Abstract
Previous studies have shown that the unusually long S5-P linker lining human ether a-go-go related gene's (hERG's) outer vestibule is critical for its channel function: point mutations at high-impact positions here can interfere with the inactivation process and, in many cases, also reduce the pore's K+ selectivity. Because no data are available on the equivalent region in the available K channel crystal structures to allow for homology modeling, we used alternative approaches to model its three-dimensional structure. The first part of this article describes mutant cycle analysis used to identify residues on hERG's outer vestibule that interact with specific residues on the interaction surface of BeKm-1, a peptide toxin with known NMR structure and a high binding affinity to hERG. The second part describes molecular modeling of hERG's pore domain. The transmembrane region was modeled after the crystal structure of KvAP pore domain. The S5-P linker was docked to the transmembrane region based on data from previous NMR and mutagenesis experiments, as well as a set of modeling criteria. The models were further restrained by contact points between hERG's outer vestibule and the bound BeKm-1 toxin molecule deduced from the mutant cycle analysis. Based on these analyses, we propose a working model for the open conformation of the outer vestibule of the hERG channel, in which the S5-P linkers interact with the pore loops to influence ion flux through the pore.
INTRODUCTION
The human ether a-go-go related gene (hERG) encodes the pore-forming subunit of the rapid delayed rectifier K channels (IKr) expressed in cardiac myocytes, certain neurons, gastrointestinal smooth muscle cells, and pancreatic insulin-secreting cells (1–8). Furthermore, hERG is upregulated in a number of cancer cell lines (9,10), prompting the suggestion that an increase in hERG channel activity may be antiapoptotic in these cells. Therefore, hERG is involved in diverse cellular activities under physiological and pathological conditions. Much has been learned about the structure-function relationship of the hERG channel (11). Our focus here is the S5-P linker and P-S6 linker that line the outer vestibule of this channel. The S5-P linker in hERG is two to three times the length of its counterparts in most other K channels. Previous data have suggested that the conformation of the long S5-P linker in the hERG channel is very dynamic, and is crucial for the channel's inactivation process and K+ selectivity (12–15). Furthermore, NMR studies of peptides corresponding to hERG's S5-P linker sequence showed that the middle portion of the peptides could adopt an amphipathic helical structure when placed in detergent micelles (15,16).
Information about the three-dimensional (3-D) structure of the S5-P linker and its relationship to other channel domains is needed to understand how conformational changes here can affect the inactivation process and K+ selectivity. Crystal structures of five different K channels are available (17–21). Of these, the sequence of KvAP is the most similar to that of hERG for the pore-forming domain. Unfortunately, the “S5-P linker equivalents” (or turrets) of all five crystal structures are much shorter and bear no sequence similarity to that of hERG. We thus sought alternative approaches to model the 3-D structure of hERG's outer mouth region. Peptide toxin “footprinting” had been used to probe the structures of outer vestibules of Kv channels before any K channel crystal structures were available (22–25). Peptide toxins purified from scorpions and several other species can bind to Kv channels with high affinities. These peptide toxins have disulfide bond-stabilized compact structures that can be determined using NMR spectroscopy (26,27). They likely retain their rigid structures upon binding to target channels' receptor sites. Spatial information about some channels' residues have been obtained by using “mutant cycle analysis” to identify channel residues that interact with specific toxin residues in a toxin/channel complex (22–25). Therefore, we used the mutant cycle analysis to study which residues on hERG's outer mouth region can interact with specific residues on BeKm-1, a scorpion peptide toxin with a high binding affinity to the hERG channel (under appropriate conditions, dissociation constant, Kd, in low nM) (28–30) and known NMR structure (Protein Data Bank code, 1J5J) (31). The second approach was molecular modeling. The transmembrane region of hERG's pore domain was modeled using the available KvAP crystal structure as the template. The extracellular S5-P linker was modeled based on secondary structure information from the NMR experiments, previous experimental observations, and a series of modeling criteria. These structures were docked to the transmembrane region of the pore domain in different orientations. The models were further restrained based on mutant cycle analysis of BeKm-1/hERG interactions. These models were then subjected to molecular dynamics (MD) simulations to test their stability. We present the most favorable structure here and discuss its implications for the outer mouth structure of the hERG channel.
MATERIALS AND METHODS
Toxin preparation
The expression and purification of BeKm-1 and its alanine mutants was performed as described previously (31). Briefly, toxins were expressed in the periplasm of Escherchia coli (HB101) as a fusion protein with two IgG-binding domains (ZZ) of staphylococcal protein A. The HB101 cells were harvested and lysed by ultrasonication. After ultracentrifugation, BeKm-1 fusion proteins in the supernatant were purified by an IgG-Sepharose 6FF column (Amersham Pharmacia Biotech, Uppsala, Sweden). Wild-type (WT) and mutant BeKm-1 toxins were cleaved from the fusion protein by enterokinase. The recombinant toxins were purified from the cleavage mixture by chromatography on a reverse phase high-performance liquid chromatography column (Delta Pak C18 300-A pore, 3.9 × 300 mm; Waters, Milford, MA). Mass spectrometry verified the composition of the purified material. The toxin peptide content was determined using the bicincholinic acid method with bovine serum albumin as the standard.
Cysteine scanning mutagenesis
Wild-type hERG in a vector, pAlterMax, was used to produce cysteine-substituted mutants using the oligonucleotide-directed method and a commercial kit (Altered Site Mammalian Mutagenesis System, Promega, Madison, WI) as described previously (14). Mutants are designated by the WT residue (one-letter code), followed by the position number and “C” for cysteine.
cRNA and oocyte preparations
cRNA transcription and oocyte injection were as described previously (32). cRNA concentrations were quantified by densitometry (ChemiImager model 4000; α-Innotech, San Leandro, CA). Oocytes were isolated from Xenopus laevis (Xenopus One, Dexter, MI) and freed from follicular cell layers after mild collagenase treatment. Each oocyte was microinjected with 40 nl of cRNA solution (total cRNA 10–18 ng). After incubating the oocytes for 2–4 days at 16°C in an ND96 medium supplemented with horse serum (4%) and antibiotics (penicillin 50 units/ml and streptomycin 50 units/ml), channels were studied in electrophysiological experiments.
Electrophysiological experiments
Before experiments, oocytes expressing cysteine-substituted mutants were incubated in DTT (reducing agent, 10 mM)-containing medium at room temperature to break disulfide bonds that might be spontaneously formed involving introduced cysteine (Cys) side chains (14). Oocytes were then thoroughly rinsed in DTT-free medium (5 ml for 1–2 min, three washes) before being placed in the tissue bath that contained 0.8 ml of low-[Cl] bath solution (to avoid interference from endogenous Cl currents). The grounding electrodes were filled with 3 M KCl (in contact with Ag/AgCl pellets) and connected to the bath solution with salt bridges made of 1% agar in bath solution ([K] = 2 or 98 mM, see below, to avoid changing [K] in bath, which could markedly affect hERG current amplitude). The oocyte was impaled with two microelectrodes, and membrane currents were recorded using an oocyte clamp amplifier (model 725B or 725C; Warner Instruments, Hamden, CT). Voltage clamp protocol generation and data acquisition were controlled by pClamp 5.5 via computer and a 12-bit D/A and A/D converter (Axon Instruments, Union City, CA).
The membrane voltage was held at −80 mV (Vh), and currents were activated by a 1-s step to +20 mV applied once per 30 or 60 s. Previous data indicated that 1-s depolarization to +20 mV was sufficient to fully activate WT and mutant hERG channels (14). After control currents (IC) were recorded, 5–10 μl of BeKm-1 stock solution (2, 20, or 200 μM in 0.1% bovine serum albumin (BSA), depending on the desired initial toxin concentration; see below) was diluted with 0.2 ml of bath solution and added to the bath. We targeted the degree of current suppression by 30–70%. To reach this goal, we set the initial [BeKm-1] at an estimated Kd value assuming no interaction between the mutated toxin and channel residues. This was calculated based on the published Kd values for WTTx:WTCh and WTTx:MutCh (28) and the Kd values for MutTx:WTCh (33):
Predicted Kd for MutTx:MutCh if no interaction 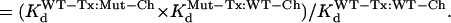
After adding the toxin stock solution, the bath solution was gently pipetted up and down repetitively to facilitate equilibration of toxin concentration in the bath. The remaining current in the presence of BeKm-1 (ITX) was recorded when the degree of current suppression reached a steady state. If the initial [BeKm-1] induced too little suppression, the toxin concentration was elevated in 5- or 10-fold steps until the desired degree of suppression was obtained or until the toxin concentration reached 5000 nM.
Solutions
The standard low-[Cl] solution contained (in mM): NaOH 96, KOH 2, CaCl2 1.8, MgSO4 1, HEPES 5, Na-pyruvate 2.5. The solution was titrated to pH 7.5 with methanesulfonic acid. In some cases when the hERG mutant channels manifested very small currents, we raised [K]o to 98 mM to increase the current amplitude. This was made by replacing NaOH with equimolar KOH and Na-pyruvate was omitted. The oocyte culture medium was made using high-[Cl] ND96 (similar to the low-[Cl] standard bath solution except that NaOH and KOH were replaced by NaCl and KCl, respectively), supplemented with 4% horse serum and antibiotics.
Lyophilized BeKm-1 toxins were dissolved in 0.1% BSA in bath solution, aliquoted, and kept at −30°C. After thawing, each aliquot was kept on ice or at 4°C and used in <2 days.
Data analysis
Data analysis was performed using the following programs: Clampfit of pClamp 6 or 8 (Axon Instruments), Excel (Microsoft, Redmond, WA), PeakFit, and SigmaPlot (Jandel Scientific, San Rafael, CA).
The amounts of charge transferred through the channels under the control conditions and at the steady state of toxin effect (QC and QTx, respectively) were estimated by integrating tail currents recorded at −80 mV over time. We chose to analyze tail currents at −80 mV because: a), the holding current at −80 mV could be used as a baseline, b), this voltage provided sufficient driving force for either outward tail currents recorded in 2 mM [K]o or inward tail currents recorded in 98 mM [K]o, and c), the rate of tail decay at −80 mV could be conveniently quantified during a 1-s step. These values were used to calculate the apparent dissociation constant (Kd). Previous data showed that the maximum degree of hERG current suppression by BeKm-1 was 90 ± 1% (28). Therefore, Kd was calculated as: QTx/QC = 0.9/(1 + [Tx]/Kd) + 0.1. The Hill coefficient is 1, consistent with our model of toxin binding to the channel (28). Perturbation of toxin:channel interaction induced by mutation of either toxin or channel (ΔΔG) was calculated by: 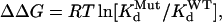 where
where 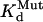 and
and 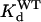 refer to Kd values for mutant toxin or channel and for WT toxin or channel, and RT = 0.59 kcal/mol. For double (toxin and channel) mutations, the coupling coefficient (Ω) was calculated as:
refer to Kd values for mutant toxin or channel and for WT toxin or channel, and RT = 0.59 kcal/mol. For double (toxin and channel) mutations, the coupling coefficient (Ω) was calculated as:
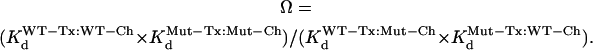 |
For clarity of data presentation, the degree of coupling was quantified using the absolute value of coupling coefficient (|Ω|). The mean ± SE of Ω (SEΩ) was calculated by:
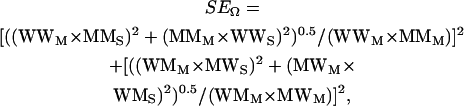 |
where WW, MM, WM, and MW refer to WTTx:WTCh, MutTx:MutCh, WTTx:MutCh, and MutTx:WTCh pairs, with subscripts “M” and “S” denoting mean and SE, respectively.
Kinetic model simulations of hERG gating and state-dependent toxin binding/unbinding was performed using ModelMaker version 4 (FamilyGenetix, Berkshire, UK).
Sequence alignment and analysis
Sequences were obtained from NCBI's nonredundant database and were aligned using ClustalW (34) followed by some manual adjustments to reduce the number of insertions and deletions within families and subfamilies using the Seqlab editor of the GCG program (Wisconsin Package Version 10.2; Accelrys, San Diego, CA). The degree of mutability, μ, for the EAG family of sequences was calculated as described in Shrivastava et al. (35).
Molecular modeling and simulations
Modeling and simulation procedures and criteria used to develop and evaluate models were similar to those described previously (35,36). Initial models of the S5, P, and S6 segments were modeled from the KvAP crystal structure (20) using the Modeler software (37). Initial models of the S5-P linkers were developed using an in-house program to generate idealized α-helices with side chains in the conformation most frequently observed in helical segments of known proteins. The PSSHOW program (38) was used to position the helical segments and to manually model the nonhelical segments. Tetrameric models were minimized with fourfold symmetry about the axis of the pore using CHARMM (39). The molecular dynamics simulations of the pore-forming domain of the hERG channel embedded in a phosphatidylethanolamine (POPE) lipid bilayer were run using the program Gromacs (http://www.gromacs.org) (see Shrivastava et al. (35) for details).
RESULTS
Using mutant cycle analysis to probe hERG residues that interact with specific BeKm-1 residues
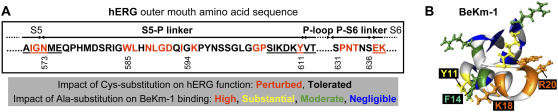
In these experiments, we used BeKm-1 toxin mutants in which surface residues were replaced by alanine (Ala) (33), and hERG channel mutants in which residues lining the outer vestibule were replaced by Cys (14). A prerequisite for the mutant cycle analysis is that the mutations should not perturb the native conformation of either the toxin or the channel. Ala and Cys have small hydrophobic side chains that are generally well tolerated in mutagenesis experiments. We have analyzed the effects of Cys substitution in the outer vestibule region of hERG (amino acids 571–613 and 631–638; Fig. 1 A) on the channel function (14). Cys substitution per se (not in disulfide bonded state) at 19 positions markedly perturbed the hERG channel function. These are “high-impact:” positions (Fig. 1 A, residues highlighted in red), and these Cys mutants were not included in the experiments. For the remaining 32 positions, Cys substitution per se had little impact on the channel function. These WT-like Cys mutants were included in the mutant cycle analysis reported here. It was shown previously that, with the exception of F21A, all Ala-substituted toxin mutants retained the native conformation of the WT toxin (33). Our preliminary experiments showed that out of the 15 Ala-substituted BeKm-1 mutants tested, only K18A, R20A, F14A, and Y11A showed coupling to hERG Cys mutants. Therefore, we focused on these four BeKm-1 mutants (see Supplemental Fig. S1 for results from the other BeKm-1 mutants). These four residues are highlighted in Fig. 1 B. To test our ability to differentiate between coupled and uncoupled toxin-channel residue pairs, we also included E9A mutant toxin in our mutant cycle analysis.
FIGURE 1.
(A) Amino acid sequence lining the outer vestibule of hERG: residues 573–611 (S5-P linker), and residues 631–636 (P-S6 linker). Segment 585–594 corresponds to a helical region determined by previous NMR studies (15,16). Red lettering highlights high-impact positions based on a previous cysteine scanning mutagenesis study (14). (B) Three-dimensional structure of BeKm-1 in ribbon format, with positions color coded for effects of alanine substitution on toxin binding to hERG: high (brown), substantial (yellow), moderate (green), and negligible (blue). BeKm-1 positions not tested are color coded light gray. Side chains for the first three classes of position are shown in ball-and-stick format. Side chains of Y11, F14, K18, and R20 are labeled.
The binding affinity of channel-blocking toxins is traditionally evaluated by the degree of suppression of current amplitude. In the case of toxin binding to the hERG channel, suppression of peak amplitudes of tail currents has been used as an estimator (28,31,40). This is because test pulse currents through the hERG channel at positive membrane voltages are small due to the fast inactivation process, whereas the peak amplitudes of tail currents are much larger due to a fast recovery from inactivation upon membrane repolarization (41). During the course of our experiments, we noticed that for some toxin:channel pairs, the peak amplitudes of tail currents were only modestly or little suppressed although the tail current decay became significantly faster. This occurred when mutation(s) introduced into toxin and/or channel weakened the binding affinity (e.g., K18A or R20A mutant toxin (33), and Q592 or S631 mutant channel (28)), so that high toxin concentrations (typically 1000–5000 nM) were needed to evaluate binding affinity. Fig. 2 A depicts three toxin:channel pairs. The left panel shows that WT toxin binding to the I583C mutant channel was strong, and 10 nM toxin suppressed both the test pulse current (time-dependent component, excluding the instantaneous current component that might arise from oocyte endogenous or leak currents) and the peak amplitude of tail current to ∼50%. The middle panels shows that WT toxin binding to the Q592C mutant channel was weaker, and in the presence of 1000 nM toxin the peak amplitude of tail current was modestly suppressed although the tail decay became much faster. Furthermore, in the presence of toxin test pulse current activation became slower, reaching the same level as control current at the end of the pulse. The right panel shows that R20A mutant toxin binding to the S631C mutant channel was very weak, so that 5000 nM toxin was needed to evaluate toxin effects. In the presence of toxin, the test pulse current and peak amplitude of tail current superimposed with the control current trace. However, the tail decay was markedly accelerated.
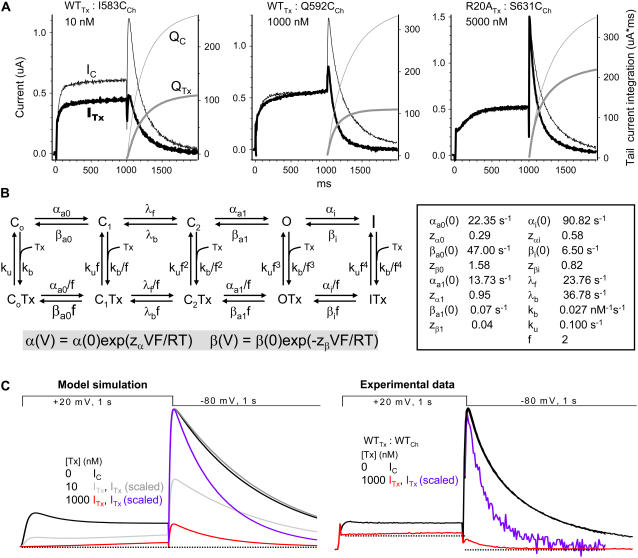
FIGURE 2.
Effects of toxin on hERG channel gating and a kinetic model of state-dependent BeKm-1 binding/unbinding. (A) Original current traces (IC and ITx) and tail current integration (QC and QTx) from three toxin:channel pairs (marked on top, with toxin concentrations denoted). The thin traces represent control data (IC and QC), whereas the thick traces represent data in the presence of toxin (ITx or QTx). For each panel, the left coordinate is for current amplitude and the right coordinate is for tail current integration. (B) (Left) State diagram of hERG channel gating and BeKm-1 binding/unbinding. (Right) Model parameter values used in the simulation shown in panel C. “Cn”, “O”, and “I” represent closed, open, and inactivated states. The upper row represents toxin-free states, whereas the lower row represents toxin-bound states. Transitions between channel gating states (except that between C1 and C2) are voltage dependent. These rate constants are described by the general equations highlighted by gray shade below the state diagram: rate(V) = rate(0)exp(zVF/RT), where rate(V) and rate(0) are rate constants at voltage “V” and 0 mV (in s−1), z is gating charge, F is Faraday constant, R is gas constant, and T is absolute temperature. Toxin binding and unbinding rate constants (kb and ku) are voltage independent, but gating state dependent as defined by the coupling coefficient (f). Therefore, with an f-value >1, toxin binding is favored with channels in closed states but disfavored as channels make transitions into open and inactivated states. To maintain the microscopic reversibility of channel gating (for any cycle in the state diagram the product of forward rate constants equals the product of backward rate constants), the forward rate constants between toxin-bound channel states are divided by the coupling coefficient, and the backward rate constants are multiplied by the coupling coefficient. (C) (Left) Simulation of BeKm-1 effects on hERG current amplitude and gating kinetics. Effects of two toxin concentrations were simulated, 10 and 1000 nM, color-coded gray and red, respectively. Tail currents of ITx traces are further scaled to match the peak amplitude of control tail current, and are color-coded dark gray and purple, respectively. (Right) Experimental data of hERG current traces recorded before and after addition of 1000 nM BeKm-1 (black and red traces, respectively). The tail current of ITx is further scaled to match the peak amplitude of IC tail current (purple trace). Dotted lines in the experimental data are drawn to help differentiate between time-dependent currents (through hERG channels expressed in the oocyte) versus oocyte endogenous or leak currents. For comparison, a dotted line is drawn in the model simulation at zero current level.
We suspect that the toxin effects on hERG gating kinetics may reflect state-dependent toxin:channel interactions. It has been shown that Bekm-1 binding to the hERG channel occurs preferentially in the closed states. Channel opening weakens toxin binding, and channel inactivation causes toxin unbinding (29). According to this scenario, BeKm-1 tends to unbind from the hERG channels upon membrane depolarization when the channels make transitions from closed to open and then to inactivated states. This can lead to an apparent slowing of hERG activation. BeKm-1 rebinds during membrane repolarization when the hERG channels recover from inactivation and begin to deactivate; that can create an apparent acceleration of hERG deactivation. To test this possibility, we set up a kinetic model of hERG gating based on a published model (42), and incorporate state-dependent BeKm-1 binding/unbinding reactions into the model. The state diagram is shown in the left panel of Fig. 2 B, whereas the model parameters are listed in the right panel. In this model, BeKm-1 binding and unbinding is voltage independent but state dependent, defined by a coupling coefficient (f) of 2. As the channel moves forward in the activation pathway from C0 to C1, C2, O, and then I states, the ratio of binding to unbinding rate constants decreases to 1:4, 1:16, 1:64, and 1:256 of that in the C0 state. Therefore, toxin binding mainly occurs in C0 and C1 states. Transition into O and I states favors toxin unbinding. Fig. 2 C, left panel, depicts model simulations of current traces elicited by the same voltage clamp protocol as used in our experiments under the control conditions and in the presence of 10 or 1000 nM toxin. In the presence of toxin, the test pulse current begins at a level lower than control, and the activation is slowed in a dose-dependent manner. These reflect toxin binding in the closed state and unbinding upon channel activation/inactivation. The peak amplitude of tail current is suppressed in a dose-dependent manner. However, although the rate of tail current decay is not altered by 10 nM toxin, the decay is markedly accelerated by 1000 nM toxin. In the presence of 1000 nM toxin, upon membrane repolarization the channels are quickly sequestered into toxin-bound closed (C0Tx and C1Tx) states, and this shift in channel state distribution accelerates channel transitions from the open (O) to closed (C2) states. In the presence of 10 nM toxin, the rebinding is too slow to manifest the change in gating kinetics. These simulations are consistent with experimental findings. The right panel of Fig. 2 C shows an example: 1000 nM BeKm-1 slowed activation and accelerated deactivation of the WT hERG channel. It is important to point out that although such a strong state dependence of BeKm-1/hERG interactions appears similar to that of “gating-modifying toxins” that bind to the S3-S4 linkers of target channels (43,44), the BeKm-1 binding site is in the outer vestibule region of the hERG channel. This is based on data from previous experiments showing that mutations in the outer vestibule region could markedly impact on BeKm-1 binding whereas mutations in the S3-S4 linker could not, and that BeKm-1 binding potency was weakened by an outer mouth blocker, tetraethylammonium (28).
Based on the above, we could choose between two methods to evaluate toxin potency: 1), suppression of peak amplitude of tail current, or 2), decrease in the amount of charges transferred through the channels during tail current (tail current integration, taking into account toxin-induced acceleration of tail decay). The examples shown in Fig. 2 A indicate that although these two methods gave the same qualitative conclusion (i.e., toxin binding was strong in WTTx:I583CCh, weaker in WTTX:Q592CCh, and weakest in R20ATx:S631CCh), quantitatively the tail integration method was more reliable than the peak amplitude method as an estimator of toxin potency. Supplemental Fig. S2 of the Supplementary Material shows a more extensive comparison of the apparent dissociation constant (Kd) values for R20A toxin binding to WT and mutant hERG channels estimated by the two methods. This comparison leads to the same conclusion. Thus, for the experiments reported here, we integrated the tail currents over time at −80 mV to estimate the amount of charge transferred under the control conditions and at the steady state of toxin effects, QC and QTx, respectively. The ratio of QTx/QC was used to calculate the apparent Kd value. In the majority of the experiments, we used a single toxin concentration to estimate Kd (see Materials and Methods), and assumed that the maximal toxin effect was a 90% suppression of charge transfer (28). For WT BeKm-1 binding to the WT hERG channel, we have shown that the maximal degree of BeKm-1 suppression of WT hERG was ∼90%, and the Kd value estimated based on a single toxin concentration (10 nM) was not different from that determined by a complete dose-response relationship (from 1 to 1000 nM) (28). The site and mechanism of BeKm-1 action are likely maintained in the interactions between mutant toxins and mutant channels, because the mutant toxins maintained the native BeKm-1 conformation (33), and we chose to use mutant channels that retained WT-like function (14). Indeed, Fig. S3 in the Supplementary Material shows that the maximal degree of current suppression induced by WT toxin in two mutant channels that exhibited markedly different toxin sensitivity than WT hERG (Q592C and S631C) was ∼90%, similar to that seen in the WT channel. Furthermore, the Kd values for toxin binding to these two mutant channels determined by dose-response relationships were similar to those estimated based on single toxin concentrations (see below, Table 1).
TABLE 1.
Summary of apparent dissociation constants (Kd) for WT or alanine-substituted BeKm-1 toxins (E9A, Y11A, F14A, K18A, and R20A, listed on top) binding to WT or cysteine-substituted hERG channels (listed on the left)
| WT
|
E9A
|
Y11A
|
F14A
|
K18A
|
R20A
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Toxin Channel | Mean | SE | n | Mean | SE | n | Mean | SE | n | Mean | SE | n | Mean | SE | n | Mean | SE | n |
| WT | 8.1 | 0.5 | 14 | 6.2 | 0.9 | 3 | 2441 | 29 | 4 | 53.8 | 8.4 | 3 | 1031 | 155 | 3 | 2358 | 419 | 4 |
| M574C | 16.7 | 5.5 | 4 | 9.4 | 4.2 | 3 | 186 | 61 | 5 | 177.7 | 56.9 | 3 | 854 | 256 | 4 | 2746 | 506 | 4 |
| E575C | 4.7 | 1.1 | 4 | 3.3 | 1.3 | 4 | 413 | 151 | 3 | 44.8 | 15.9 | 3 | 1348 | 701 | 4 | 788 | 67 | 3 |
| Q576C | 16.6 | 4.7 | 6 | 11.4 | 3.2 | 3 | 300 | 53 | 3 | 140.2 | 52.6 | 4 | 1614 | 503 | 4 | 2578 | 534 | 3 |
| P577C | 8.4 | 0.1 | 3 | 5.1 | 2.2 | 4 | 173 | 59 | 3 | 80.9 | 12.7 | 3 | 1495 | 613 | 4 | 4860 | 2036 | 4 |
| H578C | 4.6 | 1.1 | 4 | 1.5 | 0.1 | 3 | 100 | 14 | 3 | 62.2 | 16.6 | 3 | 772 | 23 | 3 | 3236 | 458 | 4 |
| M579C | 8.3 | 4.2 | 4 | 12.0 | 6.3 | 3 | 134 | 33 | 3 | 121.8 | 49.7 | 4 | 1652 | 238 | 3 | 1669 | 283 | 5 |
| D580C | 29.5 | 8.3 | 4 | 7.1 | 1.9 | 3 | 477 | 64 | 3 | 458.9 | 190.1 | 3 | 2225 | 423 | 3 | 5700 | 653 | 4 |
| S581C | 7.5 | 1.4 | 4 | 10.8 | 6.9 | 3 | 194 | 49 | 3 | 89.2 | 33.8 | 3 | 580 | 161 | 3 | 1579 | 437 | 3 |
| R582C | 22.3 | 6.7 | 5 | 30.7 | 14.6 | 4 | 728 | 256 | 5 | 1946.7 | 529.8 | 5 | 2353 | 958 | 3 | 11346 | 2231 | 7 |
| I583C | 5.6 | 1.5 | 5 | 3.4 | 0.8 | 3 | 1462 | 301 | 4 | 87.5 | 35.5 | 3 | 1823 | 429 | 4 | 2539 | 534 | 3 |
| G584C | 1.6 | 0.3 | 5 | 1.0 | 0.1 | 3 | 133 | 45 | 3 | 20.2 | 7.8 | 4 | 111 | 12 | 3 | 87 | 28 | 6 |
| H587C | 4.7 | 0.8 | 6 | 2.9 | 1.0 | 4 | 251 | 84 | 4 | 64.0 | 22.8 | 3 | 644 | 131 | 4 | 764 | 195 | 4 |
| Q592C | 438.0 | 84.8 | 3 | 153.2 | 61.0 | 5 | 4174 | 2290 | 3 | 3241.3 | 586.8 | 3 | 1.8x105 | 3 | 5662 | 631 | 4 | |
| G594C | 6.4 | 2.1 | 4 | 3.3 | 0.8 | 3 | 84 | 25 | 3 | 82.9 | 24.2 | 3 | 527 | 151 | 3 | 2295 | 503 | 4 |
| P596C | 28.1 | 12.6 | 4 | 14.9 | 3.4 | 3 | 995 | 202 | 3 | 268.9 | 36.6 | 3 | 1783 | 767 | 4 | 2335 | 156 | 4 |
| Y597C | 6.9 | 1.0 | 4 | 4.5 | 0.6 | 3 | 2036 | 198 | 6 | 1370.4 | 603.1 | 5 | 1792 | 536 | 4 | 1782 | 1100 | 3 |
| N598C | 7.5 | 1.0 | 4 | 3.4 | 0.5 | 3 | 155 | 19 | 3 | 41.1 | 9.5 | 3 | 615. | 137 | 3 | 703 | 81 | 3 |
| S599C | 5.6 | 1.6 | 4 | 2.6 | 0.6 | 4 | 91 | 10 | 3 | 36.0 | 3.7 | 3 | 517 | 33 | 3 | 504 | 73 | 3 |
| S600C | 8.1 | 1.7 | 4 | 3.8 | 1.2 | 4 | 275 | 55 | 3 | 49.3 | 11.6 | 3 | 767 | 224 | 4 | 2212 | 815 | 4 |
| G601 C | 2.8 | 0.3 | 4 | 3.4 | 0.3 | 3 | 342 | 93 | 5 | 57.5 | 9.5 | 4 | 1468 | 277 | 4 | 2995 | 473 | 3 |
| L602C | 9.3 | 3.4 | 6 | 4.4 | 1.4 | 4 | 366 | 88 | 3 | 135.9 | 81.5 | 3 | 1002 | 86 | 3 | 937 | 214 | 3 |
| G603C | 7.1 | 0.9 | 3 | 4.3 | 1.2 | 4 | 481 | 111 | 3 | 64.9 | 14.5 | 3 | 867 | 148 | 3 | 1841 | 841 | 3 |
| S606C | 5.7 | 0.9 | 4 | 6.0 | 2.8 | 3 | 543 | 91 | 3 | 61.3 | 16.2 | 3 | 1398 | 243 | 4 | 1184 | 503 | 3 |
| I607C | 5.3 | 1.0 | 3 | 6.7 | 0.8 | 3 | 262 | 45 | 5 | 126.8 | 75.2 | 4 | 1137 | 212 | 4 | 3022 | 1079 | 3 |
| K608C | 9.1 | 2.1 | 3 | 4.6 | 1.4 | 3 | 286 | 52 | 4 | 109.6 | 23.0 | 3 | 1609 | 339 | 3 | 2400 | 721 | 3 |
| D609C | 14.6 | 1.7 | 6 | 16.6 | 6.3 | 3 | 2023 | 501 | 4 | 1562.7 | 363.2 | 4 | 1363 | 201 | 3 | 1838 | 597 | 7 |
| K610C | 8.2 | 1.6 | 4 | 5.5 | 0.4 | 3 | 175 | 46 | 3 | 53.5 | 14.9 | 3 | 1081 | 326 | 3 | 832 | 59 | 3 |
| V612C | 4.5 | 0.4 | 5 | 4.9 | 2.1 | 3 | 177 | 42 | 4 | 33.9 | 14.1 | 3 | 448 | 73 | 3 | 438 | 161 | 3 |
| T613C | 50.4 | 23.4 | 3 | 64.6 | 16.9 | 3 | 2215 | 640 | 3 | 1403.7 | 198.0 | 3 | 4192 | 1440 | 6 | 16091 | 8887 | 5 |
| S631C | 119.8 | 23.1 | 6 | 91.0 | 37.5 | 3 | 1855 | 248 | 6 | 2017.9 | 389.7 | 5 | 774 | 114 | 5 | 4079 | 1485 | 4 |
| N635C | 15.8 | 6.1 | 5 | 11.2 | 1.9 | 3 | 800 | 201 | 4 | 483.7 | 176.8 | 4 | 1526 | 329 | 3 | 2692 | 595 | 4 |
| S636C | 7.0 | 2.3 | 4 | 2.3 | 0.9 | 3 | 128 | 11 | 3 | 96.8 | 34.0 | 3 | 414 | 61 | 3 | 598 | 107 | 4 |
In all experiments, channels were activated by 1-s depolarization pulses from Vh −80 mV to +20 mV (plateau level of activation) and tail currents were recorded at −80 mV. To evaluate the toxin effects, the amount of charge transferred through the channels was calculated by integrating the tail current over time at −80 mV. This was done under the control conditions (QC) and at the steady state of toxin effects (QTx). The ratio of QTx/QC was used to estimate the apparent Kd value. Our previous data showed that WT BeKm-1 could not suppress WT hERG currents. At 100-fold of IC50 (1000 nM), the remaining hERG current amounted to 10 ± 1% of control (28). Therefore, Kd was calculated as QTx/QC = 0.9/(1 + [Tx]/Kd) + 0.1. n = number of observations.
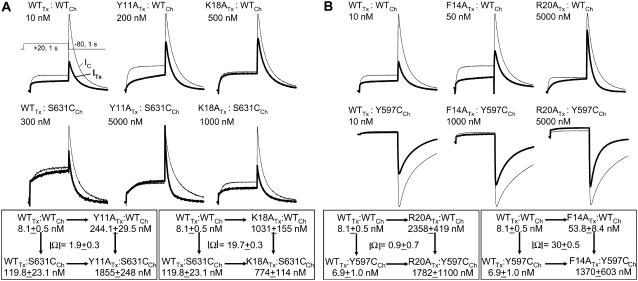
Fig. 3 A compares the effects of S631C channel mutation on toxin binding when the toxin is WT, Y11A, or K18A. Representative current traces are shown in the top panels of Fig. 3 A. The two mutant cycles, one for Y11A/S631C and the other for K18A/S631C, are shown in the lower panels of Fig. 3 A. The apparent Kd values are listed below the toxin:channel pairs. S631C reduced WT BeKm-1 toxin binding affinity (increasing the mean Kd value from 8.1 to 119.8 nM, an ∼15-fold increase). S631C also reduced the binding affinity of the Y11A mutant toxin (mean Kd value increased from 244 to 1855 nM, approximately eightfold increase). Therefore, the effects of S631C on toxin:channel interaction were not much affected by the Y11A toxin mutation, reflected in the calculated absolute value of coupling coefficient (|Ω|) 1.9 ± 0.3. On the other hand, S631C increased the binding affinity of K18A mutant toxin (mean Kd value reduced from 1031 to 774 nM). Therefore, the effect of S631C channel mutation on toxin:channel interaction depended on whether the toxin had lysine or alanine at position 18. This is reflected in the large |Ω| value of 19.7 ± 0.3.
FIGURE 3.
Detecting functional coupling between hERG channel residues and BeKm-1 toxin residues. (A) Mutant cycle analysis to compare the degree of coupling between S631 of hERG and Y11 or K18 of BeKm-1. (Top) Representative current traces from the marked toxin:channel pairs. The voltage clamp protocol is diagrammed for the WT toxin:WT channel pair. The thin and thick traces denote control current and current recorded at the steady state of toxin effect (IC and ITx, respectively), with toxin concentration marked for each pair. (Lower) Two mutant cycles, each with toxin:channel pairs marked on four corners along with apparent Kd values (mean ± SE). The absolute value of coupling coefficient (|Ω|) is marked in the center. (B) Comparison of degree of coupling between Y597 of hERG and F14 or R20 of BeKm-1. The format is the same as that in panel A. The Y597C channel currents were recorded in 98 mM [K]o. Therefore, the tail currents at −80 mV were inward.
Fig. 3 B compares the effects of Y597C channel mutation on toxin:channel interaction when the toxin was WT, F14A, or R20A. With the standard bath solution ([K]o 2 mM), the Y597C current amplitudes were very small, making it difficult to quantify toxin effects. Since we have shown previously that BeKm-1 binding to the hERG channel was insensitive to changes in ambient [K] (28), we elevated [K]o to 98 mM to boost the Y597C current amplitude (Fig. 3 B, upper panel, lower row). Y597C had little effects on the binding affinity of WT toxin (Kd values 8.1 and 6.9 nM for WT and Y597C channels), or R20A mutant toxin (Kd values 2358 and 1782 nM). The |Ω| value was ∼1. On the other hand, Y597C drastically reduced the binding affinity of F14A mutant toxin (Kd value increased from 54 to 1370 nM). This leads to a large |Ω| value of 30 ± 0.5, supporting the notion that Y597 on the channel is functionally coupled to F14 on the toxin.
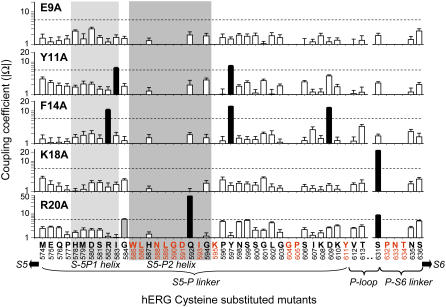
Table 1 lists the apparent Kd values for WT BeKm-1, and mutant toxins E9A, Y11A, F14A, K18A, and R20A binding to WT hERG and all 32 Cys-substituted mutant channels that maintained WT-like channel function. These values were used to calculate the absolute values of coupling coefficients (|Ω|) summarized in Fig. 4. For each of the five mutant toxins, the |Ω| values are plotted on a logarithmic scale versus the hERG channel positions where the native residues listed are replaced by Cys (high-impact positions highlighted by red were not tested). The dashed lines marked the cutoff threshold of coupling coefficient of 5.4 (equivalent to a coupling energy of 1 kcal/mol) (22,24,45). For Y11A, F14A, K18A, and R20A, there are distinct patterns of hERG residues that interact with these toxin residues (highlighted by black histogram bars). For R20A, G584C is marginally coupled (gray histogram bar). For E9A, none of the coupling coefficient reached the threshold, consistent with the notion that E9 is on the other side of the BeKm-1 interaction surface and is not involved in toxin:channel interaction. The mutant cycle analysis indicates that the critical pore-entrance residue, S631, that is an important determinant of the inactivation process of the hERG channel (12,46) (equivalent to T449 of the Shaker channel; see Lopez-Barneo et al. (47)), can interact with the two most critical toxin residues, K18 and R20. Furthermore, residues R582, I583, and G584 between the two putative S5-P linker helices (highlighted by the light gray and gray shades, described in details below), as well as Q592 and Y597 toward the C-terminus of the second S5-P helix are also coupled to specific toxin residues. Although the spatial relationship among these toxin residues is known (Fig. 1 B), this information cannot be translated directly into the spatial relationship among the channel residues that interact with them. This is because the toxin structure is asymmetric and its orientation when bound to the channel receptor site is not known. Therefore, we proceed to build structural models of the hERG pore domain based on available crystal structures (20) and NMR data (15,16), and use the mutant cycle analysis data as restraints in model building.
FIGURE 4.
Summary of absolute values of coupling coefficient (|Ω|) between 5 BeKm-1 alanine substituted mutants and 32 hERG cysteine substituted mutants. The BeKm-1 mutants are marked for each panel. The hERG native residues and position numbers are listed along the abscissa. The structural domains are also labeled. The high-impact positions are color coded red, and excluded from the analysis. Light gray shade denotes the putative S5-P1 helix that is stable during molecular dynamics simulation (see Fig. 7 B), whereas gray shade denotes the S5-P2 helix implicated by NMR spectroscopy. Dashed lines denote the cutoff |Ω| value of 5.4 (equivalent to a coupling energy of 1 kcal/mol). Histogram bars above the cutoff value are shown in black. G584 in the R20A panel is shown as a gray histogram bar because the coupling coefficient is marginally above the cutoff.
Creating a homology model for the transmembrane region of the pore domain of hERG channel
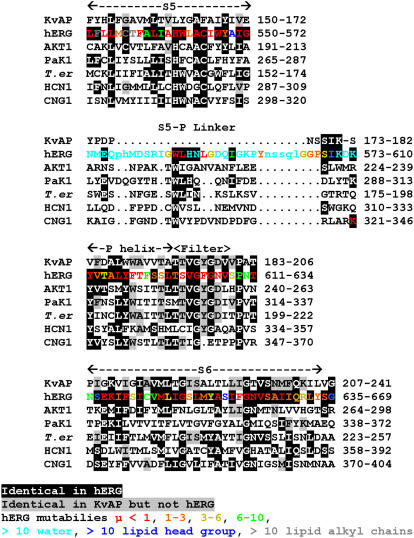
Our initial models of the transmembrane (TM) segments of hERG's pore-forming domain, S5/P/S6, were developed using homology modeling methods based on the crystal structure of KvAP for the open state conformation (20). A prerequisite for homology modeling is an accurate alignment of amino acid sequence of the target (hERG) to that of the template (KvAP). Because the two channels are only remotely related with <30% amino acid identity, sequence alignment alone was prone to errors. To aid the sequence alignment between hERG and KvAP, we employed a sequence profile alignment of hERG with >100 members of the EAG family, followed by profile alignment among KvAP, hERG (containing a cyclic nucleotide binding domain in its cytoplasmic C-terminus and thus a CNBD-containing channel), and other CNBD-containing channels. The results are shown in Fig. 5. Fig. 6 illustrates a schematic representation of our current model for the open conformation of hERG's pore-forming domain. The rationale for modeling the extracellular S5-P linkers is detailed below.
FIGURE 5.
Multisequence alignment of pore-forming domain of KvAP with hERG (ERG1, accession No. Q12809) and CNBD-containing channel homologs in plants (AKT1, accession No. NP_180233), in paramecium (PaK1, No. U19907), in Trichodesmium erythraeum bacteria ((T. er) No. YP_724331), cation nonselective HCN (No. NP_034538), and CNG (No. NP_776703) channels. The following regions are labeled on top: S5, S5-P linker, P helix, (selectivity) filter, and S6. Black background indicates residues that are identical to those of hERG, and gray background indicates residues identical to those of KvAP but not hERG. Residues of hERG are color coded according to their mutability (35) calculated from an alignment of more than 100 members of the EAG family: red μ < 1, orange μ = 1–3, yellow μ = 3–6, green μ = 6–10, cyan μ > 10 for residues predicted to be aqueous (water) exposed, blue μ > 10 for residues that occur frequently in lipid headgroup region (K, R, W, Y), gray μ > 10 for hydrophobic and ambivalent residues predicted to be exposed to lipid alkyl chains. Lower case letters in hERG S5-P linker sequence indicate positions deleted in some members of the EAG family.
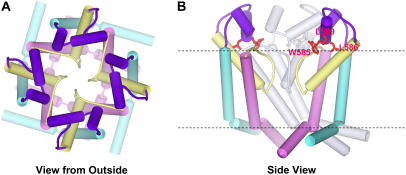
FIGURE 6.
Schematic representation of our model for hERG's pore-forming domain. Cylinders represent α-helices. Color code, S5 = cyan, S5-P linker = purple, P segment = yellow, S6 = pink. (A) View from the extracellular side through the pore. (B) Side view with two subunits colored by segment. The subunit at the back is colored light gray, and the subunit nearest the viewer has been removed. Highly conserved residues of the S5-P linker (W585, L586, and L589) are shown and colored red. Each S5-P linker is postulated to possess two helices (S5-P1 helix, H578-I583; and S5-P2 helix, W585-G594) that are connected by a hinge G584.
Experimental basis for modeling the hERG S5-P linkers
The crystal structures provide little information about the structure of hERG residues 573–611 that link S5 to the P helix (S5-P linker). We searched for the simplest models that are consistent with experimental data and a number of modeling criteria. The key experimental observations are summarized below, whereas the modeling criteria have been discussed in our recent publication (35).
Residues 583–597 in the middle of the S5-P linker are functionally important (14): cysteine (Cys)-substitution per se (in free thiol state without disulfide bond formation) or MTS modification of Cys side chains introduced here could markedly affect current amplitude and/or the channel's ability to inactivate and discriminate between K+ and Na+ ions. The pattern of functional perturbation by Cys substitution in this region suggested a helical regularity.
NMR spectroscopy studies confirmed that residues 585–594 could adopt an amphipathic α-helical structure when hERG S5-P linker peptides were placed in detergent micelles, but not in aqueous solutions (15,16). This helix will be called S5-P2 helix, to differentiate it from a putative S5-P1 helix in the current hERG model (described below).
Hydrophobic residues on the S5-P2 helix all occupied high-impact positions, consistent with the notion that they are engaged in protein-protein interactions critical for hERG channel function (14). On the other hand, most hydrophilic residues on the other face of the S5-P2 helix occupied less critical, “intermediate-impact” positions.
The G584C mutant had a unique property (14). Under reducing conditions, it had a wild-type channel phenotype (fast inactivation and high K+ selectivity). However, when G584C was exposed to either positively charged MTSET or negatively charged MTSES, it adopted a mutant behavior in which both inactivation and K+ selectivity were drastically altered. The only other cysteine mutants with this property were T613C (in the P helix) and S631C (at the pore entrance) (14). This similarity suggests a spatial proximity of G584, T613, and S631 near the entrance of the selectivity filter.
Cys introduced into positions 583–597 could form intersubunit disulfide bonds, producing dimer bands on Western blots (15). Furthermore, Cys side chains introduced into at least four positions here, 548, 585, 588, and 589, could form disulfide bonds with their counterparts from other subunits.
It is unlikely that all of the residue pairs between which disulfide bridges can form are near each other most of the time in the native conformation. However, they may occasionally come into contact if the structure is dynamic. Disulfide bonds are formed more frequently or more stable if the two thiol groups are in close proximity. Therefore, the ratio of dimer/monomer band intensities on Western blots of Cys mutants of the 583–597 segment could be used as an index for spatial proximity between partner Cys side chains. There was a general trend of higher dimer/monomer ratio when Cys was introduced into the N-terminus of the 583–597 segment and lower ratio when Cys was introduced into more C-terminal positions. This trend is consistent with the view that the N-terminus of the 583–597 segment is close to the central axis of the pore (experimental observations No. 4 above), and thus to counterparts from other subunits. Indeed, experimental data suggested that 584C could form intersubunit disulfides with 584C from an adjacent subunit, leaving the other two 584C free thiol groups capable of forming a high-affinity Cd bridge (15).
There are data suggesting that putting a charged group at position 588 or 592 can uniquely perturb the voltage dependence of hERG inactivation, although the mechanism is not clear (48).
Together these data suggest that the middle part of the S5-P linker (585–594) can form an amphipathic helix with its hydrophobic face interacting with other channel domains and the N-terminus close to the pore entrance. Furthermore, the S5-P linkers are capable of dynamic conformational changes so that G584 and N588 on the hydrophilic face, and W585 and L589 on the hydrophobic face, of the S5-P2 helix may come very close to counterparts from other subunits at certain gating conformations of the channel. When residues at these positions were replaced by Cys side chains, such occasional contacts allow intersubunit disulfide bond formation between counterparts.
Modeling the extracellular S5-P linkers
Key experimental observations listed above indicate that certain residues of the S5-P linker are likely to be located near the extracellular entrance to the pore and are likely to form an amphipathic α-helix. Thus the first modeling constraint was to assume that this linker interacts with the outer portions of the S5, P, and S6 segments that have been modeled after analogous segments in the crystal structure of KvAP channel. In docking the S5-P linkers onto the outer vestibule, we first assumed that most of the hydrophobic side chains will be buried (facing other channel domains or within the S5-P linkers) and that most of the hydrophilic side chains will be exposed to the extracellular aqueous solution. This assumption is reasonable based on key experimental observation No. 3 above. We also assumed that most of the interactions between side chains are energetically favorable; i.e., we selected models with high numbers of hydrophobic interactions, salt bridges, and hydrogen bonds (all charged residues except R582 form salt bridges and most polar side-chain groups form hydrogen bonds; see Supplemental Table S1). Furthermore, based on the multisequence alignment of the EAG family and of this family with CNBD-containing channels (Fig. 5), most of the residues in the S5-P linker are poorly conserved at both levels. However, W585, L586, and L589 are absolutely conserved within the EAG family. W585 and to a lesser extent L586 are further conserved among all CNBD-containing channel families. We constrained the models so that these hydrophobic, high-impact residues interact with other highly conserved residues of the S5, P, and S6 segments.
We wanted to use molecular dynamic simulations to analyze the stability of our models. MD simulations requires one to specify the coordinates for all of the atoms at the beginning of the simulation; i.e., we had to develop preliminary models of the segments that link the putative α-helix of the linker to the S5 and P segments that were modeled from the KvAP structure. To make preliminary models simple, we favor regular secondary structures except when secondary structure prediction methods (http://www.predictprotein.org/) (49) strongly predict otherwise. In this case we have extended the S5 α-helix to residue E575 (two residues past the S5 C-terminus in KvAP), made segments 577–583 and 585–594 helical (termed “S5-P1” and “S5-P2” helices in the following text), and began the P helix at either G604 or S606 (six or four residues before the N-terminus of the P helix in KvAP). Residues Q576, K595 and G603/G604 were given nonhelical conformations because they each precede a proline and because insertions and deletions (indels) occur in the vicinity of Q576 in an alignment of EAG channel sequences (Fig. 5). G584 was assigned a nonhelical conformation because we needed to introduce a break between the two helices and because glycines often have nonhelical conformations. The two putative helical segments of the S5-P linker are each amphipathic; i.e., one face of the helix is composed of hydrophobic residues (M579 and I583 for the S5-P1 helix and W585, L586, L589, and I593 for the S5-P2 helix) and the opposite face is composed of hydrophilic residues. Such amphipathic patterns are typical of surface α-helices. Segment 594-606 (GKPYNSSGLGGPS) is strongly predicted to form coils or turns (10 of 13 residues have high propensities for coils and turns and low propensities for α-helices and β-strands). This segment is poorly conserved among close homologs to hERG, with indels occurring in an alignment of the EAG family, and is deleted in other CNBD-containing channels (Fig. 5). G604/P605 are high-impact positions (Fig. 1 A) and are moderately conserved within the EAG family (Fig. 5), suggesting that they are functionally important.
Initial models of the transmembrane and extracellular portion of hERG were minimized with fourfold symmetry about the axis of the pore. These models included the voltage-sensing domain. Models of the voltage-sensing domain and activation gating mechanism will be presented elsewhere. However, the models of the outer vestibule region described here were not affected substantially by different models of the voltage-sensing domain, consistent with the observation that the crystal structures of the outer vestibule and selectivity filter regions of Kv1.2 (21), KvAP (20), and KcsA (17) were quite similar even though the structures of the voltage-sensing domains were very different in the Kv1.2 and KvAP, and is absent in KcsA. These models were constructed with backbone torsion angles and side-chain conformations frequently observed in known protein structures. Some initial atomic overlaps were eliminated by manual adjustments and invacu minimization. Next MD simulations were performed with the protein embedded in a lipid bilayer, with water on each side of the membrane and in the pore, and with K+ ions in the selectivity filter. The backbone atoms of the S5, P, and S6 segments were restrained during the first nanosecond of simulation. This allowed the more speculative portions of the model (the S5-P linker and voltage-sensing domain) to relax in a manner that did not substantially perturb the portion of the pore that was modeled from the KvAP crystal structure. The restraint was removed for the second nanosecond of simulation. The average structure during the unrestrained portion of the simulation was examined visually and the root-mean-square deviation (RMSD) was calculated for different portions of the model to determine how well the structure of the initial model was maintained. This procedure was performed using numerous starting models that all had fourfold symmetry. When positions and/or conformations of a given residue changed in the same manner in most of the subunits, new models with fourfold symmetry were constructed that preserved these changes while also preserving energetically favorable interactions among the residues, and the simulation was repeated. This process of simulation followed by remodeling was repeated until consistent changes were no longer observed during the simulations.
The S5-P linker was relatively dynamic during all of these MD simulations (typical RMSD of ∼3–4 Å). Some of this dynamic nature may be due to modeling errors, because this region was not modeled after any known crystal structure. However, mutagenesis experiments suggest that the S5-P linker is inherently dynamic (key experimental observations No. 5 and No. 6 above). As mentioned above, the most conserved S5-P linker residues within the EAG family are W585, L586, and L589 (Fig. 5). Coincidentally, the region containing L586 and L589 was the most stable portion of the linker in MD simulations of our models. The helical secondary structure of residues 585–589 was well maintained in the simulations. However, the helix sometimes bent or broke at residues G590 and D591. In most simulations the helical structure of the S5-P1 segment (amino acids 577–583) was maintained, although in some cases portions of it adopted a 310 helical conformation. The additional helical structures at the end of S5 and beginning of the P helix that were not present in the KvAP template structure were also well maintained in most simulations; however, residues 606–611 adopted a nonhelical conformation in a few simulations. As anticipated, the most dynamic portion of the models was the putative coiled segment that links the S5-P2 helix to the P segment; i.e., segment 594–606. Its conformation always changed substantially, and differently, in each of the four subunits during the MD simulations. The presence of this long flexible segment, which is absent in other CNBD-containing channel families (Fig. 5), likely increases the dynamic nature of the entire S5-P linker. We have little confidence that our models of residues 594–606 are correct, but this segment has been included to illustrate its general location and make this portion of the model complete for the MD simulations.
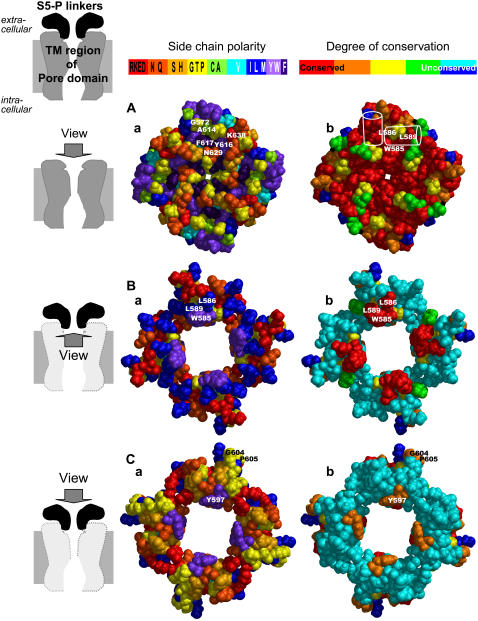
The outer surface of S5, P, and S6 segment modeled directly from the KvAP crystal structure is illustrated in Fig. 7 A. With few exceptions, most of the residues in the central core are highly conserved within the EAG family (Figs. 5 and 7 Ab), and that most of the residues on the outer surface are either hydrophobic (cyan, blue, and purple, tend to be buried in proteins) or ambivalent (green and yellow, can be either buried or exposed) (Fig. 7 Aa). Fig. 7 B illustrates the inner surface of the four S5-P linkers, which interacts with the outer surface of the pore-forming domain shown in Fig. 7 A. Although most of this segment is poorly conserved among EAG channels, the conserved W585, L586, and L589 side chains reside on this inner surface (Fig. 7 Bb). These residues interact in our models with highly conserved residues of the pore-forming domain: W585 interacts with the aromatic cuff residues Y616 and F617, and the selectivity filter residue N629. L586 interacts with G572, A614, and F617. L589 interacts with K638. The poorly conserved hydrophobic M574 and I583 residues of the putative S5-P1 helix are buried in our models and interact primarily with other poorly conserved residues of the S5-P and P-S6 linkers. In our models the upper, water-exposed, surface of the S5-P linkers are poorly conserved (Fig. 7 Cb) and relatively hydrophilic (Fig. 7 Ca). The major exception is Y597, which was modeled at the entrance of the outer vestibule to be consistent with the observation that cysteine side chains introduced here could form weak intersubunit disulfide bonds (experimental findings No. 6 above), and to better fit the BeKm-1 results as described below.
FIGURE 7.
Putative interactions between the S5-P linkers and the outer surface of the transmembrane (TM) region of hERG's pore domain in our model. (Right column) Diagrams of channel structures (TM region of pore domain and extracellular S5-P linkers) and directions of view in rows A, B, and C. (Central column) Space-filled models of channel structures viewed as indicated on the left with amino acid side chains color coded according to polarity (color scheme shown on top with one-letter amino acid codes), red = charged, orange = hydrophilic, yellow/light green = ambivalent, cyan/blue = hydrophobic, and purple = aromatic. (Right column) Same views as in central column with amino acid side chains color coded according to the degree of conservation within the EAG family. The color scheme is as illustrated in Fig. 4. Panels Aa and Ab illustrate outer surface view of the TM region of hERG pore domain. Labeled in the top subunit of Aa are residues with which highly conserved S5-P linker residues (W585, L586, and L589) may interact. Labeled in the top subunit of Ab are the approximate backbone locations of S5-P1 and S5-P2 helices (white outline of cylinders) and approximate locations of W585, L586, and L589 side chains. Panels Ba and Bb illustrate inner surface view of the S5-P linkers. Note that most of the hydrophobic residues of the S5-P linkers reside on this surface (Ba), and that some residues form a highly conserved cluster (red area in Bb). W585, L586, and L589 residues interact with highly conserved residues of the S5/P/ S6 segments shown in Aa and Ab (see details in text). Ca and Cb illustrate outer surface view of S5-P linkers. Most of these residues are hydrophilic (Ca) and poorly conserved (Cb), consistent with the notion that they are exposed to the extracellular aqueous phase.
We favor models in which the backbone portions of segments that can be aligned unambiguously would be valid for all members of the family or superfamily. Deletions can be accommodated without altering the surrounding structure substantially if the residues at the beginning and end of the deleted segment are near each other. The long poorly conserved segment that links the S5-P2 helix to the P helix is deleted in most other CNBD-containing channel families (Fig. 5). This large deletion can be accommodated in our model because G590, which is located just beyond the conserved portion of the S5-P2 helix, is near S605, which is at the beginning of the P helix. A short (two- to three-residue) deletion that occurs in most CNBD families and some members of the EAG family at the junction of S5 and the S5-P1 helix (Fig. 5) can also be accommodated because E575 at the end of S5 is near M579 in the first part of the S5-P1 helix.
Docking of BeKm-1 to the hERG's outer vestibule
The modeling described above still leaves much ambiguity about the structure of the S5-P linker; e.g., most of the criteria described above can also be satisfied by models in which the S5-P2 helices form a parallel bundle of helices that are nearer the axis of the channel than shown in Fig. 6. Our next step was to identify models of the outer vestibule of hERG developed as described above onto which NMR-determined structures of the BeKm-1 toxin could be docked in a manner consistent with two sets of experimental data. Korolkova et al. (33) analyzed the effects of alanine substitutions of surface residues of BeKm-1 on its binding affinity to the hERG channel. These mutations can be classified into four categories; very strong effects (binding affinity 52–86 times less for K18A, R20A, and F21A, although F21A may have caused a distortion of the toxin conformation), substantial effects (binding affinity ∼15 times less for Y11A and K23A), modest effects (binding affinity six to nine times less for R1A, F14A, and R27A), and negligible effects (binding affinity changed <3.3 times for P2A, D4A, K6A, E9A, Q12A, V29A, F32A, D34A, and F36A). All of the high-impact residues are on one side of the toxin, whereas all of the low impact residues are on the other side (Fig. 1 B). The toxin was docked so that the high-impact residues interact with the channel and low impact residues do not (Fig. 8 A).
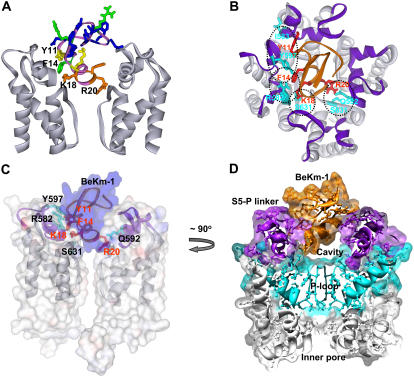
FIGURE 8.
Docking of BeKm-1 in the outer mouth of hERG. (A) Side view of two hERG pore-forming domain subunits shown in gray. BeKm-1 (top center) is colored according to the effect of alanine substitution on the binding affinity (same color scheme as in Fig. 1 B). Residues K18, R20, F14, and Y11 are labeled. (B) View from the extracellular side toward the outer entrance of hERG pore-forming domain with BeKm-1 bound. Ribbons represent the backbones of channel and toxin. Color code: light gray = S5, P, and S6 segments, purple = S5-P linker, orange = BeKm-1. Residues that interact (based on the mutant cycle analysis) are shown in red for BeKm-1 and cyan for hERG. Specific pairwise interactions are encircled with dashed lines. The figure is the averaged structure during the second half of an MD simulation of the protein embedded in a POPE lipid bilayer. The exact fourfold symmetry of the starting model (before the MD simulations) is not maintained, and the helices of the S5-P linker became distorted in two subunits. (C and D) Side views of the same model using transparent surfaces to show the tight fit of the BeKm-1 toxin in the opening between the S5-P linkers (C) but a cavity between the toxin and the entrance to the selectivity filter (D). In panel C, only two of the hERG subunits are shown. The toxin surface is shown in blue, the channel surface shown in gray, and the ribbons for toxin and S5-P linkers shown in purple, with some of the interacting side chains shown as in panel B and labeled. The view in panel D represents an ∼90° clockwise rotation of the view in panel C around the central axis, with the following key elements labeled: BeKm-1 in orange, S5-P linkers in purple, P-loops in cyan, S5 and S6 segments in gray, and inner pore. Clipping planes were used to illustrate only a central cross section through all four subunits.
The second set of data is the mutant cycle analysis presented above. In docking the toxin, we first concentrated on interactions of S631 with K18 and R20. These interactions were selected for five reasons: 1), the location of S631 near the end of the P selectivity filter is defined fairly precisely by the KvAP template used to model the pore domain; 2), the serine to cysteine mutation is very conservative and preserves the channel phenotype (12); 3), the K18A and R20A mutations have the strongest effects on BeKm-1 binding to WT channels (33); 4), the S631C channel mutation has the second strongest effect on binding of WT BeKm-1 (28); and 5), the K18-S631 pair has the second highest |Ω| value (Fig. 4). Models in which the opening of the outer vestibule was too small to allow binding of K18 and R20 to S631 were eliminated.
The strongest coupling was observed between R20 on the toxin and Q592 on the channel. Requiring the side-chain oxygens of both S631 and Q592 to bind to the guanidinium side-chain group of R20 constrains the position and orientation of the putative S5-P2 helix of the S5-P linker, and models in which both interactions could not occur were eliminated.
The mutant cycle analysis also suggests that Y11 and F14 on the toxin interact with I583 and R582 on the channel, respectively. These two hERG residues are at the C-terminus of the putative S5-P1 helix. In our favored models the axis of this helix points directly toward the pore with its C-terminus positioned in the outer vestibule just extracellular to the selectivity filter (Figs. 6 A and 7 Ab). The orientations of backbone amide groups in an α-helix produce a dipole with a partial negative charge at the C-terminus of the helix (50). Electrostatic interactions of the C-termini of these four S5-P1 helices around the outer mouth of the pore entrance with K18 and R20 may contribute to the binding of BeKm-1. The BeKm-1 structure was docked so that the side chain of F14 can make contact with the side chain of R582. In all of our models, I583 was buried beneath the S5-P1 helix, and we found it difficult to develop models in which its side chain interacts directly with that of Y11. However, in several of our models the Y11 side chain does interact with the S581 and R582 residues located at the C-terminus of the S5-P1 helix. Thus, the coupling could be due to a small shift in the location of the C-terminus of this helix that occurs when I583 is mutated to cysteine.
Weak coupling between R20A and G584C was also observed in the mutant cycle analysis. G584 links the two putative S5-P helices and was near S631, and thus R20, in most of our models, consistent with the observed functional coupling between the two.
The mutant cycle analysis also suggests that the two aromatic toxin residues, Y11 and F14, interact with an aromatic hERG residue, Y597, which lies within the latter portion of the S5-P linker that we suspect has little regular secondary structure. We developed models of this segment to allow the interaction between Y11/F14 and Y597 only for models in which the putative helical portions and their interactions with BeKm-1 had been modeled. In these models, the S5-P2 helix terminates at K595 and the P596 and Y597 residues extend back over the axis of this helix so that the Y597 side chain resides between the Y11 and F14 side chains.
The only apparent coupling interaction indicated by the mutant cycle analysis that is inconsistent with all of our models occurs between F14 and D609. In our models, D609 resides on the face of the P helix that is oriented toward the pore. However, it lies beneath the S5-P2 helix and was relatively far from F14 in our models. Because D609 interacts directly with the S5-P2 helix in the vicinity of G590 in our model, and the S5-P2 helix can adopt dynamic conformations, it is possible that the D609C mutation alters the position of some of the S5-P linker residues that interact directly with F14. Such anomalies are not unprecedented; e.g., coupling was observed between an aspartate on the P helix of Shaker (D431) and S11 of Agitoxin 2 (25); however, modeling based on crystal structures and NMR studies of the toxin-channel complex indicate that these residues do not interact directly.
Five similar models for the outer vestibule interactions with BeKm-1 were identified in which most of the interactions described above were maintained during an invacu minimization procedure. Next MD simulations of these models of the channel-toxin complex were performed. The pore domain was embedded in a POPE lipid bilayer with water on each side. During the first nanosecond of the MD simulations, restraints were imposed to 1), maintain interactions between the K18 amine group and a S631 hydroxyl group; 2), maintain interactions between the R20 guanidinium group and a Q592 amide group and a S631 hydroxyl group; and 3), maintain the backbone atoms of the inner portions of the S5 and S6 helices to within an angstrom of their starting position. These restraints allowed the position of the toxin and conformation of the channel outer vestibule to relax without dramatically altering the conformation of the pore or losing the strongest interactions between the toxin and channel. These restraints were removed during the second nanosecond of the simulations. The average structures of the complex during the last nanosecond were then examined visually to determine how well the interactions suggested by the experiments were maintained, and how the position and conformation of the toxin and the S5-P linker were altered. In most of the simulations, some of the experimentally determined pairwise interactions present in the initial models were not maintained. However, in one simulation, they all were, as illustrated in Fig. 8 (the pairwise interactions are marked in Fig. 8 B). Supplemental Fig. S4 illustrates the other models for which the interactions were not maintained as well. In the model presented in Fig. 8, which we consider our best, some of the initial fourfold symmetry of the outer vestibule was lost during the simulation. In two subunits, residues 577–583 of the S5-P1 helix and 585–594 of the S5-P2 helix maintained their secondary structure; however, the helical conformation was lost in a third subunit for residues 577, 578, 585, and 594, and in a fourth subunit for residues 585 and 590–594. The S5-P helices also shifted slightly relative to the transmembrane helices, but the direction of the shift differed among the four subunits. Some of this loss of symmetry could be due to interactions with the asymmetric BeKm-1 toxin; e.g., the two subunits in which the helical structure of the S5-P linker was completely maintained had the least contact with the toxin. However, some of the symmetry was also lost in most simulations in which the toxin was not present. This result is consistent with our hypothesis that the structure of the S5-P linker is highly dynamic, and can be easily perturbed by mutations, toxin binding, or conformational changes in other parts of the protein.
DISCUSSION
The structural model matches the functional data well
The structural model presented here not only matches seven out of eight pairs of interacting residues identified in the mutant cycle analysis, but also provides mechanistic insights into the unique features of BeKm-1/hERG interaction. The model showed that BeKm-1 is stuck above the pore entrance by the S5-P1 and S5-P2 helices that crowd the outer vestibule of the channel. The S5-P helices make contacts with Y11 and F14 on one end of the toxin's α-helix. The other end of the toxin α-helix is pointing downward, toward the pore entrance, with K18 and R20 making contacts with S631 side chains on two adjacent subunits. The observation in the model that BeKm-1 is bound above the pore entrance and none of its side chains penetrate deep into the pore explains why BeKm-1 is not sensitive to [K+] inside the pore (28). This is distinctly different from charybdotoxin (ChTx) or agitoxin2 (AgTx2) binding to the Shaker channel. In the latter case, ChTx or AgTx2 uses a critical lysine residue on their interaction surfaces to protrude into the pore and bind in the vicinity of the selectivity filter (25). Therefore, increasing K+ ion occupancy inside the pore or elevating K+ ion concentration around the outer mouth can destabilize ChTx or AgTx2 binding to the Shaker channel (25). The positive charges of the K18 and R20 side chains around outer entrance likely hinder K+ ion flux through the pore. Although electrostatic forces do not play as a major role in directing BeKm-1 (net +2.73 charges at neutral pH) binding to the hERG outer vestibule, as is the case for ChTx or AgTx2 binding to the Shaker channel (+4.89 and +5.89 at neutral pH), increasing the amount of positive charges on BeKm-1 (changing the bath solution pHo in the range of 8.5–6.5) could modestly increase its potency (28).
BeKm-1 does not totally suppress hERG currents: even in the presence of 1000 nM BeKm-1 (100-fold its IC50) there remained some residual hERG current amounting to ∼10% of the control amplitude (28). This residual conductance could indicate that BeKm1 never completely blocks the entrance to the pore. The fit of BeKm-1 in the opening between the S5-P linkers is relatively tight in this model, as illustrated in Fig. 8 C; however, there is a cavity between the toxin and entrance to the selectivity filter (Fig. 8 D). Our models are not sufficiently precise and stable to exclude the possibility that ions can pass into this cavity through gaps between the toxins and the linkers or between the linkers and the P segments at a rate sufficient to cause the residual current. It is also possible that the dynamic nature of the binding region allows flickering between blocked and unblocked states. This possibility is supported by our kinetic model simulations (Fig. 2): membrane depolarization that enhances hERG inactivation induces BeKm-1 dissociation, and toxin rebinds upon membrane repolarization. This is also consistent with experimental findings: elevating temperature markedly reduces BeKm-1 binding potency (IC50 increased from 9 nM at room temperature to 63 nM at 37°C), as if more vigorous motions of the S5-P linkers around the outer mouth at the higher temperature dislodge BeKm-1 more easily (29). Therefore, at positive voltages we used to monitor the currents, dynamic conformational changes of the S5-P linkers could temporarily dislodge BeKm-1 from the pore entrance, allowing K+ currents.
A recent report suggests that, as has been described for the P-type inactivation in the Shaker (51), Kv1.5 (52) and Kv2.1 (53) channels, the inactivation process in the hERG channel is accompanied by a transient increase in Na+ permeability through the pore (54). In our model, residues at high-impact positions on the S5-P linker interact with residues of the pore loop. For example, W585 interacts with the aromatic cuff residues Y616 and F617, as well as the selectivity filter residue N629, and L586 interacts with A614 and F617 (Fig. 7). These intimate interactions may explain why many mutations in the S5-P linker disrupt not only the inactivation process but also the ability of the hERG pore to select for K+ ions (12–14).
Limitations
The Guy laboratory at National Institutes of Health uses a long-term iterative approach to modeling. Initial models that are not highly restrained by experimental data, such as those we proposed shortly after the first voltage-gated channel sequences were determined (55), are often relatively imprecise and highly speculative. Even so, these models were valuable in designing and interpreting experiments that confirmed most of our major predictions. As more data were obtained and as methods of modeling improved, we developed successive generations of models that are less speculative and are probably more correct. Although the models presented here are less speculative and more constrained than were our early models of Na+ and K+ channels, they deal with a very difficult problem of modeling dynamic loops for which no homologous template structure has been determined. Thus these models should still be considered as tentative approximations of the loop structure of the open conformation. The most speculative aspects of the models concern segments where indels occur in alignments of closely related sequences, that are predicted not to have regular secondary structures, and for which there are no experimental data that support the proposed models. The rationale for modeling these regions is that they are required for MD simulations. Here these regions correspond to the first and last part of the S5-P linker. Thus, the proposed extension of the S5 helix and the S5-P1 helix are speculative and are supported only by the observation that they remain intact during MD simulations and allow hydrophobic residues to be buried while hydrophilic residues are exposed. However, the observation that the S5-P1 segment is poorly conserved and that some of its residues are deleted or replaced by helix breaking residues such as proline in some homologs makes it unlikely that this segment is helical in all CNBD channel families, and raises questions about the validity of this aspect of the model for hERG. The proposed structure of the last part of the loop that connects the S5-P2 helix to the P helix is even more speculative. It is even less well conserved with rather large indels in alignments of even closely related channels of the EAG family, is predicted to have a coiled structure, and is very dynamic in the simulations. The most firmly based aspect of the model is the central region that contains the high-impact residues. NMR studies support the helical nature of this segment, its hydrophobic residues are highly conserved, these interact with highly conserved residues of the P segment, and mutations of these residues alter the selectivity and gating properties of the channel. W585 is the only loop residue that is completely conserved among all CNBD families. It is also the residue that interacts with some of the most highly conserved residues of the P segment; i.e., with the two aromatic residues of the P helices that form the aromatic cuff that surrounds the selectivity filter in most K+ channels and with residues at the outer entrance to the selectivity filter. Thus, whereas some precise details of our models are likely to be incorrect, we believe that the general location of these highly conserved S5-P linker residues at the outer entrance of the selectivity filter is valid. Fortunately, this is likely to be the most important part of the linker, and errors in modeling the more peripheral parts of the linker are probably not as important.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Acknowledgments
This work was supported in part by HL 67840 and HL 46451 from National Institutes of Health/National Heart, Lung, and Blood Institute (to G.N.T.), by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (to H.R.G.), by a grant from the Russian Foundation of Basic Research (to Y.V.K.), by Ministry of Education and Science of Russian Federation and the Program of Cellular Biology, Russian Academy of Sciences (to E.V.G.), and by A. D. Williams Trust Funds (to G.N.T. and Y.V.K.).
Kailas D. Sonawane's present address is Dept. of Biochemistry, Shivaji University, Kolhapur, India.
References
- 1.Sanguinetti, M. C., C. Jiang, M. E. Curran, and M. T. Keating. 1995. A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell. 81:299–307. [DOI] [PubMed] [Google Scholar]
- 2.Emmi, A., H. J. Wenzel, P. A. Schwartzkroin, M. Taglialatela, P. Castaldo, L. Bianchi, J. M. Nerbonne, G. A. Robertson, and D. Janigro. 2000. Do glia have heart? Expression and functional role for ether-a-go-go currents in hippocampal astrocytes. J. Neurosci. 20:3915–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou, W., F. S. Cayabyab, P. S. Pennefather, L. C. Schlichter, and T. E. DeCoursey. 1998. HERG-like K+ channels in microglia. J. Gen. Physiol. 111:781–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer, C. K., I. Wulfsen, R. Schafer, G. Glassmeier, S. Wimmers, J. Flitsch, D. K. Ludecke, and J. R. Schwarz. 2003. HERG K+ currents in human prolactin-secreting adenoma cells. Pflugers Arch. 445:589–600. [DOI] [PubMed] [Google Scholar]
- 5.Faravelli, L., A. Arcangeli, M. Olivotto, and E. Wanke. 1996. A HERG-like K+ channel in rat F-11 DRG cell line: pharmacological identification and biophysical characterization. J. Physiol. 496:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang, X., E. R. Reynolds, P. Deak, and L. M. Hall. 1997. The seizure locus encodes the Drosophila homolog of the HERG potassium channel. J. Neurosci. 17:882–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Overholt, J. L., E. Ficker, T. Yang, H. Shams, G. R. Bright, and N. R. Prabhakar. 2000. HERG-like potassium current regulates the resting membrane potential in glomus cells of the rabbit carotid body. J. Neurophysiol. 83:1150–1157. [DOI] [PubMed] [Google Scholar]
- 8.Rosati, B., P. Marchetti, O. Crociani, M. Lecchi, R. Lupi, A. Arcangeli, M. Olivotto, and E. Wanke. 2000. Glucose- and arginine-induced insulin secretion by human pancreatic β-cells: the role of HERG K+ channels in firing and release. FASEB J. 14:2601–2610. [DOI] [PubMed] [Google Scholar]
- 9.Wang, H., Y. Zhang, L. Cao, H. Han, J. Wang, B. Yang, S. Nattel, and Z. Wang. 2002. HERG K+ channel, a regulator of tumor cell apoptosis and proliferation. Can Res. 62:4843–4848. [PubMed] [Google Scholar]
- 10.Crociani, O., L. Guasti, M. Balzi, A. Becchetti, E. Wanke, M. Olivotto, R. S. Wymore, and A. Arcangeli. 2003. Cell cycle-dependent expression of HERG1 and HERG1B isoforms in tumor cells. J. Biol. Chem. 278:2947–2955. [DOI] [PubMed] [Google Scholar]
- 11.Chadwick, D. J., and J. Goode. 2005. The hERG Cardiac Potassium Channel: Structure, Function and Long QT Syndrome. Novartis Foundation Symposium 266. John Wiley & Sons, Chichester, UK.
- 12.Fan, J.-S., M. Jiang, W. Dun, T. V. McDonald, and G.-N. Tseng. 1999. Effects of outer mouth mutations on hERG channel function: a comparison with similar mutations in Shaker. Biophys. J. 76:3128–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dun, W., M. Jiang, and G.-N. Tseng. 1999. Allosteric effects of mutations in the extracellular S5-P loop on the gating and ion permeation properties of hERG. Pflugers Arch. 439:141–149. [DOI] [PubMed] [Google Scholar]
- 14.Liu, J., M. Zhang, M. Jiang, and G.-N. Tseng. 2002. Structural and functional role of the extracellular S5-P linker in the HERG potassium channel. J. Gen. Physiol. 120:723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang, M., M. Zhang, I. V. Maslennikov, J. Liu, D. M. Wu, Y. V. Korolkova, A. S. Arseniev, E. V. Grishin, and G.-N. Tseng. 2005. Dynamic conformational changes of extracellular S5-P linkers in the hERG channel. J. Physiol. 569:75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torres, A. M., P. Bansal, M. Sunde, C. E. Clarke, J. A. Bursill, D. J. Smith, A. Bauskin, S. N. Breit, T. J. Campbell, P. F. Alewood, P. W. Kuchel, and J. I. Vandenberg. 2003. Structure of the HERG K+ channel S5P extracellular linker: role of an amphipathic α-helix in C-type inactivation. J. Biol. Chem. 278:42136–42148. [DOI] [PubMed] [Google Scholar]
- 17.Doyle, D. A., J. M. Cabral, R. A. Pfuetzner, A. Kuo, J. M. Gulbis, S. L. Cohen, B. T. Chait, and R. MacKinnon. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77. [DOI] [PubMed] [Google Scholar]
- 18.Jiang, Y., A. Lee, J. Chen, M. Cadene, B. T. Chait, and R. MacKinnon. 2002. The open pore conformation of potassium channels. Nature. 417:523–526. [DOI] [PubMed] [Google Scholar]
- 19.Kuo, A., J. M. Gulbis, J. F. Antcliff, T. Rahman, E. D. Lowe, J. Zimmer, J. Cuthbertson, F. M. Ashcroft, T. Ezaki, and D. A. Doyle. 2003. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science. 300:1922–1926. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, Y., A. Lee, J. Chen, V. Ruta, M. Cadene, B. T. Chait, and R. MacKinnon. 2003. X-ray structure of a voltage-dependent K+ channel. Nature. 423:33–41. [DOI] [PubMed] [Google Scholar]
- 21.Long, S. B., E. B. Campbell, and R. MacKinnon. 2005. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 309:897–903. [DOI] [PubMed] [Google Scholar]
- 22.Hidalgo, P., and R. MacKinnon. 1995. Revealing the architecture of a K+ channel pore through mutant cycles with a peptide inhibitor. Science. 268:307–310. [DOI] [PubMed] [Google Scholar]
- 23.Gross, A., and R. MacKinnon. 1996. Agitoxin footprinting the Shaker potassium channel pore. Neuron. 16:399–406. [DOI] [PubMed] [Google Scholar]
- 24.Imredy, J. P., and R. MacKinnon. 2000. Energetic and structural interactions between δ-dendrotoxin and a voltage-gated potassium channel. J. Mol. Biol. 296:1283–1294. [DOI] [PubMed] [Google Scholar]
- 25.Ranganathan, R., J. H. Lewis, and R. MacKinnon. 1996. Spatial localization of the K+ channel selectivity filter by mutant cycle-based structure analysis. Neuron. 16:131–139. [DOI] [PubMed] [Google Scholar]
- 26.Miller, C. 1995. The charybdotoxin family of K+ channel-blocking peptides. Neuron. 15:5–10. [DOI] [PubMed] [Google Scholar]
- 27.Tytgat, J., K. G. Chandy, M. L. Garcia, G. A. Gutman, M.-F. Martin-Eauclaire, J. J. van der Walt, and L. D. Possani. 1999. A unified nomenclature for short-chain peptides isolated from scorpion venoms: α-KTx molecular subfamilies. Trends Pharmacol. Sci. 20:444–447. [DOI] [PubMed] [Google Scholar]
- 28.Zhang, M., Y. V. Korolkova, J. Liu, M. Jiang, E. V. Grishin, and G.-N. Tseng. 2003. BeKm-1, an HERG-blocking scorpion toxin, shares the global structure with charybdotoxin but the mechanism of action with ErgTx. Biophys. J. 84:3022–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milnes, J. T., C. E. Dempsey, J. M. Ridley, O. Crociani, A. Arcangeli, J. C. Hancox, and H. J. Witchel. 2003. Preferential closed channel blockade of HERG potassium currents by chemically synthesized BeKm-1 scorpion toxin. FEBS Lett. 547:20–26. [DOI] [PubMed] [Google Scholar]
- 30.Yao, J.-A., X. Du, D. Lu, R. L. Baker, E. Daharsh, and P. Atterson. 2006. Estimation of potency of HERG channel blockers: impact of voltage protocol and temperature. J. Pharmacol. Toxicol. Methods. 52:146–153. [DOI] [PubMed] [Google Scholar]
- 31.Korolkova, Y. V., S. A. Kozlov, A. V. Lipkin, K. A. Pluzhnikov, J. K. Hadley, A. K. Filippov, D. A. Brown, K. Angelo, D. Strobaek, T. Jespersen, S.-P. Olesen, B. S. Jensen, and E. V. Grishin. 2001. An ERG channel inhibitor from the Scorpion Buthus eupeus. J. Biol. Chem. 276:9868–9876. [DOI] [PubMed] [Google Scholar]
- 32.Tseng-Crank, J. C. L., G.-N. Tseng, A. Schwartz, and M. A. Tanouye. 1990. Molecular cloning and functional expression of a potassium channel cDNA isolated from a rat cardiac library. FEBS Lett. 268:63–68. [DOI] [PubMed] [Google Scholar]
- 33.Korolkova, Y. V., E. V. Bocharov, K. Angelo, I. V. Maslennikov, O. V. Grinenko, A. V. Lipkin, E. D. Nosireva, K. A. Pluzhnikov, S.-P. Olesen, A. S. Arseniev, and E. V. Grishin. 2002. New binding site on the old molecular scaffold provides selectivity of HERG-specific scorpion toxin BeKm-1. J. Biol. Chem. 277:43104–43109. [DOI] [PubMed] [Google Scholar]
- 34.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shrivastava, I. H., S. R. Durell, and H. R. Guy. 2004. A model of voltage gating developed using the KvAP channel crystal structure. Biophys. J. 87:2255–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durell, S. R., I. H. Shrivastava, and H. R. Guy. 2004. Models of the structure and voltage-gating mechanism of the Shaker K+ channel. Biophys. J. 87:2116–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sali, A., and T. L. Blundell. 1993. Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815. [DOI] [PubMed] [Google Scholar]
- 38.Swanson, E. 1995. PSSHOW version 1.9. University of Washington. Seattle, WA.
- 39.Brooks, B. R., R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan, and M. Karplus. 1983. CHARMM: a program for macromolecular energy minimization and dynamic calculations. J. Comput. Chem. 4:187–217. [Google Scholar]
- 40.Pardo-Lopez, L., M. Zhang, J. Liu, M. Jiang, L. D. Possani, and G.-N. Tseng. 2002. Mapping the binding site of a HERG-specific peptide toxin (ErgTx) to the channel's outer vestibule. J. Biol. Chem. 277:16403–16411. [DOI] [PubMed] [Google Scholar]
- 41.Tseng, G.-N. 2001. IKr: the hERG channel. J. Mol. Cell. Cardiol. 33:835–849. [DOI] [PubMed] [Google Scholar]
- 42.Wang, S., S. Liu, M. J. Morales, H. C. Strauss, and R. L. Rasmusson. 1997. A quantitative analysis of the activation and inactivation kinetics of HERG expressed in Xenopus oocytes. J. Physiol. 502:45–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swartz, K. J., and R. MacKinnon. 1997. Mapping the receptor site for Hanatoxin, a gating modifier of voltage-dependent K+ channels. Neuron. 18:675–682. [DOI] [PubMed] [Google Scholar]
- 44.Winterfield, J. R., and K. J. Swartz. 2000. A hot spot for the interaction of gating modifier toxins with voltage-dependent ion channels. J. Gen. Physiol. 116:637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schreiber, G., and A. R. Fersht. 1995. Energetics of protein-protein interactions: analysis of the barnase-barstar interface by single mutations and double mutant cycles. J. Mol. Biol. 248:478–486. [DOI] [PubMed] [Google Scholar]
- 46.Smith, P. L., T. Baukrowitz, and G. Yellen. 1996. The inward rectification mechanism of the HERG cardiac potassium channel. Nature. 379:833–836. [DOI] [PubMed] [Google Scholar]
- 47.Lopez-Barneo, J., T. Hoshi, S. H. Heinemann, and R. W. Aldrich. 1993. Effects of external cations and mutations in the pore region on C-type inactivation of Shaker potassium channels. Receptors Channels. 1:61–71. [PubMed] [Google Scholar]
- 48.Clarke, C. E., A. P. Hill, J. Zhao, M. Kondo, R. N. Subbiah, T. J. Campbell, and J. I. Vandenberg. 2006. Effect of S5P α-helix charge mutants on inactivation of hERG K+ channels. J. Physiol. 573:291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rost, B., G. Yachdav, and J. Liu. 2003. The PredictProtein server. Nucleic Acids Res. 32:W321–W326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hol, W. G. 1985. Effects of the alpha-helix dipole upon the functioning and structure of proteins and peptides. Adv. Biophys. 19:133–165. [DOI] [PubMed] [Google Scholar]
- 51.Starkus, J. G., L. Kuschel, M. D. Rayner, and S. H. Heinemann. 1997. Ion conduction through C-type inactivated Shaker channels. J. Gen. Physiol. 110:539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, Z., J. C. Hesketh, and D. Fedida. 2000. A high Na+ conduction state during recovery from inactivation in the K+ channel Kv1.5. Biophys. J. 79:2416–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kiss, L., J. LoTurco, and S. J. Korn. 1999. Contribution of the selectivity filter to inactivation in potassium channels. Biophys. J. 76:253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gang, H., and S. Zhang. 2006. Na+ permeation and block of hERG potassium channels. J. Gen. Physiol. 128:55–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guy, H. R., and P. Seetharamulu. 1986. Molecular model of the action potential sodium channel. Proc. Natl. Acad. Sci. USA. 83:508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]