-cell gastric lymphomas of mucosa-associated lymphoid tissue to Helicobacter pylori. Lancet 1993, 342:571-574 [DOI] [PubMed] [Google Scholar]
- 3.Greiner A, Marx A, Heesemann J, Leebmann J, Schmausser B, Müller-Hermelink HK: Idiotype identity in a MALT-type lymphoma and B cells in Helicobacter pylori-associated chronic gastritis. Lab Invest 1994, 70:572-578 [PubMed] [Google Scholar]
- 4.Levine EG, Arthur DC, Machnicki J, Frizzera G, Hurd D, Peterson B, Gajl-Peczalska KJ, Bloomfield CD: Four new recurring translocations in non-Hodgkin lymphoma. Blood 1989, 74:1796-1800 [PubMed] [Google Scholar]
- 5.Griffin CA, Zehnbauer BA, Beschorner WE, Ambinder R, Mann R: t(11;18)(q21;q21) is a recurrent chromosome abnormality in small lymphocytic lymphoma. Genes Chromosomes Cancer 1992, 4:153-157 [DOI] [PubMed] [Google Scholar]
- 6.Horsman D, Gascoyne R, Klasa R, Coupland R: t(11;18)(q21;q21.1): a recurring translocation in lymphomas of mucosa-associated lymphoid tissue (MALT)? Genes Chromosomes Cancer 1992, 4:183-187 [DOI] [PubMed] [Google Scholar]
- 7.Leroux D, Seité P, Hillion J, Le Marc’hadour F, Pégourié-Bandelier B, Jacob MC, Larsen CJ, Sotto JJ: t(11;18)(q21;q21) may delineate a spectrum of diffuse small B-cell lymphoma with extranodal involvement. Genes Chromosomes Cancer 1993, 7:54-56 [DOI] [PubMed] [Google Scholar]
- 8.Ott G, Katzenberger T, Greiner A, Kalla J, Rosenwald A, Heinrich U, Ott MM, Müller-Hermelink HK: The t(11;18)(q21;q21) chromosome translocation is a frequent and specific finding in low-grade, but not high-grade malignant non-Hodgkin’s lymphomas of the mucosa-associated lymphoid tissue (MALT-) type. Cancer Res 1997, 57:3944-3948 [PubMed] [Google Scholar]
- 9.Auer IA, Gascoyne RD, Connors JM, Cotter FE, Greiner TC, Sanger WG, Horsman DE: t(11;18)(q21;q21) is the most common translocation in MALT lymphomas. Ann Oncol 1997, 8:979-985 [DOI] [PubMed] [Google Scholar]
- 10.Wotherspoon AC, Pan L, Diss TC, Isaacson PG: Cytogenetic study of B-cell lymphoma of mucosa-associated lymphoid tissue. Cancer Genet Cytogenet 1992, 58:35-38 [DOI] [PubMed] [Google Scholar]
- 11.Clark HM, Jones DB, Wright DH: Cytogenetic and molecular genetic studies of the t(14;18) and t(14;19) in nodal and extranodal B-cell lymphoma. J Pathol 1992, 166:129-137 [DOI] [PubMed] [Google Scholar]
- 12.Whang-Peng J, Knutsen T, Jaffe E, Raffeld M, Zhao WP, Duffey P, Longo DL: Cytogenetic study of two cases with lymphoma of mucosa-associated lymphoid tissue. Cancer Genet Cytogenet 1994, 77:74-80 [DOI] [PubMed] [Google Scholar]
- 13.Dierlamm J, Pittaluga S, Wlodarska I, Stul M, Thomas J, Boegaerts M, Michaux L, Driessen A, Mecucci C, Cassiman JJ, De Wolf-Peeters C, Van den Berghe H: Marginal zone B-cell lymphomas of different sites share similar cytogenetic and morphologic features. Blood 1996, 87:299-307 [PubMed] [Google Scholar]
- 14.Wotherspoon AC, Finn TM, Isaacson PG: Trisomy 3 in low-grade B-cell lymphomas of mucosa-associated lymphoid tissue. Blood 1995, 85:2000-2004 [PubMed] [Google Scholar]
- 15.Dierlamm J, Michaux L, Wlodarska I, Pittaluga S, Zeller W, Stul M, Criel A, Thomas J, Boogaerts M, Delaere P, Cassiman JJ, De Wolf-Peeters C, Mecucci C, Van den Berghe H: Trisomy 3 in marginal zone B-cell lymphoma: a study based on cytogenetic analysis and fluorescence in situ hybridization. Br J Haematol 1996, 93:242-249 [DOI] [PubMed] [Google Scholar]
- 16.Brynes RK, Almaguer PD, Leathery KE, McCourty A, Arber DA, Medeiros JM, Nathwani BN: Numerical cytogenetic abnormalities of chromosomes 3, 7 and 12 in marginal zone B-cell lymphomas. Mod Pathol 1996, 9:995-1000 [PubMed] [Google Scholar]
- 17.Ott G, Kalla J, Ott MM, Schryen B, Katzenberger T, Müller JG, Müller-Hermelink HK: Blastoid variants of mantle cell lymphoma: frequent bcl-1 rearrangements at the MTC region and tetraploid chromosome clones. Blood 1997, 89:1421-1429 [PubMed] [Google Scholar]
- 18.Visakorpi T, Hyytinen E, Kallioniemi A, Isola J, Kallioniemi OP: Sensitive detection of chromosome copy number aberrations in prostate cancer by fluorescence in situ hybridization. Am J Pathol 1994, 145:624-630 [PMC free article] [PubMed] [Google Scholar]
- 19.Hopman AHN, Van Hooren E, Van de Kaa CA, Vooijs GP, Ramaekers FCS: Detection of numerical chromosome aberrations using in situ hybridization in paraffin sections in routinely processed bladder cancers. Mod Pathol 1991, 4:503-513 [PubMed] [Google Scholar]
- 20.Hey MM, Feller AC, Kirchner T, Müller JM, Müller-Hermelink HK: Genomic analysis of T-cell receptor and immunoglobulin antigen receptor genes and breakpoint cluster regions in gastrointestinal lymphomas. Hum Pathol 1990, 21:1283-1287 [DOI] [PubMed] [Google Scholar]
- 21.Kerrigan DP, Irons J, Chen IM: bcl-2 gene rearrangement in salivary gland lymphoma. Am J Surg Pathol 1990, 14:1133-1138 [DOI] [PubMed] [Google Scholar]
- 22.Pan L, Diss TC, Cunningham D, Isaacson PG: The bcl-2 gene in primary B-cell lymphoma of mucosa-associated lymphoid tissue (MALT). Am J Pathol 1989, 135:7-11 [PMC free article] [PubMed] [Google Scholar]
- 23.Wotherspoon AC, Pan L, Diss TC, Isaacson PG: A genotypic study of low grade B-cell lymphomas, including lymphomas of mucosa associated lymphoid tissue (MALT). J Pathol 1990, 162:135-140 [DOI] [PubMed] [Google Scholar]
- 24.Diss TC, Wotherspoon AC, Speight P, Pan L, Isaacson PG: B-cell monoclonality, Epstein Barr virus, and t(14;18) in myoepithelial sialadenitis and low-grade B-cell MALT lymphoma of the parotid gland. Am J Surg Pathol 1995, 19:531-536 [DOI] [PubMed] [Google Scholar]
- 25.Schlegelberger B, Zhang Y, Weber-Matthiesen K, Grote W: Detection of aberrant clones in nearly all cases of angioimmunoblastic lymphadenopathy with dysproteinemia-type T-cell lymphoma by combined interphase and metaphase cytogenetics. Blood 1994, 84:2640-2648 [PubMed] [Google Scholar]
- 26.Oscier DG, Matutes E, Gardiner A, Glide S, Mould S, Brito-Babapulle V, Ellis J, Catovsky D: Cytogenetic studies in splenic lymphoma with villous lymphocytes. Br J Haematol 1993, 85:487-491 [DOI] [PubMed] [Google Scholar]
- 27.Mirro J, Kitchingman G, Williams D, Lauzon GJ, Lin CC, Callihan T, Zipf TF: Clinical and laboratory characteristics of acute leukemia with the 4;11 translocation. Blood 1986, 67:689-697 [PubMed] [Google Scholar]
- 28.Bjerrum OW, Philip P, Müller-Berat N, Hertz H, Killman SA: Acute lymphocytic leukemia with t(4;11): a clinical subentity. Scand J Haematol 1985, 35:96-101 [DOI] [PubMed] [Google Scholar]
- 29.Dierlamm J, Wlodarska I, Michaux L, Vermeesch JR, Meeus P, Stul M, Criel A, Verhoef G, Thomas J, Delannoy A, Louwagie A, Cassiman JJ, Mecucci C, Hagemeijer A, Van den Berghe H: FISH identifies different types of duplications with 12q13–15 as the commonly involved segment in B-cell lymphoproliferative malignancies characterized by partial trisomy 12. Genes Chromosomes Cancer 1997, 20:155-166 [PubMed] [Google Scholar]
- 30.Yunis JJ, Oken MM, Kaplan ME, Ensrud KM, Howe RR, Theologides A: Distinctive chromosomal abnormalities in histologic subtypes of non-Hodgkin’s lymphoma. N Engl J Med 1982, 307:1231-1236 [DOI] [PubMed] [Google Scholar]
- 31.Bloomfield CD, Arthur DC, Frizzera G, Levine EG, Peterson BA, Gajl-Peczalska KJ: Nonrandom-chromosome abnormalities in lymphoma. Cancer Res 1983, 43:2975-2984 [PubMed] [Google Scholar]
- 32.Levine EG, Arthur DC, Frizzera G, Peterson BA, Hurd DD, Bloomfield CD: There are differences in cytogenetic abnormalities among histologic subtypes of the non-Hodgkin’s lymphomas. Blood 1985, 66:1414-1422 [PubMed] [Google Scholar]
- 33.Cabanillas F, Pathak S, Trujillo J, Manning J, Katz R, McLaughlin P, Velasquez WS, Hagemeister FB, Goodacre A, Cork A, Butler JJ, Freireich EJ: Frequent nonrandom chromosome abnormalities in 27 patients with untreated large cell lymphoma and immunoblastic lymphoma. Cancer Res 1988, 48:5557-5564 [PubMed] [Google Scholar]
- 34.Monni O, Joensuu H, Franssila K, Knuutila S: DNA copy number changes in diffuse large B-cell lymphoma: comparative genomic hybridization study. Blood 1996, 87:5269-5278 [PubMed] [Google Scholar]
- 35.Chan WY, Wong N, Chan AB-W, Chow JH-S, Lee JC-K: Consistent copy number gain in chromosome 12 in primary diffuse large cell lymphomas of the stomach. Am J Pathol 1998, 152:11-16 [PMC free article] [PubMed] [Google Scholar]
- 36.Barth TFE, Döhner H, Werner CA, Stilgenbauer S, Schlotter M, Pawlita M, Lichter P, Möller P, Bentz M: Characteristic pattern of chromosomal gains and losses in primary large B-cell lymphomas of the gastrointestinal tract. Blood 1998, 91:4321-4330 [PubMed] [Google Scholar]
- 37.Bardsley WG, McMurray BP, Robson A, D’Souza S, Taylor GM: Analysis of gene-dosage effects on the expression of CD18 by trisomy 21 lymphoblastoid cell lines using a statistical model to fit flow cytometry profiles. Hum Genet 1990, 86:181-186 [DOI] [PubMed] [Google Scholar]
- 38.Ferrari S, Grande A, Zucchini P, Manfredini R, Tagliafico E, Rossi E, Temperani P, Torelli G, Emilia G, Torelli U: Overexpression of c-kit in a leukemic cell population carrying a trisomy 4 and its relationship with the proliferative capacity. Leuk Lymph 1993, 9:495-501 [DOI] [PubMed] [Google Scholar]
- 39.Hogemann I, Bock S, Heppner P, Petrides PE: Cytogenetic and growth factor gene analysis of a renal carcinoma cell line. Cancer Genet Cytogenet 1994, 78:175-180 [DOI] [PubMed] [Google Scholar]
- 40.Ohno H, Fukuhara S, Takahashi R, Mihara KI, Sugiyama T, Doi S, Uchino H, Toyoshima K: c-yes and bcl-2 genes located in 18q21.3 in a follicular lymphoma cell line carrying a t(14;18) chromosomal translocation. Int J Cancer 1987, 39:785-788 [DOI] [PubMed] [Google Scholar]
- 41.Henthorn P, Kildedjian M, Kadesch T: Two distinct transcription factors that bind the immunoglobulin enhancer μE5/kE2 motif. Science 1990, 247:467-470 [DOI] [PubMed] [Google Scholar]
- 42.Haaf T, Schmid M: Chromosome topology in mammalian interphase nuclei. Exp Cell Res 1991, 192:325-332 [DOI] [PubMed] [Google Scholar]
- 43.Vourc’h C, Taruscio D, Boyle AL, Ward DC: Cell cycle-dependent distribution of telomeres, centromeres, and chromosome-specific subsatellite domains in the interphase nucleus of mouse lymphocytes. Exp Cell Res 1993, 205:142-151 [DOI] [PubMed] [Google Scholar]