Abstract
Transcription controlled by Steroid Hormone Receptors (SHRs) plays a key role in many important physiological processes like organ development, metabolite homeostasis, and response to external stimuli. Understandably, the members of this family have drawn a lot of attention from the scientific community since their discovery, four decades ago. Still, after many years of research we are only beginning to unravel the complex nature of these receptors. The pace at which we do has improved significantly in recent years with the discovery of genetically encoded fluorescent probes, and the accompanying revival of biophysical approaches that allow more detailed study of SHRs. Here, we will look into the different aspects of SHR signalling, and discuss how biophysical techniques have contributed to visualizing their function in their native context, the living cell.
Steroid hormone receptors
As early as 1896, Thomas Beatson described that removal of the ovaries in advanced breast cancer patients often resulted in remarkable improvement [Beatson, 1896]. With that he had revealed the stimulating effect of the female ovarian hormone estrogen on breast cancer, even before the hormone itself was discovered. His work provided a foundation for the modern use of hormone therapy in treatment and prevention of breast cancer. Only much later was the cellular counterpart that mediated the described effects revealed, the estrogen receptor (ER) [Green et al., 1986; Jensen, 1962].
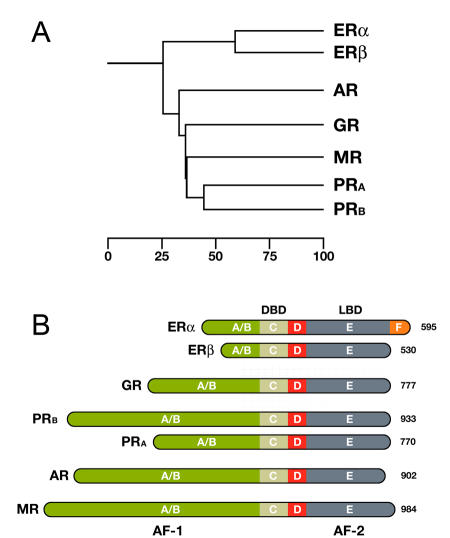
As it turned out, this receptor plays a key role in the development and maintenance of the sexual reproductive tissues, and therefore, as Beatson had discovered, in breast cancer as well. We now know that the estrogen receptor is part of the nuclear receptor superfamily, and comes in two forms, ERα [NR3A1] and ERβ [NR3A2]. More specifically, both receptors are members of the subgroup of Steroid Hormone Receptors (SHRs), to which the cortisol binding glucocorticoid receptor (GR) [NR3C1], the aldosterone binding mineralocorticoid receptor (MR) [NR3C2], the progesterone receptor (PR) [NR3C3], and the dihydrotestosterone (DHT) binding androgen receptor (AR) [NR3C4] also belong (Figure 1A).
Figure 1. Overview of the steroid hormone receptor family.
A. Phylogenetic tree of the Steroid Hormone Receptor (SHR) family showing the evolutionary interrelationships and distance between the various receptors. Based on alignments available at The NucleaRDB [Horn et al., 2001]. B. All steroid receptors are composed of a variable N-terminal domain (A/B) containing the AF-1 transactivation region, a highly conserved DNA Binding Domain (DBD), a flexible hinge region (D), and a C-terminal Ligand Binding Domain (LBD, E) containing the AF-2 transactivation region. The estrogen receptor α is unique in that it contains an additional C-terminal F domain. Numbers represent the length of the receptor in amino acids.
In addition, the SHR subgroup contains three orphan receptors closely related to ER: the estrogen related receptors α (ERRα) [NR3B1], β (ERRβ) [NR3B2], and γ (ERRγ) [NR3B3], for which a natural ligand remains to be identified. All SHRs function as nuclear transcription factors whose activity is regulated by small lipid-soluble ligands, and each member plays an important role in key physiological processes like reproduction, glucose metabolism, salt balance, and stress response.
Structural overview
The members of the Steroid Hormone Receptor family share a similar, modular architecture, consisting of a number of independent functional domains (Figure 1B). Most conserved is the centrally located DNA binding domain (DBD) containing the characteristic zinc-finger motifs. The DBD is followed by a flexible hinge region and a moderately conserved Ligand Binding Domain (LBD), located at the carboxy-terminal end of the receptor. The estrogen receptor α is unique in that it contains an additional F domain of which the exact function is unclear. The LBD is composed of twelve α-helices (H1-H12) that together fold into a canonical α-helical sandwich. Besides its ligand binding capability, the LBD also plays an important role in nuclear translocation, chaperone binding, receptor dimerization, and coregulator recruitment through its potent ligand-dependent transactivation domain, referred to as AF-2. A second, ligand independent, transactivation domain is located in the more variable N-terminal part of the receptor, designated as AF-1. To date, no crystal structure of a full-length SHR exists, though structures of the DBD and LBD regions of most SHRs are available. These have helped significantly in understanding the molecular aspects of DNA and ligand binding, but have to some extent also led to biased attention to these parts of the receptor only. For example, many coregulator interaction studies are still performed with the LBD only, while numerous studies have demonstrated that the AF-2 domain often tells only part of the story. With the help of biophysical techniques, however, it is feasible to study the full-length receptor in its native environment (Figure 2).
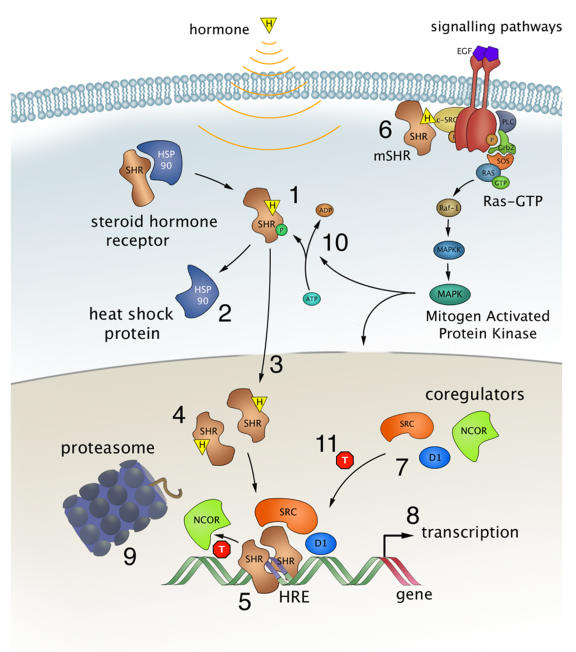
Figure 2. Steroid hormone receptor signalling.
Steroid Hormone Receptors (SHR) act as hormone dependent nuclear transcription factors. Upon entering the cell by passive diffusion, the hormone (H) binds the receptor, which is subsequently released from heat shock proteins, and translocates to the nucleus. There, the receptor dimerizes, binds specific sequences in the DNA, called Hormone Responsive Elements or HREs, and recruits a number of coregulators that facilitate gene transcription. This latter step can be modulated by receptor antagonists like tamoxifen (T), and cellular signalling pathways. Examples of processes studied using biophysical techniques and discussed in this review include: hormone binding (1), chaperone interaction (2), nuclear translocation (3), receptor dimerization (4), DNA binding (5), putative membrane-bound receptors (6), coregulator recruitment (7), transcription (8), proteasomal degradation (9), modulation by cellular signalling pathways (10), and antagonist resistance (11).
Ligand binding
Steroid Hormones (SHs) reach their target cells via the blood, where they are bound to carrier proteins. Because of their lipophilic nature it is thought that they pass the cell membrane by simple diffusion, although some evidence exists that they can also be actively taken up by endocytosis of carrier protein bound hormones [Hammes et al., 2005]. For a long time it has been assumed that binding of the ligand resulted in a simple on/off switch of the receptor (Figure 2, step 1). While this is likely the case for typical agonists like estrogen and progesterone, this is not always correct for receptor antagonists. These antagonists come in two kinds, so-called partial antagonists (for the estrogen receptors known as SERMs for Selective Estrogen Receptor Modulators) and full antagonists. The partial antagonist can, depending on cell type, act as a SHR agonist or antagonist. In contrast, full antagonists (for ER known as SERDs for Selective Estrogen Receptor Downregulators) always inhibit the receptor, independent of cell type, in part by targeting the receptor for degradation. Binding of either type of antagonist results in major conformational changes within the LBD and in release from heat shock proteins that thus far had protected the unliganded receptor from unfolding and aggregation (Figure 2, step 2). This process was nicely visualized for the estrogen receptor by Devin-Leclerc et al., who showed using fluorescence microscopy that the nuclear ER-HSP90 complex dissociates after addition of either agonist or antagonist, followed by rapid relocation of the heat shock protein to the cytoplasm [Devin-Leclerc et al., 1998].
Nuclear translocation
The constitutive nuclear localisation of ER is a unique feature of this SHR only. Although family members, SHRs are located differently in cells. The subcellular localisation of SHRs in living cells has been extensively studied using fusion constructs of green fluorescent protein (GFP). This showed that SHRs can be divided in three groups based on their unliganded distribution: ERα and ERβ are found predominantly in the nucleus [Htun et al., 1999], GR and AR are found primarily in the cytoplasm [Georget et al., 1997; Htun et al., 1996], while MR and PR have a mixed distribution over both cytoplasm and nucleus [Fejes-Toth et al., 1998; Lim et al., 1999]. The progesterone receptor is of particular interest as it exists in two forms with different ratios of nuclear versus cytoplasmic localization of the unliganded receptor. In most cell contexts, the PRA isoform is a repressor of the shorter PRB isoform, and without hormone induction it is mostly located in the nucleus, whereas PRB distributes both in the nucleus and in the cytoplasm. PRB accumulates in the nucleus after progesterone binding, a process that directly correlates with PR mediated transcription [Li et al., 2005; Lim et al., 1999].
Rapid and almost complete nuclear translocation following ligand addition is a common behavior observed for almost all SHRs (Figure 2, step 3). This translocation coincides with a striking alteration in receptor distribution within the nucleus, most apparent in the case of the already nuclear ER. Htun et al., observed that GFP-ER’s uniform distribution changes into a punctate pattern upon the addition of either agonistic or antagonistic ligands [Htun et al., 1999]. A few years earlier the same group had already made similar observations for GFP-GR [Htun et al., 1996]. Other groups confirmed similar behavior for the other SHRs, some directly by tagging two receptors with different variants of GFP and following both at the same time [Nishi et al., 2001]. Fejes-Toth and colleagues demonstrated that hormone-activated MRs accumulated in dynamic, discrete clusters in the cell nucleus, a phenomenon that only concurred with agonistic mineralocorticoids and not with full antagonists [Fejes-Toth et al., 1998]. Further work on MR and AR showed that the accumulation of these receptors in about 250-400 foci requires both the DBD and LBD regions, and is possibly influenced by AF-1 function [Farla et al., 2005; Goto et al., 2003; Pearce et al., 2002; Tomura et al., 2001; Tyagi et al., 2000]. The exact nature of these foci is still unclear and multiple roles have been proposed, including storage depots and sites of transcription, splicing, aggregation or degradation. What is clear, however, is that nuclear and subnuclear translocation of SHRs is ligand and concentration dependent. Martinez et al., recently made use of this finding to devise a molecular screen for ER ligands based on a fluorescent GR-ER chimera [Martinez et al., 2005]. Instead of a constitutive nuclear localization, this chimeric receptor adapted the cytoplasmic localization of unliganded GR, and translocates to the nucleus upon ER ligand addition. Interestingly, the GR-ER chimera retained the (anti-) estrogen binding properties, and could thus be used to screen for new ER ligands.
Dimerization
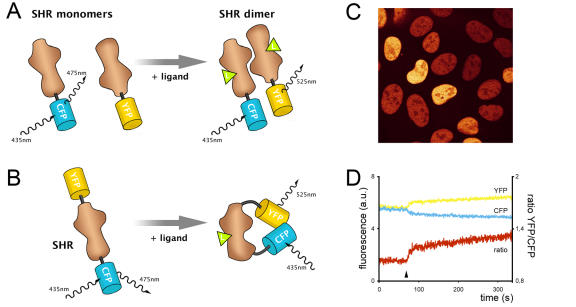
Nuclear receptors that bind steroid hormones typically form homodimers (Figure 2, step 4). Dimer formation is facilitated mainly through interactions between the LBDs of both receptors, and is essential for their function, as mutations in the dimerization domain typically render the receptor inactive. ER has been reported to exist as a dimer even in the absence of ligand, although it is important to note that these studies have again been performed with the LBD domain only [Salomonsson et al., 1994]. Biophysical in vitro studies, again with the LBD only, have confirmed these data and show slow dissociation of unliganded dimers, which is further retarded by ligand binding [Tamrazi et al., 2002]. Recent in vivo studies suggested that this might not hold for full-length receptors though, at least not for AR. Schaufele et al., used Fluorescence Resonance Energy Transfer (FRET) to study AR dimer formation [Schaufele et al., 2005]. FRET is the radiationless transfer of energy from an excited donor fluorophore to a suitable acceptor fluorophore [Förster, 1948]. Importantly, FRET is extremely sensitive to the distance between the fluorophores (its efficiency decays with the distance to the sixth power), and will therefore only occur when two proteins are on average no more than one molecule in distance apart, but usually they interact directly (Figure 3A). Schaufele and colleagues measured FRET between CFP and YFP labeled AR receptors, and their results suggest that dimerization only takes place after ligand binding, and predominantly in the nucleus. It should be noted that absence of FRET does not imply absence of protein-protein interaction since the relative orientation of two fluorophores is also critical for FRET to occur [Förster, 1948]. Further study is required to confirm these findings and to determine whether this behavior is unique for AR or also applies to other SHRs.
Figure 3. Fluorescence resonance energy transfer (FRET).
A. Principle of FRET to measure intermolecular interaction. Exciting the blue variant of GFP (CFP) linked to one Steroid Hormone Receptor (SHR) monomer at 435nm results in emission at 475nm, unless energy is transferred to a SHR monomer coupled to the yellow variant of GFP (YFP). This phenomenon only occurs when both monomers physically interact as a dimer, and results in increased YFP emission at 525nm at the cost of CFP emission at 475nm. B. A similar protocol is followed to measure intramolecular FRET. A single SHR monomer is tagged with two variants of GFP. Ligand binding induces conformational changes within the receptor and alters the relative orientation and distance between the two fluorophores, leading to changes in FRET efficiency. C. A confocal microscopy image showing U2OS cells expressing nuclear YFP-ER-CFP to monitor conformational changes through intramolecular FRET. Fluorescence is depicted in false colors. D. An example FRET trace, measuring intramolecular FRET between CFP and YFP within a single nucleus of a YFP-ER-CFP expressing cell as shown in 3C. The fluorescent signal of YFP (yellow) and CFP (cyan) is arbitrarily set to the same level such that the ratio (red) is 1, and followed in time. After 80 seconds, tamoxifen is added to the cells (arrowhead) and a conformational change is observed in the form of a change in FRET; the YFP signal increases at the cost of CFP fluorescence.
Dimers of SHRs are only efficiently formed between closely related receptors. In this light the previously mentioned two isoforms of the progesterone receptor and the two estrogen receptors are of particular interest. In both cases one of the two seems to exhibit a repressive function on the other. ERβ efficiently dimerizes with ERα and mixed dimers show identical subnuclear distribution as homodimers [Matsuda et al., 2002]. However, binding of ERβ suppresses ERα mediated gene transcription, and accordingly, the mouse knockouts of either receptor show completely opposing phenotypes [Couse and Korach, 1999].
DNA binding
Upon binding of ligand and translocation to the nucleus, SHRs bind to specific regions in the DNA called Hormone Responsive Elements (HREs) through zinc-finger motifs present in the DBD (Figure 2, step 5). The exact mode of binding has been characterised in detail with help of available crystal structures and extensive biophysical in vitro measurements. Consensus nucleotide binding sequences have been determined for all SHRs, but these show a significant amount of ambiguity, making it hard to pinpoint true target HREs in the genome. A HRE is made up of two so-called half-sites that each bind one monomer of the SHR dimer. Interestingly, single half-sites have also been found in genes that clearly respond to hormone, hinting at a possible role for receptors in their monomeric configuration.
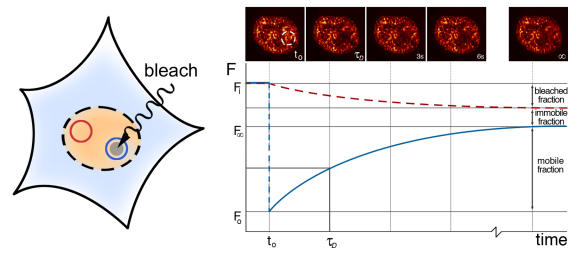
Immobilization of SHRs on DNA and other nuclear structures has been studied with photobleaching techniques like Fluorescence Recovery After Photobleaching (FRAP). By bleaching fluorescent molecules in a region of interest in a living cell and measuring recurrence of fluorescence levels in the bleached area, the mobility of the tagged molecules can be determined (Figure 4). Using this technique several groups were able to demonstrate a clear correlation between receptor immobilization in the nucleus and the appearance of the typical punctate receptor distribution, which was most convincingly demonstrated by Schaaf and colleagues who compared 13 GR ligands [Schaaf et al., 2005]. FRAP measurements show that fluorescently tagged SHRs such as ER, GR, and AR are highly mobile and dynamic in the unliganded state, whereas ligand-bound forms are less mobile [Farla et al., 2004; Sprague et al., 2004; Stenoien et al., 2001b]. Stenoien et al., further showed that in the case of ERα, FRAP could discriminate between ligands with potential agonistic properties and full antagonists on the basis of receptor immobilization in the nucleus [Stenoien et al., 2001b]. The nature of the substrate on which the receptor immobilizes remains uncertain, but almost certainly includes DNA. Carefully controlled FRAP measurements from Sprague et al., show that in free form GR is bound to a single type of substrate, most probably DNA, with each molecule binding on average 65 sites per second [Sprague et al., 2004]. This rapid sampling of GR is likely to be important in finding a specific HRE. Upon ligand binding, the residence time on DNA is significantly increased. According to Farla et al., on average one out of five ARs is immobilized in the presence of ligand, each individual AR being immobile for 1-2 min. This immobilization is dependent on DNA binding since GFP-ARs mutated in the DNA-binding domain were not immobilized [Farla et al., 2004]. Likewise, FRAP analysis by Kino et al., on several GR mutant receptors showed a significantly increased nuclear motility and decreased chromatin retention, which correlated with impaired transcriptional activity [Kino et al., 2004].
Figure 4. Fluorescence recovery after photobleaching (FRAP).
By bleaching the indicated (blue) region in the fluorescent area (here the nucleus of a cell expressing ERα fused to GFP) at time t0, fluorescence decreases from the initial fluorescence Fi to F0. The fluorescence recovers over time by diffusion. The characteristic diffusion time τD indicates the time at which half of the fluorescence has recovered. The mobile fraction can be calculated by comparing the fluorescence in the bleached region after full recovery (Fω) with the fluorescence in a distant region in the nucleus (red, dashed line).
DNA binding and transcription has been visualized directly by using cells that have stably integrated a tandem array of HREs. Pioneering work in this area has been performed by the Hager lab, which used this approach to study the interaction of GR with a natural promoter [McNally et al., 2000]. The promoter array allows significant amounts of GFP-GR to accumulate for direct detection under the microscope. The recruitment of GFP-GR leads to gross alterations in chromatin structure of the array that correlate with gene transcription [Muller et al., 2001]. Interestingly, FRAP analysis on the array again shows a rapid exchange of receptors between chromatin and the nucleoplasmic compartment. Further analysis demonstrated that following binding of GR to the promoter, the receptor is actively displaced from the template during a chromatin remodeling reaction facilitated by the hSWI/SNF complex [Nagaich et al., 2004]. Further evidence comes from work on PR by the same group, which showed that the exchange of PR-GFP on the array was slowed down (but still in the order of seconds) upon agonist addition, and even further slowed down after addition of a partial antagonist [Rayasam et al., 2005]. Strikingly, addition of a full-antagonist showed the opposite effect, with ongoing exchange at a rate faster than for an agonist bound receptor. In contrast to an agonist or partial antagonist bound receptor, addition of a full-antagonist does not lead to recruitment of the SWI/SNF chromatin remodeling complex, which may partly explain the above results. Together, these findings have led to the so-called hit-and-run model. In contrast to static binding of the receptor to a HRE and the subsequent build up of the transcription complex, this model suggests a receptor continuously probes the DNA for potential binding sites. Transcriptional activation reflects the probability that all components required for activation will meet at a certain chromatin site.
Besides binding to Hormone Responsive Elements, SHRs can also exert their effects by binding directly to other transcription factors. For example, ERα is able to bind to fos/jun, and thus regulate AP-1 mediated transcription of genes like cyclin D1. Similarly, ERα can bind Sp1 proteins and regulate transcription of genes that contain a Sp1/ER binding site. Interestingly, antagonists often have agonistic effects in this setting, which may be important when it comes to resistance to antagonistic compounds. This is illustrated by work from Kim et al., who used FRET to visualize the interaction between ERα and Sp1 [Kim et al., 2005]. Addition of the full anti-estrogen ICI 182,780 inhibits normal ERα mediated transcription, yet like agonist estradiol induced a FRET signal between ERα and Sp1 that correlated with Sp1 mediated transcription of a reporter construct.
Recently a number of groups have claimed a role for SHRs in non-genomic, extranuclear signalling events (Figure 2, step 6). Targeting ERα artificially to the plasma membrane has a marked influence on ERK1/2 signalling, which was not affected by full anti-estrogens [Rai et al., 2005]. Similar effects on the Mitogen Activated Protein Kinase (MAPK) and Protein Kinase A (PKA) pathways have also been attributed to the wildtype receptor [Levin, 2005; Razandi et al., 2004]. However, most studies are based on biochemical approaches where post-lysis artefacts are hard to exclude. Moreover, convincing microscopic pictures of SHR membrane localization are still lacking. Nevertheless, accumulating evidence seems to point to possible functions for SHRs other than those mediated by DNA binding.
Coregulator recruitment
The classical mode of action of SHRs involves ligand and DNA binding. For transcription to occur the subsequent recruitment of coregulator proteins is absolutely required (Figure 2, step 7). These regulatory proteins come in two types, coactivators and corepressors that respectively enhance or diminish transactivation activity through various enzymatic activities, including acetylating, deacetylating, methylating, ubiquitinating, and kinase activity. Ligand dependent recruitment of coregulators occurs through a hydrophobic cleft formed by helices 3, 4 and 12 in the AF-2 domain of the receptor [Gronemeyer et al., 2004]. In free receptors this pocket is shielded by a short amphipathic α-helix (H12) located at the carboxy-terminal end of the receptor that prevents AF-2 mediated coregulator binding in the absence of ligand. Upon hormone binding, this helix is repositioned, which opens a functional interface for coregulator recruitment through conserved LXXLL motifs in the cofactor. Antagonists exert their function by inducing a different conformational change of H12 that blocks or modulates the recruitment of these essential coregulators. However, not all coregulator binding occurs through the AF-2 region. Other conformational changes within the receptor and events like dimerization are also likely to be involved in coregulator recruitment. Moreover, the AF-1 region of SHRs plays an important role in ligand independent binding of coregulators. The exact coregulator requirements for transcription are dependent on cell type, and probably also on ligand and promoter context. This explains why partial antagonists can have antagonistic properties in one tissue, while exhibiting agonistic properties in another.
The most well studied coactivators are of the Steroid Receptor Coactivator (SRC) family, which includes SRC-1 (or NcoA-1), SRC-2 (also known as TIF-2 or GRIP1, NcoA-2) and SRC-3 (also known as RAC3, ACTR, AIB1, P/CIP and TRAM). Llopis et al., were the first to directly visualize the interaction between SRC-1 and a SHR in living cells using FRET [Llopis et al., 2000]. They showed that the ERα LBD exhibited some basal interaction with coactivators in unstimulated cells that was increased upon agonist addition and abolished by receptor antagonists. A large number of publications have since confirmed these findings, also with full-length ER constructs. Interestingly, these studies clearly show that the receptor adopts a slightly different conformation for various ligands, and this conformation significantly influences the binding of specific coregulatory proteins [Schaufele et al., 2000; Tamrazi et al., 2005; Weatherman et al., 2002]. Likewise, the small structural differences between the LBD of ERα and ERβ can result in profound differences in SRC-1 recruitment with the same ligand [Margeat et al., 2003]. We have visualized these conformational changes by fusing full-length ERα with YFP to its N- and CFP to its C-terminus [Michalides et al., 2004]. This allowed us to monitor conformational alterations of the receptor after ligand binding in the form of a change in FRET between CFP and YFP (Figure 3B-D). Indeed, subtle FRET differences between the various anti-estrogens tested were observed, showing that the receptor had adopted ligand specific conformations.
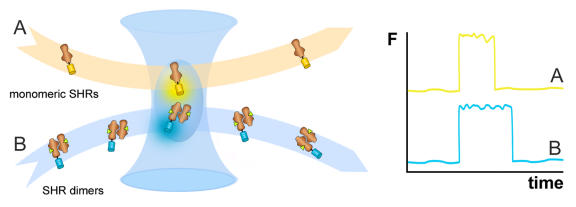
The stoichiometry of interaction between ERα and SRC-1 was studied using fluorescence correlation spectroscopy (FCS). By measuring the fluorescence signal from a very small excitation volume only, this technique allows precise determination of the diffusion coefficient of fluorescently labeled proteins, which is in part dependent on protein complex formation (Figure 5). In this way, Margeat et al., could show that the ERα dimer binds a single SRC-1 coactivator molecule [Margeat et al., 2001].
Figure 5. Fluorescence correlation spectroscopy (FCS).
By using the microscope objective lens to focus the laser beam, a diffraction limited excitation volume is created. The emitted fluorescence signal (F) from this observation volume fluctuates as labeled molecules diffuse in and out, and the duration of the fluctuations are related to the average time individual models reside within the volume. One such an event is depicted in the graph. The residence time can be used to determine the diffusion coefficient for the fluorescent molecules, which is dependent on their size and interaction with other proteins. For example, it can be used to discriminate monomeric Steroid Hormone Receptors (SHRs, A) from dimeric complexes (B).
One caveat of the above experiments is the use of only the AF-2 binding part of SRC-1. This may give false impressions when conclusions are extrapolated to the full-length receptor. Work from others and unpublished data from our lab have demonstrated that full-length SRC-1 binds the receptor through its AF-1 domain in a ligand independent manner [Dutertre and Smith, 2003] (Zwart et al., manuscript in preparation). AF-2 binding still functions as a switch to invoke full transactivation activity, but in contrast to what the above studies suggest, SRC-1 is already bound to unliganded receptors. This was also demonstrated by Stenoien et al., who used a fusion construct of CFP-ERα with a lac repressor domain to artificially target the receptor to an integrated lac operator array [Stenoien et al., 2001a]. Even in the absence of ligand, significant levels of SRC-1 were already present on the array, which further increased upon addition of agonist, and decreased after addition of antagonist. Further work by this group suggested that agonist binding predominantly stabilized SRC-1 binding, which translated in identical mobility of both proteins [Stenoien et al., 2001b]. Similar results were reported for other SHRs like GR and AR, and for other coregulators including CREB binding protein (CBP), glucocorticoid receptor (GR) interacting protein 1 (GRIP-1), and RIP140 [Becker et al., 2002; Carascossa et al., 2006; Marcelli et al., 2006]. Interestingly, in all cases dynamic DNA binding of SHRs was observed even in the presence of agonist and coregulators.
Transcription
Our view on SHR mediated transcription has more and more shifted from one in which a static holoenzyme of transcription factors is steadily built up after initial binding of the receptor to a response element, into a highly dynamic picture where different factors rapidly move in and out to perform temporary and local functions (Figure 2, step 8). In this so-called hit-and-run model, transcription only takes place when all factors coincidentally meet at the same time at the same location. Factors like DNA binding on specific HREs, and ligand dependent coregulator recruitment simply increase the odds that a successful transcriptional unit is formed and a gene is transcribed. FRAP analysis shows that indeed the majority of nuclear proteins are highly mobile [Phair and Misteli, 2000], except for Polymerase II, which once recruited has a residence time on the DNA in the order of minutes.
Whichever model is closer to reality, it is clear that transcription is a complex process, requiring dozens of proteins. Large-scale Chromatin IP assays have shown the recruitment of at least 46 factors to an empty promoter before continued transcription can take place [Metivier et al., 2003]. Interestingly, these experiments revealed a striking ATP-dependent periodicity in the recruitment of these factors, which was confirmed using FRAP analysis [Metivier et al., 2003; Reid et al., 2003]. The observed cycling time was in the order of 1h, much slower than the rapid exchange of SHRs on the template described earlier. Receptor degradation by the proteasome plays an important role in this process, as a block of proteasomal function halts the cycle after one round of transcription (Figure 2, step 9). This corresponds to previous observations that GR and ERα completely immobilize in the nucleus upon proteasome inhibition [Stavreva et al., 2004; Stenoien et al., 2001b]. However, since proteasome inhibition rapidly de-ubiquitinates histones, these effects may also be indirect, and the result of chromatin alterations [Dantuma et al., 2006]. An important role to maintain proper cycling has also been suggested for heat shock proteins like HSP-90 [Stavreva et al., 2004].
The exact role of this cyclic recruitment of transcription factors to the promoter is still unclear. It is also difficult to interpret how this slower cycle relates to the rapid exchange of SHRs that forms the basis of the hit-and-run model. Interestingly, the cyclic pattern of transcription factor presence on the promoter does suggest that some form of order in the build up of a functional transcriptional unit must exist, resulting in a so-called transcription factory. This may very well represent the summation of all rapid exchange events over a longer period of time, which suggests that both models may not necessarily be mutually exclusive, and might act in subsequent steps of transcription initiation.
Note
The literature on steroid hormone receptors is extensive and only selected studies were cited in this review due to space limitations. We sincerely apologize to all investigators whose work was not mentioned.
Abbreviations
- ER
estrogen receptor
- GR
glucocorticoid receptor
- PR
progesterone receptor
- AR
androgen receptor
- MR
mineralocorticoid receptor
References
- Beatson G.W. On the treatment of inoperable cases of carcinoma of the mamma: suggestions for a new method of treatment with illustrative cases. Lancet. 1896;2:104–107. [PMC free article] [PubMed] [Google Scholar]
- Becker M., Baumann C., John S., Walker D. A., Vigneron M., McNally J. G., Hager G. L. Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 2002;3:1188–94. doi: 10.1093/embo-reports/kvf244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carascossa S., Gobinet J., Georget V., Lucas A., Badia E., Castet A., White R., Nicolas J. C., Cavailles V., Jalaguier S. Rip140 Is a Repressor of the Androgen Receptor Activity. Mol Endocrinol. 2006;20:1506–1518. doi: 10.1210/me.2005-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couse J. F., Korach K. S. Estrogen receptor null mice: what have we learned and where will they lead us? Endocr Rev. 1999;20:358–417. doi: 10.1210/edrv.20.3.0370. [DOI] [PubMed] [Google Scholar]
- Dantuma N. P., Groothuis T. A., Salomons F. A., Neefjes J. A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J Cell Biol. 2006;173:19–26. doi: 10.1083/jcb.200510071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devin-Leclerc J., Meng X., Delahaye F., Leclerc P., Baulieu E. E., Catelli M. G. Interaction and dissociation by ligands of estrogen receptor and Hsp90: the antiestrogen RU 58668 induces a protein synthesis-dependent clustering of the receptor in the cytoplasm. Mol Endocrinol. 1998;12:842–54. doi: 10.1210/mend.12.6.0121. [DOI] [PubMed] [Google Scholar]
- Dutertre M., Smith C. L. Ligand-independent interactions of p160/steroid receptor coactivators and CREB-binding protein (CBP) with estrogen receptor-alpha: regulation by phosphorylation sites in the A/B region depends on other receptor domains. Mol Endocrinol. 2003;17:1296–314. doi: 10.1210/me.2001-0316. [DOI] [PubMed] [Google Scholar]
- Farla P., Hersmus R., Trapman J., Houtsmuller A. B. Antiandrogens prevent stable DNA-binding of the androgen receptor. J Cell Sci. 2005;118:4187–98. doi: 10.1242/jcs.02546. [DOI] [PubMed] [Google Scholar]
- Farla P., Hersmus R., Geverts B., Mari P. O., Nigg A. L., Dubbink H. J., Trapman J., Houtsmuller A. B. The androgen receptor ligand-binding domain stabilizes DNA binding in living cells. J Struct Biol. 2004;147:50–61. doi: 10.1016/j.jsb.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Fejes-Toth G., Pearce D., Naray-Fejes-Toth A. Subcellular localization of mineralocorticoid receptors in living cells: effects of receptor agonists and antagonists. Proc Natl Acad Sci U S A. 1998;95:2973–8. doi: 10.1073/pnas.95.6.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster T. Zwischenmolekulare energiewandering und fluoreszenz. . Annalen Physik. 1948;6:55–75. [Google Scholar]
- Georget V., Lobaccaro J. M., Terouanne B., Mangeat P., Nicolas J. C., Sultan C. Trafficking of the androgen receptor in living cells with fused green fluorescent protein-androgen receptor. Mol Cell Endocrinol. 1997;129:17–26. doi: 10.1016/s0303-7207(97)04034-3. [DOI] [PubMed] [Google Scholar]
- Goto K., Zhao Y., Saito M., Tomura A., Morinaga H., Nomura M., Okabe T., Yanase T., Takayanagi R., Nawata H. Activation function-1 domain of androgen receptor contributes to the interaction between two distinct subnuclear compartments. J Steroid Biochem Mol Biol. 2003;85:201–8. doi: 10.1016/s0960-0760(03)00196-1. [DOI] [PubMed] [Google Scholar]
- Green S., Walter P., Kumar V., Krust A., Bornert J. M., Argos P., Chambon P. Human oestrogen receptor cDNA: sequence, expression and homology to v-erb-A. Nature. 1986;320:134–9. doi: 10.1038/320134a0. [DOI] [PubMed] [Google Scholar]
- Gronemeyer H., Gustafsson J. A., Laudet V. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov. 2004;3:950–64. doi: 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- Hammes A., Andreassen T. K., Spoelgen R., Raila J., Hubner N., Schulz H., Metzger J., Schweigert F. J., Luppa P. B., Nykjaer A., Willnow T. E. Role of endocytosis in cellular uptake of sex steroids. Cell. 2005;122:751–62. doi: 10.1016/j.cell.2005.06.032. [DOI] [PubMed] [Google Scholar]
- Htun H., Holth L. T., Walker D., Davie J. R., Hager G. L. Direct visualization of the human estrogen receptor alpha reveals a role for ligand in the nuclear distribution of the receptor. Mol Biol Cell. 1999;10:471–86. doi: 10.1091/mbc.10.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htun H., Barsony J., Renyi I., Gould D. L., Hager G. L. Visualization of glucocorticoid receptor translocation and intranuclear organization in living cells with a green fluorescent protein chimera. Proc Natl Acad Sci U S A. 1996;93:4845–50. doi: 10.1073/pnas.93.10.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen E. V. On the mechanism of estrogen action. Perspect Biol Med. 1962;6:47–59. doi: 10.1353/pbm.1963.0005. [DOI] [PubMed] [Google Scholar]
- Kim K., Barhoumi R., Burghardt R., Safe S. Analysis of estrogen receptor alpha-Sp1 interactions in breast cancer cells by fluorescence resonance energy transfer. Mol Endocrinol. 2005;19:843–54. doi: 10.1210/me.2004-0326. [DOI] [PubMed] [Google Scholar]
- Kino T., Liou S. H., Charmandari E., Chrousos G. P. Glucocorticoid receptor mutants demonstrate increased motility inside the nucleus of living cells: time of fluorescence recovery after photobleaching (FRAP) is an integrated measure of receptor function. Mol Med. 2004;10:80–8. doi: 10.2119/2005-00026.Kino. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin E. R. Integration of the extranuclear and nuclear actions of estrogen. Mol Endocrinol. 2005;19:1951–9. doi: 10.1210/me.2004-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Fidler M. L., Lim C. S. Effect of initial subcellular localization of progesterone receptor on import kinetics and transcriptional activity. Mol Pharm. 2005;2:509–18. doi: 10.1021/mp0500418. [DOI] [PubMed] [Google Scholar]
- Lim C. S., Baumann C. T., Htun H., Xian W., Irie M., Smith C. L., Hager G. L. Differential localization and activity of the A- and B-forms of the human progesterone receptor using green fluorescent protein chimeras. Mol Endocrinol. 1999;13:366–75. doi: 10.1210/mend.13.3.0247. [DOI] [PubMed] [Google Scholar]
- Llopis J., Westin S., Ricote M., Wang Z., Cho C. Y., Kurokawa R., Mullen T. M., Rose D. W., Rosenfeld M. G., Tsien R. Y., Glass C. K. Ligand-dependent interactions of coactivators steroid receptor coactivator-1 and peroxisome proliferator-activated receptor binding protein with nuclear hormone receptors can be imaged in live cells and are required for transcription. Proc Natl Acad Sci U S A. 2000;97:4363–8. doi: 10.1073/pnas.97.8.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelli M., Stenoien D. L., Szafran A. T., Simeoni S., Agoulnik I. U., Weigel N. L., Moran T., Mikic I., Price J. H., Mancini M. A. Quantifying effects of ligands on androgen receptor nuclear translocation, intranuclear dynamics, and solubility. J Cell Biochem. 2006;98:770–788. doi: 10.1002/jcb.20593. [DOI] [PubMed] [Google Scholar]
- Margeat E., Bourdoncle A., Margueron R., Poujol N., Cavailles V., Royer C. Ligands differentially modulate the protein interactions of the human estrogen receptors alpha and beta. J Mol Biol. 2003;326:77–92. doi: 10.1016/s0022-2836(02)01355-4. [DOI] [PubMed] [Google Scholar]
- Margeat E., Poujol N., Boulahtouf A., Chen Y., Muller J. D., Gratton E., Cavailles V., Royer C. A. The human estrogen receptor α dimer binds a single SRC-1 coactivator molecule with an affinity dictated by agonist structure. J Mol Biol. 2001;306:433–42. doi: 10.1006/jmbi.2000.4418. [DOI] [PubMed] [Google Scholar]
- Martinez E. D., Rayasam G. V., Dull A. B., Walker D. A., Hager G. L. An estrogen receptor chimera senses ligands by nuclear translocation. J Steroid Biochem Mol Biol. 2005;97:307–21. doi: 10.1016/j.jsbmb.2005.06.033. [DOI] [PubMed] [Google Scholar]
- Matsuda K., Ochiai I., Nishi M., Kawata M. Colocalization and ligand-dependent discrete distribution of the estrogen receptor (ER)alpha and (ER)beta. Mol Endocrinol. 2002;16:2215–30. doi: 10.1210/me.2002-0110. [DOI] [PubMed] [Google Scholar]
- McNally J. G., Muller W. G., Walker D., Wolford R., Hager G. L. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–5. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- Metivier R., Penot G., Hubner M. R., Reid G., Brand H., Kos M., Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–63. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Michalides R., Griekspoor A., Balkenende A., Verwoerd D., Janssen L., Jalink K., Floore A., Velds A., van't Veer L., Neefjes J. Tamoxifen resistance by a conformational arrest of the estrogen receptor alpha after PKA activation in breast cancer. Cancer Cell. 2004;5:597–605. doi: 10.1016/j.ccr.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Muller W. G., Walker D., Hager G. L., McNally J. G. Large-scale chromatin decondensation and recondensation regulated by transcription from a natural promoter. J Cell Biol. 2001;154:33–48. doi: 10.1083/jcb.200011069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaich A. K., Walker D. A., Wolford R., Hager G. L. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol Cell. 2004;14:163–74. doi: 10.1016/s1097-2765(04)00178-9. [DOI] [PubMed] [Google Scholar]
- Nishi M., Ogawa H., Ito T., Matsuda K. I., Kawata M. Dynamic changes in subcellular localization of mineralocorticoid receptor in living cells: in comparison with glucocorticoid receptor using dual-color labeling with green fluorescent protein spectral variants. Mol Endocrinol. 2001;15:1077–92. doi: 10.1210/mend.15.7.0659. [DOI] [PubMed] [Google Scholar]
- Pearce D., Naray-Fejes-Toth A., Fejes-Toth G. Determinants of subnuclear organization of mineralocorticoid receptor characterized through analysis of wild type and mutant receptors. J Biol Chem. 2002;277:1451–6. doi: 10.1074/jbc.M105966200. [DOI] [PubMed] [Google Scholar]
- Phair R. D., Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–9. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- Rai D., Frolova A., Frasor J., Carpenter A. E., Katzenellenbogen B. S. Distinctive actions of membrane-targeted versus nuclear localized estrogen receptors in breast cancer cells. Mol Endocrinol. 2005;19:1606–17. doi: 10.1210/me.2004-0468. [DOI] [PubMed] [Google Scholar]
- Rayasam G. V., Elbi C., Walker D. A., Wolford R., Fletcher T. M., Edwards D. P., Hager G. L. Ligand-specific dynamics of the progesterone receptor in living cells and during chromatin remodeling in vitro. Mol Cell Biol. 2005;25:2406–18. doi: 10.1128/MCB.25.6.2406-2418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razandi M., Pedram A., Merchenthaler I., Greene G. L., Levin E. R. Plasma membrane estrogen receptors exist and functions as dimers. Mol Endocrinol. 2004;18:2854–65. doi: 10.1210/me.2004-0115. [DOI] [PubMed] [Google Scholar]
- Reid G., Hubner M. R., Metivier R., Brand H., Denger S., Manu D., Beaudouin J., Ellenberg J., Gannon F. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol Cell. 2003;11:695–707. doi: 10.1016/s1097-2765(03)00090-x. [DOI] [PubMed] [Google Scholar]
- Salomonsson M., Haggblad J., O'Malley B. W., Sitbon G. M. The human estrogen receptor hormone binding domain dimerizes independently of ligand activation. J Steroid Biochem Mol Biol. 1994;48:447–52. doi: 10.1016/0960-0760(94)90192-9. [DOI] [PubMed] [Google Scholar]
- Schaaf M. J., Lewis-Tuffin L. J., Cidlowski J. A. Ligand-selective targeting of the glucocorticoid receptor to nuclear subdomains is associated with decreased receptor mobility. Mol Endocrinol. 2005;19:1501–15. doi: 10.1210/me.2005-0050. [DOI] [PubMed] [Google Scholar]
- Schaufele F., Chang C. Y., Liu W., Baxter J. D., Nordeen S. K., Wan Y., Day R. N., McDonnell D. P. Temporally distinct and ligand-specific recruitment of nuclear receptor-interacting peptides and cofactors to subnuclear domains containing the estrogen receptor. Mol Endocrinol. 2000;14:2024–39. doi: 10.1210/mend.14.12.0572. [DOI] [PubMed] [Google Scholar]
- Schaufele F., Carbonell X., Guerbadot M., Borngraeber S., Chapman M. S., Ma A. A., Miner J. N., Diamond M. I. The structural basis of androgen receptor activation: intramolecular and intermolecular amino-carboxy interactions. Proc Natl Acad Sci U S A. 2005;102:9802–7. doi: 10.1073/pnas.0408819102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague B. L., Pego R. L., Stavreva D. A., McNally J. G. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys J. 2004;86:3473–95. doi: 10.1529/biophysj.103.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavreva D. A., Muller W. G., Hager G. L., Smith C. L., McNally J. G. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol Cell Biol. 2004;24:2682–97. doi: 10.1128/MCB.24.7.2682-2697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenoien D. L., Patel K., Mancini M. G., Dutertre M., Smith C. L., O'Malley B. W., Mancini M. A. FRAP reveals that mobility of oestrogen receptor-alpha is ligand- and proteasome-dependent. Nat Cell Biol. 2001a;3:15–23. doi: 10.1038/35050515. [DOI] [PubMed] [Google Scholar]
- Stenoien D. L., Nye A. C., Mancini M. G., Patel K., Dutertre M., O'Malley B. W., Smith C. L., Belmont A. S., Mancini M. A. Ligand-mediated assembly and real-time cellular dynamics of estrogen receptor alpha-coactivator complexes in living cells. Mol Cell Biol. 2001b;21:4404–12. doi: 10.1128/MCB.21.13.4404-4412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamrazi A., Carlson K. E., Rodriguez A. L., Katzenellenbogen J. A. Coactivator proteins as determinants of estrogen receptor structure and function: spectroscopic evidence for a novel coactivator-stabilized receptor conformation. Mol Endocrinol. 2005;19:1516–28. doi: 10.1210/me.2004-0458. [DOI] [PubMed] [Google Scholar]
- Tamrazi A., Carlson K. E., Daniels J. R., Hurth K. M., Katzenellenbogen J. A. Estrogen receptor dimerization: ligand binding regulates dimer affinity and dimer dissociation rate. Mol Endocrinol. 2002;16:2706–19. doi: 10.1210/me.2002-0250. [DOI] [PubMed] [Google Scholar]
- Tomura A., Goto K., Morinaga H., Nomura M., Okabe T., Yanase T., Takayanagi R., Nawata H. The subnuclear three-dimensional image analysis of androgen receptor fused to green fluorescence protein. J Biol Chem. 2001;276:28395–401. doi: 10.1074/jbc.M101755200. [DOI] [PubMed] [Google Scholar]
- Tyagi R. K., Lavrovsky Y., Ahn S. C., Song C. S., Chatterjee B., Roy A. K. Dynamics of intracellular movement and nucleocytoplasmic recycling of the ligand-activated androgen receptor in living cells. Mol Endocrinol. 2000;14:1162–74. doi: 10.1210/mend.14.8.0497. [DOI] [PubMed] [Google Scholar]
- Weatherman R. V., Chang C. Y., Clegg N. J., Carroll D. C., Day R. N., Baxter J. D., McDonnell D. P., Scanlan T. S., Schaufele F. Ligand-selective interactions of ER detected in living cells by fluorescence resonance energy transfer. Mol Endocrinol. 2002;16:487–96. doi: 10.1210/mend.16.3.0813. [DOI] [PubMed] [Google Scholar]