Abstract
The transmembrane surface of a multi-helix membrane protein will be rough with cavities of various sizes between the transmembrane α-helices. Efficient solvation of the surface by the lipid molecules that surround the protein in a membrane requires that the lipid fatty acyl chains be able to enter the cavities. This possibility has been investigated using fluorescence quenching methods. Trp residues have been introduced into lipid-facing sites in the first transmembrane α-helix (M1) of the mechanosensitive channel of large-conductance MscL; lipid-facing residues at the N-terminal end of M1 are buried below the transmembrane surface of the protein. Fluorescence emission maxima for lipid-facing Trp residues in M1 vary with position in the bilayer comparably to those for Trp residues in the second transmembrane α-helix (M2) despite the fact that lipid-facing residues in M2 are on the surface of the protein. Fluorescence emission spectra for most Trp residues on the periplasmic sides of M1 and M2 fit well to a model proposing a trough-like variation of dielectric constant across the membrane, but the relationship between location and fluorescence emission maximum on the cytoplasmic side of the membrane is more complex. The fluorescence of Trp residues in M1 is quenched efficiently by phospholipids with bromine-containing fatty acyl chains, showing that the lipid chains must be able to enter the Trp-containing cavities on the surface of MscL, resulting in efficient solvation of the surface.
INTRODUCTION
Solvation of a water-soluble protein by water is important in defining the structure and function of the protein (1). Some water molecules are bound in cavities on the protein surface; others interact less specifically with rather featureless regions of the surface. The overall result is that water molecules in the hydration shell around a protein have properties that differ significantly from those of the bulk water, the effects of the protein extending only to one or two shells of water molecules around the protein (1,2). Solvation of a membrane protein is more complex than solvation of a water-soluble protein because membrane proteins are solvated both by water molecules and by lipid molecules.
The transmembrane surface of a multi-helix membrane protein is rough, containing cavities of various shapes and sizes, ranging from small gaps to large crevices. In a biological membrane this surface is covered by a lipid bilayer, the lipid molecules in contact with the protein surface being commonly referred to as boundary or annular lipids, as they form an annular shell around the protein (3). In the same way that water molecules in the hydration shell around a protein are perturbed by the protein, so lipid molecules in the annular shell will be perturbed by the membrane protein. X-ray crystal structures (3,4) and molecular dynamics simulations (5) show that the annular lipid molecules make close contact with the surface of the protein, which results in the conformations adopted by the annular lipids being different from those adopted by lipids in the absence of membrane proteins; lipid molecules close to a membrane protein will have very different interactions from a lipid molecule in a protein-free lipid bilayer. Nevertheless, the rates of exchange of lipid molecules on and off a membrane protein surface are only slightly slower than the rates of exchange of lipid molecules in a bulk lipid bilayer (6), in the same way that mobilities of most water molecules on the surface of a protein are only marginally lower than those in bulk water (2). Further, ranges of interaction energies between a lipid molecule and a membrane protein are, in general, large because the total interaction energy is the sum of many weak van der Waals and charge interactions; there is no single deep energy well into which the lipids fall to give a single favored conformation, and lipid molecules are not frozen in a single long-lived conformation on the protein surface (7). Molecular dynamics simulations also suggest that effects of proteins on lipids are short range, affecting principally those lipids bound to the protein (reviewed by Lee (8)). This is consistent with experimental data with spin-labeled lipids that show two populations of lipids present in a protein-containing lipid bilayer, a relatively immobilized component corresponding to annular lipid and a more mobile component corresponding to bulk lipid (9).
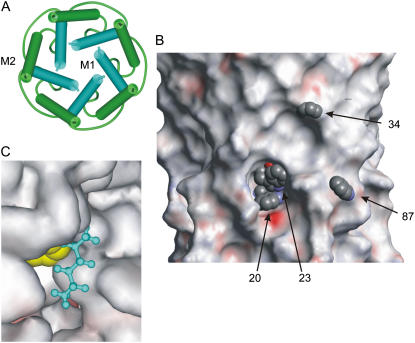
The question addressed here is to what extent lipid fatty acyl chains can enter cavities on the transmembrane surface of a membrane protein, resulting in efficient solvation of the surface. A good example of a rough transmembrane surface is provided by MscL from Mycobacterium tuberculosis. In MscL, each monomer in the homopentameric structure contains two transmembrane α-helices, M1 and M2. In the closed channel (10), a ring of five M1 helices is contained within a ring of five M2 helices so that most contacts with the surrounding lipid bilayer are made by residues in M2 (Fig. 1 A). Nevertheless, although many residues in M1 face toward the center of the channel or are involved in helix-helix contacts, some also face outward toward the lipid bilayer. The way that the M1 helices are tilted with respect to the bilayer normal means that those lipid-facing residues that are located toward the C-terminal end of M1 are relatively close to the protein surface, whereas those at the N-terminal end are buried in crevices between the M2 helices (see Fig. 1 B).
FIGURE 1.
Structure of MscL. (A) View of the transmembrane α-helices of the homopentameric structure, from the cytoplasmic side of the membrane. The first (M1) and second (M2) helices are colored blue and green, respectively. (B) View of the lipid-facing surface of MscL with Trp residues at positions 20, 23, and 34 in M1 and position 87 in M2 shown in space-fill format. The surface is colored according to electrostatic potential. The periplasmic surface is at the top, and the cytoplasmic surface is at the bottom. (C). A Trp residue at position 23 (yellow; space-fill format) is shown in van der Waals contact with hexane (blue; ball-and-stick format) modeled into the cavity in the protein surface around position 23.
The approach adopted to test the extent of chain penetration into the MscL surface is based on the ability of phospholipids with bromine-containing fatty acyl chains to quench Trp fluorescence. Phospholipids such as dibromostearoylphosphatidylcholine (di(Br2C18:0)PC) are made by bromination of the corresponding phospholipid with two cis-unsaturated fatty acyl chains and behave much like a conventional phospholipid with unsaturated fatty acyl chains because the bulky bromine atoms have effects on lipid packing that are similar to those of a cis double bond (11). Wild-type MscL lacks Trp residues so that single Trp residues can be introduced into regions of choice in MscL to study lipid binding close to the introduced Trp residue. The efficiency of quenching of Trp fluorescence by a brominated phospholipid shows a sixth-power dependence on distance between the bromine and Trp moieties, with 50% quenching at a distance of ca 8 Å (12). Thus, a bromine-containing phospholipid can efficiently quench the fluorescence of a Trp residue in M1 located within a deep cavity only if lipid fatty acyl chains are able to penetrate into the cavity to locate the chains close to the Trp residue.
EXPERIMENTAL PROCEDURES
Dioleoylphosphatidylcholine (di(C18:1)PC) was obtained from Avanti Polar Lipids (Alabaster, AL), and dimyristoylphosphatidylcholine (di(C14:0)PC) and dipalmitoylphosphatidylcholine (di(C16:0)PC) were obtained from Sigma (St. Louis, MO). Di(Br2C18:0)PC was prepared as described by East and Lee (11). A plasmid containing the M. tuberculosis mscL gene with a poly-His epitope at the N-terminus was the generous gift of Professor D. C. Rees. Site-directed mutagenesis was performed using the Quick-change protocol from Stratagene. After PCR mutagenesis, the native methylated DNA templates were digested with DpnI (Promega) for 2 h at 37°C. The mutations were confirmed by DNA sequencing. Escherichia coli BL21(λDE3)pLysS transformants carrying the pET-19b plasmid (Novagen) with the mscL gene were generally grown in 6 L Luria broth to mid-log phase (OD600 = 0.6) and then induced for 3 h in the presence of isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM). MscL was purified as described by Powl et al. (12) and stored at −80°C until use.
Test for gain of function mutations
To test for gain of function mutations, rates of growth of E. coli expressing MscL were measured. Gain of function mutants show a reduced rate of growth compared to wild-type after induction of MscL production with IPTG (13).
Fluorescence measurements
Purified MscL was reconstituted into lipid bilayers by mixing lipid and MscL in cholate, followed by dilution into buffer to decrease the concentration of cholate below its critical micelle concentration, as described by Powl et al. (12). Lipid (0.47 μmol) was dried down from solution in chloroform onto the walls of a thin glass vial. Buffer (400 μl; 20 mM Hepes, 100 mM KCl, 1 mM EGTA, pH 7.2) containing 15 mM potassium cholate was added, and the sample was then mixed on a vortex and sonicated to clarity in a bath sonicator (Ultrawave, Cardiff, UK). MscL (0.44 mg) was added to the lipid solution, and the sample was then incubated at 25°C for 15 min, followed by incubation on ice until use. For fluorescence measurements, 250 μl of the sample was diluted into 2.75 ml buffer (20 mM Hepes, 100 mM KCl, 1 mM EGTA, pH 7.2) in a stirred fluorescence cuvette at 25°C. The final protein concentration was 1 μM, based on a molecular mass of 93 kDa for the MscL pentamer, and the molar ratio of lipid to MscL pentamer was 500:1. Fluorescence measurements were recorded using a SLM-8000C fluorimeter with excitation at 280 nm. Fluorescence emission spectra were corrected for light scatter by subtraction of a blank consisting of lipid alone in buffer. Fluorescence intensities were stable for at least 20 min. Experimental values of fluorescence intensity are averages of duplicate measurements from three separate reconstitutions.
To obtain accurate values for wavelengths of maximum fluorescence emission intensity (λmax) and spectral widths (ωλ), fluorescence spectra were fitted to skewed Gaussian curves
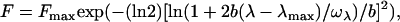 |
(1) |
where F and Fmax are the fluorescence intensities at wavelengths λ and λmax, respectively, b is the skew parameter, and ωλ is the peak width at half-height (14).
Molecular modeling
Molecular modeling was performed using WebLab ViewerPro from Molecular Simulations (San Diego, CA), using the published coordinates for wild-type MscL (10). Residues in MscL were mutated to Trp, which were then positioned to remove any interatomic distances <70% of the sum of the covalent radii while locating the Trp residue as close to the surface of the protein as possible. The depth of a Trp residue below the protein surface was determined by docking a molecule of lipid with all-trans fatty acyl chains onto the protein surface, with the lipid headgroups adjacent to the hydrophobic membrane-spanning region of MscL, defined by Leu-69 and Leu-92 on the periplasmic and cytoplasmic sides of the membrane, respectively (15). The lipid chosen, dilauroylphosphatidylethanolamine, gives a bilayer with a hydrophobic thickness, when the chains are all-trans chains, similar to the hydrophobic thickness of MscL, which is ∼25 Å (15). The lipid molecule was located on the surface of MscL to minimize the distance between the outer edge of the Trp ring and the carbon backbone of a lipid fatty acyl chain while avoiding clashes between the lipid and MscL. The coordinates for dilauroylphosphatidylethanolamine were taken from Elder et al. (16).
The hydrophobic thickness of a bilayer of di(Br2C18:0)PC in the liquid crystalline phase is ∼27 Å (17) with the two Br atoms per chain, at the 9 and 10 positions, being located at an average distance of ∼7.2 Å from the glycerol backbone region. Because the hydrophobic thickness of MscL has been estimated to be 25 Å (15), the hydrophobic thickness of the di(Br2C18:0)PC bilayer actually in contact with the MscL surface would be expected to be very similar to that for the bulk bilayer. The distance along the bilayer normal between the α-carbon of a residue mutated to Trp and the bromine atoms of di(Br2C18:0)PC was calculated from the crystal structure of MscL assuming a symmetric location for the transmembrane region of MscL within the lipid bilayer.
Analysis of fluorescence results
Quenching of Trp fluorescence in mixtures of di(Br2C18:0)PC and di(C18:1)PC was fitted to the following equation to give the value of n, the number of lipid-binding sites on MscL from which the fluorescence of the Trp residue can be quenched (18,19):
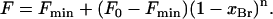 |
(2) |
Here, Fo and Fmin are the fluorescence intensities for MscL in di(C18:1)PC and in di(Br2C18:0)PC, respectively, and F is the fluorescence intensity in the phospholipid mixture when the mole fraction of di(Br2C18:0)PC is xBr. Values of n for lipid-facing residues in M1 were ∼2.5, comparable to the values obtained previously for Trp residues in M2 (12).
Quenching of Trp fluorescence by dibrominated quenchers fits to an equation with a sixth-power dependence on the distance of separation between the Trp residue and the quencher, as in Forster energy transfer (20,21) with a value for Ro, the distance at which energy transfer is 50% efficient, of 8 Å (12). The efficiency E of energy transfer from Trp to brominated lipid chains in the presence of x chains all at the same distance r from the Trp, is given by
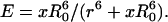 |
(3) |
For MscL in di(Br2C18:0)PC with a value for n of 2.5, the value for x is 5. This equation is valid for Trp residues in MscL located toward the cytoplasmic and periplasmic sides of MscL but not to centrally located residues that could be quenched by brominated lipids in either half of the lipid bilayer.
RESULTS
Testing for gain of function mutations
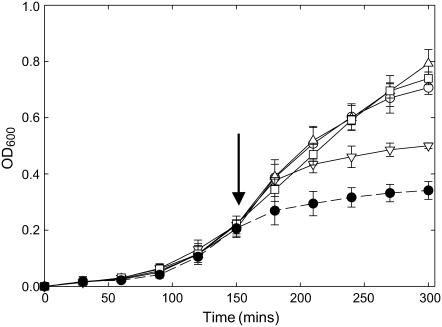
The five lipid-facing residues in transmembrane α-helix M1 listed in Table 1 were mutated to Trp. Because MscL undergoes large changes in conformation on opening (22,23), it is important to confirm that these mutations do not result in gain of function mutants, which would be in an open conformation in the absence of a membrane tension. An in vivo phenotype assay has been developed to test for gain of function mutations of MscL, gain of function mutants being detected by a reduced rate of growth after induction of MscL production with IPTG (24). Expression of a gain of function mutant such as V22W, where the Trp substitution has been made at a point of helix-helix contact in the closed channel, results in very low levels of cell growth (Fig. 2). Mutation of four of the five lipid-exposed residues in M1 resulted in no effect on growth rate, and although the mutation A20W resulted in a reduced growth rate, the effect was less than that observed for V22W (Fig. 2). We conclude that mutation of the lipid-facing Trp residues in M1 does not result in a channel that will be open at zero membrane tension. It was similarly observed that mutation of lipid-facing residues in M2 to Trp also did not result in gain of function mutants (15).
TABLE 1.
Fluorescence properties of Trp mutants of MscL
| Mutant | λmax (nm) in di(C18:1)PC | Distance along bilayer normal from L69 (Å) |
|---|---|---|
| A20W | 329.3 ± 0.2 | 24.3 |
| I23W | 325.2 ± 0.1 | 20.1 |
| L30W | 327.6 ± 0.1 | 11.6 |
| V31W | 323.9 ± 0.1 | 11.6 |
| F34W | 326.2 ± 0.4 | 7.1 |
Wavelengths of maximum fluorescence emission intensity (λmax) were obtained by fitting fluorescence spectra recorded in di(C18:1)PC to Eq. 1 for a skewed Gaussian. Distances along the bilayer normal were calculated as described in the text.
FIGURE 2.
Growth of E. coli in liquid medium expressing mutant mscL genes. E. coli BL21(DE3)pLysS transformants carrying the pET-19b plasmid with the mscL gene were grown at 37°C: (○), WT; (Δ), I23W; (□), L30W; (∇), A20W; and (•), V22W. Cells were induced with 1 mM IPTG after 150 min (arrow). Data points are the average of three colonies. Growth curves for V31W and F34W were very similar to those for WT. The mutants V22W is a gain of function mutant.
Fluorescence properties of Trp residues in M1
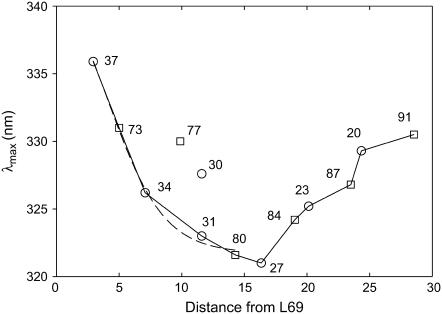
Fluorescence emission maxima for Trp mutants at lipid-exposed sites in M1 reconstituted into bilayers of di(C18:1)PC are listed in Table 1 and are plotted in Fig. 3 as a function of the distance along the bilayer normal between the α-carbon of the mutated residue and that of Leu-69, a marker for the location of the periplasmic side of the membrane. Also plotted in Fig. 3 are the corresponding data (15) for Trp residues in M2. As shown, fluorescence emission maxima for Trp residues in M1 and M2 depend on distance from the bilayer surface in a similar way, except for mutants L30W and I77W, whose emission maxima are distinct from those in the other mutants studied (Fig. 3).
FIGURE 3.
Fluorescence emission maxima for Trp mutants of MscL. The figure shows fluorescence emission maxima for Trp mutants in M1 (○) and M2 (□) of MscL reconstituted into bilayers of di(C18:1)PC plotted as a function of the distance (Å) of the α-carbon of the residue from that of Leu-69, measured along the bilayer normal. The dotted line shows a fit of the data on the periplasmic side of the membrane to Eq. 5 describing a trough-like polarity profile in the membrane. As shown, fluorescence emission maxima for mutants L30W and I77W do not fit the general trends observed with the other mutants.
The emission spectrum of Trp is polarity dependent, the wavelength of maximum fluorescence emission increasing with increasing polarity of the surrounding medium; for many fluorescent dyes the relationship between emission maximum and solvent polarity is close to linear (25,26). The dielectric constant of the hydrophobic core of a lipid bilayer is ∼2, and that of water is 78 at 25°C (27), so that the dielectric constant of a lipid bilayer must vary very markedly over the glycerol backbone and lipid headgroup regions. Flewelling and Hubbell (27) proposed that the dielectric constant varies in a smooth and continuous way over the interface, described by a sigmoidal dependence of dielectric constant on position in the interface region. They therefore described the dielectric constant of a bilayer by the following expression, giving a trough-like profile across the membrane:
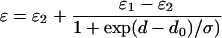 |
(4) |
Here, ɛ is the dielectric constant at a distance d from the bilayer center, and ɛ1 and ɛ2 are the dielectric constants in the bilayer center and in water, respectively. do is the value of d at the point of maximum gradient, corresponding to the point where ɛ = (ɛ1 + ɛ2)/2.0, and σ is an exponential decay constant that reflects the width of the transition region. If the emission maximum of Trp depends linearly on dielectric constant, then Eq. 4 can be recast in the following form to describe the dependence of fluorescence emission maximum on position within a bilayer (28):
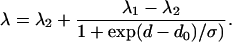 |
(5) |
Here, λ is the wavelength of maximum emission for a Trp residue at a distance d from the bilayer center, and λ1 and λ2 are limiting values for the wavelengths of maximum emission for a Trp residue in the bilayer center and in the polar headgroup region, respectively. The equation is restricted to the hydrocarbon core and glycerol backbone regions of the bilayer. With MscL located symmetrically in the bilayer, the bilayer midplane is 15.9 Å from the α-carbon of Leu-69. As shown in Fig. 3, this equation gives a good fit to the data on the periplasmic side of the membrane, except for mutants L30W and I77W, with values of do = 11.8 ± 2.6 Å and σ = 2.2 ± 1.2 Å. The value for do corresponds to a hydrophobic thickness of 23.6 ± 5.2 Å, in good agreement with the previous estimate for the hydrophobic thickness of MscL of 25 Å (15). The value of σ, an estimate of the width of the transition region, can be compared to estimates of 1–2 Å obtained from analysis of spin-label data by Marsh (29). Fluorescence emission maxima on the cytoplasmic side of the membrane do not show a trough-like dependence on distance (Fig. 3) and so cannot be fitted to Eq. 5.
Quenching by lipids with bromine-containing chains
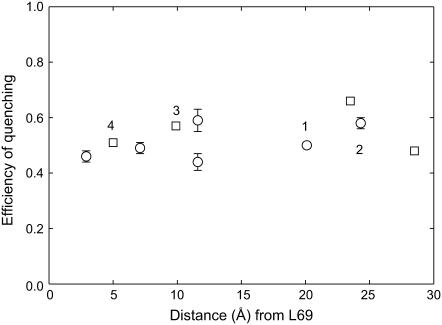
Fig. 4 compares efficiencies of fluorescence quenching for lipid-facing Trp mutants in M1 and M2 in bilayers of di(Br2C18:0)PC; values for the efficiency of quenching are plotted as a function of the distance of the residue along the bilayer normal from the interfacial residue Leu-69. As shown, the efficiencies of fluorescence quenching are similar for Trp residues in M1 and M2 at similar positions within the bilayer. For example, the efficiency of quenching of the buried residue A20W on the cytoplasmic side of MscL is similar to that of the surface exposed Trp residue in I77W at a similar depth in the bilayer on the periplasmic side. Similarly, comparable efficiencies of quenching are observed for the buried Trp residue in I23W and the surface exposed Trp residue in L73W, located at similar depths in the membrane on the cytoplasmic and periplasmic sides, respectively (Fig. 4). These results suggest that buried Trp residues in M1 and surface-exposed Trp residues in M2 are equally accessible to lipid fatty acyl chains. This conclusion can be expressed in more quantitative terms, as follows.
FIGURE 4.
Quenching of Trp fluorescence in MscL by di(Br2C18:0)PC. Efficiencies of fluorescence quenching are plotted for Trp residues in M1 (○) and M2 (□) as a function of the distance (Å) of the α-carbon of the residue from that of Leu-69, measured along the bilayer normal. Efficiencies of quenching were calculated as 1-F/F0, where F and F0 are fluorescence intensities in di(Br2C18:0)PC and di(C18:1)PC, respectively. Residues in the center of the membrane that will be quenched by brominated lipid molecules in both halves of the bilayer have not been included. Data points labeled 1, 2, 3, and 4 correspond to I23W, A20W, I77W, and L73W, respectively. Experimental values of fluorescence intensity are averages of duplicate measurements from three separate reconstitutions. The data for residues in M2 are from Powl et al. (15).
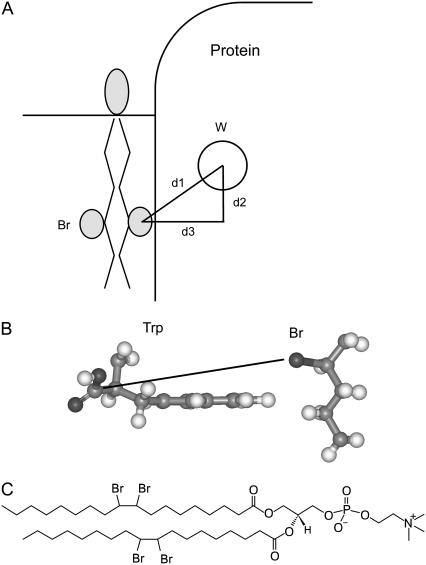
If the distance between the Trp residue in MscL and the bromine atoms of a neighboring molecule of di(Br2C18:0)PC is known (d1 in Fig. 5), then the efficiency of fluorescence quenching can be calculated from Eq. 3. The distance along the bilayer normal between a Trp residue and the bromine atoms in a bilayer of di(Br2C18:0)PC (d2 in Fig. 5) can be calculated as described in Experimental procedures; values are listed in Tables 2 and 3. The value of d3 (Fig. 5) cannot be determined purely from the structure of MscL because it requires a knowledge of the location in the Trp residue from which distances should be measured for calculations of fluorescence quenching. The value of d3 was therefore determined experimentally from the quenching efficiencies of surface-exposed residues in M2. From the experimentally determined values of F/Fo, d1 was calculated using Eq. 3, which, together with the values for d2 given in Table 2, allowed the calculation of d3 (Table 2). Values of d3 are very similar for all three mutants, with an average value of 9.6 Å. This is very close to the minimum α-carbon-to-Br distance of 9.4 Å calculated at the point of van der Waals contact between the edge of a Trp ring and a bromine-containing alkyl chain (Fig. 5). With a value of 9.6 Å for the value of d3 for a surface-exposed Trp residue (Fig. 5), the corresponding values of d3 for buried Trp residues can then be calculated from the distances between Trp residues and a surface located lipid molecule given in Table 3. These values of d3 were then used, together with the corresponding values of d2, to calculate the level of fluorescence quenching expected if lipid molecules could not penetrate into the protein surface (Table 3). For the buried Trp residues, the efficiencies of fluorescence quenching calculated in this way are very much smaller than those observed experimentally (Table 3), confirming that the experimentally observed levels of quenching require that the lipid fatty acyl chains be able to penetrate into cavities in the protein surface.
FIGURE 5.
Modeling fluorescence quenching in MscL. (A) The efficiency of quenching of the Trp fluorescence in a membrane protein by a neighboring lipid molecule with bromine-containing fatty acyl chains depends on the distance d1 between them. The distance d2 can be calculated from the position of the Trp residue in the protein and the location of the bromines in the bilayer. The distance d3 depends on the depth of the Trp residue below the protein surface and the extent of penetration of the lipid fatty acyl chains into the surface, as described in the text. (B) The figure shows a Trp residue and a bromine-containing alkane with van der Waals contact between the edge of the Trp ring and the alkyl chain. The Trp α-carbon-to-Br distance marked is 9.4 Å. (C) Structure of di(Br2C18:0)PC.
TABLE 2.
Analysis of quenching data for Trp mutants in M2
| Mutant | d1 (Å) | Trp-Br separation along bilayer normal (d2) | Trp-Br separation d3 |
|---|---|---|---|
| L73W | 10.4 | 4.5 | 9.4 |
| I77W | 9.9 | 0.4 | 9.9 |
| Y87W | 9.4 | 1.2 | 9.3 |
TABLE 3.
Quenching of fluorescence of Trp mutants of MscL by di(Br2C18:0)PC
| Mutant | Distance between Trp edge and chain on surface (Å) | Trp-Br separation along bilayer normal (d2) | Trp-Br distance d3 | Calculated efficiency of quenching with no chain penetration | Experimental efficiency of quenching |
|---|---|---|---|---|---|
| A20W | 7.2 | 2.0 | 13.3 | 0.16 | 0.58 |
| I23W | 7.0 | 2.2 | 13.1 | 0.21 | 0.50 |
| L30W | 4.8 | 2.1 | 10.9 | 0.41 | 0.59 |
| V31W | 5.2 | 2.1 | 11.3 | 0.36 | 0.44 |
| F34W | 4.6 | 2.4 | 10.7 | 0.45 | 0.49 |
| L73W | 3.5 | 4.5 | 9.6 | 0.49 | 0.51* |
| I77W | 3.5 | 0.4 | 9.6 | 0.63 | 0.57* |
| Y87W | 3.5 | 1.2 | 9.6 | 0.61 | 0.66* |
Distances between Trp residues and a lipid molecule docked on the surface of the protein were calculated as described in the text. Distances d2 and d3 are defined in Fig. 5. The efficiencies of quenching expected if lipid fatty acyl chains cannot penetrate into the protein surface were calculated as described in the text. Residues 20–34 are in M1, and residues 73–87 are in M2.
Data from Powl et al. (12).
DISCUSSION
The short distance range over which bromine can quench Trp fluorescence makes it possible to study interactions with lipid molecules in the immediate vicinity of a Trp residue introduced into a site of interest in a membrane protein. Here we use this approach to investigate the possibility of chain penetration into the cavities that are likely to exist in the transmembrane surface of a multi-helix membrane protein such as MscL. In the homopentameric MscL structure, the first transmembrane α-helices M1 from each monomer are buried within a ring made up of the second transmembrane α-helices M2 from each monomer (Fig. 1 A). As a result, a number of lipid-facing residues in M1 are buried in cavities below the transmembrane surface, whereas lipid-facing residues in M2 are located on the surface of the transmembrane domain (Fig. 1 B). Thus, lipid-facing residues in M2 will be in direct contact with the fatty acyl chains of surrounding lipid molecules even if the lipid molecules are restricted to the outer surface of the protein, whereas some of the lipid-facing residues in M1 will make contact with lipid fatty acyl chains only if the chains can penetrate into the surface cavities. This is particularly obvious for residues 20 and 23 in M1 (Fig. 1 B).
Fluorescence emission maxima
Fluorescence emission maxima for Trp residues in M1 and M2 were found to show a similar dependence on location within the membrane, with emission maxima shifting to longer wavelengths with increasing distance from the bilayer center (Fig. 3). Thus, Trp residues in M1 and M2 experience similar environments despite the fact that lipid-facing residues in M2 are fully exposed to the lipid bilayer, whereas those in M1 are to a greater or lesser extent buried between M2 helices. Fig. 3 also shows a smoother dependence of fluorescence emission maxima on location on the periplasmic side of the membrane than on the cytoplasmic side. Thus, fluorescence maxima on the periplasmic side of the membrane generally fit well to Eq. 5, describing a trough shaped dependence of dielectric constant across the membrane (Fig. 3). However, this is not true for Trp residues on the cytoplasmic side of the membrane, where the distance dependence of the fluorescence emission maxima is not of the right form to fit to Eq. 5. In particular, residues close to the bilayer center on the cytoplasmic side of the membrane have emission maxima at longer wavelengths than expected from Eq. 5, corresponding to more polar environments, in both M1 and M2. This would be the result if Trp residues, which have a preference for an interfacial location, were free to snorkel toward the membrane surface on the cytoplasmic side of the membrane but not on the periplasmic side. Inspection of the crystal structure of MscL (10) suggests that lipid-facing residues on the cytoplasmic side of the membrane are less closely packed than those on the periplasmic side, possibly allowing more freedom to move. A similar argument could apply to the Trp residues in mutants L30W and I77W that have fluorescence emission maxima distinctly different from their neighbors (Fig. 3); these residues occupy relatively open sites on the protein surface.
Quenching by bromine-containing lipids
Lipid-facing residues at the N-terminal end of M1 are buried in crevices between M2 helices, this being particularly clear for residues 20 and 23 (Fig. 1 B). An estimate of how deeply buried these residues are below the outer surface of the protein was made by docking a lipid molecule with rigid, all-trans fatty acyl chains onto the surface. The minimum distance was measured between the backbone of the lipid chain and a Trp residue oriented to extend as far as possible toward the surface. As expected, for surface-exposed Trp residues in M2, this minimum distance (3.5 Å; Table 3) corresponded to the distance of closest approach between a Trp ring and an acyl chain. By this same procedure, minimum Trp-to-chain distances of ∼7 Å were found for mutants A20W and I23W (Table 3), suggesting that in these mutants the Trp residues were buried ∼3.5 Å below the surface. Because quenching of Trp fluorescence by bromine exhibits a sixth-power dependence on distance, as in Forster energy transfer, with a value for Ro, the distance at which quenching is 50% efficient of 8 Å (12), an increase in distance between Trp and Br of 3.5 Å would result in a large decrease in the efficiency of fluorescence quenching, as shown in Table 3. The fact that the efficiencies of quenching of the buried residues A20W and I23W are much greater than would be expected if lipid fatty acyl chains were restricted to lying on the protein surface (Table 3) confirms that the lipid fatty acyl chains must be able to penetrate into cavities in the protein surface. Modeling shows that the cavity around the buried Trp residues in A20W and I23W is large enough to accommodate small sections of alkyl chain with the chain being in van der Waals contact with the Trp residues (Fig. 1 C). Modeling suggests that contact between Ile-23 and the lipid fatty acyl chains will be very similar to that observed for the Trp residue in the mutant I23W, consistent with the similar sizes of the Ile and Trp residues. Although it is possible that the smaller Ala residue will make less contact with the lipid chains than the Trp reside in A20W, the fact that the Ala residue at position 20 is exposed at the bottom of the cleft suggests that solvation of this residue could also be efficient.
The importance of solvation by lipid molecules
Solvation of buried residues by lipid chains requires flexibility in the fatty acyl chains, this flexibility being a characteristic of a lipid bilayer in the liquid crystalline phase. Solvation of buried residues will not be possible if the lipid bilayer is in the gel phase, and it has been observed that many membrane proteins are excluded from domains of gel phase lipid, an effect attributed to poor solvation of the protein surface by rigid chains (11). Results of molecular dynamics simulations of rhodopsin in bilayers of lipids with polyunsaturated chains suggest that the efficiency of solvation in the liquid crystalline phase could increase with increasing unsaturation in the fatty acyl chains (5).
Penetration of lipid chains into the protein surface could be important for protein function. For example, packing of the transmembrane α-helices of Ca2+-ATPase is distinctly different in the different conformational states of the Ca2+-ATPase (30), and consequent differences in the pattern of lipid solvation will contribute to the energetic differences between the different conformational states of the Ca2+-ATPase. A specific example is provided by the inhibitor thapsigargin, which binds between the transmembrane α-helices of Ca2+-ATPase, preventing the movement of the helices required before Ca2+ can bind to the Ca2+-ATPase (31,32). In the case of MscL, interactions between MscL and the surrounding lipid bilayer are important in linking channel opening to an increase in tension in the membrane, and the extensive rearrangement of the transmembrane α-helices involved in channel opening (22,23) will inevitably lead to changes in solvation of MscL by the lipid molecules.
Acknowledgments
We thank Professor D. C. Rees for the gift of the MscL construct and the Biotechnology and Biological Sciences Research Council for a studentship (to J.C.).
References
- 1.Raschke, T. M. 2006. Water structure and interactions with protein surfaces. Curr. Opin. Struct. Biol. 16:152–159. [DOI] [PubMed] [Google Scholar]
- 2.Modif, K., E. Liepinsh, G. Otting, and B. Halle. 200. Dynamics of protein and peptide hydration. J. Am. Chem. Soc. 126:102–114. [DOI] [PubMed]
- 3.Lee, A. G. 2003. Lipid-protein interactions in biological membranes: a structural perspective. Biochim. Biophys. Acta. 1612:1–40. [DOI] [PubMed] [Google Scholar]
- 4.Gonen, T., Y. F. Cheng, P. Sliz, Y. Hiroaki, Y. Fujiyoshi, S. C. Harrison, and T. Walz. 2005. Lipid-protein interactions in double-layered two-dimensional AQPO crystals. Nature. 438:633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grossfield, A., S. E. Feller, and M. C. Pitman. 2006. A role for direct interactions in the modulation of rhodopsin by ω-3 polyunsaturated lipids. Proc. Natl. Acad. Sci. USA. 103:4888–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.East, J. M., D. Melville, and A. G. Lee. 1985. Exchange rates and numbers of annular lipids for the calcium and magnesium ion dependent adenosinetriphosphatase. Biochemistry. 24:2615–2623. [DOI] [PubMed] [Google Scholar]
- 7.Woolf, T. B., and B. Roux. 1996. Structure, energetics, and dynamics of lipid-protein interactions: A molecular dynamics study of the gramicidin A channel in a DMPC bilayer. Proteins. 24:92–114. [DOI] [PubMed] [Google Scholar]
- 8.Lee, A. G. 2005. How lipids and proteins interact in a membrane: a molecular approach. Mol. Biosyst. 1:203–212. [DOI] [PubMed] [Google Scholar]
- 9.Brotherus, J. K., O. H. Griffith, M. O. Brotherus, P. C. Jost, and J. R. Silvius. 1981. Lipid-protein multiple binding equilibria in membranes. Biochemistry. 20:5261–5267. [DOI] [PubMed] [Google Scholar]
- 10.Chang, G., R. H. Spencer, A. T. Lee, M. T. Barclay, and D. C. Rees. 1998. Structure of the MscL homolog from Mycobacterium tuberculosis: A gated mechanosensitive ion channel. Science. 282:2220–2226. [DOI] [PubMed] [Google Scholar]
- 11.East, J. M., and A. G. Lee. 1982. Lipid selectivity of the calcium and magnesium ion dependent adenosinetriphosphatase, studied with fluorescence quenching by a brominated phospholipid. Biochemistry. 21:4144–4151. [DOI] [PubMed] [Google Scholar]
- 12.Powl, A. M., J. M. East, and A. G. Lee. 2003. Lipid-protein interactions studied by introduction of a tryptophan residue: the mechanosensitive channel MscL. Biochemistry. 42:14306–14317. [DOI] [PubMed] [Google Scholar]
- 13.Powl, A. M., J. M. East, and A. G. Lee. 2005. Heterogeneity in the binding of lipid molecules to the surface of a membrane protein: hot-spots for anionic lipids on the mechanosensitive channel of large conductance MscL and effects on conformation. Biochemistry. 44:5873–5883. [DOI] [PubMed] [Google Scholar]
- 14.Rooney, E. K., and A. G. Lee. 1986. Fitting fluorescence emission spectra of probes bound to biological membranes. J. Biochem. Biophys. Methods. 120:175–189. [DOI] [PubMed] [Google Scholar]
- 15.Powl, A. M., J. N. Wright, J. M. East, and A. G. Lee. 2005. Identification of the hydrophobic thickness of a membrane protein using fluorescence spectroscopy: studies with the mechanosensitive channel MscL. Biochemistry. 44:5713–5721. [DOI] [PubMed] [Google Scholar]
- 16.Elder, M., P. Hitchcock, R. Mason, and G. G. Shipley. 1977. A refinement analysis of the crystallography of the phospholipid 1,2-dilauroyl-dl-phosphatidylethanolamine, and some remarks on lipid-lipid and lipid-protein interactions. Proc. R. Soc. Lond. A. 354:157–170. [Google Scholar]
- 17.Lewis, B. A., and D. M. Engelman. 1983. Lipid bilayer thickness varies linearly with acyl chain length in fluid phosphatidylcholine vesicles. J. Mol. Biol. 166:211–217. [DOI] [PubMed] [Google Scholar]
- 18.London, E., and G. W. Feigenson. 1981. Fluorescence quenching in model membranes. 2. Determination of local lipid environment of the calcium adenosinetriphosphatase from sarcoplasmic reticulum. Biochemistry. 20:1939–1948. [DOI] [PubMed] [Google Scholar]
- 19.O'Keeffe, A. H., J. M. East, and A. G. Lee. 2000. Selectivity in lipid binding to the bacterial outer membrane protein OmpF. Biophys. J. 79:2066–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolen, E. J., and P. W. Holloway. 1990. Quenching of tryptophan fluorescence by brominated phospholipid. Biochemistry. 29:9638–9643. [DOI] [PubMed] [Google Scholar]
- 21.Mall, S., R. Broadbridge, R. P. Sharma, J. M. East, and A. G. Lee. 2001. Self-association of model transmembrane α-helices is modulated by lipid structure. Biochemistry. 40:12379–12386. [DOI] [PubMed] [Google Scholar]
- 22.Betanzos, M., C. S. Chiang, H. R. Guy, and S. Sukharev. 2002. A large iris-like expansion of a mechanosensitive channel protein induced by membrane tension. Nat. Struct. Biol. 9:704–710. [DOI] [PubMed] [Google Scholar]
- 23.Perozo, E., D. M. Cortes, P. Sompornpisut, and B. Martinac. 2002. Open channel structure of MscL and the gating mechanism of mechanosensitive channels. Nature. 418:942–948. [DOI] [PubMed] [Google Scholar]
- 24.Yoshimura, K., A. Batiza, M. Schroeder, P. Blount, and C. Kung. 1999. Hydrophilicity of a single residue within MscL correlates with increased channel mechanosensitivity. Biophys. J. 77:1960–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lakowicz, J. R. 1999. Principles of Fluorescence Spectroscopy. Kluwer Academic/Plenum Press, New York.
- 26.Ren, B., F. Gao, Z. Tong, and Y. Yan. 1999. Solvent polarity scale on the fluorescence spectra of a dansyl monomer copolymerizable in aqueous media. Chem. Phys. Lett. 307:55–61. [Google Scholar]
- 27.Flewelling, R. F., and W. L. Hubbell. 1986. The membrane dipole potential in a total membrane potential model. Applications to hydrophobic ion interactions with membranes. Biophys. J. 49:541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koehorst, R. B. M., R. B. Spruijt, F. J. Vergeldt, and M. A. Hemminga. 2004. Lipid bilayer topology of the transmembrane α-helix of M13 major coat protein and bilayer polarity profile by site-directed fluorescence spectroscopy. Biophys. J. 87:1445–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marsh, D. 2001. Polarity and permeation profiles in lipid membranes. Proc. Natl. Acad. Sci. USA. 98:7777–7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Obara, K., N. Miyashita, C. Xu, L. Toyoshima, Y. Sugita, G. Inesi, and C. Toyoshima. 2005. Structural role of countertransport revealed in Ca2+ pump crystal structure in the absence of Ca2+. Proc. Natl. Acad. Sci. USA. 102:14489–14496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, A. G. 2004. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta. 1666:62–87. [DOI] [PubMed] [Google Scholar]
- 32.Toyoshima, C., and H. Nomura. 2002. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature. 418:605–611. [DOI] [PubMed] [Google Scholar]