Abstract
Several cytokines have been implicated in the pathogenesis of human lymphomas. Among them, interleukin-10 (IL-10) is a pleiotropic cytokine with various biological effects on B and T lymphocytes. Its expression has been essentially studied in B-cell lymphomas, where it appears to act as an autocrine growth factor. BCRF1 (also called viral IL-10), an open reading frame of Epstein-Barr virus, exhibits extensive sequence and functional homologies with human IL-10. Some entities belonging to T- or natural killer (NK)-cell lymphomas are characterized by a frequent association with Epstein-Barr virus. We analyzed 39 cases of peripheral T-cell lymphoma, as well as 7 cases of nasal NK-cell lymphoma, for the presence of IL-10 transcripts by in situ hybridization, to see whether this cytokine was expressed in these tumors and whether its expression could be related to their histological subtype and to the presence of Epstein-Barr virus. Because the riboprobe used for in situ hybridization recognizes both human and viral IL-10, 12 cases were also analyzed by reverse transcription-polymerase chain reaction to verify the human or viral origin of IL-10. It was found that 8 of 11 (73%) anaplastic large cell lymphomas (ALCLs), 2 of 11 (18%) pleomorphic T-cell lymphomas, and 3 of 7 (43%) nasal NK-cell lymphomas exhibited a large number of IL-10-expressing cells, whereas only rare scattered cells were detected in angioimmunoblastic (11 of 11) and in γδ T-cell lymphomas (6 of 6). In ALCLs, the pattern of IL-10 mRNA-expressing cells showed an overlapping with the CD30 staining and preferential localization in sinusal and perifollicular areas, thereby suggesting that IL-10-expressing cells were tumor cells. Furthermore, IL-10 transcripts were detected in the SU-DHL-1 anaplastic lymphoma cell line. No correlation with Epstein-Barr virus profile was found, because all cases of ALCL were negative for EBER 1 and 2 genes by in situ hybridization. We confirmed the presence of human IL-10 mRNA by reverse transcription-polymerase chain reaction in ALCLs as well as in NK-cell lymphomas, whereas viral IL-10 was not detected. Thus, human and not viral IL-10 is frequently expressed by tumor cells in ALCLs and nasal NK-cell lymphomas. In view of its function in the proliferation and the differentiation of cytotoxic T and NK cells, and its immunosuppressive properties, IL-10 may have a role in the pathogenesis of these lymphomas.
There is accumulating evidence that cytokines may play important roles in the pathogenesis of human lymphomas. 1,2 Cytokines can be produced by tumor and/or reactive cells, and their secretion can provide a growth advantage for tumor cells in either an autocrine or a paracrine fashion. Among human lymphomas, peripheral T-cell lymphomas (PTCLs) constitute a heterogeneous group of malignancies with respect to clinical, immunological, genotypic, and pathogenetic features. 3 Most of them express the αβ T-cell receptor (TCR), whereas a small subset of them express the γδ TCR. 4 The possible involvement of cytokines in the pathogenesis of PTCL has been investigated in a few studies, and evidence has been provided for an association between cytokine profile of tumor cells and histological subtype of the tumor. 5-7 In this respect, the following associations have been recorded: human T-cell lymphotrophic virus 1+ adult T-cell leukemia with interleukin (IL)-2, 1 angioimmunoblastic lymphadenopathy (AILD) with tumor necrosis factor-α and lymphotoxin, 5 and CD30+ anaplastic large cell lymphomas (ALCLs) with IL-9 and IL-6. 1,6,7 In addition, some other cytokines, such as IL-4, interferon-γ, and IL-7, were also investigated, but no clear association between cytokine profile of tumor cells and histotype was reported. 7 IL-10 was first characterized as a T-cell-derived cytokine able to block interferon-γ production by T-helper-1 cells and as a B cell-derived thymocyte growth factor. 8,9 IL-10 is produced by B cells, T cells, monocytes/macrophages, and keratinocytes. 10 The cloning and sequencing of the murine and human IL-10 (hIL-10) genes revealed an extensive homology with BCRF1 gene, an open reading frame of Epstein-Barr virus (EBV). 11 The product of the BCRF1 gene, also called viral IL-10 (vIL-10), exhibits partial IL-10 activity in vitro and may play a role during EBV infection. 11 IL-10 is a pleiotropic cytokine known to be an important regulator of lymphoid and myeloid functions. 10 It is a growth factor for T lymphocytes in the mouse model and a growth and differentiation factor for human activated B cells. 12,13 IL-10 participates in the promotion of IL-2-activated mouse cytotoxic T-lymphocyte precursor to differentiate into effector cytotoxic cells. 14 In addition, IL-10 exerts immunosuppressive activities: it inhibits antigen-specific T-cell activation and blocks cytokine production by monocytes and macrophages in both human and mouse models. 10,15 IL-10 has been described to induce natural killer (NK) cell proliferation and increase of cytotoxic activity of these cells. 16
To date, IL-10 has been described in various types of B-cell lymphoproliferations, and the biological properties of IL-10 have raised questions of the potential relevance of this cytokine in the pathogenesis of B-cell lymphoproliferations. Indeed, IL-10 was suggested to be a proliferation factor for B cells, which could act in an autocrine pathway, in acquired immunodeficiency syndrome-related lymphomas and Burkitt lymphomas. 17-19 On the other hand, although IL-10 can be produced by normal T cells, only a few cases of PTCL, mostly human T-cell lymphotrophic virus 1-positive leukemia, were studied. 20 For this reason, we have investigated the expression of IL-10 mRNA by in situ hybridization (ISH) in 39 cases of PTCL as well as in 7 cases of NK-cell lymphoma to see whether this cytokine was expressed in these tumors. We have particularly addressed the question of whether this expression could be related to the histological subtype of the tumor and to the expression of αβ TCR, γδ TCR, or NK-cell antigens. In addition, because IL-10 gene has extensive homology with the BCRF1 gene of the EBV, we have investigated in parallel the cases of this series for the presence of EBV.
Materials and Methods
Tissue Specimens and Cell Lines
Tissues from 46 patients with lymphomas of T-cell or NK-cell types mainly retrieved from the files of the Département de Pathologie, Hôpital Henri Mondor (Créteil, France) were included in this study. One part of the material was routinely fixed, and the other one was snap frozen in liquid nitrogen and stored at −80°C. The diagnosis was based on histological and immunohistological data available on both fixed and frozen tissue sections. PTCLs were divided into αβ and γδ or TCR-silent T-cell lymphomas according to the expression (or the nonexpression) of the TCR proteins using the βF1 or δTCR1 antibodies as previously described. 4,21 NK-cell lymphomas had the TCR-silent, CD2+, CD5−, CD56+ phenotype, characteristic of normal NK cells and NK-cell lymphomas, and no clonal TCRγ rearrangement was detected. 22 Lymphomas were classified according to the updated Kiel classification 23 and the Revised European-American Lymphoma classification 3 (Tables 1 to 3) ▶ ▶ ▶ . Lymphoblastic lymphomas and mycosis fungoides were excluded. Because T-cell and null-cell ALCLs are now considered as a unique lymphoma entity, five null-cell ALCLs were also selected in the present study. The cytotoxic protein expression in 27 of these cases has been recently reported elsewhere. 21
Table 1.
IL-10 and EBER mRNA Expression in Nonanaplastic T-cell Lymphomas: Correlation with Cytotoxic Protein Expression by Immunohistochemistry
| Cases | Histology (Kiel/REAL) | Cell origin | Site | IL-10 mRNA expression (positive cells/mm2) | TIA-1 | Granzyme B | EBER | |
|---|---|---|---|---|---|---|---|---|
| αβ | γδ | |||||||
| 1 | AILD | − | − | Node | <1 | − | − | +* |
| 2 | AILD | + | − | Node | 4.1 | − | − | +* |
| 3 | AILD | − | − | Node | 9.7 | − | − | +* |
| 4 | AILD | + | − | Node | 6.9 | − | − | +* |
| 5 | AILD | + | − | Node | 22.2 | − | − | +* |
| 6 | AILD | + | − | Node | 19.8 | − | − | +* |
| 7 | AILD | + | − | Node | 1.4 | − | − | +* |
| 8 | AILD | ND | ND | Node | 15.5 | ND | ND | ND |
| 9 | AILD | + | − | Node | 6.25 | ND | ND | +* |
| 10 | AILD | + | − | Node | <1 | − | − | +* |
| 11 | AILD | + | − | Node | 2.4 | − | ND | +* |
| 12 | PML/Uns | + | − | Node | 12.0 | − | − | + |
| 13 | PML/Uns | + | − | Node | 23.1 | − | − | + |
| 14 | PML/Uns | + | − | Skin | 1.0 | − | − | − |
| 15 | PML/Uns | + | − | Node | 4.1 | + | + | − |
| 16 | PML/Uns | + | − | Node | <1 | + | ++ | − |
| 17 | PML/EATCL | + | − | Stomach | <1 | +++ | + | − |
| 18† | PML/Uns | − | − | Node | 50.9 | − | − | ++ |
| 19† | PML/Uns | + | − | Node | 83.3 | ++ | + | − |
| 20 | PML/EATCL | + | − | Intestine | 1.2 | + | + | − |
| 21 | PML/Uns | − | − | Node | 2.5 | − | − | − |
| 22 | PML/Uns | + | − | Node | 4.6 | − | − | − |
| 23 | −/HS γδ | − | + | HS | 2.1 | + | − | − |
| 24 | −/HS γδ | − | + | HS | 1.0 | +++ | − | − |
| 25 | PML/SPan | − | + | Skin | 1.2 | +++ | +++ | − |
| 26 | PML/AL | − | + | Larynx | 8.3 | +++ | +++ | +++ |
| 27 | PML/ITL | − | + | Stomach | 1.2 | +++ | +++ | + |
| 28 | PML/ITL | − | + | Small bowel | 1.2 | +++ | +++ | +++ |
For IL-10, the number of positive cells was estimated per mm 2 of tissue sections. For TIA-1, granzyme B, and EBER, the results are expressed as follows: −, virtually all tumor cells negative; +, <20% positive; ++, 20 to 50% positive, +++, >50% positive. For AILD cases: *A few scattered EBER + cells were observed for which the B- or T-cell origin could not be determined.
†For these two PML cases, a proportion of cells were CD30 positive, whereas ALK-1 was negative. Abbreviations: AL, angiocentric lymphoma; EATCL, enteropathy-associated T-cell lymphoma; HS γδ, hepatosplenic γδ T-cell lymphoma; ITL, intestinal T-cell lymphoma; SPan, subcutaneous panniculitic cell lymphoma, Uns (unspecified); ND, not done. REAL, Revised European-American Lymphoma classification.
Table 2.
IL-10 and EBER mRNA Expression in Anaplastic T-Cell Lymphomas: Correlation with Cytotoxic Protein Expression by Immunohistochemistry
| Cases | Age/Sex | Histological variant (Kiel/REAL) | Site of biopsy | Other sites of involvement (clinical stage) | IL-10 mRNA expression (positive cells/mm2) | TIA-1 | Granzyme | EBER | EMA | ALK1 | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 29 | 66 /M | ALCL T | Node | NA | 37.5 | − | − | − | − | − | NA |
| 30 | 20 /F | ALCL T | Node | IV | 1.4* | ++ | + | − | + | + | CR, AWD (24 months) |
| 31 | 40 /M | ALCL T | Node | III | 98.8 | ++ | ++ | − | − | − | DOD |
| 32 | 40 /M | ALCL T | Node | IV | 105/8.3† | +++ | +++ | − | + | + | CR, AWD (36 months) |
| 33 | 20 /F | ALCL T | Node | II | 80.9 | +++ | +++ | − | + | + | NA |
| 34 | 37 /M | ALCL T | Node | I | 9 | ++ | + | − | + | + | CR |
| 35 | 82 /F | ALCL null | Node | IV | 13.3 | +++ | + | − | − | − | DOD |
| 36 | 38 /M | ALCL null | Node | IV | 87.5/5.5† | + | − | − | ND | − | DOD, 2 weeks |
| 37 | 16 /F | ALCL null | Node | NA | 133.3 | + | +++ | − | ND | + | DOD |
| 38 | 21 /M | ALCL null | Node | II | 46.1 | ++ | +++ | − | ND | − | CR, AWD (16 months) |
| 39 | 11 /M | ALCL null | Node | I | 119.4 | ND | ND | − | + | + | Relapse, 2nd CR Alive (48 months) |
For IL-10, the number of positive cells was estimated per mm 2 of tissue sections. For TIA-1, granzyme B, and EBER, the results were expressed as in Table 1 ▶ .
Abbreviations: REAL, Revised European-American Lymphoma classification; NA, not available; ND, not done; M, male; F, female; AWD, alive without disease; DOD, dead of disease; and CR, complete remission.
*This lymphohistiocytic variant showed only scattered positive cells that could correspond to scattered neoplastic cells.
†Two patterns of expression were observed in the same tissue section; counts were done in two distinct areas of the tissue.
Table 3.
IL-10 and EBER mRNA expression in NK-Cell Lymphomas: Correlation with Cytotoxic Protein Expression by Immunohistochemistry
| Cases | Age/Sex | Histology Kiel/REAL | Site of biopsy | Other sites of involvement | IL-10 mRNA expression (positive cells/mm2) | TIA-1 | Granzyme B | EBER | Outcome |
|---|---|---|---|---|---|---|---|---|---|
| 40 | 28 /F | PML/AL | Nasal | I | 3.6 | +++ | +++ | +++ | NA |
| 41 | 83 /M | PML/AL | Nasal | I | 1.4 | +++ | +++ | +++ | DOD, 27 months |
| 42 | NA | PML/AL | Nasal | NA | 11.1 | +++ | +++ | +++ | CR, alive (36 months) |
| 43 | 35 /F | PML/AL | Nasal | I | 198.6 | +++ | +++ | +++ | DOD, 9 months |
| 44 | 25 /M | PML/AL | Nasal | I | 165.0 | +++ | +++ | +++ | CR after local relapse |
| 45 | 58 /M | PML/AL | Nasal | I | 34.7 | +++ | +++ | +++ | DOD, 4 months |
| 46 | 43 /F | PML/AL | Nasal | I | 2.8 | +++ | +++ | +++ | DOD, 15 months |
For IL-10, the number of positive cells was estimated per mm 2 of tissue sections. For TIA-1, granzyme B, and EBER, the results are expressed as in Table 1 ▶ . Abbreviations: REAL, Revised European-American Lymphoma classification; AL, angiocentric lymphoma; M, male; F, female; AWD, alive without disease; DOD, dead of disease; and CR, complete remission.
The SU-DHL-1 cell line, a lymphoma cell line with the t(2;5) translocation (kindly provided by M. L. Cleary, Stanford, CA) and B95-8 cell line, a marmoset lymphoblastoid B-cell line (a kind gift of I. Joab, Institut G. Roussy, Villejuif, France) were used in the present study, as well as peripheral blood mononuclear cells obtained from a healthy donor. Peripheral blood mononuclear cells were purified on a Ficoll gradient and grown for 24 hours with 1 μg/ml lipopolysaccharide. 8 Cells were then cytocentrifuged. Slides of tissue sections and cytospins were stored at −80°C before ISH.
ISH for the Detection of IL-10 Transcripts
A pBS plasmid containing a 640-bp ApaI-AccI fragment of BCRF1/IL-10 cDNA was kindly provided by I. Joab. 19 Sense and antisense riboprobes were synthesized using 35S-labeled UTP (1000 Ci/mmol, Amersham, Les Ulis, France) with the Riboprobe in vitro transcription system (Promega, Madison, WI). Fifty μCi of 35S-labeled UTP were added to the following reaction mixture: 1 μl of each adenosine, cytosine, and guanosine 5′-triphosphate (10 mmol/L each), 2 μl of 100 mmol/L dithiothreitol (DTT), 1 μl of RNase inhibitor, 4 μl of 5× transcription buffer (200 mmol/L Tris-HCl, pH 7.5, 30 mmol/L MgCl2, 10 mmol/L spermidine, and 50 mmol/L NaCl), 0.5 μg of linearized plasmid, and 15 U of T3 or T7 RNA polymerase, in a final volume of 20 μl. The reaction was allowed to proceed for 60 minutes at 37°C. The plasmid DNA was digested with 1 μl of DNase I (1 U/μl) for 10 minutes at 37°C. The size of 35S-labeled probes was adjusted to about 200 bases in length by alkaline hydrolysis in 80 mmol/L NaHCO3, 120 mmol/L Na2CO3, and 10 mmol/L DTT at 60°C for 15 minutes. After neutralization in 0.2 mol/L sodium acetate, pH 5.2, 1% acetic acid, and 10 mmol/L DTT, RNA probes were eluted on a Sephadex column (Pharmacia, Uppsala, Sweden) using a buffer containing 2 mol/L Tris-HCl, pH 7.5, 0.5 mol/L ethylenediaminetetraacetic acid, 10% sodium dodecyl sulfate, and 1 mol/L DTT and precipitated in ethanol.
Hybridization with sense and antisense probes were performed in parallel. Cryostat sections were fixed for 20 minutes in 4% paraformaldehyde and acetylated. For cytospins, cells were permeabilized in Triton 1/1000 in PBS for 15 minutes before acetylation. Slides were then dehydrated and air dried. Thirty μl of a hybridization mixture containing 50% deionized formamide, 10% dextran sulfate, 10 mmol/L DTT, 0.3 mol/L NaCl, 20 mmol/L Tris-HCl, pH 7.5, 5 mmol/L ethylenediaminetetraacetic acid, 10 mmol/L NaHPO4, pH 8, 0.4 mg/ml Denhardt’s solution, 0.5 mg/ml yeast tRNA, and 1.5 × 10 6 cpm of 35S-labeled probe were applied on each section. Hybridization was performed overnight at 50°C. Excess of probe was removed by washing in 5× standard saline citrate (SSC) and 1 mmol/L DTT for 30 minutes at 42°C and then in 2× SSC, 50% formamide, and 1 mmol/L DTT for 20 minutes at 60°C. Slides were then subjected to digestion with 30 μg/ml RNase A for 30 minutes at 37°C and washed in 2× SSC for 15 minutes at 37°C and in 0.1× SSC for 15 minutes, dehydrated, and air dried before autoradiography (Hypercoat LM1; Amersham). Slides were stored at 4°C in the dark for 15 and 35 days and were developed in Kodak D19 developer (Kodak, Hemel, Hampstead, United Kingdom), rinsed, and fixed. Slides were finally counterstained in hematoxylin and mounted. Evaluation of the number of IL-10 mRNA-containing cells was performed under light microscope by quantification of positive cells. Results were expressed as the number of positive cells per mm 2 of tissue sections. For quantification of IL-10-positive cases, we considered a case as positive when >35 positive cells/mm 2 were observed. Sense probes were used as control in each experiment and gave no significant positive signal.
Three cases (cases 29, 32, and 37) of ALCL with available formalin-fixed tissues and showing a significant number of IL-10-containing cells were examined by ISH with IL-10-digoxigenin probe on paraffin sections, to better identify the IL-10-positive cells. Probe was transcribed in vitro in the presence of digoxigenin-UTP according to the manufacturer’s procedure (Boehringer Mannheim, Mannheim, Germany) and sheared by alkaline hydrolysis to yield RNA of about 200 nucleotides in length. Sections were dewaxed and hybridized as previously described, 24 with some modifications. Tissue sections were treated with 15 μg/ml protease IV (Sigma Chemical Co., St. Louis, MO) for 15 minutes at 37°C and acetylated. Hybridization was performed overnight at 52°C in a solution containing 50% formamide, 4× SSC, 1× Denhardt’s solution, 10% dextran sulfate, 250 μg/ml yeast RNA, and 5 ng/μl of digoxigenin-labeled probe. Unbound labeled RNA probe was removed by an RNase treatment for 30 minutes at 37°C (20 μg/ml) and by washing in 2× SSC and 1× SSC for 30 minutes each at 52°C. Bound probe was detected with antidigoxigenin antibody Fab fragments conjugated with alkaline phosphatase (Boehringer Mannheim), and color reaction was performed with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (Sigma).
RNA Extraction and Reverse Transcription-Polymerase Chain Reaction (RT-PCR) for Human and Viral IL-10
Total RNAs were extracted from frozen tissues using Trizol reagent (Life Technologies, Inc., Gaithersburg, MD). RNAs were extracted in chloroform, precipitated in isopropanol, and resuspended in diethylpyrocarbonate-treated water. To avoid contamination with genomic DNA, RNA samples were treated with 1 U RNase-free RQI-DNase (Promega) for 10 minutes at 37°C and ethanol precipitated. One μg of DNase-treated RNAs was reverse transcribed using 300 ng of random hexanucleotides (Life Technologies), 200 U of SuperScript II reverse transcriptase (Life Technologies), 1 μl of 10 mmol/L deoxynucleotide triphosphate mixture, 2 μl of 0.1 mol/L DTT, 10 U of RNAsin (Life Technologies), and 4 μl of 5× first-strand buffer (250 mmol/L Tris-HCl, pH 8.3, 375 mmol/L KCl, 15 mmol/L MgCl2, and 0.1 mol/L DTT) in a final volume of 20 μl for 50 minutes at 42°C. Thirty μl of water was added at the end of the reaction.
PCR amplification of S14 ribosomal protein cDNA was performed on each sample as a control for efficient cDNA synthesis. Negative controls, which were performed without cDNA adjunction in the reagent mix, were included for every PCR analysis. Positive controls for IL-10 and BCRF1 were the cDNA of the SU-DHL-1 cell line and the B95-8 cell line, respectively. Specific primers were custom synthesized (Genomic, Collonges-sous-Salève, France). They were: IL-10 sense, 5′-ACCAAGACCCAGACATCAAG-3′; IL-10 antisense, 5′-GAGGTACAATAAGGTTTCTCAAG-3′; BCRF1 sense, 5′ATGGAGCGAAGGTTAGTGGT-3′; BCRF1 antisense, 5′-CACCCAAAGAATTGACATGGT-3′; S14 sense, 5′-GGCAGACCGAGATGAATCCTCA-3′; and S14 antisense, 5′- CAGGTCCAGGGGTCTTGGTCC-3′. One μl of the cDNA was amplified in 1× PCR buffer (10 mmol/L Tris-HCl, pH 8.3, and 50 mmol/L KCl) containing 0.4 μl of 10 mmol/L deoxynucleotide triphosphate mixture, 0.4 μl of each specific primer (10 pmol/μl), 0.5 U of Taq Gold polymerase (Perkin-Elmer Corp., Norwalk, CT), and 1.5 mmol/L MgCl2 for S14 or 2 mmol/L MgCl2 for IL-10 and BCRF1, in a total volume of 20 μl. The reaction mixture was amplified in a 2400 Perkin-Elmer thermal cycler, as follows: 10 minutes at 95°C followed by 28 (S14) or 33 (IL-10 and BCRF1) cycles of 30 seconds each at 94°C, 30 seconds at 58°C, and 30 seconds at 72°C. An autoextension of 2 seconds was added to each 72°C extension cycle. The amplified products were electrophoresed in ethidium bromide-stained 2% agarose gel.
EBV ISH
The procedure for the detection of EBER transcripts by ISH has been previously described. 25 Probes used were fluorescein-conjugated oligonucletides complementary to the nuclear RNA portions of the EBER 1 and 2 genes (Dako, Glostrup, Denmark). The hybridization was detected using an immunohistological detection system, with a mouse anti-fluorescein antibody, rabbit anti-mouse immunoglobulins, and the alkaline phosphatase-anti-alkaline phosphatase complexes (Dako). The visualization of the reaction was carried out using nitroblue tetrazolium and bromochloroindolyl phosphate.
Results
Expression of IL-10 mRNA by ISH
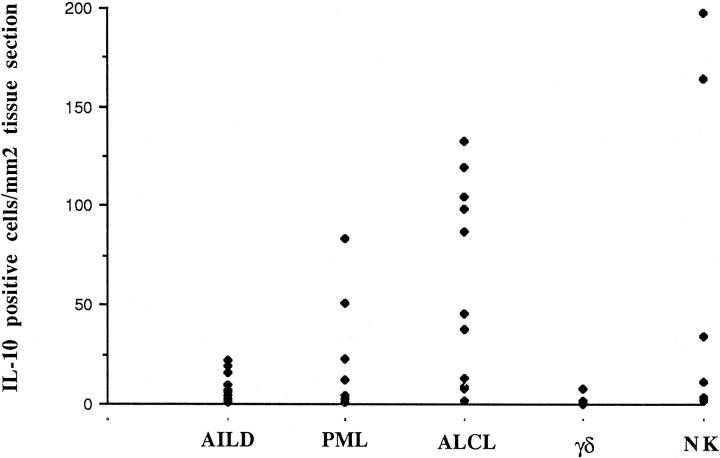
In all of the 39 cases of T-cell lymphoma and in the 7 cases of NK-cell lymphoma, cells containing IL-10 transcripts could be demonstrated, with a heterogenous pattern of expression from case to case. Moreover, a strong association with morphological subtypes was shown. Results are summarized in Tables 1 to 3 ▶ ▶ ▶ and Figure 1 ▶ . In 3 of the 7 cases of nasal NK-cell lymphoma, IL-10 transcripts were detected in a substantial proportion of cells. In one of these cases, the positive cells were scattered throughout the tissue section, thus preventing identification of the cells. In the other two cases of NK-cell lymphoma, the IL-10-positive cells were restricted to some areas, in which immunohistological studies performed on serial sections demonstrated the presence of CD30+/LMP1+ tumor cells (Figure 2, A and B) ▶ . Eight of the 11 (73%) anaplastic null-cell or T-cell lymphomas displayed a high number of IL-10-expressing cells (>35 cells/mm2). In all cases, the positivity, usually strong, seemed to be localized in the large atypical cells. In eight cases, positive cells were predominantly found in sinusal and perifollicular areas (Figure 3A) ▶ . In two other cases with partial involvement (cases 32 and 36), the localization of the positive cells was restricted to the neoplastic areas. Among the three cases with a low number of IL-10-expressing cells, a case of lymphohistiocytic variant (case 30), with only a few scattered neoplastic cells, displayed a similar pattern of IL-10-positive cells. Histological and immunohistochemical analysis performed on serial sections demonstrated an overlapping between CD30+ tumor areas and IL-10-positive cells (Figure 3B) ▶ , whereas no clear overlapping with CD68 could be observed. ISH with a digoxigenin-labeled probe, performed on paraffin sections of three ALCL IL-10 mRNA-expressing cases, confirmed the presence of IL-10 in the cytoplasm of large cells with characteristic cytological features of anaplastic cells (Figure 3C) ▶ . Five of the seven IL-10-positive ALCLs (one case was not available) expressed the cytotoxic protein granzyme B in a large proportion of tumor cells, whereas the last three cases, with only a few IL-10-expressing cells, showed granzyme B expression in less than 20% of the tumor cells. In all cases of AILD (11 of 11), only a few scattered IL-10-positive cells were observed (always less than 25 cells/mm2), but their nature could not be determined. Only 2 of 11 pleomorphic medium and large cell lymphomas (PMLs; cases 18 and 19) showed an important number of IL-10-positive cells. Hepatosplenic and nonhepatosplenic γδ T-cell lymphomas with monomorphic medium-sized or PML histology displayed IL-10 expression in only rare scattered cells.
Figure 1.
IL-10 mRNA expression detected by ISH in NK-cell lymphomas and in T-cell lymphomas of different histological subtypes. Results are shown as IL-10-positive cells/mm 2 of tissue sections.
Figure 2.

Nasal NK-cell lymphoma (case 44). Numerous cells strongly expressed IL-10 mRNA by ISH (A), with a similar pattern of latent membrane protein (LMP) expression by immunohistochemistry (alkaline phosphatase-anti-alkaline phosphatase technique; B).
Figure 3.

IL-10 mRNA expression in ALCL and the SU-DHL-1 cell line. Overlapping pattern in a sinusal area (case 32) was shown for IL-10 mRNA expression by ISH with a radiolabeled probe (A) and CD30 staining by immunohistochemistry (alkaline phosphatase-anti-alkaline phosphatase technique; B). C: IL-10 mRNA expression by ISH with a digoxigenin-labeled probe on paraffin sections in the same case demonstrates IL-10 mRNA staining detected in the cytoplasm of large anaplastic cells with kidney-shaped nuclei. D: IL-10 transcripts were detected in cells of the SU-DHL-1 cell line by ISH with a radiolabeled probe.
IL-10 transcripts were detected in cells of the SU-DHL-1 cell line (Figure 3D) ▶ . Lipopolysaccharide-stimulated peripheral blood mononuclear cells, used as positive control, showed a higher frequency of labeling of individual cells than unstimulated peripheral blood mononuclear cells (10% versus 1%).
ISH for EBV-encoded EBER 1 and 2 Transcripts
The presence of the EBV-encoded EBER 1 and 2 transcripts was determined by ISH on paraffin sections. Results are reported in Tables 1 to 3 ▶ ▶ ▶ . All NK-cell lymphomas gave a signal in most, if not all, tumor cells. Almost all cases of AILD were EBER 1 and 2 positive in some scattered cells (9 of 10 tested). EBV could be detected in some tumor cells in 3 of 11 PMLs but not in ALCLs. EBV was found in tumor cells of three nonhepatosplenic γδ T-cell lymphomas.
RT-PCR Analysis for Human and Viral IL-10
Total RNAs were extracted from frozen tissues of 12 cases of T- or NK-cell non-Hodgkin’s lymphoma (including 2 ALCLs, 2 AILDs, 3 PMLs, and 5 EBV-associated nasal NK-cell lymphomas). These cases were analyzed by RT-PCR to determine human or viral origin of the IL-10 detected by ISH (Figures 4 and 5) ▶ ▶ . In all lymphoma samples, a 141-bp S14-specific band of quite similar intensity was detected and used as a control of cDNA synthesis. The intensity of the 351-bp hIL-10 band was variable from case to case. Overall, there was a good correlation between RT-PCR results and the amount of IL-10 mRNA found by ISH despite some differences in band intensity from case to case (Figures 4 and 5 ▶ ▶ ; Tables 1 to 3 ▶ ▶ ▶ ). In addition, all analyzed samples, including the five nasal NK-cell non-Hodgkin’s lymphomas, which were strongly EBV positive, were negative for BCRF1/vIL-10 mRNA expression.
Figure 4.
RT-PCR analysis for the expression of hIL-10 and vIL-10/BCRF1 in NK-cell lymphomas (lanes 3 to 7). SU-DHL-1 and B95-8 cell lines were used as positive controls for hIL-10 and vIL-10/BCRF1, respectively (lanes 1 and 2). Omission of cDNA in all reverse transcription reactions was used as negative control. The S14 gene was used as a control for the quality and quantity of total RNAs.
Figure 5.
RT-PCR analysis for the expression of hIL-10 and vIL-10/BCRF1 in PTCLs: PML (lanes 3 to 5), AILD (lanes 6 and 7), and ALCL (lanes 8 and 9). The SU-DHL-1 and B95-8 cell lines were used as positive controls for hIL-10 and vIL-10/BCRF1, respectively (lanes 1 and 2). Omission of cDNA in all reverse transcription reactions was used as a negative control. The S14 gene was used as control for the quality and quantity of total RNAs.
Discussion
In the present study, we have investigated by ISH the expression of IL-10 transcripts in 46 lymphomas of T- or NK-cell origin. We have shown a strong association between IL-10 mRNA expression and ALCL. Indeed, in 8 of 11 cases (73%), many cells with the characteristic distribution of CD30 neoplastic cells displayed IL-10 mRNA expression. By contrast, among the remaining 28 cases of PTCL of various histotypes, only 2 cases (18%) of PMLs showed IL-10 mRNA expression in a proportion of cells, whereas only rare scattered cells were detected in cases of AILD and in the 6 γδ-PTCLs. We have also found a correlation between IL-10 mRNA expression and NK-cell lymphomas, because 3 of 7 (43%) of these tumors contained many IL-10-positive cells.
The finding that most systemic ALCLs expressed IL-10 mRNA is consistent with a recent report of IL-10 mRNA in cutaneous CD30+ lymphoproliferations. 26 Despite differences in their clinical and biological aspects, 3 these findings suggest a role for this cytokine in the pathogenesis of both cutaneous and systemic CD30+ ALCLs. Its possible implication in an autocrine pathway might be suggested because 1) the overlapping pattern between CD30 staining and IL-10 ISH results, neoplastic appearance of the IL-10-positive cells in the three cases studied on paraffin sections, and IL-10 production by the anaplastic lymphoma cell line SU-DHL-1 gave strong evidence that anaplastic cells were the major source of IL-10 production, and 2) human T cells have been shown to constitutively express the IL-10 receptor, although activation of T cells was associated with some decrease of the level of IL-10 receptor mRNA. 27 Results concerning the role of IL-10 in the T-cell growth depend on the studied model. Indeed, IL-10 was first described as a mouse T-cell growth factor in the presence of IL-2. 12 However, in human models, most in vitro studies have shown that IL-10 down-regulates mitogen or anti-CD3-induced T-cell proliferation in the presence or absence of accessory cells, via inhibition of endogenously produced IL-2. 28,29 The autocrine role of IL-10 might be supported by preliminary experiments showing a moderate but reproducible decrease of proliferation of SU-DHL-1 cell line in the presence of anti-IL-10 antibody (20%) using in vitro [3H]thymidine proliferation assays (data not shown). Besides, IL-10 has been implicated in inhibition of apoptosis of human T cells. 30 It could be hypothesized that IL-10 may rescue CD30+ ALCL cells from spontaneous apoptosis, thus allowing survival of tumor cells. Further support for an implication of IL-10 in the pathogenesis of CD30+ ALCLs can be provided by in vitro data concerning its involvement in the differentiation of mouse cytotoxic T cells. 14 This could be of particular interest because, in keeping with previous reports, 21,31-33 most cases of ALCL of this series expressed the cytotoxic proteins TIA-1 and/or granzyme B. It is noteworthy that a parallel was found between IL-10 expression and activated cytotoxic antigen profile.
IL-10 has also been shown to have pleiotropic immunosuppressive effects on T cells and macrophages, including inhibition of antigen presentation function and cytokine synthesis. 15,34 Tumor cells, melanoma, or B cell lines pretreated with IL-10 or transfected with IL-10 gene are protected from lysis by tumor-specific cytotoxic T cells. 35 These data suggest a role of IL-10 on the microenvironment resulting in the diminution of the immune surveillance and its contribution to lymphoma progression.
Taken together, our findings and those of the literature may suggest a role of IL-10 in the survival of tumor cells in CD30+ ALCL. In addition to the production of IL-6 and IL-9, as already reported, this indicates a T-helper-2 profile for anaplastic lymphomas. 6,7 Besides ALCL, only two cases of PML showed substantial levels of IL-10 mRNA. These two cases expressed the CD30 antigen in a proportion of cells (data not shown), but the relationship between IL-10 and CD30 expression needs further investigation. None of the cases of AILD showed significant levels of IL-10 mRNA. These lymphomas are characterized by tumor necrosis factor-α/lymphotoxin association 5 without IL-10 involvement. This finding in combination with the association between IL-6/IL-9/IL-10 and CD30+ ALCL 1,6,7 suggests histotype-dependent differences in the cytokine profile of PTCL. Furthermore, this particular cytokine profile reinforces the individualization of ALCL and AILD as distinct entities. In the present study, we have also investigated six cases of γδ PTCL, but IL-10 mRNA expression was found in only very rare scattered cells. Previous studies did not demonstrate IL-10 in normal γδ T cells, 36,37 and only in vitro γδ intraepithelial lymphocytes could be stimulated to express IL-10. 37 In our study, whatever the site, tumor γδ T cells were found to keep their basal IL-10 level.
In this series, three of seven NK-cell lymphomas were found to express IL-10 mRNA in a substantial proportion of tumor cells. By RT-PCR, we have demonstrated the human origin of IL-10, although all nasal NK-cell lymphomas were EBV positive. To the best of our knowledge, IL-10 production by NK cells was analyzed in only one study by enzyme-linked immunosorbent assay, and no hIL-10 was detected in resting or activated NK cells in vitro. 38 Although the direct effect of IL-10 on human NK cells has not yet been fully investigated, these cells are known to express abundant IL-10 receptor, suggesting responsiveness to IL-10. 38 Indeed, there is circumstantial evidence that IL-10 directly stimulates or potentiates NK-cell functions: hIL-10 can directly induce NK cytotoxic activity and increase IL-2-induced proliferation, cytotoxicity, and cytokine production. 16 Thus, it might be suggested that IL-10 synthesized by tumor NK cells could act in an autocrine manner on the proliferation of the tumor cells and/or on the increase of cytotoxic activities in view of the expression of cytotoxic proteins by NK-cell lymphomas. 21,33
In this study, no striking correlation was found between IL-10 expression and EBV detection irrespective of the histotype, the TCR expression, or the site of origin of the tumors. Only 4 of 22 EBV-positive T/NK-cell lymphomas, including 3 of 7 EBV-associated nasal NK-cell lymphomas, displayed significant hIL-10 expression in the absence of vIL-10/BCRF1, whereas IL-10-expressing ALCLs were EBV negative. This absence of clear correlation between IL-10 expression and presence of EBV differs from studies reported on acquired immunodeficiency syndrome-related B-cell lymphomas and Hodgkin’s disease, in which presence of EBV was associated with increased hIL-10 levels. 19,39
In summary, our findings show a frequent association between CD30+ ALCLs and hIL-10 mRNA expression by neoplastic cells of these tumors. IL-10 may rescue tumor cells from apoptosis and may be inhibitory to the generation of antitumor cytotoxic T-cell activities. In parallel with CD30+ ALCLs, a proportion of NK-cell lymphomas shows expression of cellular IL-10, suggesting an involvement of this cytokine in the pathogenesis of some NK-cell tumors. It is interesting that the level of IL-10 expression, ie, the number of IL-10-expressing cells, differs from case to case. In the present series, no clear association could be established between IL-10 expression and clinical features and outcome. However, clinical trials on larger series are needed to investigate the clinical and/or the prognostic significance of IL-10 expression, especially in ALCLs. In this respect, whatever the mechanism by which IL-10 could be involved in the pathogenesis of a proportion of CD30+ ALCLs and NK-cell lymphomas, the identification of IL-10 as a cytokine potentially involved in some T/NK-cell lymphomas could be useful for the development of new therapeutic strategies, as recently developed in B-cell lymphomas.
Acknowledgments
The authors thank the following pathologists: Drs. A-C. Baglin, M. Bernier, N. Brousse, A-M Chesneau, J-F. Emile, B. Fabiani, A. Lavergne, J. Quillard, and M. Wassef for providing samples; C. Haioun and L. Lacotteh for their contribution of clinical data; I. Joab for providing B95-8 cell line and IL-10 probe; and F. Conti for her technical assistance.
Footnotes
Address reprint requests to Dr. Philippe Gaulard, Département de Pathologie, Hôpital Henri Mondor, 94010 Créteil Cedex, France. E-mail: philippe.gaulard@hmn.ap-hop-paris.fr.
Supported by grants from Association pour la Recherche sur le Cancer and Comité des Deux-Sèvres de la Ligue contre le Cancer.
References
- 1.Hsu SM, Waldron JW, Hsu PL, Hough AJ: Cytokines in malignant lymphomas: review and perspective evaluation. Hum Pathol 1993, 24:1040-1057 [DOI] [PubMed] [Google Scholar]
- 2.Pistoia V: Production of cytokines by human B cell in health and disease. Immunol Today 1997, 18:343-350 [DOI] [PubMed] [Google Scholar]
- 3.Harris NL, Jaffe ES, Stein H, Banks PM, Chan JKC, Cleary ML, Delsol G, De Wolf-Peters C, Falini B, Gatter KC, Grogan TM, Isaacson PG, Knowles DM, Mason DY, Müller-Hermelink HK, Pileri SA, Piris MA, Ralfkiaer E, Warnke RA: A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood 1994, 84:1361-1392 [PubMed] [Google Scholar]
- 4.Gaulard P, Bourquelot P, Kanavaros P, Haioun C, Le Couedic JP, Divine M, Goossens M, Zafrani ES, Farcet JP, Reyes F: Expression of the α/β and γ/δ T-cell receptors in 57 cases of peripheral T-cell lymphomas: identification of a subset of γδ T-cell lymphomas. Am J Pathol 1990, 137:617-628 [PMC free article] [PubMed] [Google Scholar]
- 5.Foss HD, Anagnostopoulos I, Herbst H, Grebe M, Ziemann K, Hummel M, Stein H: Patterns of cytokine gene expression in peripheral T-cell lymphoma of angioimmunoblastic lymphadenopathy type. Blood 1995, 10:2862-2869 [PubMed] [Google Scholar]
- 6.Merz H, Houssiau FA, Orscheschek K, Renauld JC, Fliedner A, Herin M, Noel H, Kadin M, Mueller-Hermelink HK, Van Snick J, Feller AC: Interleukin-9 expression in human malignant lymphomas: unique association with Hodgkin’s disease and large cell anaplastic lymphoma. Blood 1991, 5:1311-1317 [PubMed] [Google Scholar]
- 7.Merz H, Fliedner A, Orscheschek K, Binder T, Sebald W, Müller-Hermelink HK, Feller AC: Cytokine expression in T-cell lymphomas and Hodgkin’s disease: its possible implication in autocrine or paracrine production as a potential basis for neoplastic growth. Am J Pathol 1991, 139:1173-1180 [PMC free article] [PubMed] [Google Scholar]
- 8.Fiorentino DF, Bond MW, Mosmann TF: Two types of mouse helper T-cells: Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. J Exp Med 1989, 170:2081-2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suda T, O’Garra A, MacNeil I, Fischer MD, Bond M, Zlotnik A: Identification of a novel thymocyte growth promoting factor from B-cell lymphomas. Cell Immunol 1990, 129:228-240 [DOI] [PubMed] [Google Scholar]
- 10.Howard M, O’Garra A, Ishida H, de Waal Malefyt R, de Vries J: Biological properties of interleukin-10. J Clin Immunol 1992, 12:239-247 [DOI] [PubMed] [Google Scholar]
- 11.Vieira P, De Waal-Malefyt R, Dang MN, Johnson KE, Kastelein R, Fiorentino DF, De Vries JE, Roncarolo MG, Mosmann TR, Moore KW: Isolation and expression of human cytokine synthesis inhibitory factor cDNA clones: homology to Epstein-Barr virus open reading frame BCRFI. Proc Natl Acad Sci USA 1991, 88:1172-1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacNeil IA, Suda T, Moore KW, Mosmann TR, Zlotnik A: IL-10, a novel growth cofactor for mature and immature T cells. J Immunol 1990, 145:4167-4173 [PubMed] [Google Scholar]
- 13.Rousset F, Garcia E, Defrance T, Péronne C, Vezzio N, Hsu DH, Kastelein R, Moore KW, Banchereau J: Interleukin 10 is a potent growth, and differentiation factor for activated human B lymphocytes. Proc Natl Acad Sci USA 1992, 89:1890-1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen WF, Zlotnik A: IL-10: a novel cytotoxic T cell differentiation factor. J Immunol 1991, 147:528-534 [PubMed] [Google Scholar]
- 15.de Waal Malefyt R, Abrams J, Bennett B, Figdor CG, de Vries JE: Interleukin 10 (IL-10) inhibits cytokine synthesis by human monocytes: an autoregulatory role of IL-10 produced by monocytes. J Exp Med 1991, 174:1209-1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warren HS, Kinnear BF, Kastelein RL, Lanier LL: Analysis of the costimulatory role of IL-2 and IL-15 in initiating proliferation of resting (CD56dim) human NK cells. J Immunol 1996, 156:3254-3259 [PubMed] [Google Scholar]
- 17.Benjamin D, Knobloch TJ, Dayton MA: Human B-cell interleukin-10: B-cell lines derived from patients with acquired immunodeficiency syndrome and Burkitt’s lymphoma constitutively secrete large quantities of interleukin-10. Blood 1992, 80:1289-1298 [PubMed] [Google Scholar]
- 18.Masood R, Zhang Y, Bond MW, Scadden DT, Moudgil T, Law RE, Kaplan MH, Jung B, Espina BM, Lunardi-Iskandar Y, Levine AM, Gill PS: Interleukin-10 is an autocrine growth factor for acquired immunodeficiency syndrome-related B-cell lymphoma. Blood 1995, 85:3423-3430 [PubMed] [Google Scholar]
- 19.Emilie D, Touitou R, Raphael M, Peuchmaur M, Devergne O, Rea D, Coumbraras J, Crevon MC, Edelman L, Joab I, Galanaud P: In vivo production of interleukin-10 by malignant cells in AIDS lymphomas. Eur J Immunol 1992, 22:2937-2942 [DOI] [PubMed] [Google Scholar]
- 20.Mori N, Gill PS, Moudgil T, Murakawi S, Eto S, Prager D: Interleukin-10 gene expression in adult T-cell leukemia. Blood 1996, 88:1035-1045 [PubMed] [Google Scholar]
- 21.Boulland ML, Kanavaros P, Wechsler J, Casiraghi O, Gaulard P: Cytotoxic protein expression in natural-killer cell lymphomas and in αβ and γδ peripheral T-cell lymphomas. J Pathol 1997, 183:432-439 [DOI] [PubMed] [Google Scholar]
- 22.Emile JF, Boulland ML, Haouin C, Kanavaros P, Petrella T, Delfau-Larue MH, Bensussan A, Farcet JP, Gaulard P: CD5-, CD56+ T-cell receptor silent peripheral T-cell lymphomas are natural killer cell lymphomas. Blood 1996, 87:1466-1473 [PubMed] [Google Scholar]
- 23.Stansfeld AG, Diebold J, Noel H, Kapanci Y, Rilke F, Kelenyi G, Sundstrom C, Lennert K, van Unnick J, Mioduszewska O, Wright D: Updated Kiel classification for lymphomas. Lancet 1988, 1:292-293 [DOI] [PubMed] [Google Scholar]
- 24.Lechner F, Vogt H-R, Seow H, Bertoni G, Cheevers WP, von Bodungen U, Zurbriggen A, Peterhans E: Expression of cytokine mRNA in lentivirus-induced arthritis. Am J Pathol 1997, 151:1053-1065 [PMC free article] [PubMed] [Google Scholar]
- 25.Kanavaros P, Lescs MC, Brière J, Divine M, Galateau F, Joab I, Bosq J, Farcet JP, Reyes F, Gaulard P: Nasal T-cell lymphoma: a clinicopathologic entity associated with peculiar phenotype and with Epstein-Barr virus. Blood 1993, 81:2688-2695 [PubMed] [Google Scholar]
- 26.Yagi H, Tokura Y, Furukawa F, Takigawa M: Th2 cytokine mRNA expression in primary cutaneous CD30-positive lymphoproliferative disorders: successful treatment with recombinant interferon-γ. J Invest Dermatol 1996, 107:827-832 [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Wei SHY, Ho ASY, de Waal Malefyt R, Moore KW: Expression, cloning, and characterization of a human IL-10 receptor. J Immunol 1994, 152:1821-1829 [PubMed] [Google Scholar]
- 28.Taga K, Tosato G: IL-10 inhibits human T cell proliferation and IL-2 production. J Immunol 1992, 148:1143-1148 [PubMed] [Google Scholar]
- 29.Taga K, Mostowski H, Tosato G: Human Interleukin-10 can directly inhibit T-cell growth. Blood 1993, 81:2964-2971 [PubMed] [Google Scholar]
- 30.Brunetti M, Martelli N, Colasante A, Piantelli M, Musiani P, Aiello F: Spontaneous and glucocorticoid-induced apoptosis in human mature T lymphocytes. Blood 1995, 86:4199-4205 [PubMed] [Google Scholar]
- 31.Foss HD, Anagnostopoulos I, Araujo I, Assaf C, Demel G, Kummer JA, Hummel M, Stein H: Anaplastic large-cell lymphomas of T-cell and null-cell phenotype express cytotoxic molecules. Blood 1996, 88:4005-4011 [PubMed] [Google Scholar]
- 32.Krenacs L, Wellmann A, Himmelmann AW, Bagdi E, Jaffe ES, Raffeld M: Cytotoxic cell antigen expression in anaplastic large cell lymphomas of T-and null-cell type and Hodgkin’s disease: evidence for distinct cellular origin. Blood 1997, 89:980-989 [PubMed] [Google Scholar]
- 33.Felgar RE, Macon WR, Kinney MC, Roberts S, Pasha T, Salhany KE: TIA-1 expression in lymphoid neoplasms: identification of subsets with cytotoxic T lymphocyte or natural killer cell differentiation. Am J Pathol 1997, 150:1893-1900 [PMC free article] [PubMed] [Google Scholar]
- 34.de Waal Malefyt R, Haanen J, Spits H, Roncarolo MG, te Velde A, Figdor CG, Johnson K, Kastelein R, Yssel H, de Vries JE: Interleukin 10 (IL-10), and viral IL-10 strongly reduce antigen-specific human T cell proliferation by diminishing the antigen-presenting capacity of monocytes via downregulation of class II major histocompatibility complex expression. J Exp Med 1991, 174:915-924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuda M, Salazar F, Petersson M, Masucci G, Hansson J, Pisa P, Zhang QJ, Masucci M, Kiessling R: Interleukin 10 pretreatment protects target cells from tumor-, and allo-specific cytotoxic T cells, and down regulates HLA class I expression. J Exp Med 1994, 180:2371-2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlaak JF, Hermann E, Gallati H: Meyer zum Büschenfelde K-H, Fleischer B: Differential effects of IL-10 on proliferation and cytokine production of human γ/δ and α/β T cells. Scand J Immunol 1994, 39:209-215 [DOI] [PubMed] [Google Scholar]
- 37.Lundqvist C, Melgar S, Mo-Wai Yeung M, Hammarström S, Hammarström M-L: Intraepithelial lymphocytes in human gut have lytic potential and a cytokine profile that suggest T helper 1 and cytotoxic functions. J Immunol 1996, 157:1926-1934 [PubMed] [Google Scholar]
- 38.Carson WE, Lindemann MJ, Baiocchi R, Linett M, Tan JC, Chou CC, Narula S, Caligiuri MA: The functional characterization of interleukin-10 receptor expression on human natural killer cells. Blood 1995, 85:3577-3585 [PubMed] [Google Scholar]
- 39.Herbst H, Foss HD, Samol J, Araujo I, Klotzbach H, Krause H, Agathanggelou A, Niedobhitek G, Stein H: Frequent expression of interleukin-10 by Epstein-Barr virus harboring tumor cells of Hodgkin’s disease. Blood 1996, 87:2918-2929 [PubMed] [Google Scholar]