Abstract
The properties of PSE-2 beta-lactamase have been examined by using two new PSE-2-producing plasmids, pMG33 and pMG74, as well as plasmid R151, found in Pseudomonas aeruginosa. PSE-2 beta-lactamase resembled other PSE enzymes in activity against carbenicillin, but it also resembled OXA enzymes, such as OXA-1, in rapid hydrolysis of oxacillin, cloxacillin, and methicillin and in inhibition by sodium chloride but not by cloxacillin. Antisera that inactivated TEM-1, TEM-2, OXA-1, or PSE-1 and PSE-4 beta-lactamase failed to cross-react with PSE-2, which thus appears to be immunologically distinct. The plasmids determining PSE-2 varied in geographical origin, size, transfer proficiency, and incompatibility specificity, but all determined resistance to carbenicillin, gentamicin, kanamycin, streptomycin, spectinomycin, sulfonamide, and tobramycin. From a pUZ8-R151 recombinant plasmid in Escherichia coli, the PSE-2 beta-lactamase gene could be transposed to a second plasmid in a 6.4-megadalton unit together with resistance to gentamicin, kanamycin, streptomycin, spectinomycin, sulfonamide, and tobramycin. Transposition was recA independent. We propose the designation Tn1404 for this unit, which, like transposons carrying OXA-1, PSE-1, PSE-4, and some transposons determining TEM-1, includes genes for beta-lactam, aminoglycoside, and sulfonamide resistance.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D. E., Rutherford E. L. Basic characterization of a lipid-containing bacteriophage specific for plasmids of the P, N, and W compatibility groups. Can J Microbiol. 1975 Feb;21(2):152–163. doi: 10.1139/m75-023. [DOI] [PubMed] [Google Scholar]
- Bryan L. E., Shahrabadi M. S., van den Elzen H. M. Gentamicin resistance in Pseudomonas aeruginosa: R-factor-mediated resistance. Antimicrob Agents Chemother. 1974 Aug;6(2):191–199. doi: 10.1128/aac.6.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood S. B., Gardella A., Philippon A. M., Jacoby G. A., Moellering R. C., Jr Effects of azlocillin in combination with clavulanic acid, sulbactam, and N-formimidoyl thienamycin against beta-lactamase-producing, carbenicillin-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1982 Aug;22(2):266–271. doi: 10.1128/aac.22.2.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee J. N., Datta N., Hedges R. W. R factors from Proteus rettgeri. J Gen Microbiol. 1972 Oct;72(3):543–552. doi: 10.1099/00221287-72-3-543. [DOI] [PubMed] [Google Scholar]
- DAVIS B. D., MINGIOLI E. S. Mutants of Escherichia coli requiring methionine or vitamin B12. J Bacteriol. 1950 Jul;60(1):17–28. doi: 10.1128/jb.60.1.17-28.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N., Barth P. T. Compatibility properties of R483, a member of the I plasmid complex. J Bacteriol. 1976 Mar;125(3):796–799. doi: 10.1128/jb.125.3.796-799.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J. B., Olsen R. H. Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids pMG1 and pMG5. J Bacteriol. 1978 Jul;135(1):227–238. doi: 10.1128/jb.135.1.227-238.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges R. W., Datta N., Kontomichalou P., Smith J. T. Molecular specificities of R factor-determined beta-lactamases: correlation with plasmid compatibility. J Bacteriol. 1974 Jan;117(1):56–62. doi: 10.1128/jb.117.1.56-62.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
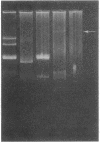
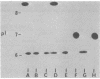
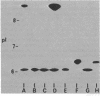
- Hedges R. W., Matthew M. Acquisition by Escherichia coli of plasmid-borne beta-lactamases normally confined to Pseudomonas spp. Plasmid. 1979 Apr;2(2):269–278. doi: 10.1016/0147-619x(79)90045-3. [DOI] [PubMed] [Google Scholar]
- Jacoby G. A., Matthew M. The distribution of beta-lactamase genes on plasmids found in Pseudomonas. Plasmid. 1979 Jan;2(1):41–47. doi: 10.1016/0147-619x(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Jacoby G. A. Properties of R plasmids determining gentamicin resistance by acetylation in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1974 Sep;6(3):239–252. doi: 10.1128/aac.6.3.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A., Sutton L. Restriction-modification systems determined by Pseudomonas plasmids. Plasmid. 1982 Sep;8(2):141–147. doi: 10.1016/0147-619x(82)90052-x. [DOI] [PubMed] [Google Scholar]
- Kabins S., Nathan C., Cohen S. Gentamicin-adenylyltransferase activity as a cause of gentamicin resistance in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1974 Jun;5(6):565–570. doi: 10.1128/aac.5.6.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsu K., Inoue M., Mitsuhashi S. Plasmid-mediated carbencillin hydrolyzing beta-lactamases of Proteus mirabilis. J Antibiot (Tokyo) 1981 Nov;34(11):1504–1505. doi: 10.7164/antibiotics.34.1504. [DOI] [PubMed] [Google Scholar]
- Katsu K., Inoue M., Mitsuhashi S. Transposition of the carbenicillin-hydrolyzing beta-lactamase gene. J Bacteriol. 1982 May;150(2):483–489. doi: 10.1128/jb.150.2.483-489.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak A., Philippon A., Chardon H., Labia R., Le Goffic F. Constantes enzymatiques (Km et Vmax) des beta-lactamases mesurées par une méthode micro-acidimétrique couplée a l'ordinateur. Ann Microbiol (Paris) 1973 Oct;124(3):259–268. [PubMed] [Google Scholar]
- Kopecko D. J., Brevet J., Cohen S. N. Involvement of multiple translocating DNA segments and recombinational hotspots in the structural evolution of bacterial plasmids. J Mol Biol. 1976 Dec;108(2):333–360. doi: 10.1016/s0022-2836(76)80124-6. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Labia R., Barthélémy M. L'enzymogramme des beta-lactamases: adaptation en cel de la méthode iodométrique. Ann Microbiol (Paris) 1979 Oct;130B(3):295–304. [PubMed] [Google Scholar]
- Labia R., Barthélémy M., Masson J. M. Multiplicité des beta lactamases: un probléme d'isoenzymes. C R Acad Sci Hebd Seances Acad Sci D. 1976 Nov 29;283(14):1597–1600. [PubMed] [Google Scholar]
- Labia R., Kazmierczak A., Philippon A., Le Goffic F., Faye J. C., Goldstein F. W., Acar J. F. beta-Lactamases de Pseudomonas aeruginosa et résistance a la carbénicilline. Ann Microbiol (Paris) 1975 May-Jun;126A(4):449–459. [PubMed] [Google Scholar]
- Labia R., Le Goffic F., Faye J. C., Philippon A. Deux céphalosporinases de Pseudomonas aeruginosa. Biochimie. 1974;56(10):1333–1337. [PubMed] [Google Scholar]
- Low B. Formation of merodiploids in matings with a class of Rec- recipient strains of Escherichia coli K12. Proc Natl Acad Sci U S A. 1968 May;60(1):160–167. doi: 10.1073/pnas.60.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew A., Harris A. M., Marshall M. J., Ross G. W. The use of analytical isoelectric focusing for detection and identification of beta-lactamases. J Gen Microbiol. 1975 May;88(1):169–178. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- Matthew M. Plasmid-mediated beta-lactamases of Gram-negative bacteria: properties and distribution. J Antimicrob Chemother. 1979 Jul;5(4):349–358. doi: 10.1093/jac/5.4.349. [DOI] [PubMed] [Google Scholar]
- Medeiros A. A., Hedges R. W., Jacoby G. A. Spread of a "Pseudomonas-specific" beta-lactamase to plasmids of enterobacteria. J Bacteriol. 1982 Feb;149(2):700–707. doi: 10.1128/jb.149.2.700-707.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melling J., Scott G. K. Preparation of gram quantities of a purified R-factor-mediated penicillinase from Escherichia coli strain W3310. Biochem J. 1972 Nov;130(1):55–62. doi: 10.1042/bj1300055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Callaghan C. H., Acred P., Harper P. B., Ryan D. M., Kirby S. M., Harding S. M. GR 20263, a new broad-spectrum cephalosporin with anti-pseudomonal activity. Antimicrob Agents Chemother. 1980 May;17(5):876–883. doi: 10.1128/aac.17.5.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle P. L., Holloway B. W. Relationship between R and FP plasmids in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1980 Mar;17(3):293–297. doi: 10.1128/aac.17.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair M. I., Holloway B. W. A chromosomally located transposon in Pseudomonas aeruginosa. J Bacteriol. 1982 Aug;151(2):569–579. doi: 10.1128/jb.151.2.569-579.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers A. O., Jacoby G. A. Plasmid-determined resistance to tellurium compounds. J Bacteriol. 1977 Jan;129(1):276–281. doi: 10.1128/jb.129.1.276-281.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. M., Grinsted J. Physical and genetic analysis of the Inc-W group plasmids R388, Sa, and R7K. Plasmid. 1982 May;7(3):239–250. doi: 10.1016/0147-619x(82)90005-1. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Tanaka M., Nohara C., Fukunaga Y., Yamagishi S. Transposition of the oxacillin-hydrolyzing penicillinase gene. J Bacteriol. 1981 Feb;145(2):808–813. doi: 10.1128/jb.145.2.808-813.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]