Abstract
Background
Most asexual eukaryotic lineages have arisen recently from sexual ancestors and contain few ecologically distinct species, providing evidence for long-term advantages of sex. Ancient asexual lineages provide rare exceptions to this rule and so can yield valuable information relating to the evolutionary forces underlying the maintenance of sex. Microsporidia are parasitic, unicellular fungi. They include many asexual species which have traditionally been grouped together into large, presumably ancient taxonomic groups. However, these putative ancient asexual lineages have been identified on the basis of morphology, life cycles and small subunit ribosomal RNA (16S rRNA) gene sequences, all of which hold questionable value in accurately inferring phylogenetic relationships among microsporidia.
Results
The hypothesis of a single, ancient loss of sex within the Nosema/Vairimorpha group of microsporidia was tested using phylogenetic analyses based on alignments of rRNA and RPB1 gene sequences from sexual and asexual species. Neither set of gene trees supported ancient asexuality, instead indicating at least two, recent losses of sex.
Conclusion
Sex has been lost on multiple, independent occasions within the Nosema/Vairimorpha group of microsporidia and there is no evidence for ancient asexual lineages. It appears therefore that sex confers important long-term advantages even upon highly simplified eukaryotes such as microsporidia. The rapid evolution of microsporidian life cycles indicated by this study also suggests that even closely related microsporidia cannot be assumed to have similar life cycles and the life cycle of each newly discovered species must therefore be completely described. These findings are relevant to the use of microsporidia as biological control agents, since several species under consideration as potential agents have life cycles that have been incompletely described.
Background
Unravelling the interplay of forces that underlie the evolution of sex in eukaryotes remains one of the most elusive goals of evolutionary biology. Sexual reproduction entails considerable short-term costs in comparison with asexual reproduction, yet most eukaryotic organisms remain capable of sex. A large number of competing hypotheses have been proposed to explain this phenomenon (see [1-5] for reviews), all of which postulate long-term selective advantage of sex over asex. The hypothesis that sex has long-term benefits coupled with short-term costs has also been used to explain the observation that most asexual eukaryotic lineages are of recent origin and contain relatively few species [6]. This general pattern has been observed so frequently among animals and plants that ancient asexual lineages containing ecologically diverse species are regarded as highly exceptional [7].
Unlike plants and animals, most sexual fungi are isogamous and so do not suffer from the "two-fold cost" of producing male gametes that do not contribute resources to the offspring [8]. All sexual fungi are also able to reproduce asexually, allowing the direct cost of sexual reproduction to be minimised by engaging in sex only when conditions are optimal [9]. Furthermore, experiments have demonstrated that sexual strains of the yeast Saccaromyces cerevisiae are able to out-compete asexual strains under a range of environmental conditions [10,11]. Given the demonstrable benefits and apparently low costs of sex in fungi, one might reasonably predict that fungi should demonstrate a high incidence of sexuality. Indeed population genetics studies using molecular markers have revealed genetic signatures of sex in most fungal species that have been tested, several of which were previously thought to be asexual [12]. However, ancient asexual lineages also exist among the fungi. In the arbuscular mycorrhizal (AM) fungi, an ancient and diverse group of plant symbionts, ancient asexuality was indicated by the relaxation of concerted evolution acting upon multi-copy ribosomal RNA (rRNA) genes [13,14].
The existence of ancient asexual lineages has also been implied in the microsporidia, a diverse group of intracellular fungal parasites that infect a wide range of vertebrate and invertebrate animal hosts. Some of the most intensively studied microsporidia belong to the genus Nosema, parasites of arthropods that have attracted attention both as causes of disease in honey bees and silk moths and as potential biological control agents for insect pests. Microsporidian life cycles typically involve the alternation of diplokaryotic (binucleate, diploid) stages and monokaryotic (uninucleate, haploid) stages (Figures 1 and 2). Members of the genus Nosema appear to have lost the monokaryotic cycle, remaining diploid throughout their life cycles, and are therefore considered to be asexual. Molecular evidence supporting asexuality in Nosema has been provided by variable rRNA gene sequences amplified from single spores of N. bombi [15], indicating relaxation of concerted evolution, a finding similar to that supporting asexuality in the AM fungi [13]. Nosema contains over 100 described species, with several new species added every year, and its host range spans the Arthropoda, including insects, arachnids and crustaceans. Furthermore, many of the physiological interactions between Nosema parasites and their hosts are complex and host-specific, indicating a high degree of host-parasite coevolution. These include many incidences of transovarial transmission [16] and, most dramatically, the feminization of genetically male crustacean hosts by the parasite N. granulosis [17]. Given the assumption of ancient asexuality within Nosema, the high level of species diversity and high degree of host-specialisation within this genus presents a major challenge to the doctrine of limited evolution within asexual eukaryotic lineages.
Figure 1.
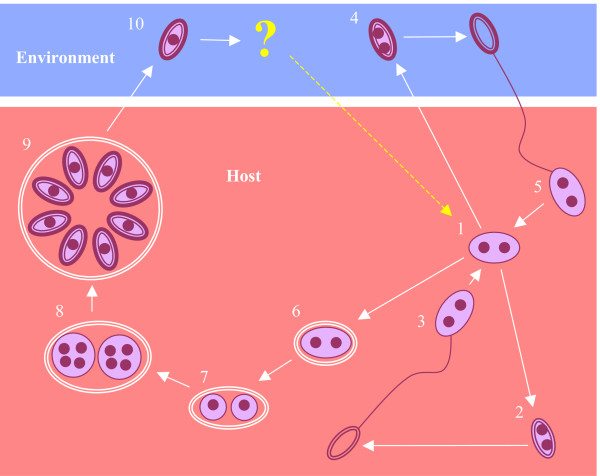
Nosema/Vairimorpha life cycles. 1. Diplokaryotic meront 2. Primary (autoinfective) diplokaryotic spore 3. Within-host transmission or vertical transmission 4. Secondary (environmental) diplokaryotic spore (not N. granulosis, N. empoascae) 5. Horizontal transmission 6. Formation of sporophorous vesicle (Vairimorpha only) 7. Meiosis and karyokinesis 8. Nuclear division to form plasmodia 9. Monokaryotic sporogony 10. Monokaryotic environmental spore. Sexual species undergo stages 1–10, producing diplokaryotic mitospores and monokaryotic meiospores. The fate of meiospores has never been directly observed in Nosema/Vairimorpha but is assumed to be similar to that within the Amblyosporidae (Figure 2), in which monokaryotic spores differentiate into gametes and undergo cytoplasmic fusion, restoring the diplokaryotic state and completing the life cycle. In V. imperfecta monokaryotic sporogony is aborted at stage 9. Asexual lepidopteran Nosema species such as N. bombycis retain only the diplokaryotic cycles (stages 1–5). The life cycle of N. granulosis is further reduced, retaining only the primary diplokaryotic cycle (stages 1–3).
Figure 2.
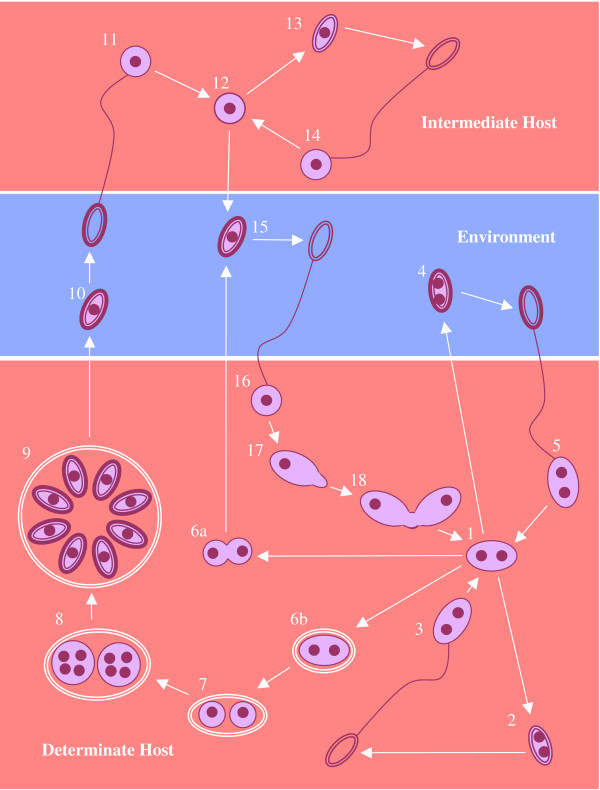
Amblyosporidae life cycles. 1. Diplokaryotic meront 2. Primary (autoinfective) diplokaryotic mitospore 3. Within-host transmission or vertical transmission (not Hyalinocysta) 4. Secondary (environmental) diplokaryotic spore (Culicosporella only) 5. Horizontal transmission 4a. Nuclear dissociation and karyokinesis (Culicospora and Edhazardia only) 4b. Formation of sporophorous vesicle (not Culicospora) 5. Meiosis and karyokinesis 6. Nuclear division to form plasmodia 7. Monokaryotic sporogony (abortive in Culicosporella and Edhazardia) 8. Monokaryotic meiospore 9. Transmission to intermediate host 10. Monokaryotic meront 11. Primary monokaryotic mitospore 12. Within-host transmission 13. Secondary monokaryotic mitospore (not Culicosporella) 14. Transmission to determinate host 15. Gametogenesis 16. Plasmogamy In the sexual Amblyospora, Duboscqia, Hyalinocysta and Parathelohania species, monokaryotic meiospores infect an intermediate host. Monokaryotic mitospores released by the intermediate host then infect the determinate host, where they differentiate into gametes and undergo plasmogamy to form diplokaryotic cells. In Edhazardia aedis and Culicosporella lunata, meiosis is abortive and any meiospores produced are non-functional while in Culicospora magna, meiosis is completely absent. E. aedis and C. magna produce monokaryotic mitospores by nuclear dissociation, eliminating meiosis. These monokaryotic spores are functionally equivalent to the spores produced by the intermediate host of Amblyospora, capable of infecting the determinate host and of undergoing gametogenesis and plasmogamy. In C. lunata, diplokaryotic mitospores are produced for horizontal transmission between larval hosts, eliminating both gametogenesis and plasmogamy.
Morphologically based taxonomies of the microsporidia have generally assumed that changes of life cycle, such as the loss of the monokaryotic cycle, correspond to major evolutionary transitions [18-21]. It was on this basis that all microsporidian species that remained diplokaryotic throughout their life cycles and lacked a sporophorous vesicle were placed within Nosema [19]. Recently, the taxonomy of Nosema has been extensively revised on the basis of phylogenetic analyses using the sequence of the small subunit ribosomal RNA (16S rRNA) gene. These molecular phylogenies have shown "Nosema" species scattered widely throughout the Phylum Microspora [22], suggesting that the monokaryotic cycle has been lost on many separate occasions. On the basis of 16S rRNA phylogenies, several microsporidian species have been transferred from Nosema to newly created genera (e.g. Antonospora locustae, Paranosema grylli, Brachiola algerae), while several species formerly assigned to the genus Vairimorpha (including Vairimorpha's type species, V. necatrix) have been tentatively placed within Nosema [23]. Following the notation of Baker et al. [23], I will henceforth refer to the monophyletic group containing the Nosema type species N. bombycis and the Vairimorpha type species V. necatrix as Nosema/Vairimorpha. Most of the "Vairimorpha" species within this group possess complete sexual life cycles, with alternating monokaryotic and diplokaryotic stages. The exception is V. imperfecta, in which meiosis is followed by an abortive monokaryotic sporogony, indicating an intermediate phase in the loss of sex [24]. The presence of these sexual species within Nosema/Vairimorpha suggests that the sexual, monokaryotic cycle has been lost since the origin of the genus, perhaps on several separate occasions.
Phylogenetic analyses based solely upon 16S rRNA sequences are, however, of limited use when comparing closely related microsporidian species. This is because the microsporidian small ribosomal RNA subunit is substantially shorter than those of other fungi and lacks many of the more variable regions [25-27]. In fact, both the small and large ribosomal RNA subunits of microsporidia are even shorter than those of most prokaryotes and consist of little more than a core of highly conserved sequences. Consequently, microsporidian 16S rRNA sequence alignments rarely contain sufficient phylogenetically informative sites to unambiguously assign a topology to trees consisting of closely related species [24,28-30]. However, despite their short length and low sequence variability, the rRNA genes of Nosema/Vairimorpha vary exceptionally in the order in which they occur within the ribosomal repeat unit. In non-microsporidian fungi, and most other eukaryotes, the order of rRNA genes from 5' to 3' is 18S, 5.8S, 25S, with internal transcribed spacers between the 18S and 5.8S subunit genes (ITS1) and between the 5.8S and 25S subunit genes (ITS2) (Figure 3a). In the Nosema species N. apis [31] and N. bombi [15] and in all known non-Nosema microsporidian species [32-36] the 18S subunit is reduced in size to 16S, the 5.8S and 25S subunit genes are reduced and the ITS2 spacer is absent, producing the gene order 5'-16S, 18S-3' (Figure 3b). In N. bombycis an additional rearrangement has occurred, placing the 18S subunit gene upstream of the 16S subunit gene [37]. A 5S subunit is also positioned downstream of the 16S subunit of N. bombycis to give the order 5'-18S, 16S, 5S-3' (Figure 3c) [37]. This deviates from the arrangement seen in the model microsporidium Encephalitozoon cuniculi, in which no 5S subunits occur in the vicinity of the larger ribosomal repeat unit [38]. The unusual ribosomal subunit gene order demonstrated by N. bombycis also occurs in N. spodopterae [39], N. plutellae (direct submission to Genbank: AY960987) and N. antheraeae [40], which, like N. bombycis, are parasites of Lepidoptera and which fall close to N. bombycis in phylogenetic trees based on the 16S rRNA gene sequence [29]. Nosema/Vairimorpha species possess multiple copies of the ribosomal RNA repeat unit which can show intragenomic variation in sequence [15,31] and some isolates of N. bombycis also possess fragmented copies of rRNA genes (Figure 3d, e) [41] which are transcriptionally active and coexist with intact rRNA copies within the same genome. The fact that, within Nosema/Vairimorpha, the rRNA repeat unit exists in multiple copies with intragenomic variation in gene order, integrity and sequence is a potential source of confusion in phylogenies based entirely upon 16S rRNA gene sequences.
Figure 3.
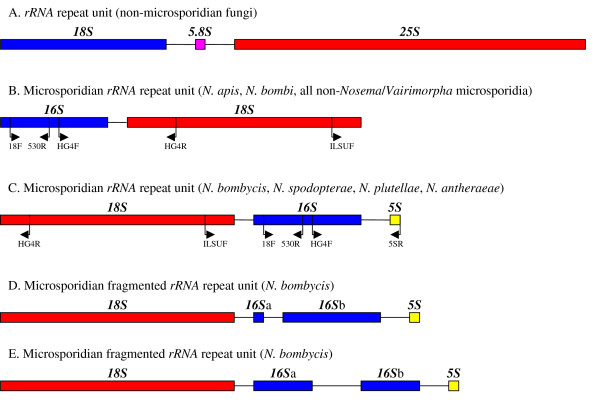
Different types of rRNA repeat unit in microsporidia and in non-microsporidian fungi. Figures 3B and 3C show the annealing sites of the primers used to determine the order of genes within microsporidian rRNA repeat units. Figures 3D and 3E show the fragmented 16S rRNA genes discovered in N. bombycis by Iiyama et al. [41].
In order to improve the resolution of the Nosema rRNA gene phylogeny, the 18S rRNA gene was sequenced from a representative group of sexual and asexual species to produce an alignment including both 16S and 18S rRNA gene sequences. As an independent test of the phylogeny indicated by the rRNA genes, the largest subunit of the single-copy protein coding gene RNA polymerase II (RPB1) was also sequenced from each of the Nosema/Vairimorpha species. These two phylogenetic analyses supported incompatible hypotheses for speciation within Nosema, casting doubt upon the reliability of the rRNA gene sequence phylogeny. However, both phylogenetic hypotheses supported multiple, independent losses of sex within the Nosema genus. The implications of these results are discussed with regard to the evolution and maintenance of sex in microsporidia and to the evaluation of microsporidia as agents of biological control.
Results
rRNA Phylogeny
In order to improve the resolution of the Nosema/Vairimorpha rRNA phylogeny, both the 16S and 18S ribosomal genes were sequenced from a representative group of two sexual and five asexual Nosema/Vairimorpha species, including parasites of insect and crustacean hosts. To these were added six published sequences, obtained from Genbank (See Additional File 1). Phylogenetic analysis of the resulting DNA sequence alignment using maximum likelihood and Bayesian inference yielded identical consensus trees, both of which supported a phylogeny in which Nosema/Vairimorpha is divided into two well-supported clades (Figure 4). These two major clades correspond to the two different rRNA gene orders found within Nosema/Vairimorpha. Sexual species occur within both clades, indicating at least two independent losses of sex.
Figure 4.

Consensus phylogenetic tree for the 16S and 18S rRNA genes of Nosema/Vairimorpha. Maximum likelihood and Bayesian inference criteria produced identical consensus tree topologies. Support for each internal node is given as maximum likelihood bootstrap value/Bayesian posterior probability of partition. Branch lengths are drawn proportionally to the number of changes assigned to each branch. The notation following each species name indicates host (Hy = Hymenoptera, L = Lepidoptera, C = Crustacea, He = Hemiptera), life cycle (A = asexual, S = sexual) and rRNA gene order.
RPB1 Phylogeny
RPB1 occurs as a single copy in V. necatrix with a length of 1,606 codons (4818 bp), uninterrupted by introns [42]. A 1979 bp fragment of the RPB1 gene was sequenced from a representative group of two sexual and six asexual Nosema/Vairimorpha species, including parasites of insect and crustacean hosts. To these was added the published sequence of N. tyriae, obtained from Genbank (See Additional File 1). Phylogenetic analysis of the resulting DNA sequence alignment using maximum likelihood and Bayesian inference yielded identical consensus trees (figure 5). The topology of this RPB1 gene tree was incongruent with that of the consensus rRNA tree, indicating no correspondence between rRNA gene order and RPB1 gene sequence. However, despite its incongruence with the rRNA tree, the topology of the consensus RPB1 tree still indicated at least two independent losses of sex.
Figure 5.
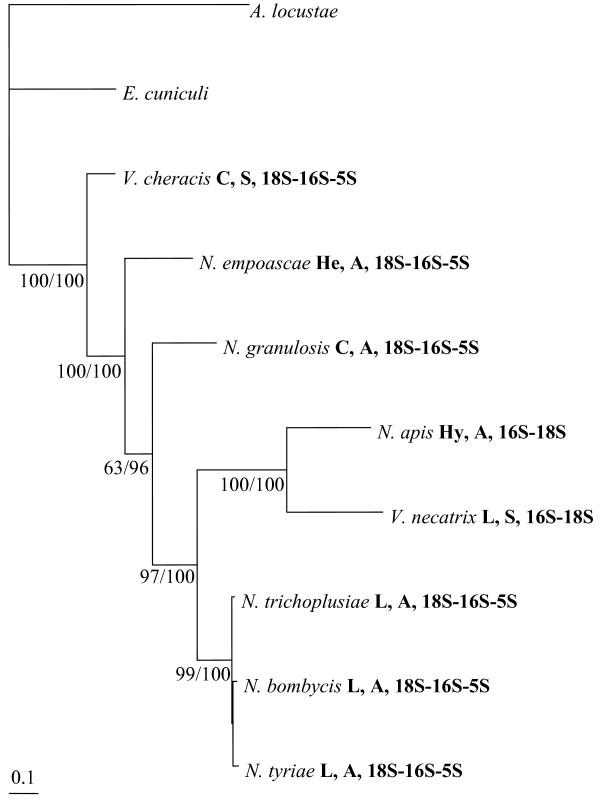
Consensus phylogenetic tree for the RPB1 gene of Nosema/Vairimorpha. Maximum likelihood and Bayesian inference criteria produced identical consensus tree topologies. Support for each internal node is given as maximum likelihood bootstrap value/Bayesian posterior probability of partition. Branch lengths are drawn proportionally to the number of changes assigned to each branch. The notation following each species name indicates host (Hy = Hymenoptera, L = Lepidoptera, C = Crustacea, He = Hemiptera), life cycle (A = asexual, S = sexual) and rRNA gene order.
Hypothesis Testing
Approximately unbiased (AU) and weighted Shimodaira-Hasegawa (WSH) tests [43,44] were performed upon the rRNA and RPB1 alignments in order to compare the likelihoods of three different phylogenetic hypotheses for Nosema/Vairimorpha speciation. This was accomplished by comparing trees generated according to maximum likelihood criteria, under topological constraints corresponding to a priori hypotheses of 1) host-parasite co-speciation (insect parasites are monophyletic), 2) ancient asexuality (asexual species are monophyletic), 3) a single fixation of the 5'-18S, 16S, 5S-3' gene order (species with the 5'-18S, 16S, 5S-3' gene order are monophyletic) (Figure 6). Results of the tests are given in table 1.
Figure 6.
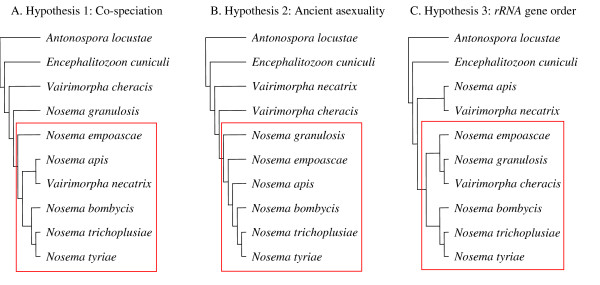
Phylogenetic hypotheses for speciation within Nosema/Vairimorpha. Hypotheses were generated according to maximum criteria, using the following topological constraints: A. co-speciation: parasites of insects are constrained to be monophyletic, B. ancient asexuality: asexual species are constrained to be monophyletic, C. rRNA gene order: species with the 5'-18S, 16S, 5S-3' gene order are constrained to be monophyletic.
Table 1.
Results of approximately unbiased (AU) and weighted Shimodaira-Hasegawa (WSH) tests, comparing three phylogenetic hypotheses for Nosema/Vairimorpha speciation.
| Gene | Constraint (monophyly) | -ln L | Diff in -ln L | AU test P | WSH test P |
| RPB1 | Insect parasites | 13940.79 | |||
| rRNA gene order | 14004.82 | 64.03 | 0.001 | 0.001 | |
| Asexuality | 14097.82 | 157.03 | <0.0001 | <0.0001 | |
| rRNA | rRNA gene order | 16981.85 | |||
| Insect parasites | 17052.06 | 70.21 | 0.0002 | <0.0001 | |
| Asexuality | 17969.55 | 987.70 | <0.0001 | <0.0001 |
The constrained rRNA tree produced by the hypothesis of a single change in rRNA gene order was completely congruent with the maximum likelihood rRNA consensus tree (Figure 4). The comparison of constrained rRNA trees indicated that the hypothesis of a single change in gene order was significantly more likely to reflect the true phylogeny than either the ancient asexuality hypothesis or the host-parasite co-speciation hypothesis (Table 1). In contrast, the comparison of constrained RPB1 trees indicated that the host-parasite co-speciation hypothesis was significantly more likely to reflect the true phylogeny than either the ancient asexuality hypothesis or the hypothesis of a single change in gene order. The host-parasite co-evolution tree and the maximum likelihood consensus tree were incongruent only with regard to the position of the crustacean parasite N. granulosis, which was weakly supported in the consensus tree. Both sets of gene trees indicated that the hypothesis of ancient asexuality was the least likely of the three alternative hypotheses (Table 1).
Discussion
Incongruence of rRNA and RPB1 Phylogenies
The consensus phylogenetic trees produced for the rRNA and RPB1 genes of Nosema/Vairimorpha are clearly incongruent. The RPB1 tree tentatively supports monophyly of the insect parasites and thereby supports the prediction that parasites should co-speciate with their hosts, especially in the case of parasites such as Nosema/Vairimorpha in which the relationship between host and parasite is very intimate [45]. In contrast, the rRNA tree indicates paraphyly for the insect parasites, with the crustacean and hemipteran parasites forming a sister clade to the lepidopteran group containing N. bombycis. These odd bedfellows share the unusual 5'-18S, 16S, 5S-3' gene order and so the rRNA tree appears to support a single origin for this gene order. However, this conclusion requires the dubious assumption that the sequences of rRNA genes are independent of the rRNA gene order.
The Nosema/Vairimorpha rRNA repeat unit occurs as multiple copies [15,31] and there is evidence for intragenomic variation in the sequence of the rRNA genes, indicating relatively low levels of concerted evolution [15,41]. It is therefore possible that rRNA repeat units with both gene orders coexisted in the Nosema/Vairimorpha genome for some time. If the genome of the common ancestor of Nosema/Vairimorpha contained some rRNA repeat units with the 5'-18S,16S,5S-3' gene order and others with the 5'-16S, 18S-3' gene order, it would have been possible for the 5'-18S, 16S, 5S-3' gene order to have been lost or fixed on several independent occasions within the genomes of different Nosema/Vairimorpha species (Figure 7). Nucleotide substitutions that occurred after the rearrangement that produced the 5'-18S, 16S, 5S-3' gene order but before speciation from the common ancestor would, in this case, be shared across species by all 5'-18S, 16S, 5S-3' rRNA repeat units. Such shared nucleotide polymorphisms would then be fixed or lost along with the gene order with which they were associated. Under this scenario, Nosema/Vairimorpha phylogenies based upon the rRNA sequence would falsely support monophyly of all species sharing the 5'-18S, 16S, 5S-3' gene order. In contrast, RPB1 is a single-copy gene and so its phylogeny is not likely to have been confused by the presence of different copies with different evolutionary histories.
Figure 7.

Multiple, independent fixations of one rRNA copy can produce an rRNA phylogenetic tree that is incongruent with the species tree. A-B: two copies of the rRNA repeat unit accumulate differences in nucleotide sequence and in gene order. C: speciation occurs and one rRNA repeat unit copy is fixed within each new species. D-E: each rRNA repeat unit copy is fixed twice, independently, resulting in an rRNA gene tree that is incongruent with the species tree.
Multiple Losses of Sex within Nosema/Vairimorpha
Despite the differences in topology between the rRNA and RPB1 phylogenies, Shimodaira-Hasegawa and Kishino-Hasegawa tests of both phylogenies conclusively rejected monophyly of the asexual Nosema/Vairimorpha species. The closest phylogenetic group to Nosema/Vairimorpha (according to 16S rRNA trees) contains three species of microsporidian parasites of freshwater crayfish, erroneously placed within the genus Thelohania [46-48]. The life cycles of all three "Thelohania" species are similar to that of V. necatrix and V. cheracis (Figure 1), containing both monokaryotic and diplokaryotic phases, with evidence of meiosis. This suggests that the most recent common ancestor of Nosema/Vairimorpha and the "Thelohania" parasites of crayfish was sexual and that the monokaryotic cycle, and hence the capacity for sexual reproduction, was lost on at least two occasions within the Nosema/Vairimorpha group. It can therefore be concluded that Nosema/Vairimorpha is not an ancient asexual lineage but consists of a core of sexual species that have repeatedly given rise to asexual clones. This situation is not uncommon among the fungi [49]. Many asexual strains of otherwise sexual fungal species have been isolated from environmental and medical sources [50,51] and laboratory studies have demonstrated that the capability for sexual reproduction can be rapidly eroded when fungi are constrained to reproduce asexually [52]. When the loss of sex occurs due to the deletion or inactivation of genes necessary for meiosis or sexual interactions, reversion to sex is considered to be unlikely [7] since this would generally require multiple, specific back-mutations.
The phylogenetic group including the crayfish "Thelohania" and Nosema/Vairimorpha is one of only three microsporidian groups in which the production of sexual meiospores has been described in detail. The others are the dipteran parasites of the family Amblyosporidae [53] and the "Thelohania" parasites of Solenopsis ants [54]. According to 16S rRNA trees, the sexual "Thelohania" parasites of ants are phylogenetically distant both from the true Thelohania and from the false "Thelohania" of crayfish but form a close sister group to the asexual microsporidia of the genus Anncaliia [55] indicating that members of the latter genus may also have recently abandoned sex. Within the Amblyosporidae, life cycle evolution also appears to have followed a very similar pattern to that within Nosema/Vairimorpha, with 16S rRNA trees indicating independent losses of the sexual cycle in the species Edhazardia aedis, Culicospora magna and Culicosporella lunata [56]. Like V. imperfecta, E. aedis and C. lunata undergo abortive meiotic sporogony, suggesting relatively recent loss of the sexual cycle. Frequent losses of the sexual cycle therefore appear to be common to several microsporidian groups.
Why give up sex?
For obligate parasites such as the microsporidia sex has the potential to increase the efficiency of directional selection upon the parasite and hence the speed with which the parasite can adapt to overcome the resistance of its host [49,57]. Furthermore microsporidia are isogamous and are therefore unaffected by the "two-fold cost" of sex. Given these apparent advantages of the sexual cycle it is, perhaps, surprising that its loss appears to have occurred so frequently among the microsporidia. However, sex can carry additional costs arising from the production of gametes and the process of fertilisation. Nosema/Vairimorpha may be particularly affected by such costs because many species rely on efficient vertical transmission from female hosts to their offspring. Any additional energy costs incurred by sexual reproduction are passed on to the host, reducing its survival and/or fecundity and hence reducing vertical transmission of the parasite [58].
The situation is further complicated by the fact that the monokaryotic stages of sexual microsporidia perform other functions in addition to sex. Selection to retain monokaryotic stages of the life cycle may not, therefore, correspond directly to selection for the maintenance of sex. For example, meiospores produced by the sexual species V. necatrix and V. plodiae are very resistant to degradation in the extracellular environment. Production of meiospores in these species increases markedly at low temperatures [59,60], suggesting that meiospores may function as an overwintering stage, allowing the parasite to survive while active hosts are unavailable. In contrast, asexual Nosema parasites of Lepidoptera do not produce resistant meiospores but overwinter within the eggs, pupae or adults of the host and are vertically transmitted to the next generation. In this case, loss of the monokaryotic cycle may have been due to selection against the production of resistant spores rather than selection against sex.
Where the monokaryotic phase of the life cycle involves an intermediate host, as occurs in the Amblyosporidae [61] (Figure 2b), the absence or poor availability of this host might also select against the production of monokaryotic stages and hence against sex. The fate of meiospores produced by V. necatrix and other sexual Nosema/Vairimorpha species has never been documented and it is not known whether transmission is direct or involves an intermediate host. Similarly, the fate of meiospores is known in neither the crustacean Thelohania species nor the ant "Thelohania" species, leading to speculation that these sexual microsporidian groups might also use intermediate hosts [54,62]. If sexuality tends to be intimately associated with a multi-host life cycle in microsporidia then the loss of sex might occur as a by-product of the elimination of an intermediate host from the life cycle.
Implications for Biological Control and Pest Management
In view of their status as pests and their potential for use in biological control, it is important to understand the sexual cycles of Nosema/Vairimorpha species and of microsporidia in general. There are now several documented cases of microsporidia being accidentally introduced to new geographical areas along with their hosts [63] and further, deliberate, introductions are contemplated for the purpose of biological control. Several of the proposed introductions involve sexual microsporidian species such as V. necatrix, V. lymantriae and T. solenopsis [64]. These examples are particularly worrying because the fate of the meiospores of these species is unknown. Previous studies have investigated the host range of microsporidian isolates with regard only to their determinate hosts [65,66]. There are, however, close similarities in meiospore production between Amblyospora, Vairimorpha and Thelohania. The possibility that meiospores of the latter two genera might, like those of Amblyospora, infect non-target intermediate hosts needs, therefore, to be taken into consideration by proponents of their use in biological control. The potential for introduced microsporidian species to hybridise with native species and the effects that such gene flow would have upon the host range and pathogenicity of these organisms also requires consideration. This is particularly relevant with regard to the introduction of V. necatrix to Europe, where there exist many native Nosema/Vairimorpha species, including pests such as N. apis and parasites such as N. bombi that infect threatened insect species [67]. In order to conduct appropriate experiments to investigate these issues, a more detailed understanding of the sexual phase of microsporidian life cycles is urgently required, including knowledge of mating types and intermediate hosts.
Conclusion
Phylogenetic analysis indicates that sex has been lost within the genus Nosema/Vairimorpha on at least two separate occasions. It is therefore concluded that that this genus does not constitute or include an ancient asexual lineage. Instead, the evidence presented here suggests that the many asexual Nosema/Vairimorpha species have evolved recently from sexual ancestors.
There is currently a trend towards describing new microsporidian species on the basis of incomplete life cycle data or even solely on the basis of their 16S rRNA sequences. The evidence presented here indicates that the life cycles of Nosema/Vairimorpha species can evolve very rapidly relative to evolution at the molecular level. This means that a newly discovered species will not necessarily share life cycle characteristics with a model species, even if their DNA sequences are extremely similar. The evidence presented here also suggests that rRNA sequences are unreliable indicators of the phylogenetic relationships of closely related species and so the phylogenetic information yielded by comparison of rRNA sequences is of limited value. On this basis, it is proposed that taxonomic descriptions of microsporidian species should describe every stage of the life cycle and should include molecular phylogenetic information based on the sequences of several genes.
The finding that life cycle reduction through the loss of sex has occurred on multiple occasions both in Nosema/Vairimorpha and in the Amblyosporidae has important implications for the way in which life cycle evolution within the microsporidia, and within the fungi in general, is viewed. The majority of described Nosema/Vairimorpha species possess a reduced, asexual life cycle and so description of the genus on the basis of one or a few species might have indicated that the entire group was asexual. Many putatively asexual groups of microsporidia and other fungi are described on the basis of one or a few species. It is therefore possible that their life cycles are truncated versions of the life cycles of recent, sexual ancestors, sexual descendents of which have yet to be described.
Methods
Species used and nucleic acid extraction
Complete ribosomal repeat unit sequences, including both the 16S and the 18S rRNA genes, are published for nine Nosema/Vairimorpha species. All of these were parasites of Lepidoptera or Hymenoptera and all are asexual (See Additional File 1). The only Nosema/Vairimorpha species for which RPB1 sequences are published are V. necatrix and N. tyriae, both of which are parasites of Lepidoptera. In order to include species from a more representative range of hosts and to include multiple sexual species, the complete ribosomal repeat unit and a fragment of the RPB1 gene were sequenced from the crustacean parasites N. granulosis and V. cheracis, from the hemipteran parasite N. empoascae, from the lepidopteran parasites N. bombycis, N. trichoplusiae and V. necatrix and from the hymenopteran parasite N. apis. The genomic DNA of N. granulosis, N. apis, N. empoascae, V. cheracis and V. necatrix were extracted from infected tissue samples stored in ethanol. The genomic DNA of N. bombycis and N. trichoplusiae was extracted from purified spores stored in ethanol. Genomic DNA extraction was performed using Qiagen's DNeasy® DNA purification kit, according to the manufacturer's instructions.
Determination of rRNA gene order, amplification and sequencing of rRNA genes
The order of rRNA genes in the ribosomal repeat unit of each species was ascertained by PCR using gene order specific primer combinations. The primer set HG4F: HG4R [31] produced a 947 bp product only when rRNA genes occurred in the 5'-16S, 18S-3' order (Figure 3B), while the primer sets ILSUF: 530R [37,68] and 18F: 5SR [37,69] produced products of 870 bp and 1624 bp only when the rRNA genes occurred in the 5'-18S, 16S, 5S-3' order (Figure 3C). The ribosomal repeat unit was amplified with Invitrogen's recombinant Taq DNA polymerase, in 5 fragments with an annealing temperature of 50°C and an extension time of one minute. Direct sequencing was performed using BigDye® terminators on an ABI 3100 high throughput sequencer. PCR and sequencing were performed using the species-specific primers (see Additional File 2).
Amplification and sequencing of RPB1
RPB1 occurs as a single copy in V. necatrix with a length of 1,606 codons (4818 bp), uninterrupted by introns [42]. A large (2958 bp) fragment of RPB1 was previously sequenced from N. tyriae [70], providing a second sequence from species within the Nosema/Vairimorpha group. Alignment of these two sequences allowed conserved regions of RPB1 to be identified, facilitating the design of degenerate PCR primers with which to amplify RPB1 from additional species. These initial sequences were then used to design specific RPB1 primers (See Additional File 1) for each of the Nosema/Vairimorpha species included in the analysis. The 1979 bp RPB1 fragment used in the analysis was amplified with Invitrogen's recombinant Taq DNA polymerase, using the species specific primer combinations: N. apis:NaRPB1_1F:NaRPB1_4R, N. bombycis and N. trichoplusiae: NbRPB1_1F:NbRPB1_5R, N. empoascae: NeRPB1_1F:NeRPB1_3R, N. granulosis:NgRPB1_1F:NgRPB1_4R, V. cheracis:VcRPB1_1F:VcRPB1_4R, V. necatrix:VnRPB1_1F:VnRPB1_5R. All primer combinations used an annealing temperature of 50°C and an extension time of two minutes. Direct sequencing was performed using BigDye® terminators on an ABI 3100 high throughput sequencer, with the species specific primers (see Additional File 2).
Phylogenetic analysis
Due to the high level of DNA sequence divergence between microsporidia and other fungi [71,72], outgroup species were selected from within Phylum Microspora. The two species chosen as outgroups were Encephalitozoon cuniculi, a parasite of mammals and Antonospora locustae, a parasite of orthopteran insects. According to the phylogenetic analysis of Vossbrinck and Debrunner-Vossbrinck [28], Encephalitozoon is a close sister group to Nosema/Vairimorpha within the Class Terresporidia while Antonospora is more distantly related, falling within Class Aquasporidia.
Ribosmal RNA gene sequences were aligned using Clustal W with an equal transition: transversion ratio, a gap opening penalty of 15 and a gap elongation penalty of 6.66. Since the internal spacers of ribosomal repeat units with the 5'-18S, 16S, 5S-3' rRNA gene order are not homologous to those of units with the 5'-16S, 18S-3' order [37], the spacers were excluded from the alignment. RPB1 DNA sequences were aligned on the basis of an RPB1 amino acid sequence template, implemented in the programme DAMBE [73]. The rRNA and RPB1 alignments were tested for saturation using the methods of Steel et al. [74] and Xia et al. [75], implemented in DAMBE. In neither case was there significant evidence for saturation so all polymorphic sites were included in the phylogenetic analyses. In order to test the possibility that some RPB1 sequences were those of pseudogenes, Ka/Ks values were calculated between each new RPB1 sequence and the well-characterised RPB1 sequence of V. necatrix. All pairwise comparisons produced Ka/Ks values within the range 0.05–0.18 (Table 2), indicating strong purifying selection upon all sequences used in the alignment and hence providing a good indicator that all sequences were those of functional genes.
Table 2.
Ka/Ks values for each new Nosema/Vairimorpha RPB1 sequence, compared to that of V. necatrix.
| Species pair | Non-synonymous positions | Non-synonymous differences | Ka | Synonymous positions | Synonymous differences | Ks | Ka/Ks |
| N. apis:V. necatrix | 1437.58 | 229.25 | 0.1793 | 383.42 | 211.75 | 0.9999 | 0.1793 |
| N. bombycis:V. necatrix | 1434.67 | 259.58 | 0.2071 | 386.33 | 247.42 | 1.4426 | 0.1436 |
| N. empoascae:V. necatrix | 1427.75 | 250.17 | 0.1996 | 393.25 | 291.83 | 3.4155 | 0.0584 |
| N. granulosis:V. necatrix | 1427.58 | 262.58 | 0.2110 | 393.42 | 285.42 | 2.5655 | 0.0822 |
| N. trichoplusiae:V. necatrix | 1434.00 | 260.83 | 0.2083 | 387.00 | 249.17 | 1.4664 | 0.1420 |
| V. cheracis:V. necatrix | 1421.33 | 278.25 | 0.2269 | 399.67 | 372.75 | - | - |
| V. cheracis:N. empoascae | 1409.42 | 143.17 | 0.1091 | 411.58 | 283.83 | 1.8895 | 0.0577 |
Values of Ka/Ks considerably lower than 1 indicate relatively low levels of sequence polymorphism at non-synonymous sites, an indicator of strong purifying selection and hence of gene activity. The sequence of V. cheracis is too divergent from that of V. necatrix for Ka/Ks cannot be calculated, so an additional comparison is performed between V. cheracis and N. empoascae.
Likelihood comparison of evolutionary models, based on the Akaike information criteria in ModelTest [76], indicated that the General Time Reversible model with gamma-distributed rate variation across sites and a proportion of invariable sites (GTR+G+I) was most suitable for phylogenetic analysis of both the rRNA and RPB1 alignments. Phylogenetic analysis was performed separately upon the rRNA and RPB1 alignments using maximum likelihood, implemented in PAUP* 4b10 [77] and Bayesian inference, implemented in MrBayes [78]. For each alignment, a maximum likelihood consensus tree was generated by conducting a heuristic search and bootstrapping with 100 replicates. The rRNA tree (figure 4) indicated a relatively long branch length for the A. locustae outgroup sequence. In order to check that this long branch was not distorting the tree, additional maximum likelihood and Bayesian searches were conducted, in which A. locustae was either replaced with the alternative outgroup Heterosporis anguillarum (AF387331) or omitted entirely, leaving the single outgroup E. cuniculi. The trees produced by these additional searches had identical topologies to those produced using the original E. cuniculi and A. locustae outgroup pair and were supported by qualitatively similar bootstrap values.
Hypothesis testing
Three a priori evolutionary hypotheses (Figure 6) were compared by generating constrained phylogenetic trees in PAUP* and selecting the tree with the highest likelihood value using the approximately unbiased (AU) test of Shimodaira [43] and the more conservative weighted Shimodaira-Hasegawa (WSH) test [44], both implemented in Consel [79].
Hypothesis 1 (Co-speciation) proposes co-speciation of parasites and hosts and constrains all insect-parasitizing Nosema/Vairimorpha to form a monophyletic group.
Hypothesis 2 (Sex/Asex) proposes that sex has been lost once within Nosema/Vairimorpha and constrains all asexual Nosema to form a monophyletic group.
Hypothesis 3 (rRNA gene order) proposes that the rRNA gene order has changed once from 5'-16S, 18S-3' to 5'-18S, 16S, 5S-3' and constrains all Nosema/Vairimorpha with the 5'-18S, 16S, 5S-3' rRNA gene order to form a monophyletic group.
Supplementary Material
Sequences included in rRNA and RPB1 alignments. Species, host, life cycle, source and Genbank accession number for each microsporidian rRNA and RPB1 sequence.
Primers used for PCR and sequencing of microsporidian rRNA and RPB1 genes. Names and sequences of oligonucleotide primers
Acknowledgments
Acknowledgements
This research was funded by a research grant from the Royal Society of London. I would like to thank Rachel Down, Judith Smith, Elizabeth Moodie, Leellen Solter and Elaine O'Mahony for providing microsporidian samples for use in this project. I would also like to thank Jo Porter for advice on phylogenetic analysis and three anonymous reviewers for their comments.
References
- Barton NH, Charlesworth B. Why sex and recombination? Science. 1998;281:1986–1990. doi: 10.1126/science.281.5385.1986. [DOI] [PubMed] [Google Scholar]
- Burt A. Perspective: Sex, recombination, and the efficacy of selection - Was Weismann right? Evolution. 2000;54:337–351. doi: 10.1554/0014-3820(2000)054[0337:PSRATE]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Kondrashov AS. Classification of Hypotheses on the Advantage of Amphimixis. Journal of Heredity. 1993;84:372–387. doi: 10.1093/oxfordjournals.jhered.a111358. [DOI] [PubMed] [Google Scholar]
- Otto SP, Lenormand T. Resolving the paradox of sex and recombination. Nature Reviews Genetics. 2002;3:252–261. doi: 10.1038/nrg761. [DOI] [PubMed] [Google Scholar]
- Rice WR. Experimental tests of the adaptive significance of sexual recombination. Nature Reviews Genetics. 2002;3:241–251. doi: 10.1038/nrg760. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J. Evolution - Contemplating Life without Sex. Nature. 1986;324:300–301. doi: 10.1038/324300a0. [DOI] [PubMed] [Google Scholar]
- Normark BB, Judson OP, Moran NA. Genomic signatures of ancient asexual lineages. Biological Journal of the Linnean Society. 2003;79:69–84. doi: 10.1046/j.1095-8312.2003.00182.x. [DOI] [Google Scholar]
- Maynard Smith J. The evolution of sex. Cambridge , Cambridge University Press; 1978. [Google Scholar]
- Xu JP. The prevalence and evolution of sex in microorganisms. Genome. 2004;47:775–780. doi: 10.1139/g04-037. [DOI] [PubMed] [Google Scholar]
- Zeyl C, Bell G. The advantage of sex in evolving yeast populations. Nature. 1997;388:465–468. doi: 10.1038/41312. [DOI] [PubMed] [Google Scholar]
- Goddard MR, Charles H, Godfray J, Burt A. Sex increases the efficacy of natural selection in experimental yeast populations. Nature. 2005;434:636–640. doi: 10.1038/nature03405. [DOI] [PubMed] [Google Scholar]
- Taylor JW, Jacobson DJ, Fisher MC. The evolution of asexual fungi: Reproduction, speciation and classification. Annual Review of Phytopathology. 1999;37:197–246. doi: 10.1146/annurev.phyto.37.1.197. [DOI] [PubMed] [Google Scholar]
- Sanders IR, Alt M, Groppe K, Boller T, Wiemken A. Identification of Ribosomal DNA Polymorphisms among and within Spores of the Glomales - Application to Studies on the Genetic Diversity of Arbuscular Mycorrhizal Fungal Communities. New Phytologist. 1995;130:419–427. doi: 10.1111/j.1469-8137.1995.tb01836.x. [DOI] [Google Scholar]
- Pawlowska TE, Taylor JW. Organization of genetic variation in individuals of arbuscular mycorrhizal fungi. Nature. 2004;427:733–737. doi: 10.1038/nature02290. [DOI] [PubMed] [Google Scholar]
- Tay WT, O'Mahony EM, Paxton RJ. Complete rRNA gene sequences reveal that the microsporidium Nosema bombi infects diverse bumblebee (Bombus spp.) hosts and contains multiple polymorphic sites. Journal of Eukaryotic Microbiology. 2005;52:505–513. doi: 10.1111/j.1550-7408.2005.00057.x. [DOI] [PubMed] [Google Scholar]
- Dunn AM, Terry RS, Smith JE. Transovarial transmission in the microsporidia. Advances in Parasitology. 2001;48:57–100. doi: 10.1016/s0065-308x(01)48005-5. [DOI] [PubMed] [Google Scholar]
- Terry RS, Smith JE, Dunn AM. Impact of a novel, feminising microsporidium on its crustacean host. Journal of Eukaryotic Microbiology. 1998;45:497–501. [Google Scholar]
- Weiser J. Contribution of the classification of Microsporidia. Vestnik Ceskoslovenske Zoologicke Spolecnosti. 1977;41:308–320. [Google Scholar]
- Sprague V, Becnel JJ, Hazard EI. Taxonomy of Phylum Microspora. Critical Reviews in Microbiology. 1992;18:285–395. doi: 10.3109/10408419209113519. [DOI] [PubMed] [Google Scholar]
- Sprague V. Systematics of the Microsporidia. In: Bulla LACTC, editor. Comparative Pathobiology. Vol. 2. New York , Plenum Press; 1977. [Google Scholar]
- Issi IV. Microsporidia as a phylum of parasitic protozoa. Protozoology (Leningrad) 1986;10:1–136. [Google Scholar]
- Sokolova YY, Dolgikh VV, Morzhina EV, Nassonova ES, Issi IV, Terry RS, Ironside JE, Smith JE, Vossbrinck CR. Establishment of the new genus Paranosema based on the ultrastructure and molecular phylogeny of the type species Paranosema grylli Gen. Nov., Comb. Nov (Sokolova, Selezniov, Dolgikh, Issi 1994), from the cricket Gryllus bimaculatus Deg. Journal of Invertebrate Pathology. 2003;84:159–172. doi: 10.1016/j.jip.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Baker MD, Vossbrinck CR, Maddox JV, Undeen AH. Phylogenetic-Relationships among Vairimorpha and Nosema Species (Microspora) Based on Ribosomal-Rna Sequence Data. Journal of Invertebrate Pathology. 1994;64:100–106. doi: 10.1006/jipa.1994.1077. [DOI] [PubMed] [Google Scholar]
- Canning EU, Curry A, Cheney S, Lafranchi-Tristem NJ, Haque MA. Vairimorpha imperfecta n.sp., a microsporidian exhibiting an abortive octosporous sporogony in Plutella xylostella L. (Lepidoptera : Yponomeutidae) Parasitology. 1999;119:273–286. doi: 10.1017/S0031182099004734. [DOI] [PubMed] [Google Scholar]
- Vossbrinck CR, Maddox JV, Friedman S, Debrunnervossbrinck BA, Woese CR. Ribosomal-Rna Sequence Suggests Microsporidia Are Extremely Ancient Eukaryotes. Nature. 1987;326:411–414. doi: 10.1038/326411a0. [DOI] [PubMed] [Google Scholar]
- Neefs JM, Vandepeer Y, Derijk P, Chapelle S, Dewachter R. Compilation of Small Ribosomal-Subunit Rna Structures. Nucleic Acids Research. 1993;21:3025–3049. doi: 10.1093/nar/21.13.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartskeerl RA, Schuitema ARJ, Dewachter R. Secondary Structure of the Small Subunit Ribosomal-Rna Sequence of the Microsporidium Encephalitozoon-Cuniculi. Nucleic Acids Research. 1993;21:1489–1489. doi: 10.1093/nar/21.6.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossbrinck CR, Debrunner-Vossbrinck BA. Molecular phylogeny of the Microsporidia: ecological, ultrastructural and taxonomic considerations. Folia Parasitologica. 2005;52:131–142. doi: 10.14411/fp.2005.017. [DOI] [PubMed] [Google Scholar]
- Tsai SJ, Lo CF, Soichi Y, Wang CH. The characterization of microsporidian isolates (Nosematidae : Nosema) from five important lepidopteran pests in Taiwan. Journal of Invertebrate Pathology. 2003;83:51–59. doi: 10.1016/S0022-2011(03)00035-1. [DOI] [PubMed] [Google Scholar]
- Pieniazek NJ, daSilva AJ, Slemenda SB, Visvesvara GS, Kurtti TJ, Yasunaga C. Nosema trichoplusiae is a synonym of Nosema bombycis based on the sequence of the small subunit ribosomal RNA coding region. Journal of Invertebrate Pathology. 1996;67:316–317. doi: 10.1006/jipa.1996.0049. [DOI] [PubMed] [Google Scholar]
- Gatehouse HS, Malone LA. The ribosomal RNA gene region of Nosema apis (microspora): DNA sequence for small and large subunit rRNA genes and evidence of a large tandem repeat unit size. Journal of Invertebrate Pathology. 1998;71:97–105. doi: 10.1006/jipa.1997.4737. [DOI] [PubMed] [Google Scholar]
- Zhu XL, Wittner M, Tanowitz HB, Cali A, Weiss LM. Ribosomal-Rna Sequences of Enterocytozoon-Bieneusi, Septata-Intestinalis and Ameson-Michaelis - Phylogenetic Construction and Structural Correspondence. Journal of Eukaryotic Microbiology. 1994;41:204–209. doi: 10.1111/j.1550-7408.1994.tb01498.x. [DOI] [PubMed] [Google Scholar]
- Tsai SJ, Kou GH, Lo CF, Wang CH. Complete sequence and structure of ribosomal RNA gene of Heterosporis anguillarum. Diseases of Aquatic Organisms. 2002;49:199–206. doi: 10.3354/dao049199. [DOI] [PubMed] [Google Scholar]
- Nilsen F, Endresen C, Hordvik I. Molecular phylogeny of microsporidians with particular reference to species that infect the muscles of fish. Journal of Eukaryotic Microbiology. 1998;45:535–543. doi: 10.1111/j.1550-7408.1998.tb05113.x. [DOI] [PubMed] [Google Scholar]
- Muller A, Trammer T, Chioralia G, Seitz HM, Diehl V, Franzen C. Ribosomal RNA of Nosema algerae and phylogenetic relationship to other microsporidia. Parasitology Research. 2000;86:18–23. doi: 10.1007/PL00008501. [DOI] [PubMed] [Google Scholar]
- Gresoviac SJ, Khattra JS, Nadler SA, Kent ML, Devlin RH, Vivares CP, De La Fuente E, Hedrick RP. Comparison of small subunit ribosomal RNA gene and internal transcribed spacer sequences among isolates of the intranuclear microsporidian Nucleospora salmonis. Journal of Eukaryotic Microbiology. 2000;47:379–387. doi: 10.1111/j.1550-7408.2000.tb00064.x. [DOI] [PubMed] [Google Scholar]
- Huang WF, Tsai SJ, Lo CF, Soichi Y, Wang CH. The novel organization and complete sequence of the ribosomal RNA gene of Nosema bombycis. Fungal Genetics and Biology. 2004;41:473–481. doi: 10.1016/j.fgb.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Peyretaillade E, Biderre C, Peyret P, Duffieux F, Metenier G, Gouy M, Michot B, Vivares CP. Microsporidian Encephalitozoon cuniculi, a unicellular eukaryote with an unusual chromosomal dispersion of ribosomal genes and a LSU rRNA reduced to the universal core. Nucleic Acids Research. 1998;26:3513–3520. doi: 10.1093/nar/26.15.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SJ, Huang WF, Wang CH. Complete sequence and gene organization of the Nosema spodopterae rRNA gene. Journal of Eukaryotic Microbiology. 2005;52:52–54. doi: 10.1111/j.1550-7408.2005.3291rr.x. [DOI] [PubMed] [Google Scholar]
- Wang LL, Chen KP, Zhang Z, Yao Q, Gao GT, Zhao Y. Phylogenetic analysis of Nosema antheraeae (Microsporidia) isolated from Chinese oak silkworm, Antheraea pernyi. Journal of Eukaryotic Microbiology. 2006;53:310–313. doi: 10.1111/j.1550-7408.2006.00106.x. [DOI] [PubMed] [Google Scholar]
- Iiyama K, Chieda Y, Yasunaga-Aoki C, Hayasaka S, Shimizu S. Analyses of the ribosomal DNA region in Nosema bombycis NIS 001. Journal of Eukaryotic Microbiology. 2004;51:598–604. doi: 10.1111/j.1550-7408.2004.tb00592.x. [DOI] [PubMed] [Google Scholar]
- Hirt RP, Logsdon JM, Healy B, Dorey MW, Doolittle WF, Embley TM. Microsporidia are related to Fungi: Evidence from the largest subunit of RNA polymerase II and other proteins. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:580–585. doi: 10.1073/pnas.96.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Systematic Biology. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Molecular Biology and Evolution. 1999;16:1114–1116. [Google Scholar]
- Klassen GJ. Coevolution - a History of the Macroevolutionary Approach to Studying Host-Parasite Associations. Journal of Parasitology. 1992;78:573–587. doi: 10.2307/3283532. [DOI] [PubMed] [Google Scholar]
- Lom J, Nilsen F, Dykova I. Thelohania contejeani Henneguy, 1892: dimorphic life cycle and taxonomic affinities, as indicated by ultrastructural and molecular study. Parasitology Research. 2001;87:860–872. doi: 10.1007/s004360100436. [DOI] [PubMed] [Google Scholar]
- Moodie EG, Le Jambre LF, Katz ME. Thelohania parastaci sp nov (Microspora : Thelohaniidae), a parasite of the Australian freshwater crayfish, Cherax destructor (Decapoda : Parastacidae) Parasitology Research. 2003;91:151–165. doi: 10.1007/s00436-003-0941-3. [DOI] [PubMed] [Google Scholar]
- Moodie EG, Le Jambre LF, Katz ME. Thelohania montirivulorum sp nov (Microspora : Thelohaniidae), a parasite of the Australian freshwater crayfish, Cherax destructor (Decapoda : Parastacidae): fine ultrastructure, molecular characteristics and phylogenetic relationships. Parasitology Research. 2003;91:215–228. doi: 10.1007/s00436-003-0948-9. [DOI] [PubMed] [Google Scholar]
- Bell G. The Masterpiece of Nature. London , Croom Helm; 1982. [Google Scholar]
- Yan Z, Li XG, Xu JP. Geographic distribution of mating type alleles of Cryptococcus neoformans in four areas of the United States. Journal of Clinical Microbiology. 2002;40:965–972. doi: 10.1128/JCM.40.3.965-972.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler KB, Wang P, Cox GM, Perfect JR, Heitman J. Identification of the MATa mating-type locus of Cryptococcus neoformans reveals a serotype A MATa strain thought to have been extinct. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:14455–14460. doi: 10.1073/pnas.97.26.14455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JP. Estimating the spontaneous mutation rate of loss of sex in the human pathogenic fungus Cryptococcus neoformans. Genetics. 2002;162:1157–1167. doi: 10.1093/genetics/162.3.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazard EI, Brookbank JW. Karyogamy and Meiosis in an Amblyospora Sp (Microspora) in the Mosquito Culex-Salinarius. Journal of Invertebrate Pathology. 1984;44:3–11. doi: 10.1016/0022-2011(84)90039-9. [DOI] [Google Scholar]
- Sokolova YY, McNally LR, Fuxa JR, Vinson SB. Spore morphotypes of Thelohania solenopsae (microsporidia) described microscopically and confirmed by PCR of individual spores microdissected from smears by position ablative laser microbeam microscopy. Microbiology-Sgm. 2004;150:1261–1270. doi: 10.1099/mic.0.26838-0. [DOI] [PubMed] [Google Scholar]
- Franzen C, Nassonova FS, Scholmerich J, Issi IV. Transfer of the members of the genus Brachiola (Microsporidia) to the genus Anncaliia based on ultrastructural and molecular data. Journal of Eukaryotic Microbiology. 2006;53:26–35. doi: 10.1111/j.1550-7408.2005.00066.x. [DOI] [PubMed] [Google Scholar]
- Vossbrinck CR, Andreadis TG, Vavra J, Becnel JJ. Molecular phylogeny and evolution of mosquito parasitic microsporidia (Microsporidia : Amblyosporidae) Journal of Eukaryotic Microbiology. 2004;51:88–95. doi: 10.1111/j.1550-7408.2004.tb00167.x. [DOI] [PubMed] [Google Scholar]
- Hamilton WD, Axelrod R, Tanese R. Sexual Reproduction as an Adaptation to Resist Parasites (a Review) Proceedings of the National Academy of Sciences of the United States of America. 1990;87:3566–3573. doi: 10.1073/pnas.87.9.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn AM, Smith JE. Microsporidian life cycles and diversity: the relationship between virulence and transmission. Microbes and Infection. 2001;3:381–388. doi: 10.1016/S1286-4579(01)01394-6. [DOI] [PubMed] [Google Scholar]
- Pilley BM. New Genus, Vairimorpha (Protozoa-Microsporida), for Nosema-Necatrix Kramer 1965 - Pathogenicity and Life-Cycle in Spodoptera-Exempta (Lepidoptera-Noctuidae) Journal of Invertebrate Pathology. 1976;28:177–183. doi: 10.1016/0022-2011(76)90119-1. [DOI] [Google Scholar]
- Malone LA, Canning EU. Fine-Structure of Vairimorpha-Plodiae (Microspora, Burenellidae), a Pathogen of Plodia-Interpunctella (Lepidoptera, Phycitidae) and Infectivity of the Dimorphic Spores. Protistologica. 1982;18:503–516. [Google Scholar]
- Andreadis TG. Evolutionary strategies and adaptations for survival between mosquito-parasitic microsporidia and their intermediate copepod hosts: a comparative examination of Amblyospora connectius and Hyalinocysta chapmani (Microsporidia : Amblyosporidae) Folia Parasitologica. 2005;52:23–35. doi: 10.14411/fp.2005.004. [DOI] [PubMed] [Google Scholar]
- Valles SM, Briano JA. Presence of Thelohania solenopsae and Vairimorpha invictae in South American populations of Solenopsis invicta. Florida Entomologist. 2004;87:625–627. doi: 10.1653/0015-4040(2004)087[0625:POTSAV]2.0.CO;2. [DOI] [Google Scholar]
- Galbreath J, Smith JE, Terry RS, Becnel JJ, Dunn AM. Invasion success of Fibrillanosema crangonycis, n.sp., n.g.: a novel vertically transmitted microsporidian parasite from the invasive amphipod host Crangonyx pseudogracilis. International Journal for Parasitology. 2004;34:235–244. doi: 10.1016/j.ijpara.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Sokolova Y, Fuxa J. Development of Thelohania solenopsae in red imported fire ants Solenopsis invicta from polygynous colonies results in formation of three spore types. Journal of Eukaryotic Microbiology. 2001:85S–85S. doi: 10.1111/j.1550-7408.2001.tb00463.x. [DOI] [PubMed] [Google Scholar]
- Solter LF, Pilarska DK, Vossbrinck CF. Host specificity of microsporidia pathogenic to forest Lepidoptera. Biological Control. 2000;19:48–56. doi: 10.1006/bcon.2000.0845. [DOI] [Google Scholar]
- Briano JA, Williams DF, Oi DH, Davis LR. Field host range of the fire ant pathogens Thelohania solenopsae (Microsporida : Thelohaniidae) and Vairimorpha invictae (Microsporida : Burenellidae) in South America. Biological Control. 2002;24:98–102. doi: 10.1016/S1049-9644(02)00009-9. [DOI] [Google Scholar]
- Colla SR, Otterstatter MC, Gegear RJ, Thomson JD. Plight of the bumble bee: Pathogen spillover from commercial to wild populations. Biological Conservation. 2006;129:461–467. doi: 10.1016/j.biocon.2005.11.013. [DOI] [Google Scholar]
- Baker MD, Vossbrinck CR, Didier ES, Maddox JV, Shadduck JA. Small-Subunit Ribosomal DNA Phylogeny of Various Microsporidia with Emphasis on Aids-Related Forms. Journal of Eukaryotic Microbiology. 1995;42:564–570. doi: 10.1111/j.1550-7408.1995.tb05906.x. [DOI] [PubMed] [Google Scholar]
- Baker MD, Vossbrinck CR, Becnel JJ, Andreadis TG. Phylogeny of Amblyospora (Microsporida : Amblyosporidae) and related genera based on small subunit ribosomal DNA data: A possible example of host parasite cospeciation. Journal of Invertebrate Pathology. 1998;71:199–206. doi: 10.1006/jipa.1997.4725. [DOI] [PubMed] [Google Scholar]
- Cheney SA, Lafranchi-Tristem NJ, Bourges D, Canning EU. Relationships of microsporidian genera, with emphasis on the polysporous genera, revealed by sequences of the largest subunit of RNA polymerase II (RPB1) Journal of Eukaryotic Microbiology. 2001;48:111–117. doi: 10.1111/j.1550-7408.2001.tb00422.x. [DOI] [PubMed] [Google Scholar]
- Philippe H, Lopez P, Brinkmann H, Budin K, Germot A, Laurent J, Moreira D, Muller M, Le Guyader H. Early-branching or fast-evolving eukaryotes? An answer based on slowly evolving positions. Proceedings of the Royal Society of London Series B-Biological Sciences. 2000;267:1213–1221. doi: 10.1098/rspb.2000.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Ben Ali A, Meyer A. Microsporidia: accumulating molecular evidence that a group of amitochondriate and suspectedly primitive eukaryotes are just curious fungi. Gene. 2000;246:1–8. doi: 10.1016/S0378-1119(00)00063-9. [DOI] [PubMed] [Google Scholar]
- Xia X, Xie Z. DAMBE: Software package for data analysis in molecular biology and evolution. Journal of Heredity. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- Steel MA, Lockhart PJ, Penny D. Confidence in Evolutionary Trees from Biological Sequence Data. Nature. 1993;364:440–442. doi: 10.1038/364440a0. [DOI] [PubMed] [Google Scholar]
- Xia XH, Xie Z, Salemi M, Chen L, Wang Y. An index of substitution saturation and its application. Molecular Phylogenetics and Evolution. 2003;26:1–7. doi: 10.1016/S1055-7903(02)00326-3. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP* 4.0. Phylogenetic analysis using parsimony (*and other methods). Beta version 4.0b4a. Sunderland, Massachusetts , Sinauer Associates; 2002. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences included in rRNA and RPB1 alignments. Species, host, life cycle, source and Genbank accession number for each microsporidian rRNA and RPB1 sequence.
Primers used for PCR and sequencing of microsporidian rRNA and RPB1 genes. Names and sequences of oligonucleotide primers


