Abstract
Steroid hormones are a diverse class of structurally related molecules, derived from cholesterol, that include androgens, estrogens, progesterone and corticosteroids. They represent an important group of physiologically active signalling molecules that bind intracellular receptor proteins and regulate genes involved in developmental, reproductive and metabolic processes. The receptor proteins share structurally and functionally related ligand binding and DNA-binding domains, but possess distinct N-terminal domains (NTD) of unique length and amino acids sequence. The NTD contains sequences important for gene regulation, exhibit structure plasticity and are likely to contribute to the specificity of the steroid hormone/receptor response.
Introduction
Steroid hormone receptors (SHR) are intracellular proteins that regulate patterns of gene expression in target tissues in response to small lipophilic hormones synthesised from cholesterol. They are important for normal development, reproductive function and the regulation of metabolism and salt balance. The receptor proteins have a well-defined domain organisation and high-resolution structures are available for the C-terminal ligand binding domain (LBD), with different agonist and antagonist ligands bound, and the zinc-finger DNA-binding domain (DBD) (Figure 1). SHR have been shown to contain two transactivation functions: one is represented by a structurally defined hydrophobic groove on the surface of the LBD, formed by residues from helices 3, 4, 5 and 12 (AF2), while the other maps to the structurally flexible N-terminal domain (NTD) and is termed AF1 (Figure 1). The main determinants for transactivation map to NTD of both the androgen (AR) and glucocorticoid (GR) receptors (Figure 2A and reviewed in [Lavery and McEwan, 2005]) and while generally there is little sequence conservation between the different SHR-NTDs, in contrast to the DBD and LBD, short regions of similarity have been observed for the AR and GR. For example, a four amino acid synergy control motif [Iniguez-Lluhi and Pearce, 2000], a hydrophobic signature sequence implicated in transactivation [Betney and McEwan, 2003] and a 26 amino acid sequence, that is located within the AF1 domain, that shows 46% amino acid identity between AR (amino acids 237 to 262) and GR (amino acids 47 to 103) (IJM unpublished observation). The NTD is potentially involved in multiple protein-protein interactions [Lavery and McEwan, 2005] and the length of this domain has been positively correlated with the activity of AF1 for different members of the nuclear receptor superfamily [He et al., 2004b].
Figure 1. Androgen and glucocorticoid receptor domain organisation.
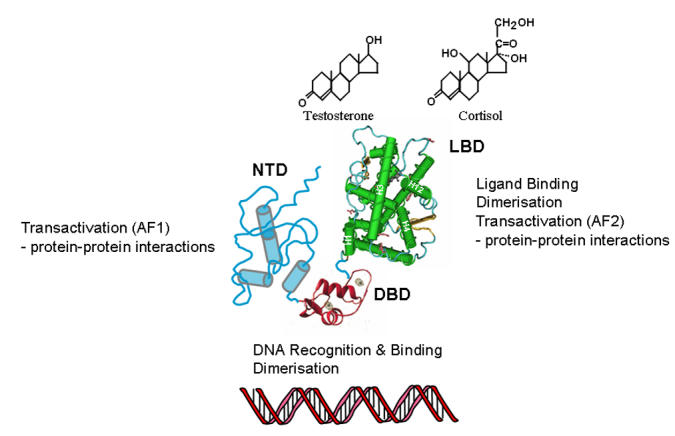
A hypothetical structural model for the full-length AR and GR based on structural information for the isolated ligand-binding (LBD) and DNA-binding (DBD) domains is shown. Note: high resolution structures are available for the isolated LBD and DBD of nearly all SHR, but no multi-domain structure has yet been reported for a member of the nuclear receptor superfamily. The LBD consists of 12 α-helices folded in a three-layer helical sandwich and is linked via a flexible hinge region to the DBD, which has a characteristic globular fold made up of two perpendicular α-helices. The NTD is unique to each SHR and has variable sequence and length [Lavery and McEwan, 2005]. The proposed helical and unfolded conformation for this domain is based in structure predictions (see below), spectroscopy analysis and site-directed mutagenesis and proteolytic sensitivity of the AR- and GR-NTD (see text for details).
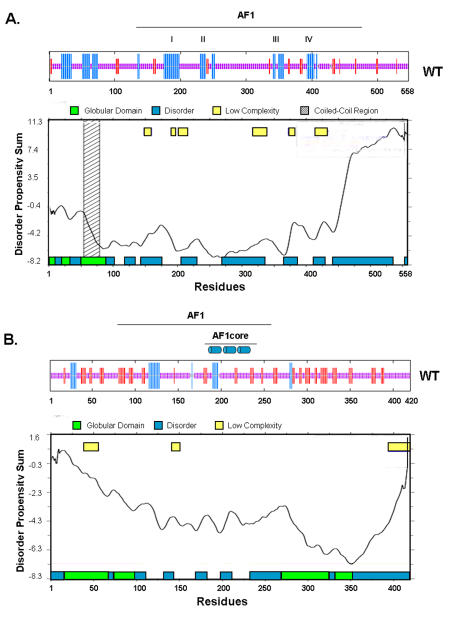
Figure 2. Consensus secondary structure and disorder predictions for AR-NTD and GR-NTD.
(A) and (B) Consensus secondary structure [Combet et al., 2000] and disorder predictions (Glob plot: Linding et al., 2003) for the AR-NTD and GR-NTD, respectively. The location of AF1 is shown above the secondary structure plot (blue and red bars represent α-helix and β-strand structure, respectively) for AR and GR. In addition the four putative helical regions in AR-AF1 are indicated (A, I-IV) and the location of three helices observed in the GR-AF1 core (B) are highlighted.
Androgen receptor-NTD (NR3C4)
Folding and function
The AR-NTD transactivation function is highly modular, with key sequences mapping to amino acids 101 to 370 and 360 to 485 and termed TAU1 and TAU5, respectively [Chamberlain et al., 1996; Jenster et al., 1995; Simental et al., 1991]. A fusion protein containing amino acids 142 to 485 (human AR), consisting of most of TAU1 and all of TAU5, retained about 70% of the activity of the full-length NTD when measured in yeast cells in vivo [Reid et al., 2002b] and also activated a reporter gene in vitro, when fused to a heterologous DBD [Choudhry et al., 2006; McEwan and Gustafsson, 1997].
The first 25 amino acids of the AR-NTD contain a FxxLF motif, that forms part of an amphipathic α-helix (residues 21 to 30) and interacts with the AR-LBD (AF2) [He et al., 2004b; He et al., 2000]. Predictions of disordered structure suggest this region is globular (Figure 2A) and recent structural studies comparing the binding of peptides containing either an FxxLF or an LxxLL motif revealed that the AR-NTD peptide forms a charge clamp with glutamic acid 897 in helix 12 and lysine 720 at the end of helix 3, and the more bulky hydrophobic residues fit better into the surface pocket of AF2 [He et al., 2004b; Hur et al., 2004]. In contrast, a LxxLL motif peptide fails to make hydrogen bond contacts with the glutamic acid residue in helix 12 and makes fewer hydrophobic contacts with the surface of the LBD [He et al., 2004b; Hur et al., 2004]. Interestingly, while other SHR interact with coactivator proteins via LxxLL motifs, which bind in the hydrophobic groove of AF2, the AR-LBD preferentially binds the AR-NTD and coactivators with more bulky hydrophobic residues in the sequence F/WxxLF/W/Y [He et al., 2000; Hsu et al., 2003], which may explain the relatively weak activity observed for AR-AF2.
The AR-AF1 (amino acids 142 to 485) is predicted to be highly disordered and has limited structure, as revealed by circular dichroism and fluorescence spectroscopy, which is lost upon urea-induced unfolding (Figure 2A and [Reid et al., 2002a]). In contrast, incubation with the natural organic osmolyte, trimethylamine-N oxide (TMAO), or the hydrophobic solvent trifluoroethanol (TFE) induces and/or stabilises a more folded conformation in the AF1 domain. This folding of AF1 is associated with a more protease resistant conformation and is concomitant with a significant increase in α-helical content of AR-AF1 (13-16% to 37-40%), at the expense of both non-ordered and β-structure [Kumar et al., 2004a; Reid et al., 2002a]. Significantly, this more folded helical conformation is induced upon binding the AR target factor TFIIF, which in turn appears to create a platform for further protein-protein interactions [Kumar et al., 2004a; Reid et al., 2002a].
Mutations affecting the structure-function of the AR-NTD
We initially predicated four helical segments within AR-AF1, which could be disrupted under native conditions by the introduction of proline residues: helix I, amino acids 175 to 201; helix II, 231 to 252; helix III, 341 to 358; and helix IV, 393 to 411 (Figure 2A: I-IV) [Reid et al., 2002a]. A 14 amino acid sequence in the first of these regions was previously shown to be important for AF1 activity and predicated to form an amphipathic α-helix [Chamberlain et al., 1996]. Recently, residues in this region were found to act as a docking site for TAB2, a component of the N-CoR corepressor complex, which acts as a sensor molecule for androgen, estrogen and cytokine signalling pathways [Zhu et al., 2006].
When the AR-NTD sequences from at least twelve different species were compared, three highly conserved regions were identified: amino acids 1 to 30, 224 to 258 and 500 to 541 [Betney and McEwan, 2003]. The second of these sequences includes helical region II identified in the AF1 domain (see above and Figure 2A) and mutation of conserved hydrophobic residues reduced transactivation activity, but had minimal, if any effect, on conformation as determined by fluorescence spectroscopy and limited proteolysis [Betney and McEwan, 2003]. Interestingly, two mutations A234T and E236G, originally identified in a mouse model for prostate cancer, also map to this sequence [Han et al., 2005; He et al., 2004a]. Structure predictions suggest no dramatic changes in secondary structure for the A234T change, but the possible increase in an adjacent β-strand structure for the E234G mutation (IJM unpublished observations). In recent molecular dynamic simulations these mutations were suggested to stabilize the predicted helical conformation [Han et al., 2005]. Garabedian and co-workers reported on a third mutation identified in a patient with prostate cancer, P340L, which may also have altered structural properties [Li et al., 2005]. This mutation is predicted to disrupt local secondary structure (IJM unpublished observations) and to alter folding of α-helices in the region of amino acids 331 to 355 [Li et al., 2005]. Interestingly, these mutations have been shown to alter different protein-protein interactions with coregulatory proteins: A234T and E236G reduced the binding of the chaperone protein CHIP [He et al., 2004a], while P340L enhanced the binding of the AR coregulatory protein ART-27 [Li et al., 2005]. Intriguing structural and functional changes were also observed when a double point mutation was introduced into a six amino acid repeat sequence in AF1, 158PSTLSL163 (serine to alanine). These changes disrupted binding of TFIIF [Reid et al., 2002b] and resulted in a more folded conformation for AF1 [Betney and McEwan, 2003]. Thus, taken together these findings describe two classes of mutation in the AR-NTD: the first have been shown and/or predicted to alter the conformational flexibility of this domain and receptor activity, while the second group alter function without apparently disrupting structure. To date, about a third of the single point mutations identified in clinical samples of prostate cancer map to the AR-NTD [Scher et al., 2004]. Overall, the structure-function studies of the AR-NTD/AF1 have emphasised a dynamic interplay between protein folding and conformation and receptor function, which may be further modulated by clinically relevant point mutations.
Glucocorticoid receptor-NTD (NR3C1); folding and function
The AF1 function of the GR was initially mapped to amino acids 77 to 262 for the human receptor. Subsequently, a 58 amino acid core function was delineated to amino acids 187 to 244 (Reviewed in [Lavery and McEwan, 2005]). Mutations altering hydrophobicity or the acidic character of both the human and rat GR transactivation domains were found to impair transactivation activity [Almlof et al., 1998; Almlof et al., 1995; Iniguez-Lluhi et al., 1997]. This suggests that such residues are important structurally and/or formed surfaces involved in protein-protein interactions. Recently, a study combining mutations in the GR AF1 or AF2 domains with gene array analysis revealed a differential requirement for the NTD (AF1) and LBD (AF2) transactivation functions [Rogatsky et al., 2003], emphasising a potential additional level of selectivity in SHR signalling.
Figure 2B shows the predictions for secondary structure and regions of disordered structure within the GR-NTD and suggests significant disorder within the AF1 domain and the region N-terminal to the DBD. Folding of the GR-AF1 (amino acids 77 to 262) or AF1 core (amino acids 187 to 244) domain into a more stable helical conformation has been observed in the presence of both the solvent TFE and the natural solute TMAO. Using NMR spectroscopy, Dahlman-Wright and co-workers originally described three helical segments within the AF1 core, in the presence of 40% TFE: amino acids 189 to 200, 216 to 226 and 235 to 241 (Figure 2B and [Dahlman-Wright et al., 1995]). Introduction of helix-breaking proline mutations resulted in disruption to both secondary structure and transactivation potential [Dahlman-Wright and McEwan, 1996]. A subsequent study by Thompson and Kumar and co-workers showed changes in tryptophan fluorescence emission spectra in the presence of TMAO and upon DNA binding, indicative of structural changes within the GR-NTD [Baskakov et al., 1999]. Folding of the NTD was also shown to enhance the binding of the coregulatory target proteins TBP, CBP and the p160 family member SRC-2 (GRIP1) [Kumar et al., 2001]. More recently, these authors showed the binding of TBP to GR-AF1 occurred in vivo and induced a coil to α-helix transition with concomitant alterations in the chemical shifts of four glycine residues present in the GR-AF1 core region in vitro [Copik et al., 2006; Kumar et al., 2004b]. Therefore, similar to the AR, the GR-AF1 appears structurally flexible and has the propensity to form an α-helical conformation. In addition to the above studies on AR and GR, Warnmark et al (2001) found that the NTD of estrogen receptor (ER) α and β were unstructured when studied by both NMR and circular dichroism spectroscopy. Significantly, the ERα, but not ERβ, -NTD adopted a more structured conformation upon binding the general transcription factor the TATA-binding protein [Warnmark et al., 2001]. Thus, in spite of the lack of high resolution structural data, a large body of evidence now exists underlining an induced structure model for both the AR and GR AF1 domains upon specific DNA and/or protein binding.
Natural disordered sequences and the SHR-NTD
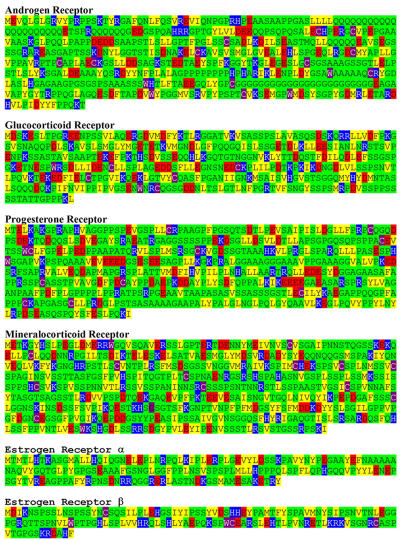
What is the nature of this structural flexibility? The concept of regions or even whole domains of proteins being naturally disordered has gained much attention recently as more evidence accumulates to support the hypothesis that such sequences are (i) very common in eukaryotic proteins and (ii) functionally important [Dunker et al., 2002; Dyson and Wright, 2005]. Intrinsically unstructured regions have been implicated in molecular recognition and assembly of protein complexes and as sites of post-translational modification [Dunker et al., 2002; Dyson and Wright, 2005]. Thus, a consideration of natural disordered sequences within steroid receptor-NTDs is highly appropriate given the available structural information, the function of this domain in multiple protein-protein interactions and as a target for post-translational modifications (phosphorylation and sumoylation) (Reviewed in [Lavery and McEwan, 2005]). While lacking any significant level of primary sequence homology, analysis of the amino acid composition of SHR-NTDs suggests some shared structural properties. The presence of a high proportion of proline, serine and glycine residues in the NTDs of steroid receptors conforms to the amino acid compositional bias associated with a signature for intrinsic disorder [Dyson and Wright, 2005]. Figure 3 shows an analysis of the primary amino acid sequence of the NTD for the six major steroid receptors, with amino acids colour coded depending on the properties of their side-chains. It is clear there are large stretches of potentially disordered structure with a compositional bias towards small (Ala, Gly), uncharged hydrophilic (Asn, Gln, Ser, Thr) and proline residues (Green shading). Interspersed are sequences of charged (Red and Blue shading) and hydrophobic (Yellow shading) residues, which may represent interaction sites for protein-protein interactions and/or more structured regions. A recent analysis [Lavery and McEwan, 2005] of individual SHR-NTDs using the trained neural network programme PONDR® and the GlobPlot predictions for the AR and GR-NTDs (Figure 2A and Figure 2B) supports the view that these domains contain significant levels of intrinsically disordered structure and is in good general agreement with the amino acid compositional bias profiling shown in Figure 3. There is also good general agreement correlating potential regions of naturally disordered structure with sequences identified as playing a role in the control of gene expression for the different SHR. Thus, structural flexibility is likely to result from the amino acid compositional bias and underpin the ability of SHR-NTD to make multiple protein-protein interactions with the cellular transcription machinery.
Figure 3. SHR-NTD: regions of intrinsic structural disorder.
The amino acid compositional bias for regions within the SHR-NTD is shown. Small (Ala, Gly), uncharged hydrophilic (Asn, Gln, Ser, Thr) and proline residues (Green shading); charged residues (Red and Blue shading); and hydrophobic residues (Yellow shading). Androgen Receptor, amino acids 1 to 558; Glucocorticoid Receptor, amino acids 1 to 420; Progesterone Receptor, amino acids 1 to 566; Mineralocorticoid Receptor, amino acids 1 to 602; Estrogen Receptor α, amino acids 1 to 184 and β, amino acids 1 to 148. Amino acid numbering and sequences are for the human receptors.
Conclusions and future perspectives
Specificity in SHR signalling is exerted at a number of key steps, notably the binding of hormone and subsequent recognition and binding to DNA response elements. While the structural basis of both hormone and DNA binding specificity are generally well described, the structural basis for AF1 activity, located in the distinct NTD, is only now beginning to be revealed. Extensive analysis of the isolated AR and GR AF1 domains emphasises the structural plasticity of this domain and the importance of flexibility for receptor function. An important question is what is the conformation of the NTD in the context of the full-length SHR? From the available experimental evidence two possible models can be proposed for SHR-NTD structure: in the first the NTD exists in an equilibrium of essentially non-ordered, random-coil conformers, while in the second, the NTD would exhibit limited structural stability consisting of regions of disorder and variable levels of secondary structure. The latter is a particularly attractive model, with the NTD in a flux of partially folded intermediate structures that collapse into a stable structure upon protein-protein interactions. In both models it seems likely that the NTD structure will be stabilised, at least partially by intramolecular interactions with the DBD and/or LBD. Indeed, studies with two domain constructs of the AR [Brodie and McEwan, 2005], GR [Kumar et al., 1999], progesterone receptor [Bain et al., 2001] and the full-length ER [Wood et al., 2001] suggest that the presence of the DBD and/or specific DNA binding leads to structural changes in the NTD. Therefore, both intra- and inter-molecular interactions will result in an induction and/or stabilisation of a more folded, active conformation in the SHR-NTD. Future analysis will have to consider whether different conformations are induced in the NTD depending on DNA response element and/or target protein binding, and also the role played by post-translational modifications (i.e. phosphorylation), which are likely to lead to additional levels of complexity and potential specificity in SHR action. Furthermore, as studies have revealed that both active and non-productive conformations may be stabilised for the AR-NTD, it is reasonable to propose that folding of SHR-NTD may serve as a future target for drug discovery for the treatment of conditions ranging from cancer to neurodegeneration.
Acknowledgments
The authors wish to thank colleagues participating in the recent FASEB Summer Research Meeting on Dynamics of the Nuclear Hormone Receptors (July 2006) for stimulating discussions on SHR-NTD structure and function. Financial support for the research in the Author’s laboratory is also gratefully acknowledged from the Association for International Cancer Research, Biotechnology and Biological Sciences Research Council, College of Life Sciences and Medicine, University of Aberdeen and the Medical Research Council.
Abbreviations
- AF
activation function
- AR
androgen receptor
- CBP
CREB binding protein
- DBD
DNA-binding domain
- GR
glucocorticoid receptor
- LBD
ligand binding domain
- N-CoR
nuclear receptor corepressor
- NTD
N-terminal domain
- SHR
steroid hormone receptor
- SRC
steroid receptor coactivator
- TAB2
transforming growth factor β-activated kinase 1 binding protein
- TAU
transactivation unit
- TBP
TATA-binding protein
- TFE
trifluorethanol
- TFIIF
Transcription Factor IIF
- TMAO
trimethylamine-N oxide
References
- Almlof T., Wright A. P., Gustafsson J. A. Role of acidic and phosphorylated residues in gene activation by the glucocorticoid receptor. J Biol Chem. 1995;270:17535–40. doi: 10.1074/jbc.270.29.17535. [DOI] [PubMed] [Google Scholar]
- Almlof T., Wallberg A. E., Gustafsson J. A., Wright A. P. Role of important hydrophobic amino acids in the interaction between the glucocorticoid receptor τ 1-core activation domain and target factors. Biochemistry. 1998;37:9586–94. doi: 10.1021/bi973029x. [DOI] [PubMed] [Google Scholar]
- Bain D. L., Franden M. A., McManaman J. L., Takimoto G. S., Horwitz K. B. The N-terminal region of human progesterone B-receptors: biophysical and biochemical comparison to A-receptors. J Biol Chem. 2001;276:23825–31. doi: 10.1074/jbc.M102611200. [DOI] [PubMed] [Google Scholar]
- Baskakov I. V., Kumar R., Srinivasan G., Ji Y. S., Bolen D. W., Thompson E. B. Trimethylamine N-oxide-induced cooperative folding of an intrinsically unfolded transcription-activating fragment of human glucocorticoid receptor. J Biol Chem. 1999;274:10693–6. doi: 10.1074/jbc.274.16.10693. [DOI] [PubMed] [Google Scholar]
- Betney R., McEwan I. J. Role of conserved hydrophobic amino acids in androgen receptor AF-1 function. J Mol Endocrinol. 2003;31:427–39. doi: 10.1677/jme.0.0310427. [DOI] [PubMed] [Google Scholar]
- Brodie J., McEwan I. J. Intra-domain communication between the N-terminal and DNA-binding domains of the androgen receptor: modulation of androgen response element DNA binding. J Mol Endocrinol. 2005;34:603–15. doi: 10.1677/jme.1.01723. [DOI] [PubMed] [Google Scholar]
- Chamberlain N. L., Whitacre D. C., Miesfeld R. L. Delineation of two distinct type 1 activation functions in the androgen receptor amino-terminal domain. J Biol Chem. 1996;271:26772–8. doi: 10.1074/jbc.271.43.26772. [DOI] [PubMed] [Google Scholar]
- Choudhry M. A., Ball A., McEwan I. J. The role of the general transcription factor IIF in androgen receptor-dependent transcription. Mol Endocrinol. 2006;20:2052–61. doi: 10.1210/me.2005-0486. [DOI] [PubMed] [Google Scholar]
- Copik A. J., Webb M. S., Miller A. L., Wang Y., Kumar R., Thompson E. B. Activation function 1 of glucocorticoid receptor binds TATA-binding protein in vitro and in vivo. Mol Endocrinol. 2006;20:1218–30. doi: 10.1210/me.2005-0257. [DOI] [PubMed] [Google Scholar]
- Dahlman-Wright K., Baumann H., McEwan I. J., Almlof T., Wright A. P., Gustafsson J. A., Hard T. Structural characterization of a minimal functional transactivation domain from the human glucocorticoid receptor. Proc Natl Acad Sci U S A. 1995;92:1699–703. doi: 10.1073/pnas.92.5.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlman-Wright K., McEwan I. J. Structural studies of mutant glucocorticoid receptor transactivation domains establish a link between transactivation activity in vivo and α-helix-forming potential in vitro. Biochemistry. 1996;35:1323–7. doi: 10.1021/bi952409k. [DOI] [PubMed] [Google Scholar]
- Dunker A. K., Brown C. J., Lawson J. D., Iakoucheva L. M., Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–82. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- Dyson H. J., Wright P. E. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Han G., Buchanan G., Ittmann M., Harris J. M., Yu X., Demayo F. J., Tilley W., Greenberg N. M. Mutation of the androgen receptor causes oncogenic transformation of the prostate. Proc Natl Acad Sci U S A. 2005;102:1151–6. doi: 10.1073/pnas.0408925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Bai S., Hnat A. T., Kalman R. I., Minges J. T., Patterson C., Wilson E. M. An androgen receptor NH2-terminal conserved motif interacts with the COOH terminus of the Hsp70-interacting protein (CHIP) J Biol Chem. 2004b;279:30643–53. doi: 10.1074/jbc.M403117200. [DOI] [PubMed] [Google Scholar]
- He B., Kemppainen J. A., Wilson E. M. FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J Biol Chem. 2000;275:22986–94. doi: 10.1074/jbc.M002807200. [DOI] [PubMed] [Google Scholar]
- He B., Gampe R. T., Jr., Kole A. J., Hnat A. T., Stanley T. B., An G., Stewart E. L., Kalman R. I., Minges J. T., Wilson E. M. Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance. Mol Cell. 2004a;16:425–38. doi: 10.1016/j.molcel.2004.09.036. [DOI] [PubMed] [Google Scholar]
- Hsu C. L., Chen Y. L., Yeh S., Ting H. J., Hu Y. C., Lin H., Wang X., Chang C. The use of phage display technique for the isolation of androgen receptor interacting peptides with (F/W)XXL(F/W) and FXXLY new signature motifs. J Biol Chem. 2003;278:23691–8. doi: 10.1074/jbc.M211908200. [DOI] [PubMed] [Google Scholar]
- Hur E., Pfaff S. J., Payne E. S., Gron H., Buehrer B. M., Fletterick R. J. Recognition and accommodation at the androgen receptor coactivator binding interface. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniguez-Lluhi J. A., Pearce D. A common motif within the negative regulatory regions of multiple factors inhibits their transcriptional synergy. Mol Cell Biol. 2000;20:6040–50. doi: 10.1128/mcb.20.16.6040-6050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniguez-Lluhi J. A., Lou D. Y., Yamamoto K. R. Three amino acid substitutions selectively disrupt the activation but not the repression function of the glucocorticoid receptor N terminus. J Biol Chem. 1997;272:4149–56. doi: 10.1074/jbc.272.7.4149. [DOI] [PubMed] [Google Scholar]
- Jenster G., van der Korput H. A., Trapman J., Brinkmann A. O. Identification of two transcription activation units in the N-terminal domain of the human androgen receptor. J Biol Chem. 1995;270:7341–6. doi: 10.1074/jbc.270.13.7341. [DOI] [PubMed] [Google Scholar]
- Kumar R., Betney R., Li J., Thompson E. B., McEwan I. J. Induced α-helix structure in AF1 of the androgen receptor upon binding transcription factor TFIIF. Biochemistry. 2004a;43:3008–13. doi: 10.1021/bi035934p. [DOI] [PubMed] [Google Scholar]
- Kumar R., Baskakov I. V., Srinivasan G., Bolen D. W., Lee J. C., Thompson E. B. Interdomain signaling in a two-domain fragment of the human glucocorticoid receptor. J Biol Chem. 1999;274:24737–41. doi: 10.1074/jbc.274.35.24737. [DOI] [PubMed] [Google Scholar]
- Kumar R., Volk D. E., Li J., Lee J. C., Gorenstein D. G., Thompson E. B. TATA box binding protein induces structure in the recombinant glucocorticoid receptor AF1 domain. Proc Natl Acad Sci U S A. 2004b;101:16425–30. doi: 10.1073/pnas.0407160101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Lee J. C., Bolen D. W., Thompson E. B. The conformation of the glucocorticoid receptor af1/tau1 domain induced by osmolyte binds co-regulatory proteins. J Biol Chem. 2001;276:18146–52. doi: 10.1074/jbc.M100825200. [DOI] [PubMed] [Google Scholar]
- Lavery D. N., McEwan I. J. Structure and function of steroid receptor AF1 transactivation domains: induction of active conformations. Biochem J. 2005;391:449–64. doi: 10.1042/BJ20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Cavasotto C. N., Cardozo T., Ha S., Dang T., Taneja S. S., Logan S. K., Garabedian M. J. Androgen receptor mutations identified in prostate cancer and androgen insensitivity syndrome display aberrant ART-27 coactivator function. Mol Endocrinol. 2005;19:2273–82. doi: 10.1210/me.2005-0134. [DOI] [PubMed] [Google Scholar]
- McEwan I. J., Gustafsson J. Interaction of the human androgen receptor transactivation function with the general transcription factor TFIIF. Proc Natl Acad Sci U S A. 1997;94:8485–90. doi: 10.1073/pnas.94.16.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid J., Kelly S. M., Watt K., Price N. C., McEwan I. J. Conformational analysis of the androgen receptor amino-terminal domain involved in transactivation. Influence of structure-stabilizing solutes and protein-protein interactions. J Biol Chem. 2002b;277:20079–86. doi: 10.1074/jbc.M201003200. [DOI] [PubMed] [Google Scholar]
- Reid J., Murray I., Watt K., Betney R., McEwan I. J. The androgen receptor interacts with multiple regions of the large subunit of general transcription factor TFIIF. J Biol Chem. 2002a;277:41247–53. doi: 10.1074/jbc.M205220200. [DOI] [PubMed] [Google Scholar]
- Rogatsky I., Wang J. C., Derynck M. K., Nonaka D. F., Khodabakhsh D. B., Haqq C. M., Darimont B. D., Garabedian M. J., Yamamoto K. R. Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc Natl Acad Sci U S A. 2003;100:13845–50. doi: 10.1073/pnas.2336092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher H. I., Buchanan G., Gerald W., Butler L. M., Tilley W. D. Targeting the androgen receptor: improving outcomes for castration-resistant prostate cancer. Endocr Relat Cancer. 2004;11:459–76. doi: 10.1677/erc.1.00525. [DOI] [PubMed] [Google Scholar]
- Simental J. A., Sar M., Lane M. V., French F. S., Wilson E. M. Transcriptional activation and nuclear targeting signals of the human androgen receptor. J Biol Chem. 1991;266:510–8. [PubMed] [Google Scholar]
- Warnmark A., Wikstrom A., Wright A. P., Gustafsson J. A., Hard T. The N-terminal regions of estrogen receptor α and β are unstructured in vitro and show different TBP binding properties. J Biol Chem. 2001;276:45939–44. doi: 10.1074/jbc.M107875200. [DOI] [PubMed] [Google Scholar]
- Wood J. R., Likhite V. S., Loven M. A., Nardulli A. M. Allosteric modulation of estrogen receptor conformation by different estrogen response elements. Mol Endocrinol. 2001;15:1114–26. doi: 10.1210/mend.15.7.0671. [DOI] [PubMed] [Google Scholar]
- Zhu P., Baek S. H., Bourk E. M., Ohgi K. A., Garcia-Bassets I., Sanjo H., Akira S., Kotol P. F., Glass C. K., Rosenfeld M. G., Rose D. W. Macrophage/cancer cell interactions mediate hormone resistance by a nuclear receptor derepression pathway. Cell. 2006;124:615–29. doi: 10.1016/j.cell.2005.12.032. [DOI] [PubMed] [Google Scholar]