Abstract
Clusterin [CLU, a.k.a. TRPM-2, SGP-2, or ionizing radiation (IR)-induced protein-8 (XIP8)] was implicated in apoptosis, tissue injury, and aging. Its function remains elusive. We reisolated CLU/XIP8 by yeast two-hybrid analyses using as bait the DNA double-strand break repair protein Ku70. We show that a delayed (2–3 days), low-dose (0.02–10 Gy) IR-inducible nuclear CLU/XIP8 protein coimmunoprecipitated and colocalized (by confocal microscopy) in vivo with Ku70/Ku80, a DNA damage sensor and key double-strand break repair protein, in human MCF-7:WS8 breast cancer cells. Overexpression of nuclear CLU/XIP8 or its minimal Ku70 binding domain (120 aa of CLU/XIP8 C terminus) in nonirradiated MCF-7:WS8 cells dramatically reduced cell growth and colony-forming ability concomitant with increased G1 cell cycle checkpoint arrest and increased cell death. Enhanced expression and accumulation of nuclear CLU/XIP8-Ku70/Ku80 complexes appears to be an important cell death signal after IR exposure.
Formation of DNA double-strand breaks (DSBs) correlates well with lethality after ionizing radiation (IR) (1). Because the ability of cancer cells to repair and recover from DSBs appears to be the most critical factor in the radiation responsiveness of a tumor, a complete understanding of DSB repair and signaling events after the creation of these lesions will be required to improve radiotherapy. Damage-inducible proteins that interact with DSB repair proteins in human cells have not been described.
DNA-dependent protein kinase (DNA-PK), a crucial component in nonhomologous DNA end joining repair of DSBs, is a multisubunit complex composed of a DNA-PK catalytic subunit (460 kDa) and the Ku autoantigen. The Ku autoantigen is a nuclear DNA end binding heterodimer composed of 70-kDa (Ku70) and 80-kDa (Ku80) proteins. Recent data suggest that DNA-PK is a DNA damage sensor, which actively recruits other components of the repair machinery to DNA lesions. Once bound to DNA lesions, this complex stimulates DNA repair and signals damage stress responses, facilitating the transcriptional activation of p53 (2) and/or NF-κB (3), which influences cell cycle arrest, apoptois, and carcinogenesis. Ku70/Ku80 may play a role in other basic cellular processes, such as telomere function(s) (4, 5).
The exact role of DNA-PK in DSB repair is still not fully understood. Additional components may include XRCC4 (6), DNA ligase IV (6), and poly(ADP-ribose) polymerase (7). To search for additional repair proteins we performed yeast two-hybrid analyses using human Ku70 as bait and a human liver cDNA library as a source for prey proteins (8).
Materials and Methods
Yeast Two-Hybrid Analyses.
The yeast two-hybrid system was generously provided by S. Elledge (Baylor College of Medicine, Houston, TX). Briefly, full-length human Ku70 cDNA (from W. Reeves, University of North Carolina, Chapel Hill) was subcloned (by PCR) in-frame into the pAS2 vector, forming a hybrid protein with the GAL4 DNA-binding domain (DBD). Cloning was verified by DNA sequencing, and expression of DBD-Ku70 hybrid protein was confirmed by Western immunoblotting using anti-DBD antibody (Santa Cruz Biotechnology). The DBD-Ku70 hybrid protein did not self-activate in recipient Y190 yeast. A yeast two-hybrid screen (using a human liver cDNA library purchased from CLONTECH, catalogue no. HL4024AH) was performed to identify Ku70 binding proteins (KUBs) (8). Positive interacting yeast colonies were grown on medium containing 25 mM 3-AT (3-amino-1,2,4-triazole, Sigma A-8056, freshly prepared), but lacking histidine (HIS3+), then tested for β-galactosidase activity using 5-bromo-4-chloro-3-indolyl β-d-galactoside staining (β-gal+). KUB plasmids were isolated from yeast, and interactions were reconfirmed by cotransfecting each KUB plasmid with pAS2-Ku70 into yeast and testing for HIS3+ and β-gal+. No yeast clones self-activated. Yeast expressing p53, cdk2, lamin B, and SNF-1 were described (8), and yeast expressing Ku70, Ku70 StuI del, and Ku80 proteins were developed by us and confirmed by Western blot analyses.
psort (Prediction of Protein Sorting Signals and Localization Sites in Amino Acid Sequence) Analyses.
The psort program was developed by Kenta Nakai (Osaka University, Japan) (web site: psort.nibb.ac.jp). Analyses of CLU/IR-induced protein-8 (XIP8) amino acids showed (i) an endoplasmic reticulum (ER)-hydrophobic signal peptide (amino acids 1–21); (ii) two simian virus 40 large T antigen-like nuclear localization signals (NLSs), KKKK (amino acids 78–81) and RKKH (amino acids 443–446), and (iii) two Xenopus nucleoplasmin bipartite motif-like NLSs, RK-10-aa spacer-KKKED (amino acids 67–83) and RR-10-aa spacer-RLTRK (amino acids 324–340). An alternative translational AUG site was located at amino acid 34, after the ER signal peptide in human clu/xip8 (Fig. 1); this site is not conserved in other species.
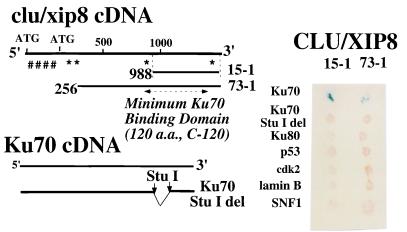
Figure 1.
Human CLU/XIP8 interacts with Ku70 by yeast two-hybrid analyses. (Left) Limited deletion analyses showed that the C terminus of CLU/XIP8 interacted with the C terminus of Ku70. Nineteen separate CLU/XIP8 clones, with 15–1 (C-terminal 120 aa) being the shortest and 73–1 the longest, bound to the C terminus of Ku70. A Ku70 deletion mutant, eliminating 276 bp, was created by StuI digestion (8). The ER-hydrophobic signal is indicated by ####. NLSs are indicated by *. (Right) CLU/XIP8 clones, 15–1 and 73–1, were used to characterize the interaction between CLU/XIP8 and Ku70. Positive interacting yeast colonies were grown on histidine(−) medium and monitored for β-galactosidase activity (blue spots, shown). Ku70 StuI deletion mutant, Ku80, p53, cdk2, lamin B, and SNF-1 did not interact with CLU/XIP8 clones, 15–1 or 73–1.
Antibodies.
A polyclonal antisera (rCY1, clone #62228), directed against a potential nuclear nonglycosylated CLU/XIP8 protein, was generated in rabbits. Briefly, clu/xip8 cDNA was obtained from M. Tenniswood (University of Notre Dame, South Bend, IN). An N-terminally truncated clu/xip8 cDNA, starting from the second start site without the ER signal peptide DNA sequence, was cloned into the pET28a vector (Novagen) by PCR. The protein was induced by isopropyl β-d-thiogalactopyranoside as per the manufacturer's instructions. Bacterial inclusion bodies were collected, and the protein was partially purified by SDS/PAGE and then injected into rabbits. The rCY1 antisera was collected after the third “boost” and used for Western immunoblot analyses at 1:1,500 dilution and immunofluorescence cell staining at 1:10 dilution. Anti-human clusterin (CLU/XIP8) antibody (clone 41D, Upstate Biotechnology, Lake Placid, NY) preferentially detected 60- and 40-kDa CLU/XIP8 forms, but not the nuclear 55-kDa protein. Commercial or private sources of CLU/XIP8 monoclonal or polyclonal antibodies were unable to detect this nuclear protein form, presumably because of the glycosylated state(s) of the CLU/XIP8 protein, with which all other antibodies were generated. A polyclonal antibody to green fluorescent protein (GFP) was obtained from CLONTECH (#8367–1).
Whole-Cell and Nuclear Lysate Preparations.
Growth and IR treatment of mycoplasma-free MCF-7:WS8 (MCF-7) cells was described (9–11). Control or irradiated MCF-7 cells were extracted for whole-cell or nuclear proteins as described (8–11). Protein concentrations were determined by Bradford assays (Bio-Rad), and 10 μg nuclear or 50 μg whole-cell lysates were analyzed by Western blotting.
Coimmunoprecipitation.
Antibodies and protein G-agarose beads (Pierce) were incubated in reaction buffer for 2 h at room temperature with constant rocking to generate an antibody-bead affinity matrix (8). Goat polyclonal antibodies raised against human Ku70 (C19) or Ku80 (C20) (Santa Cruz Biotechnology) were used. Antibody-bound beads were collected by centrifugation (550 × g, 2 min) at 4°C, washed with reaction buffer, and resuspended in the same buffer, and nuclear lysates from irradiated MCF-7 cells were added. Binding reactions were performed overnight at 4°C with constant rocking. Precipitated proteins were collected by centrifugation (550 × g, 2 min) at 4°C and washed three times with reaction buffer. Bound proteins were eluted by boiling for 5 min in SDS/PAGE loading buffer (8) and analyzed by Western blotting (Fig. 3B and ref. 8).
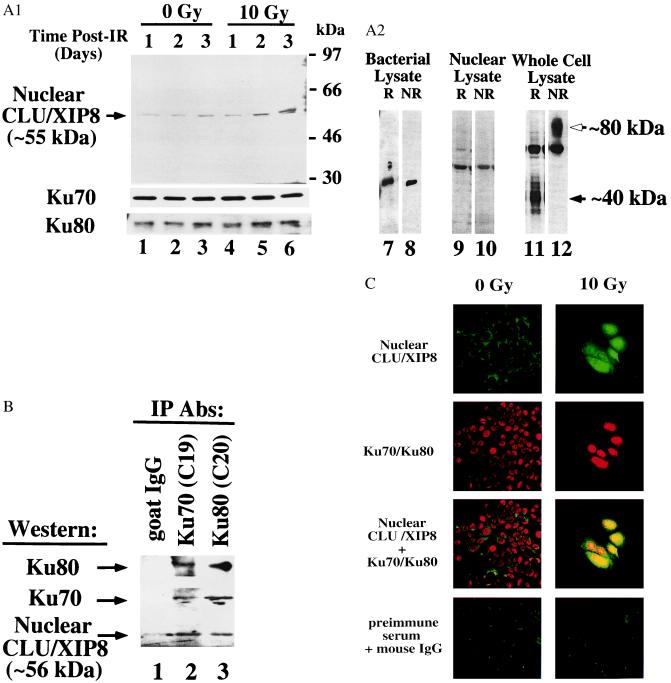
Figure 3.
Analyses of Ku70, Ku80, and CLU/XIP8 in nuclear lysates of MCF-7 cells after IR. (A) Log-phase MCF-7 cells were treated with or without 10 Gy and nuclear lysates prepared at various times (lanes 1–6). Ku70 and Ku80 levels served as loading controls. The 55-kDa form of nuclear CLU/XIP8, Ku70, and Ku80 were detected by Western blot analyses using anti-CLU/XIP8 (rCY1), Ku70 (N3H10), or Ku80 (LL1) antibodies, respectively. CLU/XIP8 produced in bacteria (lanes 7 and 8) or found in IR-exposed MCF-7 nuclei (lanes 9 and 10) was not altered under reducing (R, with β-mercaptoethanol) or nonreducing (NR, without β-mercaptoethanol) conditions. In contrast, secretory CLU/XIP8 protein, detected by the 41D antibody, was converted from its native 80-kDa form (open arrow) to 40-kDa α and β glycopeptides (closed arrow) as indicated (lanes 11 and 12). (B) Nuclear lysates prepared from MCF-7 cells 3 days after 10 Gy were immunoprecipitated by using Ku70 (C19) or Ku80 (C20) antibodies. Coimmunoprecipitates were detected by Western blotting using anti-CLU/XIP8 (rCY1), Ku70 (N3H10), or Ku80 (LL1) antibodies. Goat IgG was used as a negative control. (C) Colocalization of CLU/XIP8 with Ku70/Ku80 in the nuclei of MCF-7 cells treated with IR by confocal microscopy. MCF-7 cells were treated with or without IR (10 Gy) and fixed 3 days post-IR. Cells were doubly stained by using rabbit anti-CLU/XIP8 (rCY1) and mouse anti-Ku70/Ku80 heterodimer (162) antibodies. Secondary antibodies were FITC-anti-rabbit (green) and Texas red-anti-mouse. Colocalization of CLU/XIP8 (green) and Ku70/Ku80 (red) was shown by yellow in “giant” irradiated cells. Untreated MCF-7 cells showed no colocalization of proteins. Digital images were taken at ×400 magnification. Preimmune serum and normal mouse IgG were used as negative controls. Experiments in A–C were repeated three or more times.
Immunocytochemistry and Confocal Microscopy.
MCF-7 cells were grown on 6-well Teflon-coated microscope slides (Erie Scientific, Erie, PA) and irradiated with 10 Gy or left untreated. RPMI medium was changed daily. Three days postirradiation, control or IR-treated cells were fixed in 3.7% formaldehyde at room temperature for 10 min. Fixed cells were permeabilized by methanol at −20°C for 10 min and air-dried. Combinations of primary antibodies (see below) were diluted in antibody dilution buffer (1% BSA/0.005% saponin in PBS, pH 7.2) and incubated with fixed cells at room temperature for 1 h. Dilutions of primary antibodies were as follows: CLU/XIP8 polyclonal antisera (rCY1, 1:10 dilution) was added with a mouse mAb (Ab 162, 1:20), which specifically detected the Ku70/Ku80 heterodimer. Slides then were immersed in washing buffer (0.005% saponin in PBS, pH 7.2) for 1 h (8). Fluorochrome-conjugated secondary antibodies [FITC (green)-anti-rabbit and Texas red-anti-mouse, used at 1:50 (Vector Laboratories)] were incubated with samples at room temperature for 1 h. Slides then were washed for 1 h as above, then mounted with cover slips in Vectashield solution (Vector Laboratories). Preimmune serum and normal mouse IgG (Santa Cruz Biotechnology) were used as negative controls at 1:10 dilution. Confocal microscopy was analyzed by using a Bio-Rad MRC 1024 Confocal Laser Scanning microscope.
Construction of GFP Fusion Proteins, Fluorescence-Activated Cell Sorting (FACS), Cell Growth, and Survival Assays.
Various GFP constructs were generated by using GFP fusion TOPO cloning kits, as per the manufacturer's instructions (Invitrogen). GFP fusion cDNAs then were subcloned into the retroviral vector, pLNCX (CLONTECH), to enhance fusion protein expression. Cloning was confirmed by DNA sequencing. In pLNCX, GFP fusion protein expression was controlled by a cytomegalovirus promoter, and the neomycin (neo) resistance gene was used for selection. Overexpression of GFP fusion proteins was confirmed by Western analyses using a polyclonal anti-GFP antibody (CLONTECH, #8367–1). MCF-7 cells were grown in phenol red free RPMI medium and transiently transfected with GFP-clu/xip8 or GFP-alone cDNA constructs using Effectene (Qiagen, Valencia, CA). Twenty-four hours after transfection, cells overexpressing GFP or various GFP-CLU/XIP8 fusion proteins (at least 30-fold higher fluorescence intensity than nontransfected cells) were isolated by using FACS (Coulter, EPICS ELITE ESP). For cell growth assays, 1,000 sorted cells/well were seeded into 96-well dishes. Cells were incubated for 14 days and relative growth rates were measured (12). Data were normalized to cells overexpressing GFP alone (100%). For cell survival assays, 104 sorted cells/well (as described above) were seeded into 60-mm tissue culture plates. Seven days after seeding, 500 μg/ml G418 (neomycin) was added to select stable transfectants. After 7 days of selection, live colonies were examined by phase and fluorescence microscopy using a GFP filter (Chroma Technology, Brattleboro, VT, #31019). GFP-positive colonies (100 or more) were counted and scored as viable. Survival was determined by the ratio of GFP positive to total colonies. All tissue culture plates were coated with 0.2% gelatin (Sigma, G-1890 in PBS) at least 15 min before seeding to enhance plating efficiency and eliminate micrometastasis.
Flow Cytometry.
Control or IR-treated MCF-7 cells were transiently transfected with GFP constructs. Cells were harvested 24–48 h later, fixed in 1.9% formaldehyde, permeabilized by methanol at −20°C for 1 h, washed in phosphate citric acid buffer (13) for 15 min, and stained with propidium iodide containing 2% BSA and RNase A. Samples were analyzed by flow cytometry (Coulter, EPICS XL-MCC). Changes in cell cycle distribution were recorded at the same time. Cell death was measured as described (9–11). Data were analyzed by modfit lt (Verity Software House, Topsham, ME). Statistical evaluations (P values) were calculated by using the Student's t test.
Results
Ku70 and CLU/XIP8 Interactions.
We isolated CLU/XIP8 as KUB1 and Ku80 as KUB2 in a yeast two-hybrid screen using Ku70 as “bait” (8). Sequences of 19 clu/xip8 (KUB1) cDNA clones indicated that the shortest clone (15–1, also referred to as C-120) contained only 360 bp (≈120 aa) of C terminus and was sufficient for Ku70 binding (Fig. 1). Clu/xip8 did not interact with Ku80, p53, cdk2, lamin B, or SNF-1 (Fig. 1), showing that its association with Ku70 was specific. Deletion of a 276-bp StuI fragment near the Ku7 C terminus eliminated binding of all 19 clu/xip8 cDNA clones (i.e. clones 15–1 and 73–1, Fig. 1), but not its association with Ku80 or other KUBs (8).
X-Ray Induction of clu/xip8 mRNA in MCF-7 Cells.
Clu cDNA was isolated by us as x-ray-induced gene transcript leading to protein 8 (xip8) by using differential hybridization (14). Its steady-state mRNA levels were induced ≈5- to 10-fold, 3 days after as little as 2.5 Gy in MCF-7 cells (Fig. 2A), in agreement with earlier kinetics (14). No further induction of clu/xip8 mRNA levels was noted at higher IR doses (5–20 Gy, Fig. 2A). RNase protection analyses, using a probe that spanned a region between the middle of two AUG start sites in the clu mRNA and extended 100 bases after the second AUG (bases 35–235, between two AUGs in clu/xip8 cDNA, Fig. 1), indicated that a single transcript of ≈1.8 kb was induced in MCF-7 cells after IR (not shown).
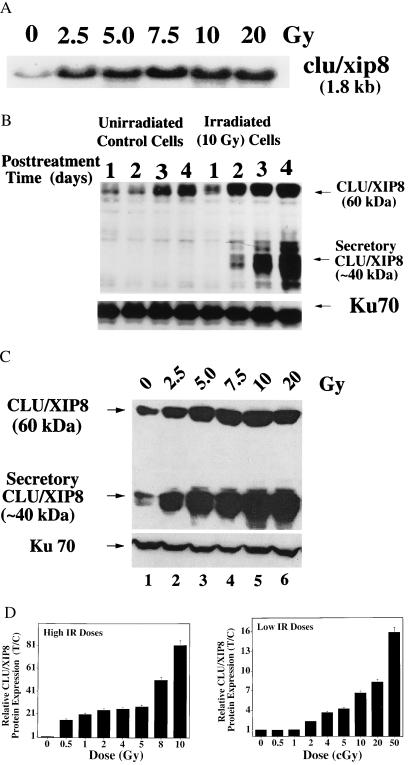
Figure 2.
Induction of CLU/XIP8 after IR. Log-phase MCF-7 cells were treated with various doses of IR. (A) Increased steady-state expression of clu/xip8 transcripts in MCF-7 cells, 3 days after IR. Equivalent total RNA loading was monitored by ethidium bromide staining and 36B4 transcript levels as described (14). (B) Time-course induction of CLU/XIP8 proteins in MCF-7 cells after IR. Log-phase MCF-7 cells were treated with 10 Gy and whole-cell lysates were prepared. CLU/XIP8 proteins (60- and 40-kDa forms) were monitored by Western blot assays and detected by using the 41D antibody. Ku70 protein was detected by the N3H10 antibody and served as a loading control. (C) Increased steady-state levels of the 60-kDa precursor and ≈40-kDa secretory forms of CLU/XIP8 after various doses of IR. Whole-cell lysates from MCF-7 cells were prepared 3 days after IR. Ku70 levels remained unaltered. (D) Dose-response of CLU/XIP8 protein induction in MCF-7 cells after IR. Data in Fig. 3B, as well as other experiments using lower IR doses were compiled. Induction of nuclear CLU/XIP8 protein was not apparent until 1 Gy (not shown). Western blots of CLU/XIP8 compared with Ku70 or α-tubulin protein levels in control or IR-treated MCF-7 cells were quantified by densitometric scans and normalized for loading. Values for treated/control protein levels for each dose of IR then were graphed (mean ± SE). Data represent the results of experiments performed at least three times, each in duplicate.
IR Induction of CLU/XIP8 Protein.
Multiple CLU/XIP8 protein forms are expressed in human cells. This protein was first discovered as an ER-targeted (Fig. 1), secreted glycoprotein (80-kDa intramolecularly cleaved glycopeptide, with two 40-kDa subunits linked by five disulfide bonds) (15, 16). An ≈60-kDa intracellular precursor is uncleaved and less glycosylated (16). The levels of both the ≈60-kDa precursor and 40-kDa secretory forms of CLU/XIP8 were strongly induced in MCF-7 cells at 2–4 days post 10-Gy treatment (Fig. 2B). The protein also was induced by UV irradiation, heat shock, and exposure to phorbol esters as described for its mRNA (14). We also noted that CLU/XIP8 levels increased (control group, Fig. 2B) when cells reached confluence arrest at 4 days, under conditions when significant increases (≈10%) in apoptosis were noted (9). In contrast, Ku70 protein levels were not altered in irradiated or confluent MCF-7 cells at various times (Fig. 2B) or doses (Fig. 2C) posttreatment. Increases in CLU/XIP8 protein levels greatly exceeded its transcriptional up-regulation (Fig. 2A), suggesting that accumulation of all forms of this protein were regulated at both transcriptional and posttranscriptional levels after IR. The lowest dose of IR found to induce CLU/XIP8 (all forms) was 0.02 Gy (2.0 cGy) (Fig. 2D).
Identification of a Nuclear Form of CLU/XIP8.
The 60-kDa precursor form of the CLU/XIP8 protein, but not its secretory form, associated with Ku70 (8). The physiological significance of such an association between a nuclear and secreted protein was unclear, even though both Ku70 (17) and DNA-PK (18) may be located outside the nucleus and near cell membranes.
Analyses of the CLU/XIP8 amino acid sequence using psort revealed several cryptic NLSs downstream from its hydrophobic secretory signal peptide, a leader sequence that normally targets the protein to the ER (see ####, Fig. 1A). Undefined signal transduction processes may induce expression of a nuclear form of the CLU/XIP8 protein from its in-frame, second translational AUG site (19); however, the existence of a nuclear protein was not shown. Expression from this second AUG initiation sequence may allow NLSs within the CLU/XIP8 protein to be exposed. We therefore predicted the IR-inducible expression of a nuclear form of CLU/XIP8 protein, which could associate with nuclear Ku70 or Ku70/Ku80 complexes.
To visualize the nuclear form of this IR-inducible protein, a polyclonal antisera directed against a bacterial-expressed, nonglycosylated CLU/XIP8 protein translated from the second AUG site was generated. This polyclonal antisera (rCY1) detected an IR-inducible 55-kDa protein band in nuclear lysates from irradiated MCF-7 cells (Fig. 3A, lanes 1–6). The antibody weakly detected the ≈60-kDa underglycosylated precursor CLU/XIP8 protein and did not detect the ≈40-kDa heavily glycosylated, secretory polypeptides (not shown). The 55-kDa protein was not detected with preimmune sera from the same rabbit, nor with rCY1 antisera that was preabsorbed with excess membrane-bound CLU/XIP8 antigen. Unlike the secretory forms of CLU/XIP8 protein, the nuclear form was not cleaved at its α/β site (19); the molecular weight of the protein was unaltered under reduced SDS/PAGE conditions (Fig. 3A, lanes 9 and 10), whereas the secretory form was converted from its native 80-kDa form to 40-kDa α and β glycopeptides (Fig. 3A, lanes 11 and 12) (16).
Coimmunoprecipitation of Nuclear CLU/XIP8 with Ku70/Ku80 After IR.
The 55-kDa nuclear form of CLU/XIP8 increased ≈3- to 5-fold in nuclear lysates derived from MCF-7 cells 3 days after 10 Gy. In contrast, nuclear Ku70 and Ku80 protein levels were unaltered (Fig. 3A, lanes 1–6). Furthermore, Ku70, the 55-kDa nuclear form of CLU/XIP8, and Ku80 coimmunoprecipitated from nuclear lysates of irradiated MCF-7 cells using Ku70 (C19) or Ku80 (C20) polyclonal antibodies (Fig. 3B), even though Ku80 did not interact with CLU/XIP8 by yeast two-hybrid analyses (Fig. 1). Goat IgG failed to precipitate Ku70, Ku80, or CLU/XIP8 (Fig. 3B, lane 1). Moreover, these complexes were not detected in nonirradiated cells, nor in IR-exposed cells 24 h posttreatment (not shown), when nuclear CLU/XIP8 protein levels were not expressed (Fig. 3A) or translocated (Fig. 3C). These data strongly suggest that CLU/XIP8 associates with the Ku70/Ku80 heterodimer in vivo by binding to Ku70, forming a trimeric protein complex as well as a CLU/XIP8-Ku70 heterodimer. These complexes form over a matter of days rather than hours. Moreover, Northern blot (Fig. 2A) and RNase protection assays indicated that only one species of clu/xip8 mRNA, (i.e., full length) was induced in MCF-7 cells after IR. We believe that increased expression of nuclear CLU/XIP8 after IR is a result of altered translation/processing from one clu/xip8 mRNA.
Colocalization of Nuclear CLU/XIP8 with Ku70/Ku80 After IR.
Indirect immunofluorescence cell staining confirmed IR-inducible nuclear CLU/XIP8 colocalization with Ku70/Ku80 by using confocal microscopy, rCY1 antisera, and Ab 162, a mAb that recognizes only the Ku70/Ku80 heterodimer (20) (Fig. 3C). Nuclear CLU/XIP8 protein accumulated both within nuclei and around perinuclear membranes of the 2- to 3-fold enlarged, IR (10 Gy)-treated, MCF-7 cells (Fig. 3C); all cells showed this increase in 2–4 days after this supralethal dose of IR. The protein was observed in the cytoplasm of untreated cells and appeared to translocate to perinuclear and nuclear regions of lethally irradiated MCF-7 cells. Increased nuclear CLU/XIP8 protein (green) colocalized (yellow) with nuclear Ku70/Ku80 heterodimer (red), whereas CLU/XIP8 at the perinuclear boundary of irradiated cells failed to colocalize with Ku70/Ku80 (Fig. 3C). Ku70/Ku80 complexes were exclusively nuclear and remained unaltered after IR. Preimmune serum and mouse IgG were used as negative controls (Fig. 3C).
Nuclear CLU/XIP8 Is A Death Signal.
CLU/XIP8 was cloned and identified in various tissues associated with aging (21), neurodegenerative diseases (Alzheimer's disease, scrapie), renal disease (glomerulonephritis), atherosclerosis, and in some cancers (22). The association between Ku70 and CLU/XIP8 may correlate with the pathophysiology of CLU/XIP8-related diseases and provides a useful clinical or environmental marker of IR, renal, or heart injury. The association of clu/xip8 expression with apoptosis (i.e., TRPM-2, a marker for apoptosis) was first made when clu/xip8 was identified and cloned as the major mRNA species induced during regression of the rat ventral prostate after castration (23). The association of this protein with apoptosis was confirmed in many other biological systems (19, 22).
Because the 55-kDa nuclear form of CLU/XIP8 did not increase until 3 days post-IR, it seemed unlikely that this protein played an immediate role in DSB repair, despite its interaction with Ku70. The repair of DSBs occurs within 2 h posttreatment (24, 25), although delayed induction and repair of DSBs have been described. Given the proposed role of CLU/XIP8 as a “marker of apoptosis” and the translocation of nuclear CLU/XIP8 in IR-treated and dying cells (Fig. 3C), we investigated the possibility that the 55-kDa CLU/XIP8 protein may play a role in cell death, important for elimination of severely damaged and/or misrepaired cells. Overexpression of nuclear CLU/XIP8 protein may mimic stress-inducible responses and reduce cell growth or survival.
We transiently transfected MCF-7 cells with various GFP-tagged clu/xip8 constructs (Fig. 4A). Attempts to produce stably transfected or inducible mammalian cell systems with nuclear clu/xip8 failed because of induced cell death. We separated cells overexpressing various GFP-CLU/XIP8 fusion proteins or GFP alone by using fluorescence-activated cell sorting (FACS) and measured changes in cell cycle distribution, growth rates (Fig. 4B), or colony-forming survival assays (Fig. 4C). Cells transiently overexpressing GFP N-terminal-tagged nuclear CLU/XIP8 (G-NC) or GFP N-terminal-tagged minimum Ku70 binding domain (i.e., G-C120, containing the C-terminal 120 aa of CLU/XIP8, same as clone 15–1 in Fig. 1) showed significant cell growth inhibition (60–65%, P < 0.005, Fig. 4B) compared with cells overexpressing GFP (G) alone. GFP C-terminally tagged nuclear CLU/XIP8 (NC-G) resulted in only moderate growth inhibition [≈25% inhibition compared with >60% inhibition for G-NC (P < 0.25) (Fig. 4B)], presumably because of steric hindrance of its interaction with Ku70 in vivo. Cell cycle distribution analyses at 48 h posttransfection revealed a 12% ± 2% increase in G1 cell cycle checkpoint arrested and sub-G0G1 cells after transfection of nonsynchronized, log-phase MCF-7 cells with G-NC or G-C120. In contrast, no alterations in G1 or G2 cell cycle arrest responses were noted after transfection with GFP alone or NC-G.
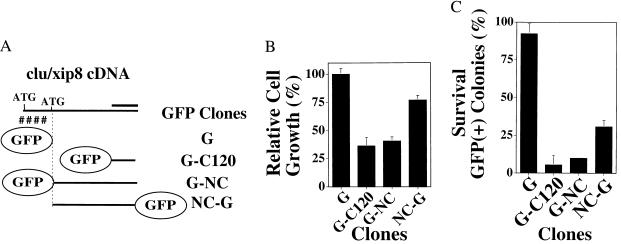
Figure 4.
Nuclear CLU/XIP8 is a cell death signal. (A) Various GFP-CLU/XIP8 fusion constructs were prepared: G, GFP alone; G-C120, 120 aa of the C terminus of CLU/XIP8 (i.e., clone 15–1 in Fig. 1, a Ku70 minimum binding domain) fused to N-terminal GFP; G-NC, nuclear CLU/XIP8, without ER-hydrophobic signal peptide, fused to N-terminal GFP; NC-G, nuclear CLU/XIP8, without ER-hydrophobic signal peptide, containing a C-terminal GFP tag. The thick solid line represents the C120 fragment, containing the CLU/XIP8 minimal Ku70 binding domain (Fig. 1). (B) MCF-7 cells overexpressing various GFP fusion proteins were isolated by FACS sorting and relative growth inhibition (T/C, with GFP transfectants serving as 100% control) monitored as described (9, 10). (C) In separate assays, MCF-7 cells overexpressing GFP fusion proteins and isolated as in B were analyzed for survival. Survival was determined by the ratio of GFP positive versus all colonies. Experiments in B and C were performed three times, each in duplicate. Graphed data represent mean ± SE. Transfection of G-C120 and G-NC resulted in statistically indistinguishable growth inhibition and lethality. Transfection of NC-G resulted in less growth inhibition (P < 0.05) and less lethality (P < 0.02) compared with G-C120 or G-NC. G-NC, but not G, bound Ku70 by yeast two-hybrid analyses.
Consistent with increases in G1 checkpoint responses and cell death, more dramatic effects on cell survival were observed in cells stably overexpressing G-NC or G-C120 (90–95% loss of survival) compared with cells containing GFP alone (0–25% loss of survival compared with nontransfected, sorted cells) (Fig. 4C). Cells stably overexpressing NC-G exhibited an intermediate loss of survival (≈70% lethality) compared with MCF-7 cells transfected with G-NC (92% lethality) (Fig. 4C). More importantly, overexpression of the minimal Ku70-binding domain of CLU/XIP8 (i.e., C120) was sufficient for dramatic loss of DNA content, colony-forming ability, as well as moderate and significant increases in G1 cell cycle arrest responses and cell death. We demonstrated coimmunoprecipitation of nuclear CLU/XIP8 and Ku70 or Ku80 proteins using anti-Ku70, and more importantly anti-Ku80 antibodies; the Ku80 protein did not interact with CLU/XIP8 by yeast two-hybrid assays (Fig. 1). These data support our hypothesis that nuclear CLU/XIP8 plays a role in delayed cell death responses, a cytotoxic response that appears to be minimally triggered by the interaction of the C terminus CLU/XIP8 with the C terminus of Ku70. We previously showed that this interaction of CLU/XIP8 with Ku70 can suppress Ku70/Ku80 heterodimeric-dependent DNA end binding activity (8).
Discussion
We speculate that formation of CLU/XIP8-Ku70/Ku80 complexes, as shown by immunocytochemical staining and confocal microscopy (Fig. 3C), may constitute a signal for killing severely damaged MCF-7 cells. Interestingly, MCF-7 cells did not undergo significant apoptosis after IR exposure (Fig. 3C). We have noted that IR is a very poor inducer of apoptosis in MCF-7 and other epithelial cancer cell lines. IR causes a variety of changes in cells, including irreversible cell cycle checkpoint blockage and delayed apoptosis (26). MCF-7 cells, in particular, lack caspase 3 and 10a (27) that may be required for immediate IR-induced apoptosis. Furthermore, the formation of free radicals may inhibit apoptosis (28), making it possible that proapoptotic and antiapoptotic signals are simultaneously induced by IR. When dissociated from all other responses induced by IR, the single event of overexpressing nuclear apoJ/XIP8 caused growth inhibition, increased G1 cell cycle arrest responses, cell death, and loss of survival. Thus, we were able to evaluate the relative cell death contribution of overexpressed nuclear CLU/XIP8 protein (via GFP-fused transient transfection and sorting) from the spectrum of IR-induced cytotoxic responses. The exact mechanism of cell death is currently under study. The formation of free radicals after IR may interfere with cell death apoptotic “clean-up” responses.
Although we do not yet know whether binding of nuclear CLU/XIP8 to Ku70 is essential for lethality, our data are consistent with this theory. We speculate that enhanced CLU/XIP8 binding to the Ku70/Ku80 heterodimer, along with the reported caspase-mediated cleavage of DNA-PK catalytic subunit (29), may cause loss of nonhomologous DNA repair and prevent unwanted (i.e., error prone) recombinational events in severely damaged cells. The induction of this protein at 24–72 h post-IR exposure clearly indicates that this protein does not influence the immediate repair of DSBs via homologous or nonhomologous recombination. Our data suggest that CLU/XIP8 may play an important role in monitoring cells with genomic instability and/or infidelity, created by translesion DNA synthesis, by facilitating removal of severely damaged and genetically unstable cells. Removal of these cells may occur at different times after treatment with various cytotoxic agents. CLU/XIP8 lethality responses occur after the initial DSB repair is over. Interestingly, delayed programmed cell death responses and simultaneous induction of CLU/XIP8 after IR or other cytotoxic agents occur in both human and rodent cells, 11–15 days post-IR (26). We speculate that IR-induced endoreduplication after G2 arrest responses create damage that induces CLU/XIP-mediated cell death responses. We are examining the radiosensitivity and carcinogenic consequences of elimination of the CLU/XIP8 gene in knockout mice after IR. The recent finding that Ku70, but not Ku80, knockout mice are cancer prone (30) may be consistent with the notion that formation of nuclear CLU/XIP8 with Ku70 may be required for elimination of carcinogenic initiated, severely damaged cells. We currently are elucidating the signal transduction pathways leading to the up-regulation of nuclear CLU/XIP8 expression, as well as the downstream mechanism(s) of cell death after formation of nuclear CLU/XIP8-Ku70/Ku80 complexes. We currently are exploring whether the interaction of nuclear CLU/XIP8 with Ku70 is necessary and sufficient for cell death. Nuclear CLU/XIP8 appears to be an important mediator of cell death after IR. Previous studies showed clu/xip8 mRNA induction after UV irradiation, heat shock, and phorbol ester (TPA) treatments (14). These data strongly suggest that this protein is a general cell death signal, monitoring the overall health of the cell. Determining the signaling pathways that induce this death protein after IR or other antineoplastic agents will be important for understanding its up-regulation in tumor compared with normal tissues. Selective modulation of the levels of nuclear CLU/XIP8 could be important for therapy against breast, prostate, colon, or other cancers. This protein appears to be an important protein for monitoring cell responses to cytotoxic drug or radiation exposures and could be used in a variety of clinical or environmental situations.
Acknowledgments
We thank Dr. S. Yeh (University of Rochester) for her assistance in yeast two-hybrid cloning, Dr. T. Pugh (University of Wisconsin) for guidance with immunocytochemistry, and Drs. J. Petrini (University of Wisconsin), J. K. Harmony (University of Cincinnati) and B. Aronow (University of Cincinnati) for their helpful discussions. This work was supported by Grant ER62723 from the Department of Energy and Grant CA78530 from the National Institutes of Health (National Cancer Institute) to D.A.B.
Abbreviations
- CLU
clusterin
- IR
ionizing radiation
- XIP8
IR-induced protein-8
- DSB
double-strand break
- DNA-PK
DNA-dependent protein kinase
- KUB
Ku70 binding protein
- ER
endoplasmic reticulum
- NLS
nuclear localization signal
- GFP
green fluorescent protein
References
- 1.Hall E J, Astor M, Bedford J, Borek C, Curtis S B, Fry M, Geard C, Hei T, Mitchell J, Oleinick N, et al. Am J Clin Oncol. 1988;11:220–252. doi: 10.1097/00000421-198806000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Woo R A, McLure K G, Lees-Miller S P, Rancourt D E, Lee P W K. Nature (London) 1998;394:700–704. doi: 10.1038/29343. [DOI] [PubMed] [Google Scholar]
- 3.Liu L, Kwak Y-T, Bex F, Garcia-Martinez L F, Li X-H, Meek K, Lane W S, Gaynor R B. Mol Cell Biol. 1998;18:4221–4234. doi: 10.1128/mcb.18.7.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Featherstone C, Jackson S P. Mutat Res. 1999;434:3–15. doi: 10.1016/s0921-8777(99)00006-3. [DOI] [PubMed] [Google Scholar]
- 5.Hsu H L, Gilley D, Blackburn E H, Chen D J. Proc Natl Acad Sci USA. 1999;96:12454–12458. doi: 10.1073/pnas.96.22.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Critchlow S E, Bowater R P, Jackson S P. Curr Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 7.Galande S, Kohwi-Shigematsu T. J Biol Chem. 1999;274:20521–20528. doi: 10.1074/jbc.274.29.20521. [DOI] [PubMed] [Google Scholar]
- 8.Yang C-R, Yeh S, Leskov K, Odegaard E, Hsu H-L, Chang C, Kinsella T J, Chen D J, Boothman D A. Nucleic Acids Res. 1999;27:2165–2174. doi: 10.1093/nar/27.10.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pink J J, Wuerzberger-Davis S M, Tagliarino C, Planchon S M, Yang X-H, Froelich C J, Boothman D A. Exp Cell Res. 2000;255:144–155. doi: 10.1006/excr.1999.4790. [DOI] [PubMed] [Google Scholar]
- 10.Pink J J, Planchon S M, Tagliarino C, Wuerzberger-Davis S M, Varnes M E, Siegel D, Boothman D A. J Biol Chem. 2000;275:5416–5422. doi: 10.1074/jbc.275.8.5416. [DOI] [PubMed] [Google Scholar]
- 11.Wuerzberger S M, Pink J J, Planchon S M, Byers K L, Bornmann W G, Boothman D A. Cancer Res. 1998;58:1876–1885. [PubMed] [Google Scholar]
- 12.Labarca C, Paigen K. Anal Biochem. 1980;102:344–352. doi: 10.1016/0003-2697(80)90165-7. [DOI] [PubMed] [Google Scholar]
- 13.Gong J, Traganos F, Darzynkiewicz Z. Anal Biochem. 1994;218:314–319. doi: 10.1006/abio.1994.1184. [DOI] [PubMed] [Google Scholar]
- 14.Boothman D A, Meyers M, Fukunaga N, Lee S W. Proc Natl Acad Sci USA. 1993;90:7200–7204. doi: 10.1073/pnas.90.15.7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aronow B J, Lund S D, Brown T L, Harmony J A, Witte D P. Proc Natl Acad Sci USA. 1993;90:725–729. doi: 10.1073/pnas.90.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burkey B F, deSilva H V, Harmony J A. J Lipid Res. 1991;32:1039–1048. [PubMed] [Google Scholar]
- 17.Prabhakar B S, Allaway G P, Srinivasappa J, Notkins A L. J Clin Invest. 1990;86:1301–1305. doi: 10.1172/JCI114838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bandyopadhyay D, Mandal M, Adam L, Mendelsohn J, Kumar R. J Biol Chem. 1998;273:1568–1573. doi: 10.1074/jbc.273.3.1568. [DOI] [PubMed] [Google Scholar]
- 19.Wilson M R, Easterbrook-Smith S B, Lakins J, Tenniswood M P R. In: Clusterin: Role in Vertebrate Development, Function, and Adaptation. Harmony J A K, editor. Austin, TX: Landes; 1995. pp. 1–300. [Google Scholar]
- 20.Wang J, Chou C H, Blankson J, Satoh M, Knuth M W, Eisenberg R A, Pisetsky D S, Reeves W H. Mol Biol Rep. 1993;18:15–28. doi: 10.1007/BF01006891. [DOI] [PubMed] [Google Scholar]
- 21.Major D E, Kesslak J P, Cotman C W, Finch C E, Day J R. Neurobiol Aging. 1997;18:523–526. doi: 10.1016/s0197-4580(97)00102-4. [DOI] [PubMed] [Google Scholar]
- 22.Rosenberg M E, Silkensen J. Int J Biochem Cell Biol. 1995;27:633–645. doi: 10.1016/1357-2725(95)00027-m. [DOI] [PubMed] [Google Scholar]
- 23.Russo P, Warner J A, Huryk R, Perez G, Heston W D. Prostate. 1994;24:237–243. doi: 10.1002/pros.2990240504. [DOI] [PubMed] [Google Scholar]
- 24.Hall E J. Radiobiology for the Radiologist. Philadelphia: Lippincott; 1994. [Google Scholar]
- 25.Longo J A, Nevaldine B, Longo S L, Winfield J A, Hahn P J. Radiat Res. 1997;147:35–40. [PubMed] [Google Scholar]
- 26.Boothman D A, Odegaard E, Yang C-R, Hosley K, Mendonca M S. Hum Exp Toxicol. 1998;17:448–453. doi: 10.1177/096032719801700809. [DOI] [PubMed] [Google Scholar]
- 27.Janicke R U, Ng P, Sprengart M L, Porter A G. J Biol Chem. 1998;273:15540–15545. doi: 10.1074/jbc.273.25.15540. [DOI] [PubMed] [Google Scholar]
- 28.Lee Y-J, Shacter E. J Biol Chem. 1999;274:19792–19798. doi: 10.1074/jbc.274.28.19792. [DOI] [PubMed] [Google Scholar]
- 29.Han Z, Malik N, Carter T, Reeves W H, Wyche J H, Hendrickson E A. J Biol Chem. 1996;271:25035–25040. doi: 10.1074/jbc.271.40.25035. [DOI] [PubMed] [Google Scholar]
- 30.Li G C, Ouyang H, Li X, Nagasawa H, Little J B, Chen D J, Ling C C, Fuks Z, Cordon-Cardo C. Mol Cell. 1998;2:1–8. doi: 10.1016/s1097-2765(00)80108-2. [DOI] [PubMed] [Google Scholar]