Abstract
The Glutathione-S-Transferase (GST) “pulldown” assay has been used extensively to assay protein interactions in vitro. This methodology has been especially useful for investigating the interactions of nuclear hormone receptors with a wide variety of their interacting partners and coregulatory proteins. Unfortunately, the original GST-pulldown technique relies on multiple binding, washing and elution steps performed in individual microfuge tubes, and requires repeated centrifugation, aspiration, and suspension steps. This type of batch processing creates a significant liquid handling bottleneck, limiting the number of sample points that can be incorporated into one experiment and producing inherently less efficient washing and elution than would a flow-through methodology. In this manuscript, we describe the adaptation of this GST-pulldown assay to a 96-well filter plate format. The use of a multi-well filter plate makes it possible to assay more samples in significantly less time using less reagents and more efficient sample processing than does the traditional single tube assay.
Introduction
Nuclear receptors (also known as nuclear hormone receptors or intracellular receptors) are ligand-regulated transcription factors that play key roles in metazoan homeostasis, development, and reproduction [Apriletti et al., 1998; Beato and Klug, 2000; Chambon, 1996; Mangelsdorf et al., 1995; Zhang and Lazar, 2000]. To regulate their target genes, nuclear receptors must participate in an interwoven network of protein-protein interactions. For example, most nuclear receptors bind to their DNA recognition sites through a combination of DNA-protein and protein-protein interactions that generate receptor dimers; the nature and composition of these protein dimers define both the target gene specificity and the transcriptional properties of the participating receptors [Glass, 1996; Mangelsdorf and Evans, 1995]. Once assembled on DNA, the receptor dimers then must recruit additional auxiliary polypeptides, denoted corepressors and coactivators, to modulate expression of their target genes [Glass and Rosenfeld, 2000; Lazar, 2003; Lee et al., 2001; Leo and Chen, 2000; McKenna and O'Malley, 2002; Ordentlich et al., 2001; Xu and Li, 2003]. Corepressors and coactivators themselves assemble into multiple protein complexes to exert their functions, which can include further interactions with chromatin proteins and with components of the general transcriptional machinery. All levels of nuclear receptor function are subject to regulation by post-translational modifiers, such as kinases and allosteric modulators, which recognize their substrates through still additional protein-protein interactions (e.g. [Rochette-Egly, 2003; Stallcup et al., 2003; Weigel, 1996]).
A variety of methodologies have been applied to dissect this web of protein-protein interactions. One of the most broadly useful techniques has been a "pull-down" approach by which an immobilized "bait" protein is assayed for the ability to bind to and retain a second "prey" protein after incubation in vitro. The most common version of this approach uses a glutathione-S-transferase (GST) bait protein fusion, expressed in bacteria and immobilized on glutathione-agarose, together with a radiolabeled prey protein synthesized by in vitro transcription and translation; after co-incubation, washing, and elution, the recovered bait and prey proteins can be analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and quantified by autoradiography or phosphorimager analysis [Guan and Dixon, 1991]. This basic protocol can also be modified in a variety of ways. For example, other fusion tags, such as six-histidines or the maltose binding protein, can be used in place of GST to immobilize the bait protein to a corresponding insoluble matrix (e.g. [Bedouelle and Duplay, 1988; Chen and Hai, 1994; di Guan et al., 1988; Janknecht et al., 1991; Maina et al., 1988]). Bait protein fusions can be expressed in yeast, in baculovirus-infected insect cells, or in mammalian cell expression systems rather than in bacteria. The prey protein need not be radioactive, but alternatively can be detected by immunoblot, enzyme assay or ELISA methodologies.
Despite their general utility, standard GST-pulldown assays are performed in individual microcentrifuge tubes and require considerable sample handling involving repeated mixing, centrifugation, aspiration, and resuspension of each sample. This batch collection/resuspension approach reduces the efficiency of the wash steps, restricts the number of samples that can be expediently analyzed in any one experiment to the capacity of a microcentrifuge rotor, and introduces a considerable time delay that can interfere with detection of unstable protein-protein interactions. We describe here a modification of the GST-protocol to a 96-well filter plate format that substantially simplifies the technique, saves in reagent costs and investigator time, and enhances the efficiency of the process. This modification permits multiple protein-protein interaction assays to be rapidly and reproducibly performed in parallel, simplifying the generation of apparent affinity curves and the determination of the effects of modifiers and reagent conditions. This general methodology should also be readily adaptable to a variety of bait protein tags, prey protein detection techniques, and automation systems.
Reagents and instruments
COMPLETE protease inhibitor, (Boehringer-Mannheim/Roche, Indianapolis, IN); Glutathione-agarose, (Sigma Chemical Company, Saint Louis, MO); TNT T7 Quick Coupled Transcription/Translation System, (Promega, Madison, WI); 96-well MultiScreen-HV plate, (Millipore, Billerica, MA); 96 V-well polystyrene plate, (Corning, Acton, MA); TemPlate adhesive film, (USA Scientific, Ocala, FL); Branson sonifier 250, (Branson Corporation, Danbury, CT); 840 Phosphorimager Storm System, (Molecular Dynamics, Sunnyvale, CA); ImageQuant software, (Amersham Biosciences, Piscataway, NJ); Roto-Shake Genie, (Scientific Industries, Bohemia, NY); IEC Centra MP4 centrifuge with microplate rotor, (Thermo Electron, Waltham, MA); Reacti-Bind glutathione-coated plate, (Pierce Biotechnology, Rockford, IL)
Methods
Plasmids
A variety of GEX bacterial vectors have been described that can express a glutathione-S-transferase (GST) fusion of the bait protein of interest; for the experiments described here, we used pGEX-KG vectors to synthesize non-recombinant GST (employed as a negative control), a GST-SMRT corepressor fusion (encoding amino acids 2077-2471 of SMRTτ) or a GST-ACTR coactivator fusion (encoding amino acids 621-821of ACTR) [Chen et al., 1997; Goodson et al., 2005]. Similarly, a variety of vectors containing T7 or analogous phage promoters can be employed for the synthesis of radiolabeled prey protein in vitro; pSG5 vectors encoding full length retinoic acid receptor (RAR)α, thyroid hormone receptor (TR)α, or farnesoid X receptor (FXR) were employed here [Chan and Privalsky, 2006; Farboud and Privalsky, 2004; Forman et al., 1995].
Standard GST-pulldown assay
The glutathione-S-transferase (GST) fusion proteins are expressed in Escherichia coli (BL-21-strain) transformed by the corresponding pGEX vectors [Guan and Dixon, 1991]. Overnight cultures are inoculated 1:100 into LB broth (50 to 1000 ml) containing 100 µg/ml ampicillin and are grown with vigorous aeration at 37°C to an O. D. 600 nm = 0.8. Expression of the GST-fusion protein is induced by addition of 1 mM isopropylthiogalactopyranoside (IPTG) and the cultures are maintained at 37°C for an additional 3 hr. prior to harvesting by centrifugation at 4000 x g for 15 min.; this protocol was followed for the GST and GST-ACTR constructs described here. For some protein fusions (such as the GST-SMRT construct), greater solubility is achieved if the cultures are induced by IPTG addition and maintained at 16°C for 16 hr. prior to harvesting by centrifugation.
All subsequent steps are either on ice or at 4°C. The bacteria are washed twice in phosphate-buffered saline (PBS), resuspended in 5 ml of PBS containing COMPLETE protease inhibitor (Boehringer-Mannheim/Roche, Indianapolis, IN), and are lysed by three 30 sec bursts in a Branson sonicator with 1/2 inch horn at the #9 setting. The lysates are then clarified by a 20 min. centrifugation at 30,000 x g and either used immediately, or quick frozen as aliquots and stored at −80°C for future use. Glutathione-agarose (Sigma Chemical Company, Saint Louis, MO) is prepared by swelling for 10 min. in PBS (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.45 mM KH2PO4), then collected by a 30 sec. centrifugation at 1,000 x g, washed twice in 20 volumes each of PBS, and suspended as a 50% slurry in PBS. For each binding reaction 50 µl of washed glutathione agarose slurry is mixed with the desired amount of bacterial lysate (containing from 50 to 1000 ng of immobilized GST fusion protein) in 500 µl of PBS + COMPLETE protease inhibitor + 1mM DTT. After a 1 hr. incubation on a rocker platform at 4°C, the GST-fusion bait protein, now immobilized on the glutathione-agarose, is recovered by a 30 sec. centrifugation at 1,000 x g, washed twice with PBS containing 0.5% Triton X-100 and 1mM DTT, and resuspended as a 50% slurry in Buffer A (25mM HEPES, pH 7.8; 50mM KCl; 5mM EDTA; 5mM MgCl2; 0.5% Triton X-100; 6% glycerol; 1mM dithiothreitol) for binding to the radiolabeled prey protein, below.
35S-radiolabeled proteins are prepared in vitro by use of a phage RNA polymerase-coupled transcription/translation kit following the manufacturer’s recommendations (e.g. TNT T7 Quick Coupled Transcription/Translation System, Promega, Madison, WI). For the traditional tube-based assay, the 35S-labeled proteins are transferred into 1.5 ml microfuge tubes, 500 µl of Buffer A containing COMPLETE proteinase inhibitor, and 5 mg/ml of bovine serum albumin are added, and the incubation is begun by addition of 50 µl/per tube of a 50% slurry of the appropriate immobilized GST fusion protein. The bovine serum albumin can be omitted in experiments utilizing hormones that may be adsorbed by this protein carrier. The microfuge tubes are capped and incubated on a rocker platform for 30 min. at 4°C. The immobilized GST-protein, and any prey protein bound to it, are then recovered by a 10 sec. centrifugation (1,000 x g). The supernatants are aspirated, the agarose matrix is resuspended in 1 ml/tube of PBS containing 0.5% Triton X-100 and 1mM DTT by a 5 sec. inversion mixing, and the centrifugation step is repeated. The wash steps are repeated 3 times, and proteins remaining bound to the glutathione-agarose matrix are eluted by incubation for 30 min. at 4°C in 50 µl/tube of 20 mM free glutathione in 100 mM Tris-Cl pH 8.0. The matrix is pelleted by a final centrifugation and the eluated proteins are resolved by SDS-PAGE and visualized/quantified by phosphorimager analysis (Molecular Dynamics Storm System, Sunnyvale, CA).
Improved 96 well filter plate format
The immobilized GST-fusion bait proteins and radiolabeled prey proteins are prepared as for the standard procedure, above. Four to five µl of glutathione agarose (containing from 50 to 1000 ng of immobilized GST fusion protein) plus 100 µl of Buffer A (containing 5 mg/ml bovine serum albumin and COMPLETE protease inhibitor) are aliquoted per well of a MultiScreen-HV plate (Millipore Billerica, MA). Aliquots of the in vitro translated prey protein (4 µl unless otherwise stated) representing 4 x 105 to 4 x 106 dpm 35S radiolabel are added per well, the filter plate is placed on top of an empty 96-well polystyrene plate (Corning, Acton, MA), and the top filter plate is sealed with TemPlate adhesive film (USA Scientific, Ocala, FL). Although not required, we also typically heat-seal the dual plate sandwich in a polyethylene bag to provide redundant radiological containment. The prey and GST-bait are then incubated together on an inverting platform at 5 to 8 rpm for 30 min. at 4°C; we typically use magnetic strips to attach the bag and plate sandwich to a rotating metal platform such as the Roto-Shake Genie (Scientific Industries, Bohemia, NY) or similar apparatus. The plates are subsequently unsealed and the prey proteins not bound to the GST-fusion are removed by a 1,000 x g centrifugation for 60 seconds at 25°C in an IEC Centra MP4 centrifuge with a microplate rotor (Thermo Electron, Waltham, MA). Plates are then washed three times by adding 200 µl of PBS containing 0.5% Triton X-100 and 1mM DTT per well with a multi-channel pipetter and centrifuging as above. Bound proteins are eluted by placing the filter plate on top of a fresh 96 well V-bottom plate, adding 50 µl of elution buffer (20 mM glutathione, 100 mM Tris-HCl pH 8.0) to each well, sealing the plate sandwich as before, and incubating it for an additional 30 min. at 4°C with continuous rotation. Protein complexes eluted by the 20 mM glutathione are then collected by centrifuging the samples into the 96-well bottom plate, and are prepared for electrophoresis by adding 20 µl of 4x SDS-PAGE sample buffer (125 mM Tris-HCl, pH 6.8, 20% Glycerol, 4% SDS, 1.4 M ß-mercaptoethanol, 0.5 mg/ml bromophenol blue) to each well. The samples are heated at 95°C on a dry block for 5 min. and analyzed by SDS-PAGE. The recovery of GST-bait proteins can be visualized by Coomassie blue staining and the recovery of prey proteins can be determined and quantified by phosphorimager analysis (Storm840 and ImageQuant software, Amersham Biosciences, Piscataway, NJ).
Surface immobilized plate format
Saturating amounts (equivalent to the amounts used for the corresponding filter plate assay) of GST or GST-fusion protein extract were bound to Reacti-Bind glutathione coated plates (Pierce Biotechnology, Rockford, IL). After a 30 min. incubation on a rocker platform at 4°C, the GST-fusion bait proteins, now immobilized on the glutathione coated plate, are washed twice with PBS containing 0.5% Triton X-100 and 1mM DTT. The in vitro translated prey proteins are prepared and bound as previously described for the filter microplate assay. After a 30 min. incubation on a rocker platform at 4°C, the prey proteins not bound to the GST-fusion are removed by aspiration. Plates are then washed three times by adding 200 µl of PBS containing 0.5% Triton X-100 and 1mM DTT per well with a multi-channel pipetter and aspirating as above. Bound proteins are eluted by adding 50 µl of elution buffer (20 mM glutathione, 100 mM Tris-HCl pH 8.0) to each well, and incubating for an additional 30 min. at 4°C with continuous agitation. Eluted protein complexes are removed from the Reacti-bind coated plates and analyzed as previously described for the filter microplate assay.
Results
Comparison of the 96-well plate and the individual tube GST-pulldown assays
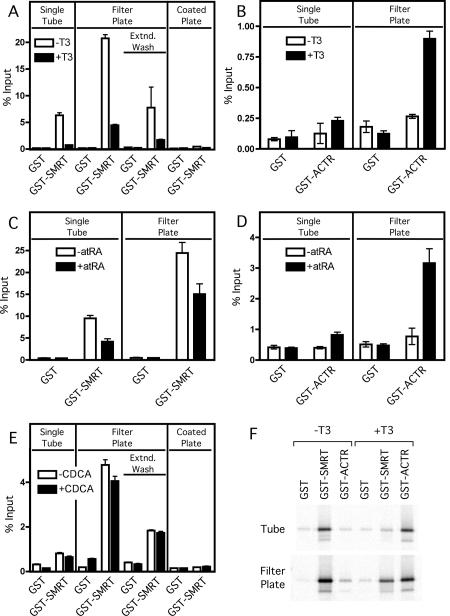
We compared the 96-well plate and the individual tube assay by examining the ability of two different nuclear receptors, thyroid hormone receptor α (TRα) and retinoic acid receptor α (RARα), to bind to two coregulatory proteins, the SMRT corepressor and the ACTR coactivator. We synthesized the radiolabeled receptors and GST-coregulators, performed the GST-pulldowns, and analyzed the eluted proteins by SDS-PAGE/phosphorimager analysis as described in Materials and Methods; the average and standard deviation of triplicate experiments are presented (Figure 1). Nuclear receptors are known to bind to corepressors in the absence of hormone agonist, but release from corepressors and bind coactivators in the presence of hormone agonist. The results of both methods paralleled these expectations: both TRα and RARα bound to the GST-SMRT corepressor construct in the absence of hormone, but this binding was significantly reduced in the presence of cognate hormone agonist (T3 or all-trans retinoic acid, respectively) (Figure 1A and Figure 1C). Reciprocally, binding of either receptor to the GST-ACTR coactivator construct was minimal in the absence of hormone, but was significantly increased in the presence of the cognate agonist (Figure 1B and Figure 1D). Both tube and plate based assays were internally reproducible, and both yielded very low background binding to the GST negative control construct (Figure 1). Representative Phosphorimager gel images showing the interaction of TRα with GST, GST-SMRT and GST-ACTR in the presence and absence of hormone for both the individual tube assay and filter microplate assay are presented in Figure 1F.
Figure 1. Comparison of GST-pulldown methodologies: microfuge tube versus filter-plate protocols.
Non-recombinant GST, GST-SMRTτ (amino acids 2077-2471; A, C, E and F), or GST-ACTR (amino acids 621-821; B, D, and F) bound to glutathione agarose were incubated with in vitro translated 35S-methionine radiolabeled TRα1 (A, B and F), RARα (C and D), or FXR (E) protein in either the absence (open bars) or presence (filled bars) of cognate agonist (1 µM T3 for TRα1, 1µM ATRA for RARα or 100 µM CDCA for FXR). Samples were bound, washed, and eluted with free reduced glutathione using the conventional individual tube assay (“Single Tube”), the modified filter microplate assay (“Filter Plate”), or the glutathione coated Reacti-Bind plate assay (“Coated Plate”). Extended wash filter microplate samples (“Extnd. Wash”) were incubated 15 min between washes. Samples were resolved on a SDS-10% PAGE gel prior to fixing, staining and scanning of the dried gel with a Molecular Dynamics Storm 840 Phosphorimager. Quantification of the radiolabeled bands was performed using ImageQuant software version 4.2. Assays were performed in triplicate; error bars indicate standard deviation. (F) Representative Phosphorimager gel images used for analysis in panels A and B.
Notably, the plate assay was consistently more efficient than was the tube assay under these conditions, displaying a higher percent of the input receptor binding to the GST-SMRT and GST-ACTR constructs than was observed with the tube-based technique. To determine if this increased interaction in the microplate was due to the significantly reduced processing time involved with the wash steps of the filter microplate assay, we added a 15 minute incubation to each of the wash steps to simulate the extended wash times of the individual tube assays (Figure 1 A, “Extnd. Wash”). Extending the wash times reduced the efficiency of the filter microplate binding reaction to nearly that observed for the individual tube assay. Because of the increased efficiency of the filter microplate assay, we wanted to determine if this assay would be suitable for analyzing weaker protein interactions, such as the interaction between the farnesoid X receptor (FXR) and the corepressor SMRT (Figure 1E). In the individual tube assay, the interaction between FXR and SMRT is only marginally above background; whereas in the filter microplate interaction, there is a readily observable interaction (Figure 1E). This interaction is slightly, but reproducibly, diminished (p<0.05) in the presence of the FXR ligand, chenodeoxycholic acid (CDCA) [Makishima et al., 1999; Parks et al., 1999]. As with TRα, the amount of FXR bound to SMRT in the filter plate assay is reduced with extended wash times, though not to the extent of the single tube assay (Figure 1E). These results suggest that the 96-well filter plate approach is equal or better in utility to the traditional individual tube method for high affinity interactions, such as between TR or RAR and their coregulatory partners. Unlike the individual tube assay, the filter microplate assay is also suitable for analyzing lower affinity interactions, such as between FXR and SMRT or other cofactors.
A different GST pulldown assay has been described using a specialized microplate with a glutathione coated surface to assay antibody-antigen interactions [Murray et al., 1998]. To compare our filter microplate assay to the coated microplate assay, we incubated identical amounts of GST-fusion bait protein (approximately 1000 ng) with glutathione agarose and the Reacti-Bind plate. This amount of protein was saturating for the coated plates, which have a reported maximum GST binding capacity of 10 ng/well (as opposed to 5 µg/µl for the glutathione agarose resin). Not unexpectedly, only a slight interaction is observed between TRα and SMRT using the coated microplates (Figure 1A) and no interaction is observed between FXR and SMRT (Figure 1E). Thus the lower binding capacity of the coated microplate appears to restrict its suitability to only very high affinity interactions.
Further characterization of the 96-well filter plate assay
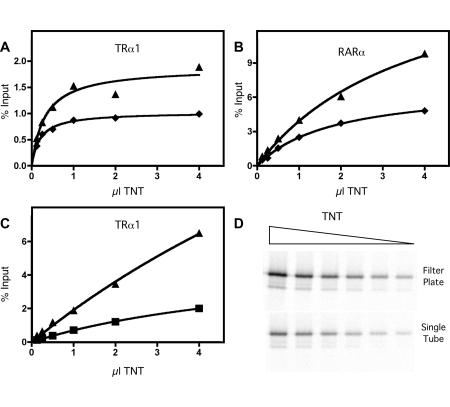
GST-pulldown methods are inherently non-equilibrium systems, and, when used with in vitro transcription/translation mixtures, contain additional proteins and other components that can potentially influence the results. Nonetheless, a useful GST-pulldown assay should mimic many of the characteristics anticipated from a Michaelis-Menten description and display well-behaved saturable binding that adequately reflects the avidity of one protein for the other. We tested this by repeating the GST-pulldown filter plate assay using a wide range of TRα or RARα proteins and two different concentrations of GST-ACTR construct (Figure 2). As hoped, both nuclear receptors displayed binding to the GST-coactivator that fit theoretic expectations for a two-component protein-protein interaction (Figure 2 and Table 1). These results confirm that the GST-pulldown microplate method is well-behaved and is a useful measure of the protein-protein interaction itself. Similar experimentally well-behaved interactions are observed between TRα and SMRT in both the single tube and filter microplate assays (Figure 2C and Figure 2D), with the overall interaction being reduced in the individual tube assays, as before.
Figure 2. Titration of the binding of regulators to nuclear receptors by the plate or single tube methods.
GST-ACTR (amino acids 621-821), either 400 ng (triangles) or 200 ng (diamonds), bound to glutathione agarose was incubated with increasing amounts of in vitro translated 35S-methionine radiolabeled TRα1 (A) or RARα (B) in the presence of cognate agonist (1 µM T3 for TRα or 1 µM ATRA for RARα). GST-SMRTτ (amino acids 2077-2471) bound to glutathione agarose was incubated with increasing amounts of in vitro translated 35S-methionine radiolabeled TRα1 (C) in the absence of cognate agonist. Samples were bound, washed, and eluted with free reduced glutathione in either the conventional individual tube assay (squares) or the modified filter microplate assay (triangles) and were analyzed as in Figure 1. Data was fit to a single-site hyperbolic binding curve using GraphPad Prism v. 4.0. (D) The Phosphorimager gel images used for analysis in panel C.
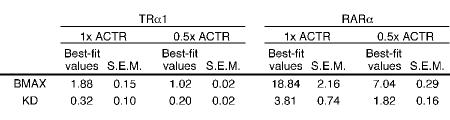
Table 1. Regression analysis of plate assay.
Best fit values and standard errors from the non-linear regression analysis of the quantification of the interaction of GST-ACTR with varying amount of in vitro translated TRα1 and RARα protein (Figure 2). The data were fit to the equation Y=Bmax*X/(Kd+X) with the constraints of Bmax<100 and Kd>0. The analysis was done using GraphPad Prism v. 4.0.
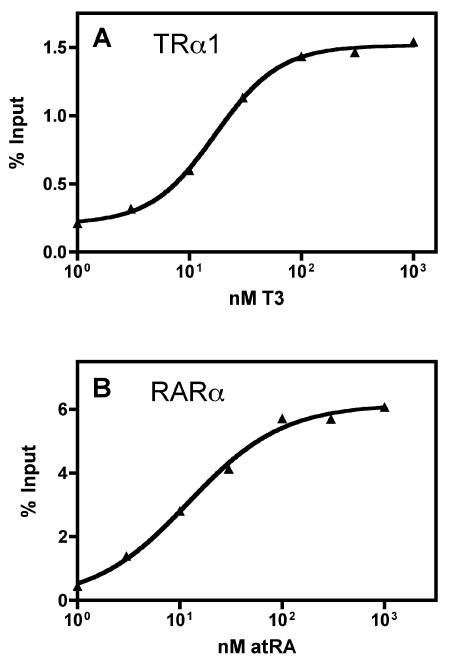
We next determined if the GST filter plate methodology could also quantify the effects of hormone ligands on the interactions of nuclear receptors with their coregulators. We tested the ability of a range of hormone agonist concentrations (T3 or all-trans retinoic acid) to induce binding of TRα or RARα to the GST-ACTR coactivator (Figure 3). These titrations produced sigmoidal curve fits fully consistent with prior, tube-based GST-pulldown assays (Figure 3). Reciprocal results were observed when testing the hormone-induced release of these receptors from the GST-SMRT corepressor construct (data not shown). We conclude that the GST-pulldown filter plate method is sufficient to allow a quantitative analysis of these phenomena.
Figure 3. Analysis of the effects of hormone agonists on the receptor/coactivator interaction using the plate method.
GST-ACTR (amino acids 621-821) bound to glutathione agarose was incubated with in vitro translated 35S-methionine radiolabeled TRα1 (A) or RARα (B) protein in presence of increasing amounts of cognate agonist. Samples were analyzed using the filter plate method as in Figure 1. Data was fit to a sigmoidal dose response curve with variable slope using GraphPad Prism v. 4.0.
Comparison of centrifugation verses vacuum manifold washing methods
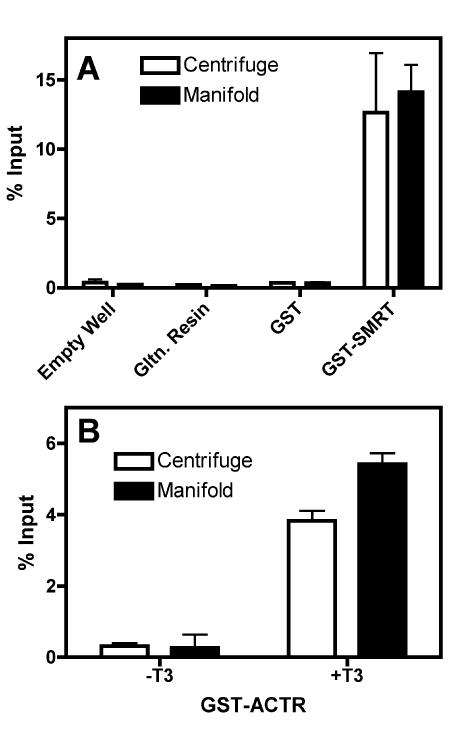
The assays detailed above employed a tabletop centrifuge equipped with microplate holders to expel each wash into a collecting tray. Our setup permits up to four 96-well filter plates (384 samples) to be analyzed in parallel; other centrifuges and configurations potentially further double these quantities. Alternatively, any of a number of commercially-available or custom-constructed manifolds can be employed to wash the plate wells under vacuum. We therefore compared these two wash methods in parallel, testing the ability of TRα to bind to GST-SMRT corepressor in the absence or presence of T3 hormone (Figure 4). Notably, the filter plate centrifugation and filter plate vacuum manifold assays yielded virtually indistinguishable results in terms of background, binding efficiency, and reproducibility. These results further extend the general applicability of this microplate methodology.
Figure 4. Comparison of centrifuge washing versus vacuum manifold washing for plate methodology.
(A) GST-SMRTτ (amino acids 2077-2471) bound to glutathione agarose was incubated with in vitro translated 35S-methionine radiolabeled TRα1 protein; empty wells, glutathione agarose alone, or glutathione agarose bound to GST were used as negative controls as indicated. Samples were washed using either a centrifuge (open bars) or vacuum manifold (filled bars), eluted and analyzed as described in Figure 1. (B) GST-ACTR (amino acids 621-821) bound to glutathione agarose was incubated with in vitro translated 35S-methionine radiolabeled TRα1 protein in either the absence or presence of cognate agonist (1 µM T3). Samples were washed using either a centrifuge (open bars) or vacuum manifold (filled bars), eluted, and analyzed using the filter plate method as described in Figure 1. Assays were performed in triplicate; error bars indicate standard deviation.
Comparison of SDS-PAGE analysis versus scintillation counting for quantification
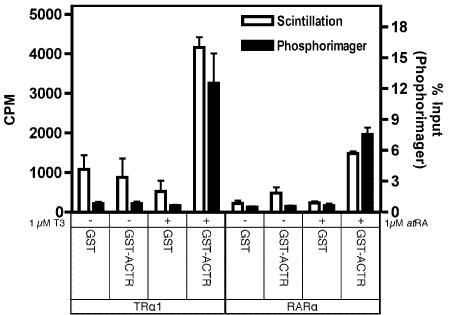
Typically, prey proteins bound and eluted from the GST-bait are then analyzed by SDS-PAGE. This permits a specific prey protein of interest to be resolved and quantified independent of any other radiolabeled translation product, and also allows the integrity and quantity of the GST-bait proteins to be determined by Coomassie blue staining of the same electrophoretogram as that used for the radioactive quantification. Notably, however, the in vitro transcription/translation system often produces a relatively pure radiolabeled prey protein with virtually no other detectable translation products; under this circumstance, SDS-PAGE analysis after the GST-pulldown might prove unnecessary. We tested this concept by analyzing the interaction of TRα with GST-SMRT using the 96-well filter plate assay, but quantifying each sample either by SDS-PAGE/phosphorimager analysis, or by subjecting the entire protein eluate to aqueous liquid scintillation counting in the absence of any further fractionation (Figure 5). No significant differences were detected between these analyses (Figure 5). We conclude that (a) when using radiologically homogeneous prey protein preparations and (b) for sufficiently robust protein-protein interactions, the filter plate GST-pulldown approach can be further simplified, and the analysis considerably accelerated, by direct scintillation analysis of the radioactive eluates without a subsequent SDS-PAGE step.
Figure 5. Comparison of plate assay quantification by SDS-PAGE/phosphorimager analysis versus analysis by liquid scintillation counting.
Non-recombinant GST or GST-ACTR (amino acids 621-821) were bound to glutathione agarose, and were incubated with in vitro translated 35S-methionine radiolabeled TRα1 or RARα protein in presence of cognate agonist (1 µM T3 or 1 µM ATRA). Samples were washed and eluted using the filter plate method as described in Figure 1. After elution, a 20 µl aliquot (out of 70 µl total) of each sample was resolved on a SDS-10% PAGE gel prior to Coomassie staining and visualization of the dried gel with a Molecular Dynamics Storm 840 Phosphorimager. A separate 5 µl aliquot of each sample was subjected to scintillation counting using BioSafe II scintillation cocktail (Research Products International, Mt. Prospect, IL) and a Beckman LS6500 liquid scintillation counter (Beckman Coulter, Fullerton, CA). Assays were performed in triplicate; error bars indicate standard deviation.
Discussion
We have adapted the existing GST-pulldown protein-protein interaction assay, traditionally performed in individual microfuge tubes, to a high-throughput 96-well filter plate format. As demonstrated here, the 96-well format displays low background, good reproducibility, and strong positive signals, and is comparable or better to the individual tube method by these criteria. The 96-well format produces good fits to expected two-component binding curves in response to varying receptor, coregulator, or hormone ligand concentrations. The speed of the filter plate method also favors the detection of relatively weak or short half-life interactions which may not be sufficiently stable to detect over the longer time course required for the single tube methodology.
The relative ease of analyzing multiple samples in parallel makes the 96-well filter plate format particularly well suited to high throughput analysis of large numbers of different receptors or their coregulators, different mutant constructs, or for detailed titrations examining the effect of hormone or receptor concentration on a protein-protein interaction. Of equal importance: the 96-well modification of the GST-pulldown method reduces the manual intervention time required for pipetting during the wash and elution steps from approximately 6 minutes per wash (assuming 20 samples) to approximately twenty seconds per wash (assuming 96 samples); thus the microplate assay represents a more than 10-fold reduction in investigator time. Using a simple tabletop centrifuge for the wash steps, 384 samples or more can readily be manually analyzed in parallel; alternatively, the same 96-well format can be utilized with a vacuum manifold to further speed the washing steps, or can be adapted to robotics work stations to further minimize investigator intervention. The micro-plates are readily stacked, manipulated, and stored, and unlike the traditional tube method, do not require the additional effort of uniquely labeling each individual sample tube.
In addition to the reductions in investigator time and effort, there are also reagent savings. A minimum of 25 µl per sample of glutathione agarose is needed when using the individual microfuge tube format to form a visible pellet and to avoid significant loss of resin during the repeated manual washing steps; in contrast, the membrane-like surface of the 96-well filter plates permits as little as 5 µl of glutathione-agarose to be used per sample. The cost of the glutathione agarose (and the cost of the protease inhibitor in the reduced volume of binding buffer used in the plate method) are therefore approximately 20% of the cost for the individual tube assay ($0.15/sample versus $0.75/sample). With the filter microplate assay there is a fixed cost of approximately $15.00 for each microplate and seal, resulting in the filter plate method being more economical for assays representing 25 samples or greater. Depending on the nature of the interaction, it may also be possible to further reduce the expense per assay by reducing the amount of in vitro translated protein used, although potentially with the risk of reduced sensitivity (Figure 2).
The 96-well filter plate assay system described here can also be modified to expand its general applicability. A variety of different agarose matrices are available that can be substituted in place of the glutathione-agarose employed here, permitting a range of bait protein fusion tags to be employed in the 96-well plate format (e.g. maltose binding protein, chitin binding protein, biotin, or 6-histidine tags). Alternatively, approaches other than radioisotopic labeling can be used to detect the prey protein. For example, a non-radioactive prey protein, either isolated from its native context or artificially generated in an over-expression system, can be used in the GST-pulldown; once eluted, the prey protein can be analyzed by SDS-PAGE and quantitative immunoblotting, or by immunological assay of the total bound fraction using a microplate formatted assay.
Acknowledgments
The authors thank Liming Liu for superb technical assistance. This work was supported by Public Health Service/National Institutes of Health award DK53528. B.F. was funded, in part, by a predoctoral training award, T32GM07377, from the National Institute of General Medical Sciences. M.L.G. was supported by a Ruth L. Kirschstein National Research Service Award, 5F32DK062654.
Abbreviations
- GST
glutathione-S-transferase
- TRα
thyroid hormone receptor-α
- RARα
retinoic acid receptor-α
- T3
3,3′,5-triiodothyronine
- ATRA
all-trans retinoic acid
- EDTA
ethylenediamine tetraacetic acid
- PAGE
polyacrylamide gel electrophoresis
- SDS
sodium dodecyl sulphate
- SMRT
silencing mediator for retinoid and thyroid hormone receptors
- ACTR
activator for thyroid hormone and retinoid receptors
References
- Apriletti J. W., Ribeiro R. C., Wagner R. L., Feng W., Webb P., Kushner P. J., West B. L., Nilsson S., Scanlan T. S., Fletterick R. J., Baxter J. D. Molecular and structural biology of thyroid hormone receptors. Clin Exp Pharmacol Physiol Suppl. 1998;25:S2–11. doi: 10.1111/j.1440-1681.1998.tb02293.x. [DOI] [PubMed] [Google Scholar]
- Beato M., Klug J. Steroid hormone receptors: an update. Hum Reprod Update. 2000;6:225–36. doi: 10.1093/humupd/6.3.225. [DOI] [PubMed] [Google Scholar]
- Bedouelle H., Duplay P. Production in Escherichia coli and one-step purification of bifunctional hybrid proteins which bind maltose. Export of the Klenow polymerase into the periplasmic space. Eur J Biochem. 1988;171:541–9. doi: 10.1111/j.1432-1033.1988.tb13823.x. [DOI] [PubMed] [Google Scholar]
- Chambon P. A decade of molecular biology of retinoic acid receptors. Faseb J. 1996;10:940–54. [PubMed] [Google Scholar]
- Chan I. H., Privalsky M. L. Thyroid hormone receptors mutated in liver cancer function as distorted antimorphs. Oncogene. 2006;25:3576–88. doi: 10.1038/sj.onc.1209389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. P., Hai T. Expression vectors for affinity purification and radiolabeling of proteins using Escherichia coli as host. Gene. 1994;139:73–5. doi: 10.1016/0378-1119(94)90525-8. [DOI] [PubMed] [Google Scholar]
- Chen H., Lin R. J., Schiltz R. L., Chakravarti D., Nash A., Nagy L., Privalsky M. L., Nakatani Y., Evans R. M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–80. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- di Guan C., Li P., Riggs P. D., Inouye H. Vectors that facilitate the expression and purification of foreign peptides in Escherichia coli by fusion to maltose-binding protein. Gene. 1988;67:21–30. doi: 10.1016/0378-1119(88)90004-2. [DOI] [PubMed] [Google Scholar]
- Farboud B., Privalsky M. L. Retinoic acid receptor-α is stabilized in a repressive state by its C-terminal, isotype-specific F domain. Mol Endocrinol. 2004;18:2839–53. doi: 10.1210/me.2004-0236. [DOI] [PubMed] [Google Scholar]
- Forman B. M., Goode E., Chen J., Oro A. E., Bradley D. J., Perlmann T., Noonan D. J., Burka L. T., McMorris T., Lamph W. W., Evans R. M., Weinberger C. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 1995;81:687–93. doi: 10.1016/0092-8674(95)90530-8. [DOI] [PubMed] [Google Scholar]
- Glass C. K. Some new twists in the regulation of gene expression by thyroid hormone and retinoic acid receptors. J Endocrinol. 1996;150:349–57. doi: 10.1677/joe.0.1500349. [DOI] [PubMed] [Google Scholar]
- Glass C. K., Rosenfeld M. G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–41. [PubMed] [Google Scholar]
- Goodson M. L., Jonas B. A., Privalsky M. L. Alternative mRNA splicing of SMRT creates functional diversity by generating corepressor isoforms with different affinities for different nuclear receptors. J Biol Chem. 2005;280:7493–503. doi: 10.1074/jbc.M411514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K. L., Dixon J. E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–7. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Janknecht R., de Martynoff G., Lou J., Hipskind R. A., Nordheim A., Stunnenberg H. G. Rapid and efficient purification of native histidine-tagged protein expressed by recombinant vaccinia virus. Proc Natl Acad Sci U S A. 1991;88:8972–6. doi: 10.1073/pnas.88.20.8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar M. A. Nuclear receptor corepressors. Nucl Recept Signal. 2003;1:e001. doi: 10.1621/nrs.01001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. W., Lee Y. C., Na S. Y., Jung D. J., Lee S. K. Transcriptional coregulators of the nuclear receptor superfamily: coactivators and corepressors. Cell Mol Life Sci. 2001;58:289–97. doi: 10.1007/PL00000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leo C., Chen J. D. The SRC family of nuclear receptor coactivators. Gene. 2000;245:1–11. doi: 10.1016/s0378-1119(00)00024-x. [DOI] [PubMed] [Google Scholar]
- Maina C. V., Riggs P. D., Grandea A. G., 3rd, Slatko B. E., Moran L. S., Tagliamonte J. A., McReynolds L. A., Guan C. D. An Escherichia coli vector to express and purify foreign proteins by fusion to and separation from maltose-binding protein. Gene. 1988;74:365–73. doi: 10.1016/0378-1119(88)90170-9. [DOI] [PubMed] [Google Scholar]
- Makishima M., Okamoto A. Y., Repa J. J., Tu H., Learned R. M., Luk A., Hull M. V., Lustig K. D., Mangelsdorf D. J., Shan B. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–5. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schutz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. The nuclear receptor superfamily: the second decade. Cell. 1995a;83:835–9. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf D. J., Evans R. M. The RXR heterodimers and orphan receptors. Cell. 1995b;83:841–50. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- McKenna N. J., O'Malley B. W. Minireview: nuclear receptor coactivators--an update. Endocrinology. 2002;143:2461–5. doi: 10.1210/endo.143.7.8892. [DOI] [PubMed] [Google Scholar]
- Murray A. M., Kelly C. D., Nussey S. S., Johnstone A. P. Production of glutathione-coated microtitre plates for capturing recombinant glutathione S-transferase fusion proteins as antigens in immunoassays. J Immunol Methods. 1998;218:133–9. doi: 10.1016/s0022-1759(98)00114-8. [DOI] [PubMed] [Google Scholar]
- Ordentlich P., Downes M., Evans R. M. Corepressors and nuclear hormone receptor function. Curr Top Microbiol Immunol. 2001;254:101–16. doi: 10.1007/978-3-662-10595-5_5. [DOI] [PubMed] [Google Scholar]
- Parks D. J., Blanchard S. G., Bledsoe R. K., Chandra G., Consler T. G., Kliewer S. A., Stimmel J. B., Willson T. M., Zavacki A. M., Moore D. D., Lehmann J. M. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–8. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- Rochette-Egly C. Nuclear receptors: integration of multiple signalling pathways through phosphorylation. Cell Signal. 2003;15:355–66. doi: 10.1016/s0898-6568(02)00115-8. [DOI] [PubMed] [Google Scholar]
- Stallcup M. R., Kim J. H., Teyssier C., Lee Y. H., Ma H., Chen D. The roles of protein-protein interactions and protein methylation in transcriptional activation by nuclear receptors and their coactivators. J Steroid Biochem Mol Biol. 2003;85:139–45. doi: 10.1016/s0960-0760(03)00222-x. [DOI] [PubMed] [Google Scholar]
- Weigel N. L. Steroid hormone receptors and their regulation by phosphorylation. Biochem J. 1996;319 ( Pt 3):657–67. doi: 10.1042/bj3190657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Li Q. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol Endocrinol. 2003;17:1681–92. doi: 10.1210/me.2003-0116. [DOI] [PubMed] [Google Scholar]
- Zhang J., Lazar M. A. The mechanism of action of thyroid hormones. Annu Rev Physiol. 2000;62:439–66. doi: 10.1146/annurev.physiol.62.1.439. [DOI] [PubMed] [Google Scholar]