Abstract
A flexible Numerical Matrices Method (NMM) for nonlinear system identification has been developed based on a description of the dynamics of the system in terms of kinetic complexes. A set of related methods are presented that include increasing amounts of prior information about the reaction network structure, resulting in increased accuracy of the reconstructed rate constants. The NMM is based on an analytical least squares solution for a set of linear equations to determine the rate parameters. In the absence of prior information, all possible unimolecular and bimolecular reactions among the species in the system are considered, and the elements of a general kinetic matrix are determined. Inclusion of prior information is facilitated by formulation of the kinetic matrix in terms of a stoichiometry matrix or a more general set of representation matrices. A method for determination of the stoichiometry matrix beginning only with time-dependent concentration data is presented. In addition, we demonstrate that singularities that arise from linear dependencies among the species can be avoided by inclusion of data collected from a number of different initial states. The NMM provides a flexible set of tools for analysis of complex kinetic data, in particular for analysis of chemical and biochemical reaction networks.
INTRODUCTION
An extensive theoretical framework has been developed for analysis of nonlinear dynamic systems, in particular, for the case of chemical reaction kinetics (1). A variety of theoretical and computational approaches for studying complex reaction networks have been developed and described (1–3). Most recently, advances in genomics have made the understanding of the complex biochemical reaction networks of metabolism, gene regulation, and cell function an active area of investigation (4,5). Many of these approaches make extensive use of a stoichiometry matrix that describes the set of reactions present in the system. Analysis of the null spaces and the span of a stoichiometry matrix provides extensive information about the system, including steady-state solutions, elementary flux modes, and conservation of mass relations (4,6–9). For cases where the stoichiometry matrix is not known, correlation methods have been applied to deduce the structure of a reaction network from time-dependent data for the chemical species (10–12). A complete understanding of metabolic networks will require methods to determine the network structure, the resulting steady states and fluxes for the network, and the transient response of a system away from the steady state.
In this work, the theoretical basis for a set of numerical tools is developed to practically address two important problems encountered in analysis of biochemical reaction networks. The approach is conceptually distinct from previous work based on the properties of the stoichiometry matrix or correlation methods (4–12). Here, we present an approach that both solves the reaction identification problem, and provides a least squares analysis to determine the values of the rate constants that best fit a given reaction structure to the time-dependent data. The tools presented here represent a powerful “bottom-up” approach to determine reaction network structure and to quantitatively analyze reaction network dynamics.
The reaction identification problem
Experimentally observed rates and concentrations can be used to identify the underlying nature of the chemical reactions present. Solving the identification problem involves finding the elementary reaction steps and kinetics that give rise to the observed data without any prior knowledge of the reaction chemistry. The basis of most of the modern methods of system identification was developed during the period 1950–1970. These methods have been successfully used to obtain the rate constants for nontrivial chemical reaction systems with several species.
A set of time-dependent data for the concentrations of the species can be represented as Xj(ti), where the index i runs from 1 to nt, which is the number of time points, and the index j runs from 1 to ns, which is the number of species. In addition, for each species and time point, there is a matrix of observed rates dXj(ti)/dt with the same indices. In practice, the rates may be either directly observed, or calculated from the observed concentration data, Xj(ti). A set of np chemical reactions is generally described by Cauchy equations in the form:
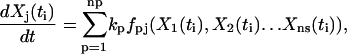 |
(1) |
where the 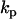 are the rate constants, and the
are the rate constants, and the 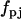 are the empirical functions that contain information about structure of the chemical reaction, for each reaction p. A number of approaches have been developed to solve sets of equations such as Eq. 1 for the rate constants, including empirical optimization (13–16), direct integral methods (13,17–22), and the Prony method for quasilinear systems (23).
are the empirical functions that contain information about structure of the chemical reaction, for each reaction p. A number of approaches have been developed to solve sets of equations such as Eq. 1 for the rate constants, including empirical optimization (13–16), direct integral methods (13,17–22), and the Prony method for quasilinear systems (23).
A direct differential approach has been developed (24–31) to obtain rate constants from observed rates of change of the species using a least squares criterion:
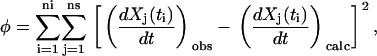 |
(2) |
where the observed value of the derivatives is obtained from directly or by numerical differentiation of observed value of concentrations and the calculated value of derivatives is obtained from Eq. 1. Differentiation of Eq. 2 with respect to each rate parameter 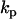 , gives a system of linear equations that can be directly solved for the rate constants
, gives a system of linear equations that can be directly solved for the rate constants 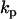 .
.
Chemical reaction network theory
A general formalism for chemical reaction network theory (32,33) has been developed based on the idea of reaction complexes, which are the combinations of species that are reaction products and reactants. Each species is represented by a unit vector 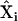 , linear combinations of species that are products or reactants are represented by complex vectors
, linear combinations of species that are products or reactants are represented by complex vectors 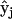 , and reactions are represented by differences between two complex vectors as vectors
, and reactions are represented by differences between two complex vectors as vectors 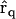 , as outlined in Appendix I in the Supplementary Material. Any network of chemical reactions can be described as a set of reaction vectors that constitute a stoichiometry matrix. The dynamics of a network of species can be described using the stoichiometry matrix, and the properties of the stoichiometry matrix can be analyzed to identify steady states and steady-state fluxes for the reaction network (6).
, as outlined in Appendix I in the Supplementary Material. Any network of chemical reactions can be described as a set of reaction vectors that constitute a stoichiometry matrix. The dynamics of a network of species can be described using the stoichiometry matrix, and the properties of the stoichiometry matrix can be analyzed to identify steady states and steady-state fluxes for the reaction network (6).
Despite the extensive developments in nonlinear system identification and chemical reaction network theory, there has been no strong connection made between these two important areas. In this article, we make such a theoretical connection by formulating the Numerical Matrices Method (NMM) for nonlinear system identification in terms of the formalism of reaction complexes, and presenting a general method for de novo determination of the stoichiometry matrix. First, the previously developed method for nonlinear system identification (34,35) has been formulated as the Kinetic Matrix Method (KMM) using the formalism of kinetic complexes that provides a key connection to the other matrix representations of the kinetic equations. Second, a variant of the KMM is described that incorporates varying degrees of prior information as the Representation Matrix Method (RMM), which result in increased accuracy of the determined rate constants. Third, the Stoichiometry Matrix Method (SMM) is presented as a general method for determination of rate constants where the reaction network structure is known. Fourth, a method for breaking the holonomic conditions arising from linear dependence among species is presented, which is essential for general application of the Numerical Matrices Method to large complex reaction networks. Finally, a general method is presented for construction of a stoichiometry matrix with the Numerical Matrices Method, using only concentration data as a starting point, without any prior knowledge of the reaction network structure. The approach outlined here provides a general and powerful set of analytical tools that will have a wide variety of applications in analysis of chemical reaction dynamics and metabolic networks.
RESULTS
The Numerical Matrices Method for nonlinear system identification
The direct integral, direct differential, and empirical optimization methods are not generally suitable for the task of investigating chemical reaction cascades with strong nonlinear dynamics, a large number of species, and unknown structure of the underlying chemical reactions. A direct differential method using a linearly quadratic kinetic model has been recently developed that can be used to solve the identification problem for these more demanding systems (34,35). Here we extend this approach to a more general set of related matrix methods that constitute a flexible and powerful set of tools for the quantitative analysis of nonlinear dynamic systems.
The linearly quadratic kinetic model
Considering a set of ns chemical species, there are a number of elementary reactions that might occur among them, including unimolecular, bimolecular, and higher order reactions, as well as mass flow in and out of the system. The vast majority of chemical and biochemical reactions can be represented as cascades of uni- and bimolecular reactions. If the unusual cases of trimolecular and higher order reactions are neglected, a general mathematical expression for the time dependence of a particular species j is given by:
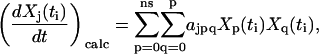 |
(3) |
where the rate of change is described as a linear sum of flux terms, which are the rate parameters, ajpq, times the appropriate concentrations of species p and q. To conveniently describe the kinetics for unimolecular reactions or mass flow in this quadratic expression, a fictitious species, X0, is introduced whose concentration is held constant. In this way, kinetics that depend linearly on a single species can be accounted for. It should be noted that the complete set of rate constants describing the kinetics should contain all of the combinations of indices p and q, but that Eq. 3 contains the additional index j. For the purpose of system identification, it is necessary to treat each of the species j individually, as described below. The linearly quadratic form of Eq. 3 is very general for common reactions and avoids the ambiguity in specification of the arbitrary functions  in Eq. 1.
in Eq. 1.
Matrix formalisms for chemical reaction dynamics
The vector formalism from chemical reaction network theory provides useful and compact expressions for chemical reaction kinetics, and provides strong connections between different representations of the kinetics. As described in Appendices I and II in the Supplementary Material, the reaction dynamics from Eqs. 1 and 3 can be equivalently expressed in the following three forms:
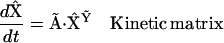 |
(4) |
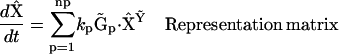 |
(5) |
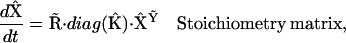 |
(6) |
where 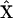 is the vector of species,
is the vector of species, 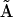 is a generalized kinetic matrix,
is a generalized kinetic matrix, 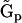 is a set of representation matrices,
is a set of representation matrices,  is a stoichiometry matrix formed from the set of reaction vectors,
is a stoichiometry matrix formed from the set of reaction vectors, 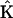 is a vector containing the rate constants
is a vector containing the rate constants 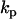 , and
, and 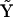 is a matrix formed from the set of complex vectors. The exponentiation of the species vector by the complex matrix,
is a matrix formed from the set of complex vectors. The exponentiation of the species vector by the complex matrix,  , is a compact expression for the vector of kinetic complexes
, is a compact expression for the vector of kinetic complexes 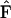 , as described in Appendices I and III in the Supplementary Material.
, as described in Appendices I and III in the Supplementary Material.
The kinetic matrix  is most useful in the absence of any knowledge about the reaction structure, the representation matrices
is most useful in the absence of any knowledge about the reaction structure, the representation matrices 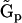 are useful when there is partial information about the reaction structure or rates, while the stoichiometry matrix
are useful when there is partial information about the reaction structure or rates, while the stoichiometry matrix  is useful when the complete structure of the reaction network is known. Each of these representations offers advantages for analysis of reaction network kinetics in particular situation, and this set of dynamic equations serves as the basis for the Numerical Matrices Method.
is useful when the complete structure of the reaction network is known. Each of these representations offers advantages for analysis of reaction network kinetics in particular situation, and this set of dynamic equations serves as the basis for the Numerical Matrices Method.
The Kinetic Matrix Method
The dynamics of a cascade of uni- and bimolecular reactions is expressed in matrix form in Eq. 4, where 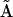 is a general kinetic matrix containing the elements
is a general kinetic matrix containing the elements 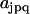 , and
, and 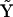 is the matrix of complexes. Because of the inclusion of the fictitious species X0 in the linearly quadratic model, there are ns + 1 species, and nk = (ns + 2)(ns + 1)/2 possible rate constants, each of which corresponds to all possible reaction among the ns species. The dimensions of matrices
is the matrix of complexes. Because of the inclusion of the fictitious species X0 in the linearly quadratic model, there are ns + 1 species, and nk = (ns + 2)(ns + 1)/2 possible rate constants, each of which corresponds to all possible reaction among the ns species. The dimensions of matrices 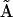 , and
, and 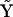 are ns rows, one for each species, and nk columns, one for each kinetic complex.
are ns rows, one for each species, and nk columns, one for each kinetic complex.
The agreement of the observed data and model data calculated with Eq. 4 is quantitated by the least squares discrepancy in analogy to Eq. 2. Differentiation of Eq. 2 with respect to each of the elements  gives a linear system of nk equations for the
gives a linear system of nk equations for the 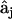 (row j of matrix
(row j of matrix 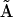 ) as described in Appendix II in the Supplementary Material. The observed rates in Eq. 2 are calculated from the concentration data using finite differences as described in Appendix II in the Supplementary Material. The set of equations can be solved by defining the elements of matrix
) as described in Appendix II in the Supplementary Material. The observed rates in Eq. 2 are calculated from the concentration data using finite differences as described in Appendix II in the Supplementary Material. The set of equations can be solved by defining the elements of matrix 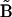 and vector
and vector 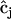 :
:
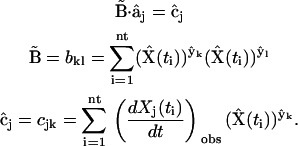 |
(7) |
The desired vector of rates 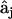 is obtained by inversion of matrix
is obtained by inversion of matrix 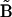 . It is necessary to solve this system of equations for each of the ns species j separately to ensure the condition of
. It is necessary to solve this system of equations for each of the ns species j separately to ensure the condition of 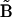 . The results of this analysis are estimates for the rate constants for each of the possible quadratic combinations of species involved in all possible reactions that affect the concentration of species j. Solving for each
. The results of this analysis are estimates for the rate constants for each of the possible quadratic combinations of species involved in all possible reactions that affect the concentration of species j. Solving for each 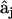 in turn allows for the construction of the kinetic matrix
in turn allows for the construction of the kinetic matrix 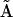 , which we term the Kinetic Matrix Method for nonlinear system identification. Many of the
, which we term the Kinetic Matrix Method for nonlinear system identification. Many of the  are zero, and the nonzero values give information about which of these many possible reactions are occurring. The elements of
are zero, and the nonzero values give information about which of these many possible reactions are occurring. The elements of 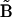 and
and  are completely defined in terms of the concentration time-series data, the rates that must be obtained by differentiation of the concentration data, and the complex matrix
are completely defined in terms of the concentration time-series data, the rates that must be obtained by differentiation of the concentration data, and the complex matrix 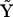 .
.
The Representation Matrix Method
An alternative approach to reducing the number of parameters to be determined from the data is to decompose the kinetic matrix 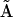 into the product of a set of representation matrix
into the product of a set of representation matrix 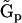 and a vector of nonzero parameters
and a vector of nonzero parameters 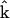 = kp that are to be determined. The representation matrices contain information about the complexes that are present in the dynamics. In this case, the dynamics takes the form of Eq. 5, and minimizing the least squares discrepancy function in Eq. 2 leads to the formulas similar to Eq. 7 for the vector of parameters
= kp that are to be determined. The representation matrices contain information about the complexes that are present in the dynamics. In this case, the dynamics takes the form of Eq. 5, and minimizing the least squares discrepancy function in Eq. 2 leads to the formulas similar to Eq. 7 for the vector of parameters 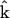 = kp:
= kp:
 |
(8) |
The Stoichiometry Matrix Method
For the case of the dynamics in terms of the stoichiometry matrix in Eq. 6, the discrepancy function in Eq. 2 is differentiated with respect to each of the elements of 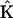 , giving a system of linear equations in direct analogy to Eq. 7. The sparse nature of the stoichiometry matrix makes it numerically tractable to treat all ns species simultaneously, and the sum over all species j is retained in analogy to Eq. 2. This linear system of equations can be readily solved for
, giving a system of linear equations in direct analogy to Eq. 7. The sparse nature of the stoichiometry matrix makes it numerically tractable to treat all ns species simultaneously, and the sum over all species j is retained in analogy to Eq. 2. This linear system of equations can be readily solved for 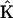 by defining the elements of matrix
by defining the elements of matrix 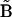 and vector
and vector 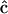 :
:
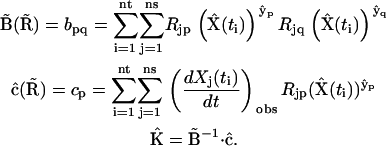 |
(9) |
An important difference in Eq. 9 compared to Eq 7 is that the matrix elements are determined by summing over all ns species. The stoichiometry matrix effectively matches the various complexes to the appropriate rate constants according to the reaction scheme, and ensures the good condition of 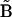 . In this way, the set of rate constants that best describes the observed data is obtained in a closed form.
. In this way, the set of rate constants that best describes the observed data is obtained in a closed form.
For all three matrix representations, the elements of 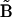 and
and 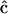 are defined in terms of the concentration data, the rates that must be obtained from the concentration data, and the complex matrix
are defined in terms of the concentration data, the rates that must be obtained from the concentration data, and the complex matrix  . These three matrix methods constitute the basic tools of the Numerical Matrices Method. Analysis of reaction kinetics and reaction structure is based on calculation of the Numerical Matrices Method described below.
. These three matrix methods constitute the basic tools of the Numerical Matrices Method. Analysis of reaction kinetics and reaction structure is based on calculation of the Numerical Matrices Method described below.
Application of the Kinetic Matrix Method
In the case of a de novo nonlinear system identification problem, where there is no prior knowledge of the reaction dynamics, the NMM is implemented using the generalized kinetic matrix 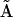 and the dynamics are expressed in Eq. 4, which we term the Kinetic Matrix Method. As an example of applying the KMM, consider the parametric nonlinear oscillator system described by the following set of differential equations:
and the dynamics are expressed in Eq. 4, which we term the Kinetic Matrix Method. As an example of applying the KMM, consider the parametric nonlinear oscillator system described by the following set of differential equations:
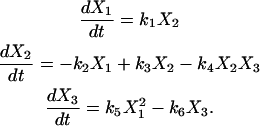 |
(10) |
A synthetic data set with N = 1000 points was constructed by numerical integration of Eq. 10, using initial concentrations X1(0) = 0; X2(0) = 0.25; X3(0) = 0.0625, the target rate values k10 = k20 = k30 = k40 = k50 = k60 = 1, and introducing noise at a level of 1%, shown in Fig. 1.
FIGURE 1.
Data set for a nonlinear oscillator. The set of differential equations in Eq. 10 was numerically integrated using rate constant values and initial conditions given in the text, and 1% random noise was added to the data.
All possible reactions among the ns species are considered, and the complete set of complex vectors is considered including the fictitious species X0. For a system of ns species, there are nk = (ns + 1)(ns + 2)/2 possible combinations of two species p and q. The set of nk possible complex vectors 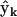 that describe all possible kinetic processes can be assembled into the complete complex matrix
that describe all possible kinetic processes can be assembled into the complete complex matrix 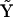 :
:
 |
(11) |
The construction of the complete matrix of complexes and the standard mapping between indices 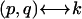 is given in Appendix III in the Supplementary Material. For the case of three species (ns = 3) the matrix
is given in Appendix III in the Supplementary Material. For the case of three species (ns = 3) the matrix 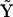 is a 3 × 10 matrix:
is a 3 × 10 matrix:
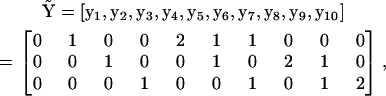 |
(12) |
which produces vector of complexes  :
:
 |
(13) |
These complexes represent all possible unimolecular and bimolecular reactions that can occur among the ns species, and includes a constant term that allows for mass to be added to or taken away from the system.
The elements of the set of 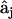 are determined using the formulas in Eq. 7 for each of the three species j, giving a set of three solutions that can be assembled into the kinetic matrix
are determined using the formulas in Eq. 7 for each of the three species j, giving a set of three solutions that can be assembled into the kinetic matrix 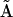 that represents the dynamics of the system in Eq. 4:
that represents the dynamics of the system in Eq. 4:
 |
(14) |
The elements of 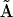 are all nonzero due to the effects of noise on the reconstruction, however, the bold elements that correspond to the processes in the dynamics are all significantly above the noise floor. A significant nonzero value for Ajp indicates that the dynamics of species j depends on complex p. Element A13 indicates that the dynamics of X1 depend on complex
are all nonzero due to the effects of noise on the reconstruction, however, the bold elements that correspond to the processes in the dynamics are all significantly above the noise floor. A significant nonzero value for Ajp indicates that the dynamics of species j depends on complex p. Element A13 indicates that the dynamics of X1 depend on complex 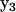 , which can be seen from inspection of
, which can be seen from inspection of 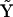 is X2, which is in turn due to the term k1X2 in Eq. 10. Element A35 indicates that the dynamics of X3 depend on complex y5, which can be seen from inspection of
is X2, which is in turn due to the term k1X2 in Eq. 10. Element A35 indicates that the dynamics of X3 depend on complex y5, which can be seen from inspection of 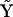 is
is 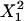 , which is due to the term k5
, which is due to the term k5  in Eq. 10. There are six significant values in
in Eq. 10. There are six significant values in 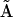 : A13, A22, A23, A29, A34, and A35 each of which directly corresponds to one of the flux terms in Eq. 10. Thus, the Kinetic Matrix Method in terms of the complexes formalism successfully extracts the structure of the dynamics of the system from the time-dependent data.
: A13, A22, A23, A29, A34, and A35 each of which directly corresponds to one of the flux terms in Eq. 10. Thus, the Kinetic Matrix Method in terms of the complexes formalism successfully extracts the structure of the dynamics of the system from the time-dependent data.
The accuracy of the reconstruction can be determined from the relative deviation of the np fitted nonzero rates to the input rates 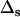 averaged over S = 40 noise realizations:
averaged over S = 40 noise realizations:
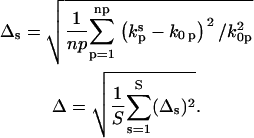 |
(15) |
For small numbers of time points (N ≤ 50), Δ is on the order of 10%, and there is rapid decrease that plateaus at 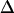 < 1% for N > 500.
< 1% for N > 500.
If an entire column p of the reconstructed matrix 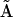 is null, this indicates that complex p does not contribute to the observed dynamics. Thus, the minimal reduced set of complexes required to reconstruct the data can be identified by inspection of
is null, this indicates that complex p does not contribute to the observed dynamics. Thus, the minimal reduced set of complexes required to reconstruct the data can be identified by inspection of 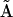 . The accuracy of determination of the rate constants for the system can be improved by subsequently neglecting the complexes that are not present in the dynamics. The complex
. The accuracy of determination of the rate constants for the system can be improved by subsequently neglecting the complexes that are not present in the dynamics. The complex 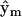 is neglected if
is neglected if  that fulfils the condition:
that fulfils the condition:
 |
(16) |
where 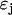 is a threshold that must be empirically determined. The reduced matrix of complexes
is a threshold that must be empirically determined. The reduced matrix of complexes 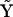 for the system of Eq. 10 can be simply obtained from the matrix
for the system of Eq. 10 can be simply obtained from the matrix 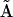 :
:
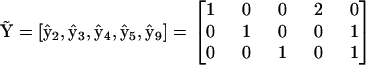 |
(17) |
Having constructed the reduced matrix 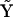 , it is now straightforward to solve again for the set of rate constant assembled into the reduced kinetic matrix
, it is now straightforward to solve again for the set of rate constant assembled into the reduced kinetic matrix 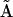 , using Eq. 7:
, using Eq. 7:
 |
(18) |
Using these reduced matrices reduces the number of possible kinetic complexes that must be identified from the data from 10 to 5, and reduces the number of kinetic parameters from 30 to 15. There is a reduction in the root mean square (rms) error (Δ) of the target rate constants by a factor of at least 2 using the reduced KMM compared to the full KMM, as shown in Fig. 2. It should be noted that if there is prior information about the nature of the complexes present in the dynamics, this information can be used to directly construct the reduced matrix of complexes.
FIGURE 2.
Comparison of the accuracy of three variants of the Numerical Matrices Method applied to the nonlinear oscillator data set.
Application of the Representation Matrix Method
Describing the kinetics in terms of a set of representation matrices provides a general and flexible way of including prior information about the structure of the dynamics. In addition, the representation matrices allow description of nonlinear dynamic systems that do not correspond to chemical reaction networks. For the system in Eq. 10, there are a total of six kinetic terms operative, and the complex matrix 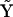 takes the form in Eq. 17, but the kinetic equations cannot be described with a standard stoichiometry matrix. However, we can use the Representation Matrix Method to determine the rate constants from the time-series data. If all of the rates are nonequal,
takes the form in Eq. 17, but the kinetic equations cannot be described with a standard stoichiometry matrix. However, we can use the Representation Matrix Method to determine the rate constants from the time-series data. If all of the rates are nonequal, 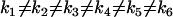 , the set of the representation matrices
, the set of the representation matrices 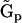 in Eq. 5 is given by:
in Eq. 5 is given by:
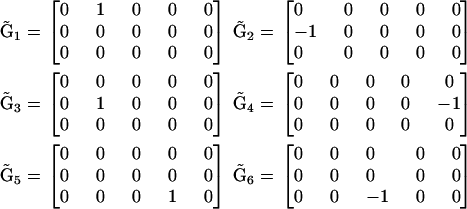 |
(19) |
The values for the rate constants 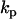 are determined using Eqs. 8 and 19, and the deviation from the data is calculated using Eq. 15. The RMM includes prior information about the reaction structure, which results in a decrease in the number of parameters to be determined, compared to the KMM, and provides an additional increase in the accuracy of determination of the rates, as shown in Fig. 2.
are determined using Eqs. 8 and 19, and the deviation from the data is calculated using Eq. 15. The RMM includes prior information about the reaction structure, which results in a decrease in the number of parameters to be determined, compared to the KMM, and provides an additional increase in the accuracy of determination of the rates, as shown in Fig. 2.
Application of the Stochiometry Matrix Method
The Stoichiometry Matrix Method can be applied to a large class of problems involving chemical reactions. The structure of the chemical reaction network and the relationship among the chemical species is given by the stoichiometry matrix, and the dynamical equations are given by Eq. 6. This work was in part motivated by the need to analyze complex kinetic data for assembly of the 30S ribosomal subunit, which is responsible for decoding the mRNA during protein synthesis in bacteria. The 30S subunit is composed of 20 small proteins and a large 16S rRNA that form a large globular structure with a dense RNA interior decorated by the ribosomal proteins (36,37). It is possible to reconstitute 30S subunits from purified components in vitro, which led to an assembly map involving a complex series of parallel and sequential protein binding events (38). Recently, quantitative kinetic data for the binding rates of 30S proteins has been collected using an isotope pulse chase method (39). There is currently no straightforward method to analyze this complex kinetic data to determine the mechanism of assembly and to extract rate constants for the binding reactions.
As a model system to demonstrate the application of the Stoichiometry Matrix Method to assembly of a ribonucleoprotein complex, we consider three RNA binding proteins, A, B, and C, that bind to an RNA, R, to form a quartenary complex RABC. We specify that A and B can bind to the RNA independently, but that binding of C requires prior binding of A (i.e., there is no RC complex), and that no protein-protein complexes are formed among A, B, or C, as shown in Fig. 3.
FIGURE 3.
Hypothetical assembly mechanism of a quarternary complex between an RNA R, and three proteins (A,B,C). Proteins A and B can bind independently to R, but binding of protein C requires prior binding of protein B.
There are 14 rate constants associated with these seven reactions (seven forward and seven reverse), and nine different species. The species vector  is given by:
is given by:
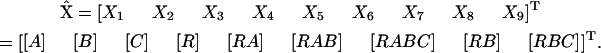 |
(20) |
For the reaction scheme shown in Fig. 3, the stoichiometry matrix 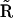 , and complex matrix
, and complex matrix 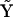 are given by:
are given by:
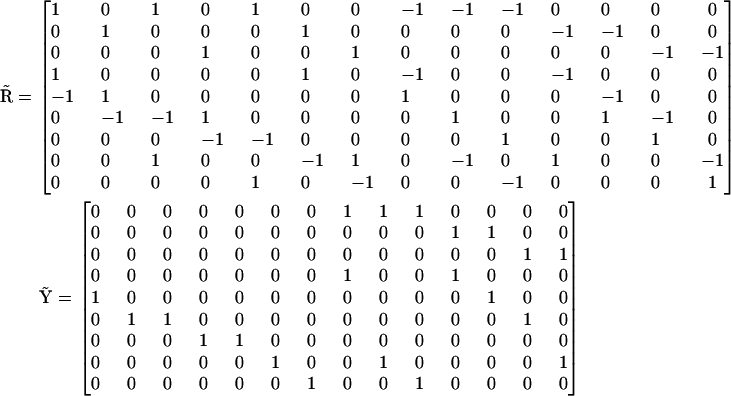 |
(21) |
To demonstrate the application of the SMM, a synthetic data set was constructed by numerical integration of Eq. 6, using the following values for the rate constants 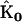 and initial concentrations
and initial concentrations 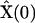 :
:
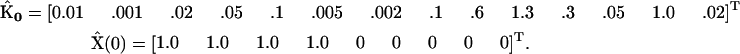 |
(22) |
Noise was introduced into the data set at a level of σ = 0.001 and the data are shown in Fig. 4. Application of Eq. 9 gives values for the set of rate parameters from the model data set shown in Fig. 4. The estimation of the rate constants for the synthetic data using Eq. 9 is shown as the solution vector 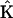 :
:
 |
(23) |
which closely matches the initial values used to generate the data, in Eq. 22.
FIGURE 4.
Model kinetic data set for assembly of the RABC quarternary complex. Data were generated by numerical integration of Eq. 6 using the matrices in Eq. 21 and the rate constants and initial concentrations in Eq. 22.
The SMM procedure was repeated for synthetic data sets with different numbers of time points, and for comparison, reconstructions were also performed using the full KMM and the reduced KMM. The accuracy of the reconstructions were quantified as the root mean square deviation of the reconstructed rate constants ( ) from the target rate constants
) from the target rate constants  , in Eq. 22. A plot of the rms error Δ for the three methods as a function of the number of time points is shown in Fig. 5. Inclusion of the stoichiometry matrix improves the accuracy of the reconstructed rates for data sets of all size.
, in Eq. 22. A plot of the rms error Δ for the three methods as a function of the number of time points is shown in Fig. 5. Inclusion of the stoichiometry matrix improves the accuracy of the reconstructed rates for data sets of all size.
FIGURE 5.
Comparison of the accuracy of three variants of the Numerical Matrices Method applied to the quarternary ribonucleoprotein complex.
Breaking holonomic constraints in the KMM by using multiple initial states
Considering the case where no information about the structure of the reaction network is known a priori, we seek to reconstruct the reaction pathways and determine the rate constants given data from the time dependence of the concentrations of the various species. Data from a real experiment such as that shown in Fig. 4 would be collected after initiating assembly of the RNA-protein complex by mixing equimolar amounts of R, A, B, and C. Using an appropriate measurement method, we would observe the presence of five new RNA-protein complexes, X5, X6, X7, X8, and X9, whose identity is not known, a priori. We assume for this exercise that we can identify the four input molecules R, A, B, and C using the measurement method.
There are several classes of mechanisms that might be operative for assembly of the final particle RABC. There might be a required sequential order to binding, there might be completely independent binding of the three proteins in any order, or, as is the case for the mechanism in Eq. 59, a combination of the two. Here we demonstrate the application of the KMM to this model system to determine the mechanism and extract the rate constants. Determining the mechanism allows the stoichiometry matrix to be constructed from the kinetic matrix 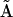 .
.
In principle, the KMM can be directly applied as described above for the nonlinear oscillator example. However, in practice, the matrix 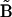 becomes singular using data in Fig. 4 due to linear dependencies among the concentration data for the system of Fig. 3. The linear dependencies arise from conservation of mass in the system of reactions:
becomes singular using data in Fig. 4 due to linear dependencies among the concentration data for the system of Fig. 3. The linear dependencies arise from conservation of mass in the system of reactions:
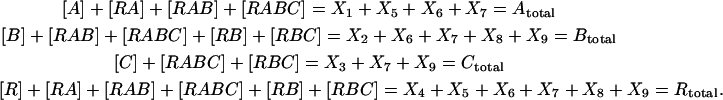 |
(24) |
These holonomic constraints are a property of the stoichiometry matrix that describes the reaction network. Analysis of the left and right null spaces and range of the stoichiometry matrix can be used to derive the steady-state fluxes and equilibrium points, and in particular, the conservation of mass relations that are the holonomic constraints on the dynamics of the network. For the purpose of applying the KMM, the structure of the stoichiometry matrix is not known, and thus the nature of the holonomic constraints cannot be known, a priori.
To apply the KMM, it is necessary to break the holonomic conditions arising from the conservation of the quantities  that make the numerical matrix
that make the numerical matrix 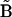 singular. One strategy to break the holonomic conditions is to collect the time-series data beginning with a sufficient number of different initial concentrations to increase the rank of
singular. One strategy to break the holonomic conditions is to collect the time-series data beginning with a sufficient number of different initial concentrations to increase the rank of  and to avoid its singularity. For the model system of Fig. 3, it is sufficient to use 16 different time series, using the initial concentrations:
and to avoid its singularity. For the model system of Fig. 3, it is sufficient to use 16 different time series, using the initial concentrations:
 |
(25) |
where  is the initial set of concentrations used to generate the time series
is the initial set of concentrations used to generate the time series 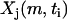 using
using  from Eq. 22, and m is the index for the time series that runs from 1 to nm = 16 data sets. Noise was introduced into the data with standard deviations
from Eq. 22, and m is the index for the time series that runs from 1 to nm = 16 data sets. Noise was introduced into the data with standard deviations 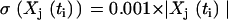 .
.
From this set of 16 synthetic data sets, the Kinetic Matrix Method of Eq. 7 can be applied to identify the complexes responsible for the kinetics and to extract the rate constants. The kinetic matrix for this system is derived from the ns = 9 species, and the nk = 55 canonical complexes 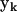 given by Eq. 11, resulting in the 9 × 55 matrix
given by Eq. 11, resulting in the 9 × 55 matrix 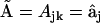 . In addition, the formulas for
. In addition, the formulas for 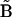 and
and 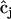 in Eq. 7 must be modified to include summation over the nm data sets recorded at different initial concentrations:
in Eq. 7 must be modified to include summation over the nm data sets recorded at different initial concentrations:
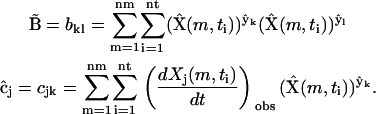 |
(26) |
Most of the elements  of the kinetic matrix
of the kinetic matrix 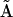 obtained from Eq. 26 are close to zero, but some
obtained from Eq. 26 are close to zero, but some 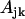 that correspond to the
that correspond to the 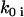 are close to the initial value used to generate the data. The quality of reconstruction of the reaction system of Fig. 3 can be estimated from the deviation of reconstructed value of
are close to the initial value used to generate the data. The quality of reconstruction of the reaction system of Fig. 3 can be estimated from the deviation of reconstructed value of 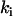 from initial
from initial 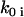 , using Eq. 15, where
, using Eq. 15, where 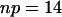 is the total number of nonzero rate constants. The value of
is the total number of nonzero rate constants. The value of  as function of number of time points used for the reconstruction is shown in Fig. 5.
as function of number of time points used for the reconstruction is shown in Fig. 5.
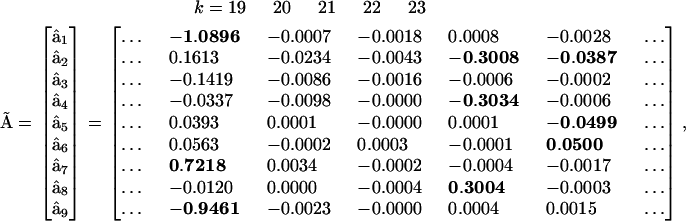
The full kinetic matrix 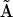 is too large to be conveniently shown, but columns k = 19….23 of matrix
is too large to be conveniently shown, but columns k = 19….23 of matrix  are shown to illustrate the essential features of the reconstructed matrix:
are shown to illustrate the essential features of the reconstructed matrix:
 |
(27) |
 |
(28) |
At this point, the details of the mechanism for assembly become clear by examining the structure of these two matrices. First, all of the irrelevant possible reactions, such as dimerization of proteins and formation of heterodimeric protein-protein complexes have been eliminated from consideration. Importantly, there is no complex that corresponds to the forward or reverse reaction R + C <-> RC. However, there are two elementary reactions that correspond to the complexes  and
and 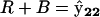 , which indicates that either A or B can bind independently to R. Inspection of columns 1 and 6 of
, which indicates that either A or B can bind independently to R. Inspection of columns 1 and 6 of 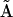 reveal that the dynamics of X5 also depends on
reveal that the dynamics of X5 also depends on  , which clearly identifies X5 as the RNA-protein complex RA. Similarly, complex
, which clearly identifies X5 as the RNA-protein complex RA. Similarly, complex  also affects the dynamics of X8, identifying it as the RNA-protein complex RB. Complex
also affects the dynamics of X8, identifying it as the RNA-protein complex RB. Complex 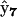 identifies X6 as RAB, complex
identifies X6 as RAB, complex 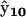 identifies X9 as RBC, and complex
identifies X9 as RBC, and complex 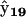 identifies X7 as RABC. It can also be seen from the structure of
identifies X7 as RABC. It can also be seen from the structure of 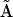 , that both forward and reverse reactions are occurring in the dynamics. A great deal of the mechanism is clear from the reconstruction, however there are some ambiguities evident from column 3 of
, that both forward and reverse reactions are occurring in the dynamics. A great deal of the mechanism is clear from the reconstruction, however there are some ambiguities evident from column 3 of 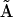 . Application of Eq. 7 to the reduced matrix of complexes gives rise to the reconstructed reduced kinetic matrix
. Application of Eq. 7 to the reduced matrix of complexes gives rise to the reconstructed reduced kinetic matrix 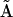 . The accuracy of the reconstruction of the rates is significantly improved using the reduced KMM, as shown in Fig. 5.
. The accuracy of the reconstruction of the rates is significantly improved using the reduced KMM, as shown in Fig. 5.
Reconstruction of the stoichiometry matrix
In the previous sections we have described the application of the KMM to identify the processes inherent in kinetic data, and to extract the rate constants for the processes. In the case of a dynamical system that corresponds to a chemical reaction network, we have shown that application of the stoichiometry matrix, as in the SMM, further improves the accuracy of the rate reconstructions. A complete and powerful synthesis of these two approaches would involve application of the KMM to identify the dynamics present, and use of this information to construct de novo a stoichiometry matrix that describes the reactions present in the system.
One of the difficulties encountered in construction of a stoichiometry matrix from the kinetic matrix  is apparent in column 2 of Eq. 28. It would appear that an apparent reaction of the form:
is apparent in column 2 of Eq. 28. It would appear that an apparent reaction of the form:
 |
(29) |
is taking place, but it is clear that such a complex reaction is a composite of two separate elementary reactions that both give the same product, RAB. It is necessary to describe several of the properties of networks of unimolecular and biomolecular reactions to develop an algorithm to resolve kinetic ambiguities in the structure of a kinetic matrix.
Consider one of the elementary chemical reaction steps from Fig. 3:
 |
(30) |
The reaction in Eq. 30 is represented by a reaction vector 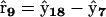 , which is a bimolecular reaction where RAB is the only possible product of the reaction of A and RB, and we define this as a “univariant” reaction. More specifically, there is only one possible product complex,
, which is a bimolecular reaction where RAB is the only possible product of the reaction of A and RB, and we define this as a “univariant” reaction. More specifically, there is only one possible product complex,  , arising from reactant complex
, arising from reactant complex 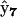 . In contrast, for the dissociation of RABC, there are two possible products:
. In contrast, for the dissociation of RABC, there are two possible products:
 |
(31) |
and we define this as a “bivariant”, or more generally a “multivariant” reaction. There are two possible product complexes that can be formed from the same reactant complex. The observed rate of dissociation of RABC is determined by the composite rate constant:
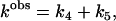 |
(32) |
as outlined in Appendix IV in the Supplementary Material. The composite reactions contained in the matrix 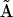 due to the presence of bivariant reactions must be decomposed into their elementary reactions.
due to the presence of bivariant reactions must be decomposed into their elementary reactions.
With these considerations in mind, we can devise a method for reconstruction of the stochiometry matrix  from the primary set of dynamical data
from the primary set of dynamical data  . The first step is to use the KMM to obtain a model of the chemical system in terms of the kinetic matrix
. The first step is to use the KMM to obtain a model of the chemical system in terms of the kinetic matrix 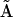 and a complete set of complexes
and a complete set of complexes 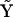 . As described above, matrices
. As described above, matrices  and
and 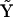 will both have dimensions of 9 × 55. Second, the reduced set of complexes is formed by elimination of the null columns of
will both have dimensions of 9 × 55. Second, the reduced set of complexes is formed by elimination of the null columns of  , and the reduced dimension of matrices
, and the reduced dimension of matrices 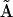 and
and  will be 9 × 12. The third step is to use the Representation Matrix Method for extraction of nonzero rates from the reduced kinetic matrix
will be 9 × 12. The third step is to use the Representation Matrix Method for extraction of nonzero rates from the reduced kinetic matrix 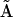 , which are then organized into a vector of rates
, which are then organized into a vector of rates 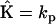 . The kinetic matrix
. The kinetic matrix 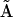 can be equivalently expressed in terms of a set of representation matrices, given by Eq. 5. Each of the resulting representation matrices
can be equivalently expressed in terms of a set of representation matrices, given by Eq. 5. Each of the resulting representation matrices 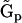 will have only one nonzero element, as in Eq. 19. Fourth, since the form of the dynamical equations using the set of representation matrices is significantly different from the form using the stochiometry matrix, as in Eq. 6, and it is necessary to generate an intermediate “collapsed” kinetic matrix
will have only one nonzero element, as in Eq. 19. Fourth, since the form of the dynamical equations using the set of representation matrices is significantly different from the form using the stochiometry matrix, as in Eq. 6, and it is necessary to generate an intermediate “collapsed” kinetic matrix 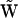 . If the dynamics of the system corresponds to a set of univariant chemical reactions, matrix
. If the dynamics of the system corresponds to a set of univariant chemical reactions, matrix 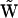 will be identical to the stoichiometry matrix
will be identical to the stoichiometry matrix 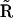 . However, in general, there are differences between
. However, in general, there are differences between 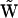 and
and 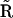 in columns that correspond to the multivariant reactions. Inspection of
in columns that correspond to the multivariant reactions. Inspection of  allows the identification of the multivariant reactions.
allows the identification of the multivariant reactions.
 |
(33) |
A final step in construction of the stoichiometry matrix 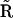 from the collapsed matrix
from the collapsed matrix 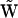 is necessary. Each column of
is necessary. Each column of 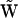 may correspond to a true reaction vector for a univariant reaction, or it may correspond to a reactant complex vector for a multivariant reaction. The formal mapping of indices to convert the kinetic matrix
may correspond to a true reaction vector for a univariant reaction, or it may correspond to a reactant complex vector for a multivariant reaction. The formal mapping of indices to convert the kinetic matrix 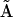 into the set of representation matrices
into the set of representation matrices 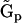 and vector of rates
and vector of rates 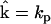 , the construction of the collapsed matrix
, the construction of the collapsed matrix  , and the decomposition of the multivariant columns of
, and the decomposition of the multivariant columns of 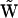 into reaction vectors is detailed in Appendix V in the Supplementary Material. Finally, the newly constructed stoichiometry matrix
into reaction vectors is detailed in Appendix V in the Supplementary Material. Finally, the newly constructed stoichiometry matrix 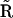 can be used in the SMM to further improve the accuracy of the rate constants for the dynamical system. The outlined procedure provides a robust method for determination of the structure of a chemical reaction network beginning only with the time series data
can be used in the SMM to further improve the accuracy of the rate constants for the dynamical system. The outlined procedure provides a robust method for determination of the structure of a chemical reaction network beginning only with the time series data 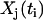 .
.
DISCUSSION
The numerical matrices method for nonlinear system identification
We have developed and implemented several variant methods of nonlinear system identification and determination of rate constants for cascades of chemical reactions. All of these methods describe the chemical kinetics in a similar manner using various numerical matrices: the matrix of complexes 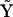 , the kinetic matrix
, the kinetic matrix 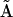 , the stochiometry matrix
, the stochiometry matrix 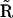 , the vector of rates
, the vector of rates 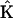 , and the set of representation matrices
, and the set of representation matrices 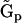 . However, these methods differ from each other in the amount of prior information about the reaction system that is used in the determination. Inclusion of prior information increases the accuracy of the reconstruction, in large part due to the decrease in the number of unknown parameters that must be determined.
. However, these methods differ from each other in the amount of prior information about the reaction system that is used in the determination. Inclusion of prior information increases the accuracy of the reconstruction, in large part due to the decrease in the number of unknown parameters that must be determined.
The most basic method is the Kinetic Matrix Method, which uses an arbitrary kinetic matrix  and the complete set of complexes
and the complete set of complexes 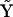 to describe all possible unimolecular and bimolecular reactions among the species present. No prior information is included about the structure of the reaction network, the input being the identity of the chemical species and the time dependence of their concentrations. The KMM identifies which reactions are taking place, and provides estimates for the rate constants for the reactions. Although the KMM is robust, it can be seen from Figs. 2 and 5 that there is a significant error in the rates that are determined.
to describe all possible unimolecular and bimolecular reactions among the species present. No prior information is included about the structure of the reaction network, the input being the identity of the chemical species and the time dependence of their concentrations. The KMM identifies which reactions are taking place, and provides estimates for the rate constants for the reactions. Although the KMM is robust, it can be seen from Figs. 2 and 5 that there is a significant error in the rates that are determined.
If the nature of the reactions are known or have been determined using the KMM, or if some possible reactions can be excluded a priori, it is possible to form a reduced matrix of complexes, which provides increased accuracy of the determined rates using the KMM. In addition, prior information about the structure of the reaction network is included by construction of the reduced matrix of complexes 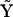 to reflect only the complexes that are present. As can be seen in Figs. 2 and 5, the reduced KMM provides a significant increase in the accuracy of the determined rates compared to the full KMM, which is presumably due to elimination of rate parameters for complexes that are irrelevant to the dynamics of the system.
to reflect only the complexes that are present. As can be seen in Figs. 2 and 5, the reduced KMM provides a significant increase in the accuracy of the determined rates compared to the full KMM, which is presumably due to elimination of rate parameters for complexes that are irrelevant to the dynamics of the system.
If the complete structure of the reaction network is known, the dynamics can be represented using the stoichiometry matrix formalism, which effectively pairs up the reactant and product complexes appropriate for each reaction in the system. The stoichiometry matrix  contains information about the structure of the reaction network, whereas the rate constants are contained in a vector
contains information about the structure of the reaction network, whereas the rate constants are contained in a vector 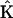 . This method requires determination of the fewest parameters from the data, and consequently provides the most accurate determination of the rate constants, as can be seen in Fig. 5.
. This method requires determination of the fewest parameters from the data, and consequently provides the most accurate determination of the rate constants, as can be seen in Fig. 5.
A particularly significant part of the NMM is the Representation Matrix Method. The RMM is the most generalized and universal of the methods, and can work with different levels of prior information, and the KMM and the SMM can be considered as specific implementations of the RMM, where the level of prior information included in the reconstruction is determined by the choice of the set of representation matrices 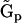 . In the complete absence of prior information, the KMM can be fully and equivalently represented and implemented by choosing the set of representation matrices
. In the complete absence of prior information, the KMM can be fully and equivalently represented and implemented by choosing the set of representation matrices 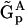 such that:
such that:
 |
(34) |
When the full structure of the reaction network is known, the SMM can be similarly represented by the choice of representation matrices 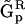 such that:
such that:
 |
(35) |
In addition, the RMM is particularly applicable when some of the rates of reactions are exactly known and others remain to be determined. In this most general form, the dynamics of the system is given by:
 |
(36) |
where the matrix  contains the information about known rate parameters (35).
contains the information about known rate parameters (35).
Determination of reaction network structure
For a given reaction mechanism, it is straightforward to construct the set of differential equations that describe the dynamics of the reaction network, as shown in the top part of Fig. 6. Given initial conditions and values for the rate constants it is also straightforward to either analytically or numerically integrate the system of differential equations to provide the time-dependent concentrations of the species in the network. The NMM described here provides a general method for the reverse process, where the time dependence of the species alone is sufficient to determine the reaction structure, as shown in the lower part of Fig. 6. From the time dependence of the species and the corresponding rates, the KMM is applied to determine the basic reaction structure and provide initial estimates for rate constants in the general kinetic matrix 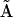 . In some cases it is possible to deduce the stoichiometry matrix
. In some cases it is possible to deduce the stoichiometry matrix 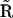 directly from
directly from 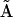 , but in particular for the case where there are multivariant reactions present in the system, additional steps are required. We have outlined general procedure for generating
, but in particular for the case where there are multivariant reactions present in the system, additional steps are required. We have outlined general procedure for generating 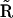 from
from 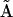 for a chemical reaction network via a set of intermediate representation matrices
for a chemical reaction network via a set of intermediate representation matrices 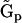 and an intermediate collapsed matrix
and an intermediate collapsed matrix 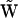 . Thus, beginning only with the time dependence of the species, the structure of the reaction network can be determined using the NMM.
. Thus, beginning only with the time dependence of the species, the structure of the reaction network can be determined using the NMM.
FIGURE 6.
Overview of the Numerical Matrices Method to nonlinear system identification. Given a reaction mechanism, it is straightforward to construct a set of differential equations that can be numerically integrated to give time dependence of the species. The NMM formally carries out the reverse of this process in a series of steps involving kinetic matrices.
Comparison to correlation metric construction
There are some strong analogies between the NMM and the previously developed correlation metric construction (CMC) method (10–12). For both methods, reaction pathways can be identified from concentration measurements of the species, and a type of correlation matrix is used for the analysis. In addition, the stochastic modulation of the inputs in the CMC is similar in spirit to the use of multiple initial states in NMM. Both the NMM and CMC have the common limitation that data for all species must be considered.
There are significant differences between the NMM and CMC as well, and the NMM has several advantages. First, an important part of NMM is that values for rate parameters are determined, but no such values are produced by CMC, which is limited to identification of the reaction structure. Second, NMM is based on correlations between kinetic complexes such as  and
and 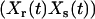 , which are analyzed using linear algebra and statistical methods. In contrast, the CMC is based on analysis of a time-lagged two-species correlation function between
, which are analyzed using linear algebra and statistical methods. In contrast, the CMC is based on analysis of a time-lagged two-species correlation function between  and
and 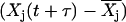 that is interpreted using a heuristic algorithm. Finally, there is no way to include prior information about reaction structure using the CMC, which is a strength of the NMM.
that is interpreted using a heuristic algorithm. Finally, there is no way to include prior information about reaction structure using the CMC, which is a strength of the NMM.
Domain of applicability
To apply the methods in the NMM, it is necessary to collect time-series data on all of the species present, and data sets with many time points are required. One advantage of the NMM is that no information is required about the initial conditions. Data collected beginning at any arbitrary time t > 0 is sufficient, and there are no boundary conditions imposed for determination of the rate constants. The only requirement is that significant changes in the concentrations occur in the data sets, with respect to the noise. The Numerical Matrices Method can be carried out using not only direct differential methods, but also the direct integral or empirical optimization methods described above. The primary application for these alternative implementations would be when the number of time points is small. For a large number of time points, the condition number of the matrix 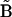 is much better for the direct differential method, which is particularly important when the number of parameters to be determined is large.
is much better for the direct differential method, which is particularly important when the number of parameters to be determined is large.
The linearly quadratic model used here for the kinetics is generally applicable to most chemical reaction networks. However, this choice is not imperative, and other more complex models for the kinetics can be used within the framework of the NMM. In addition, the NMM can be generalized to important cases with nonlinear dependence on parameters, such as the case of Michaelis-Menten enzyme kinetics. A more generalized nonlinear kinetic model of the form:
 |
(37) |
contains strong nonlinearity of parameters 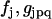 . In analogy to Eqs. 2 and 50 in Appendix II in the Supplementary Material, the least squares criterion:
. In analogy to Eqs. 2 and 50 in Appendix II in the Supplementary Material, the least squares criterion:
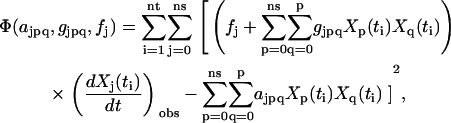 |
(38) |
can be used to solve the reaction identification problem for the nonlinear parameters using a tractable system of linear equations for all of the parameters in Eq. 37, including the nonlinear parameters 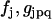 .
.
Multiple initial states
Application of the NMM is complicated by linear dependencies among the species in the time-dependent data. In the absence of knowledge about the reaction network structure or the stoichiometry matrix, the linear system of equations for the rate constants becomes singular. However, recording data sets with multiple initial states where the concentrations of the various species are changed in combination results in a numerically tractable problem. The choice of the number and the nature of the initial states will depend on the nature of the kinetics present, which cannot be known a priori for an unknown reaction structure. This consideration offers an important guiding principle for designing experiments to determine rate constants in complex kinetic systems, and it will be generally necessary to collect data using a range of concentrations of all of the initial species present.
CONCLUSION
The matrix methods described here are based on a very general kinetic framework that is applicable to a wide variety of dynamic systems, particularly chemical and biochemical reaction networks. An important and simplifying principle is that most chemical reactions can be represented by a linearly quadratic kinetic equation, making it possible to enumerate all possible uni- and bimolecular reactions. For all of the above methods, the description of the dynamics is based on the matrix of complexes 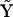 that correspond to these reactions. An important feature of the approach is that rate constants for these reactions can be obtained by solving a linear system of equations with a least squares solution to the observed data in closed form. The various methods can be readily combined with each other and can be applied in succession as the understanding of the dynamics increases. The individual matrix methods constitute important parts of a universal approach to the task of the system identification that we term the Numerical Matrices Method.
that correspond to these reactions. An important feature of the approach is that rate constants for these reactions can be obtained by solving a linear system of equations with a least squares solution to the observed data in closed form. The various methods can be readily combined with each other and can be applied in succession as the understanding of the dynamics increases. The individual matrix methods constitute important parts of a universal approach to the task of the system identification that we term the Numerical Matrices Method.
The NMM provides an opportunity to use a continuously changing amount of prior information for system identification and rate constant determination. This in turn provides the possibility of developing iterative methods for identification of large chemical and biochemical reaction networks. Information that is obtained in early steps can be subsequently included to iteratively improve the description of the dynamics. A complete implementation of such an iterative procedure will include analysis of errors at each stage to guide the reconstruction of the reaction network. Here we have described the essential elements of the NMM that will provide the basis for further development of such powerful tools for analysis of kinetic data and for system identification.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Acknowledgments
This work was supported by grants from the National Institutes of Health (TW-007478 to A.V.K. and J.R.W and GM-53757 to J.R.W.).
References
- 1.Maria, G. 2004. A review of algorithms and trends in kinetic model identification for chemical and biochemical systems. Chem. Biochem. Eng. Q. 18:195–222. [Google Scholar]
- 2.Crampin, E. J., S. Schnell, and P. E. McSharry. 2004. Mathematical and computational techniques to deduce complex biochemical reaction mechanisms. Prog. Biophys. Mol. Biol. 86:77–112. [DOI] [PubMed] [Google Scholar]
- 3.Moles, C. G., P. Mendes, and J. R. Banga. 2003. Parameter estimation in biochemical pathways: a comparison of global optimization methods. Genome Res. 13:2467–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Famili, I., and B. O. Palsson. 2003. Systemic metabolic reactions are obtained by singular value decomposition of genome-scale stoichiometric matrices. J. Theor. Biol. 224:87–96. [DOI] [PubMed] [Google Scholar]
- 5.Schuster, S., D. A. Fell, and R. Dandekar. 2000. A general definition of metabolic pathways useful for systematic organization and analysis of complex metabolic networks. Nat. Biotechnol. 18:326–332. [DOI] [PubMed] [Google Scholar]
- 6.Famili, I., and B. O. Palsson. 2003. The convex basis of the left null space of the stoichiometric matrix leads to the definition of metabolically meaningful pools. Biophys. J. 85:16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forster, J., I. Famili, P. Fu, B. O. Palsson, and J. Nielsen. 2003. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 13:244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuster, S., C. Hilgetag, J. H. Woods, and D. A. Fell. 2002. Reaction routes in biochemical reaction systems: algebraic properties, validated calculation procedure and example from nucleotide metabolism. J. Math. Biol. 45:153–181. [DOI] [PubMed] [Google Scholar]
- 9.Stelling, J., S. Klamt, K. Bettenbrock, S. Schuster, and E. D. Gilles. 2002. Metabolic network structure determines key aspects of functionality and regulation. Nature. 420:190–193. [DOI] [PubMed] [Google Scholar]
- 10.Arkin, A., and J. Ross. 1995. Statistical construction of chemical reaction mechanisms from measured time-series. J. Phys. Chem. 99:970–979. [Google Scholar]
- 11.Arkin, A., P. Shen, and J. Ross. 1997. A test case of correlation metric construction of a reaction pathway from measurements. Science. 277:1275–1279. [Google Scholar]
- 12.Vance, W., A. Arkin, and J. Ross. 2002. Determination of causal connectivities of species in reaction networks. Proc. Natl. Acad. Sci. USA. 99:5816–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bard, Y. 1974. Nonlinear Parameter Estimation. Academic Press, New York and London, UK.
- 14.Hemker, P. W. 1972. Numerical methods for differential equations in system simulation and in parameter estimation. In Analysis and Simulation of Biochemical Systems. H. C. Hemker and B. Hess, editors. North Holland, Amsterdam. 59–80.
- 15.Stortelder, W. J. H. 1998. Parameter Estimation in Nonlinear Dynamic Systems. Stichting Mathematische Centrum, Amsterdam.
- 16.Stortelder, W. J. H., P. W. Hemker, and H. C. Hemker. 1997. Mathematical Modelling in Blood Coagulation; Simulation and Parameter Estimation. Modelling, Analysis and Simulation. Report CWI, Stichting Mathematische Centrum, Amsterdam.
- 17.Bard, Y. 1970. Comparison of gradient methods for the solution of nonlinear parameter estimation problems. SIAM J. Numer. Anal. 7:157–186. [Google Scholar]
- 18.Himmelblau, D. M., C. R. Jones, and K. B. Bischoff. 1967. Determination of rate constants for complex kinetic models. Ind. Eng. Chem. Fundam. 6:539–543. [Google Scholar]
- 19.Hosten, L. H. 1979. A comparative study of shortcut procedures for parameter estimation in differential equations. Comp. Chem. Eng. 3:117–126. [Google Scholar]
- 20.Vajda, S., P. Valko, and A. Yermakova. 1986. A direct-indirect procedure for estimation of kinetic parameters. Comp. Chem. Eng. 10:49–58. [Google Scholar]
- 21.Yermakova, A., and P. Valco. 1985. A remark on parameter estimation in dynamic models. AIChE J. 31:1213. [Google Scholar]
- 22.Yermakova, A., P. Valco, and S. Vajda. 1982. Direct integral method via spline-approximation for estimating rate constants. Appl. Cat. 2:139–154. [Google Scholar]
- 23.Prony, R. 1975. Essai experimental et analyticque. J. de L'ecole Polytechnique. 1:24–76. [in French]. [Google Scholar]
- 24.Bak, K. 1963. Numerical differentiation and integration in chemical kinetics. Acta Chem. Scand. 17:985–988. [Google Scholar]
- 25.Fay, L., and A. Balogh. 1968. Determination of reaction order and rate constants on the basis of the parameter estimation of differential equations. Acta Chim. Acad. Sci. Hung. 57:391–401. [Google Scholar]
- 26.Garfinkel, D., J. D. Rutledge, and J. J. Higgins. 1961. Simulation and analysis of biochemical systems. Comm. Assoc. Computing Machinery. 4:559–562. [Google Scholar]
- 27.Levenspiel, O. 1962. Chemical Reaction Engineering. Wiley, New York.
- 28.Lindsay, K. L. 1962. Optimum time scheduling of kinetic experiments. Ind. Eng. Chem. Fundamentals. 1:241–245. [Google Scholar]
- 29.Rudakov, E. 1960. Differential methods of determination of rate constants of noncomplicated chemical reactions. Kinetics and Catalysis. 1:177–187. [Google Scholar]
- 30.Rudakov, E. 1970. Determination of rate constants. Method of support function. Kinetics and Catalysis. 11:228–234. [Google Scholar]
- 31.Steiner, R., and K. Schoenemann. 1965. Korrelationsrechnung beim Aufstellen von Geshwindigkeitsgleichungen fur Komplexe Chemische. Chem. Ing. Tech. 37:101–107. [in German]. [Google Scholar]
- 32.Feinberg, M., and F. Horn. 1977. Chemical mechanism structure and the coincidence of the stoichiometric and kinetic subspace. Arch. Rational Mech. Anal. 66:83–97. [Google Scholar]
- 33.Horn, F., and R. Jackson. 1972. General mass action kinetics. Arch. Rational Mech. Anal. 47:81–116. [Google Scholar]
- 34.Karnaukhov, A. V., and E. V. Karnaukhova. 2003. Application of a new method of nonlinear dynamical system identification to biochemical problems. Biochemistry (Mosc.). 68:253–259. [DOI] [PubMed] [Google Scholar]
- 35.Karnaukhov, A. V., E. V. Karnaukhova, and V. I. Zarnitsina. 2005. Consideration of prior information and the type of experimental error in the identification method based on minimizing square residuals. Biophysics (Moscow). 50:309–313. [Google Scholar]
- 36.Schluenzen, F., A. Tocilj, R. Zarivach, J. Harms, M. Gluehmann, D. Janell, A. Bashan, H. Bartels, I. Agmon, F. Franceschi, and A. Yonath. 2000. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 102:615–623. [DOI] [PubMed] [Google Scholar]
- 37.Wimberly, B. T., D. E. Brodersen, W. M. Clemons Jr., R. J. Morgan-Warren, A. P. Carter, C. Vonrhein, T. Hartsch, and V. Ramakrishnan. 2000. Structure of the 30S ribosomal subunit. Nature. 407:327–339. [DOI] [PubMed] [Google Scholar]
- 38.Traub, P., and M. Nomura. 1968. Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc. Natl. Acad. Sci. USA. 59:777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trevathan, M. T., G. Siuzdak, and J. R. Williamson. 2005. Assembly landscape of the 30S ribosomal subunit. Nature. 438:628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]








