Summary
Translation and mRNA degradation are affected by a key transition where eukaryotic mRNAs exit translation and assemble an mRNP state that accumulates into processing bodies (P bodies), cytoplasmic sites of mRNA degradation containing nontranslating mRNAs, and mRNA degradation machinery. We identify the decapping activators Dhh1p and Pat1p as functioning as translational repressors and facilitators of P body formation. Strains lacking both Dhh1p and Pat1p show strong defects in mRNA decapping and P body formation and are blocked in translational repression. Contrastingly, overexpression of Dhh1p or Pat1p causes translational repression, P body formation, and arrests cell growth. Dhh1p, and its human homolog, RCK/p54, repress translation in vitro, and Dhh1p function is bypassed in vivo by inhibition of translational initiation. These results identify a broadly acting mechanism of translational repression that targets mRNAs for decapping and functions in translational control. We propose this mechanism is competitively balanced with translation, and shifting this balance is an important basis of translational control.
Introduction
A key aspect of the regulation of eukaryotic gene expression is control of mRNA translation and degradation. A major pathway of mRNA decay in eukaryotes is initiated by shortening of the 3′ poly(A) tail (deadenylation), followed by cleavage of the 5′ m7GpppN cap (decapping), and ultimately, 5′ to 3′ exonucleolytic degradation (for review, see Coller and Parker, 2004). Decapping is a critical step in this decay pathway as it permits destruction of the mRNA body and is a site of numerous control inputs. Translation and mRNA decapping are intertwined. For example, decreasing translation initiation by a variety of means increases the rate of decapping. Conversely, inhibition of translation elongation significantly decreases the rate of decapping (Coller and Parker, 2004). Moreover, translation initiation factors such as the cap binding protein eIF-4E can directly inhibit decapping, and thus before an mRNA undergoes decapping, it exits translation by an unknown mechanism wherein translational initiation factors dissociate from the transcript, and an mRNA decapping complex assembles on the mRNA (Schwartz and Parker, 1999, 2000; Tharun and Parker, 2001).
The requirement for exiting translation prior to decapping implies that there is a general, and unknown, process by which the vast majority of mRNAs can exit translation and enter a translationally inactive state. In principle, exiting of mRNAs from translation could simply be a passive process occurring in the absence of ongoing recruitment of the translational apparatus. In this view, translational repression would occur as a default state. Alternatively, and more consistent with mRNA decapping being a regulated process, the cell may possess a general mechanism for actively targeting mRNAs into a translationally repressed state.
Evidence suggesting cells might have a general mechanism for translational repression comes from the observation that mRNAs that have exited translation and can be targeted for decapping, accumulate within discrete cytoplasmic foci, referred to as P bodies (also referred to as GW182 or DCP bodies). P bodies are dynamic RNA-protein aggregates containing untranslated mRNAs, the decapping machinery (Bashkirov et al., 1997; Ingelfinger et al., 2002; Lykke-Andersen, 2002; Van Dijk et al., 2002; Teixeira et al., 2005), and are sites where decapping can occur (Sheth and Parker, 2003; Cougot et al., 2004). This suggests that when mRNAs exit translation, they assemble an mRNP complex containing the decapping machinery, which can then undergo either decapping, or aggregation into P bodies. It is currently unclear whether assembly of individual mRNPs into a larger P body structure directly affects the rate of decapping or plays a different role in control of mRNA function.
An important and unresolved issue is the manner by which mRNAs exit translation and are targeted for decapping and accumulation in P bodies. Moreover, because P bodies increase under conditions that globally repress translation, it is possible that the mechanism that targets mRNAs for decapping and P body accumulation might also play an important role in the control of translation, both globally and on specific mRNAs.
Given the intertwined nature of mRNA decapping and translation, we hypothesized that a subset of the proteins involved in decapping may function by promoting translational repression. Indeed, several observations led us to hypothesize that the decapping activators, Dhh1p, and Pat1p might function in the pathway of moving mRNAs from polysomes and into the translationally inert state that accumulates in P bodies. First, both proteins show genetic interactions with translation initiation factors (Bonnerot et al., 2000; Wyers et al., 2000; Coller et al., 2001). Second, Pat1p coimmunoprecipitates with Pab1p and eIF-4E in an RNA-dependent manner (Tharun and Parker, 2001), suggesting that Pat1p can interact with the transcript while it is still translationally competent. Third, two homologs of Dhh1p, Me31b in Drosophila and the Xenopus protein Xp54, function in translational control of metazoan maternal mRNAs (Ladomery et al., 1997; Minshall et al., 2001; Nakamura et al., 2001). Finally, because strains lacking Dhh1p or Pat1p each show partial defects in mRNA decapping, we hypothesized that these proteins might work independently to repress translation, thereby activating decapping and P body formation.
In this work, we tested this hypothesis by examining the functions of Dhh1p and Pat1p with respect to mRNA decay, P body formation, and translational repression. Our results indicate that Dhh1p and Pat1p function as a general and active mechanism of translational repression, which functions independent of the cap structure and can also repress translation from a broad spectrum of IRES elements. This translational repression machinery targets mRNAs for decapping, facilitates their assembly into P bodies, and appears to be conserved throughout eukaryotes. Importantly, these results indicate that a general translation repression machinery exists within cells that is in constant competition with the translation apparatus and that the balance of this competition provides the basis for translational repression that occurs under numerous biological contexts.
Results
Dhh1p and Pat1p Have Additive Effects on mRNA Decay
As discussed above, we hypothesized that the decapping regulators, Dhh1p, and Pat1p might function independently in the pathway of moving mRNAs from polysomes and into the translationally inert state that accumulates in P bodies. To test this hypothesis, we first examined the phenotypes of dhh1Δ, pat1Δ, and dhh1Δ/pat1Δ double mutants, with respect to mRNA decay, P body formation, and translational repression. In addition, because the Lsm1-7p complex can also promote mRNA decapping, we examined the effects of combining a lsm1Δ, which inactivates the Lsm1-7p complexes function in mRNA decapping, with the pat1Δ and dhh1Δ lesions.
The effect of each mutant background on decapping was determined by using a transcriptional shut-off strategy for the MFA2pG reporter gene, which is under the control of the GAL promoter, thereby allowing repression of transcription by addition of glucose (Muhlrad et al., 1994). In addition, the MFA2pG gene contains a polyguanosine tract (pG) in its 3′UTR that blocks 5′ to 3′ exonuclease digestion and thereby traps an intermediate in mRNA degradation (Muhlrad et al., 1994).
We observed that dhh1Δ/pat1Δ double mutants had a severe block to mRNA decapping, stronger than either dhh1Δ or pat1Δ single mutants. Specifically, following glucose repression of transcription, decay rate of the MFA2pG mRNA is slowed in dhh1Δ (t1/2 = 12′; Figure S1B in the Supplemental Data available with this article online) or pat1Δ (t1/2 = 15′; Figure S1C) compared to wild-type (WT) (t1/2 = 5′; Figure S1A), whereas the dhh1Δ/pat1Δ strain shows an even slower decay rate (t1/2 = 26′; Figure S1D). Accumulation of stable, deadenylated mRNAs in dhh1Δ/pat1Δ indicates a significant defect in mRNA decapping (Figure S1D), consistent with the partial block to decapping observed in dhh1Δ or pat1Δ single mutants (Coller et al., 2001, Fischer and Weis, 2002; Tharun et al., 2000; Bouveret et al., 2000). In contrast, a pat1Δ/lsm1Δ strain behaved similar to a pat1Δ or lsm1Δ strain (Figure S1F). This is consistent with other results arguing that the Lsm1-7p complex and Pat1p work together to promote mRNA decapping (Bouveret et al., 2000). Thus, a dhh1Δ/lsm1Δ strain showed an additive effect on mRNA decay but not as strong as dhh1Δ/pat1Δ (Figure S1G and data not shown). These results suggest that Dhh1p and Pat1p can function independently, but at a similar step, in promoting mRNA decapping.
Dhh1p and Pat1p Have Additive Effects on P Body Formation
P bodies are cytoplasmic foci within which mRNA decapping can occur and presumably reflect formation of an mRNP state that has undergone translational repression and is capable of decapping (Sheth and Parker, 2003; Cougot et al., 2004; Teixeira et al., 2005). Their size and abundance is proportional to the pool of untranslating mRNAs, which can be substrates for the decapping reaction. Therefore, P body formation can be used as a microscopic assay for monitoring the flux of mRNA through different stages of the decapping pathway.
The strong block to mRNA decapping seen in dhh1Δ/pat1Δ could be a result of either an inability to exit translation and thus access the decapping machinery (decreasing P body abundance) or an inability to be processed by the decapping enzyme (increasing P body abundance). To distinguish between these two possibilities, we monitored P body formation in dhh1Δ, pat1Δ, and dhh1Δ/pat1Δ strains under glucose deprivation, which stimulates P body formation (Teixeira et al., 2005). Visualization of P bodies was achieved using a GFP fusion protein of the decapping enzyme Dcp2p (Bashkirov et al., 1997; Ingelfinger et al., 2002; Lykke-Andersen, 2002; Van Dijk et al., 2002; Sheth and Parker, 2003). In WT cells, P bodies are seen in nearly all cells visualized, with 1–4 foci per cell (Figure 1A, panel i). In both dhh1Δ and pat1Δ, the number of P bodies did not change significantly (Figure 1A, panel ii and iii). However, in dhh1Δ/pat1Δ, less than 10% of cells contained P bodies (Figure 1A, panel iv). This is in contrast to a dcp1Δ strain, which lacks a component of the decapping enzyme, thus inhibiting the decapping reaction directly and causing P bodies to accumulate (Sheth and Parker, 2003) (Figure 1A, panel v). These strains all contained similar levels of the Dcp2p-GFP protein, indicating that differences in P body abundance were not due to decreased expression of the fusion protein (Figure S2A). Additionally, changes in P body size and abundance are not reflective of growth rate differences as dhh1Δ/pat1Δ grows similarly to the dcp1Δ strain. Furthermore, similar results were obtained by monitoring P bodies using a GFP fusion to Lsm1p (Figure S3). Together, these results argue that Dhh1p and Pat1p act at an early step in mRNA decay prior to P body formation. Since translational repression is a key step in promoting mRNA decapping (Schwartz and Parker, 1999, 2000) and P bodies contain pools of nontranslating mRNPs (Sheth and Parker, 2003; Teixeira et al., 2005), we hypothesized that Dhh1p and Pat1p might function to repress translation of mRNAs, thereby promoting P body formation.
Figure 1. Dhh1p and Pat1p Have Additive Effects on P Body Formation and Are Required for Translational Repression.

(A) Using Dcp2p-GFP as a marker, P bodies were observed in wild-type (WT), dhh1Δ, pat1Δ, dhh1Δ/pat1Δ, or dcp1Δ strains.
(B) Depicted are polysome profiles (OD260nm traces) of WT, dhh1Δ, pat1Δ, dhh1Δ/pat1Δ, or dcp1Δ before and after glucose deprivation. The nontranslating region (RNP), 80S monosome (80S), and polysomes (P) are indicated above.
(C) Northern of RPL41a mRNA following polysome analysis in WT or dhh1Δ/pat1Δ before and after glucose deprivation.
(D) Incorporation of [35S]methionine in WT and dhh1Δ/pat1Δ cells before and after glucose deprivation. Values are reported as cpm incorporated. Time after label addition indicated.
(E) Polysome profiles (OD260nm traces) of WT and dhh1Δ/pat1Δ strains before and after amino acid deprivation.
Dhh1p or Pat1p Is Required for Translational Repression
To determine if Dhh1p and Pat1p function in translational repression, we examined the cell’s response to glucose deprivation. In yeast, glucose deprivation leads to repression of protein synthesis by an unknown mechanism (Ashe et al., 2000) and increases P body size and abundance (Teixeira et al., 2005). If Dhh1p and Pat1p function to move mRNAs out of polysomes, thereby allowing P body assembly, then dhh1Δ/pat1Δ cells would be unable to repress translation in response to a physiological cue.
WT, dhh1Δ, pat1Δ, and dhh1Δ/pat1Δ strains were grown in the presence of glucose until reaching mid-log phase. Cultures were shifted to media lacking glucose for 10 min and then examined for translational repression by polysome analysis. Consistent with earlier work, glucose depletion led to a rapid loss of polysomes in WT cells (Ashe et al., 2000) (Figure 1B). In a dhh1Δ or pat1Δ mutant, the ability to undergo translational repression is only slightly impaired (Holmes et al., 2004) (Figure 1B). In contrast, translation is no longer repressed in the dhh1Δ/pat1Δ mutant upon glucose deprivation (Figure 1B). Consistent with this global analysis, specific mRNAs are moved from polysomes into the mRNP fraction in WT cells but retained on polysomes in dhh1Δ/pat1Δ following glucose repression (Figure 1C). Moreover, translation rate as measured by [35S]methionine incorporation is not significantly reduced in dhh1Δ/pat1Δ as compared to WT following glucose depletion (Figure 1D). These results demonstrate that either Dhh1p or Pat1p are required for the general translational repression that occurs upon glucose deprivation. Moreover, we observed that dhh1Δ/pat1Δ were unable to repress translation in response to amino acid starvation (Figure 1E). These observations indicate that Dhh1p or Pat1p are required for global translational repression in a variety of biological contexts. A role of Dhh1p and Pat1p in promoting translational repression is consistent with the observation that dhh1Δ/pat1Δ cells have severe defects in both mRNA decapping and P body formation.
Previous results have demonstrated that dcp1Δ, dcp2Δ, and xrn1Δ strains are also defective in translational repression in response to glucose deprivation (Figure 1B) (Holmes et al., 2004). Several observations suggest that this is an indirect effect of sequestering Dhh1p and Pat1p in P bodies. First, unlike dhh1Δ/pat1Δ mutants, dcp1Δ, dcp2Δ, and xrn1Δ cells contain high levels of P bodies, which contain Dhh1p and Pat1p (Sheth and Parker, 2003; Teixeira et al., 2005) (Figure 1A panel iv versus panel v). Second, in contrast to Dhh1p and Pat1p, overexpression of Dcp1p or Dcp2p does not repress mRNA translation (see below). Third, overexpression of either Dhh1p or Pat1p restores translational repression in dcp1Δ, and dcp2Δ strains (see below and data not shown).
Overexpression of Either Dhh1p or Pat1p Drives Translational Repression and P Body Formation
Additional evidence that Dhh1p and Pat1p function to repress translation was obtained from analyzing the effects of overexpression of either Dhh1p or Pat1p from an inducible galactose promoter in WT cells. First, overexpression of Dhh1p or Pat1p, but not Dcp1p, Dcp2p, or Edc3p, inhibits cell growth (Figure 2E). Second, overexpression of either Dhh1p or Pat1p, but not Dcp1p, dramatically reduced polysomes and resulted in a concomitant increase in P body abundance (Figure 2A). Third, overexpression of either Dhh1p or Pat1p reduced the rate of translation as indicated by a decrease in [35S]methionine incorporation (Figure 2B). Finally, overexpression of either Dhh1p or Pat1p caused relocation of the PGK1, RPL41a, COX17, TIF51a, and CYH2 mRNAs from heavy polysomes to the mRNP region of the density gradient (Figure 2C and data not shown), where components of P bodies, such as Dhh1p (Figure 2D), Pat1p, Lsm1p, and Dcp1p are seen to migrate (Bonnerot et al., 2000). These results provide a second line of evidence that Dhh1p and Pat1p function to promote disassociation of mRNA from polysomes and thus facilitate assembly of mRNAs into P bodies. Interestingly, overexpression of either Dhh1p or Pat1p had no effect on the steady-state abundance of mRNA (Figure S4). This suggests that following Dhh1p and/or Pat1p function, there is an additional limiting step after assembly of a P body that is required to dictate mRNA decay.
Figure 2. Overexpression of Dhh1p or Pat1p Causes General Translational Repression and Increases P Bodies.

(A) Polysome profiles and P body accumulation in control cells (vector) or cells overexpressing Dhh1p, Pat1p, or Dcp1p. Times after induction by the addition of galactose are indicated. P bodies were visualized using Dcp2p-GFP.
(B) Incorporation of [35S]methionine in cells overexpressing Dhh1p, Pat1p, or a vector control. Values are reported as cpm incorporated. Time after addition of label indicated.
(C) Northerns of the PGK1 and RPL41a mRNA following polysome analysis, in control cells, or cells overexpressing Dhh1p or Pat1p. Minutes after induction are indicated. Nontranslating (RNP) and translating areas (polysomes) are indicated above.
(D) Western analysis indicating location of Dhh1p on a sucrose gradient.
(E) Growth of yeast cells when Dhh1p, Pat1p, Dcp1p, Dcp2p, or Edc3p are overexpressed. Cells were plated on either glucose media, or galactose.
Dhh1p and Pat1p Stimulate Translational Repression and P Body Formation Independently
To determine the basis for the overexpression lethality and the interrelationship of Dhh1p and Pat1p function, we examined the consequences of overexpression of either protein in strains lacking other components of P bodies. Overexpression of Dhh1p or Pat1p still inhibited growth in strains lacking Dcp1p, Dcp2p, and Lsm1p (Figure 3A). As strains lacking Dcp1p or Dcp2p are blocked for decapping, this indicates that overexpression lethality does not require mRNA decapping. Importantly, we observed that overexpression of Dhh1p in pat1Δ cells or overexpression of Pat1p in dhh1Δ cells still resulted in inhibition of cell growth, decreased polysomes, and increased P body abundance (Figures 3A–3C). These results indicate that Pat1p and Dhh1p stimulate translational repression in yeast independently, consistent with the observations that Dhh1p and Pat1p have additive effects on turnover, P body formation, and translational repression.
Figure 3. Dhh1p and Pat1p Stimulate Translational Repression via Parallel Pathways.

(A) Shown is growth of yeast cells when Dhh1p or Pat1p are overexpressed in strains deleted for components of P bodies. Cells were plated on glucose media or galactose media.
(B) Polysome and P body analysis of cells overexpressing Dhh1p in a pat1Δ strain or (C) Pat1p in a dhh1Δ strain. Nontranslating region (RNP), 80S monosome (80S), and polysomes (P) are indicated above first trace. P bodies were visualized using Dcp2p-GFP. Time after galactose induction is indicated.
Dhh1p and Its Human Homolog RCK/p54 Repress mRNA Translation In Vitro
The above results document that Dhh1p and Pat1p function in vivo to repress translation, thereby allowing them to assemble into the translational inactive state that can accumulate within P bodies. To determine if Dhh1p directly represses translation, we asked if recombinant Dhh1p could repress translation in vitro. In these experiments, we added purified GST-Dhh1p to yeast and rabbit reticulocyte extracts programmed with a luciferase reporter (LUC) in vitro and assayed translation by monitoring LUC activity. We analyzed translation in both yeast and mammalian systems because we anticipated that any function of Dhh1p in repressing translation would be conserved. This hypothesis is supported by the observations that Dhh1p is highly conserved, vertebrate homologs can complement a yeast dhh1Δ strain, and overexpression of the mammalian homolog, RCK/p54, in tissue culture cells represses growth (Tseng-Rogenski et al., 2003; Westmoreland et al., 2003; Akao et al., 1998). We focused on Dhh1p in these experiments because it is more conserved than Pat1p, and recombinant Pat1p has been difficult to purify to date.
Addition of recombinant Dhh1p repressed translation in a dose-dependent manner in both yeast and rabbit reticulocyte extracts (Figure 4A). Importantly, these results were not due to destabilization of the LUC reporter mRNA in either system (Figure 4A and data not shown). Furthermore, by using mutant alleles of Dhh1p, we demonstrated that repression of translation in vitro is similar to Dhh1p function in vivo. For example, the recombinant mutant protein, Dhh1-8p, was unable to stimulate translational repression in either yeast or reticulocyte extracts (Figure 4C). The dhh1-8 allele, which has alanine substitutions of Arg 345 and Gly 346, prevents a required conformational rearrangement in Dhh1p, thus inhibiting Dhh1p’s function in decapping in vivo (Cheng et al., 2005). Importantly, overexpression of Dhh1-8p in vivo is unable to induce translational repression (Figure S5). Similar effects were seen for additional mutant forms of Dhh1p, such as the dhh1-2 allele (D195A, E196A), which is predicted to be unable to hydrolyze ATP, and is null for function in vivo (Cheng et al., 2005 and data not shown). Moreover, inhibition of translation is specific to Dhh1p activity because addition of recombinant Ded1p, an ATP-dependent helicase with similarity to Dhh1p, stimulated rather than repressed translation in both yeast and reticulocyte extracts (Figure 4B).
Figure 4. Dhh1p and Its Human Homolog RCK/p54 Stimulate Translational Repression In Vitro.
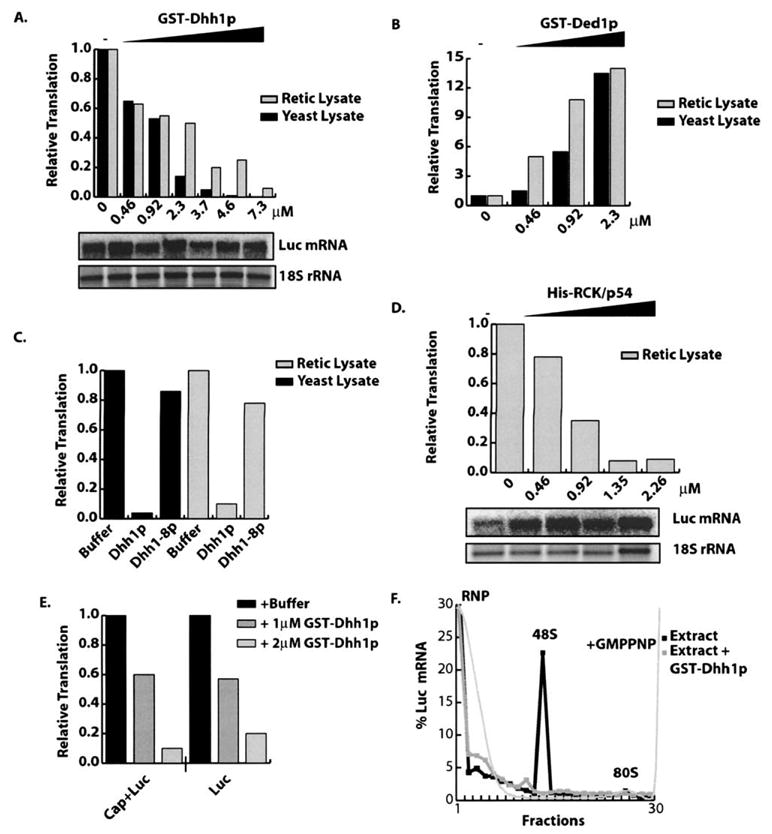
(A) GST-Dhh1p, (B) GST-Ded1p, (C) GST-Dhh1-8p, or (D) His-RCK/p54 was added to either yeast or reticulocyte extracts. LUC activity normalized to translation observed in absence of test protein. Protein concentration is indicated below graph. For (C), 7.3 μM of protein was added. Northern of LUC reporter from extracts containing Dhh1p (A) or RCK/p54 (D).
(E) Graph indicates relative LUC activity of cap plus (Cap+Luc) and cap minus (Luc) LUC reporter in the presence of buffer, 1.0 μM GST-Dhh1p, or 2.0 μM GST-Dhh1p.
(F) Dhh1p inhibited 48S preinitiation complex formation in vitro. Extracts were programmed with radiolabeled reporter and GMPPNP in the presence or absence of 7.3 μM GST-Dhh1p. Graph indicates percent of radiolabeled reporter in each fraction. Position of the RNP, 48S, and 80S complexes are indicated.
The ability of Dhh1p to repress translation in both yeast and mammalian cell extracts suggested that this function would be conserved in its homologs. To test this prediction, we purified the human homolog of Dhh1p, termed RCK/p54, and examined if it could repress translation in vitro. Similar to Dhh1p, recombinant RCK/p54 repressed LUC translation in a dose-dependent manner (Figure 4D). These results demonstrate that Dhh1p can directly repress translation and this function is conserved from yeast to mammals.
Dhh1p Inhibits the Production of a Stable 48S Preinitiation Complex
Recapitulation of Dhh1p-dependent translational repression in vitro facilitated examination of the mechanism by which Dhh1p affects translation. Translational initiation can be divided into three broad steps. First, eIF-4F recognizes and binds the 5′-cap structure. Second, eIF-4F, with eIF3, recruits 43S preinitiation complex (consisting of the 40S ribosome, eIF2, GTP, and Met-tRNAi), leading to formation of a 48S preinitiation complex. Third, 48S complex recognizes the AUG, at which point GTP is hydrolyzed and the 60S ribosomal subunit joins, initiating polypeptide synthesis.
It has previously been proposed that Dhh1p might act to dissociate eIF-4E from the 5′-cap structure and thereby promote decapping (Fischer and Weis, 2002). Such a mode of function could also explain Dhh1p’s inhibition of translation. If Dhh1p acts to dissociate eIF-4E from the cap structure than Dhh1p would be predicted to have no effect on the translation of uncapped mRNAs. However, we observed that both Dhh1p and RCK/p54 were equally effective at repressing the translation of both capped and uncapped mRNAs (Figure 4E and data not shown). This argues that Dhh1p inhibits a step in translation independent of cap recognition.
To determine if Dhh1p affects a step upstream of formation of 48S preinitiation complex, translation extracts were programmed with radiolabeled mRNA with or without Dhh1p and in the presence of various translational inhibitors. Use of substeps specific inhibitors allows translational initiation to be studied by trapping dynamic intermediates (Gray and Hentze, 1994). Accumulation of initiation complexes was monitored by sucrose density gradient sedimentation and quantification of labeled mRNA following fractionation. In WT cells, addition of a nonhydrolysable GTP analog (GMPPNP) blocks 60S subunit joining causing accumulation of 48S preinitiation complex (Figure 4F) (Gray and Hentze, 1994). Strikingly, addition of purified Dhh1p eliminated accumulation of 48S preinitiation complex (Figure 4F). This result suggests that Dhh1p represses translation in vitro by inhibiting the ability of 48S complex to form on mRNA. These effects may be a direct consequence of Dhh1p acting upstream of 48S formation, acting on the 48S complex itself, or via promotion of assembly of a repression complex that is independent of translational initiation factors.
Broad Spectrum Inhibition of mRNA Translation by Dhh1p and RCK/p54
To determine if Dhh1p or RCK/p54 affect a specific step in translation initiation or were broadly acting general translational repressors, we examined their effect on translational initiation driven by various internal ribosomal entry sites (IRES). IRES elements allow translational initiation in a manner independent of subsets of translation initiation factors (Figure 5A). This allows translation inhibitors that act on discrete translational initiation factors to have their site of action identified (Ostareck et al., 2001). We focused on RCK/p54 for these experiments as IRES elements function more efficiently in mammalian systems and used IRES elements from the encephalomycarditis (EMCV), hepatitis C (HCV), and cricket paralysis (CrPV) viruses. Strikingly, we observed that RCK/p54 inhibited translation from all three IRES elements including CrPV IRES (Figure 5B), which is known stimulate translation independent of all initiation factors by binding directly to 40S and 60S ribosomal subunits (Wilson et al., 2000). Similar results were seen for purified Dhh1p in reticulocyte extracts (data not shown). Repression was specific to RCK/p54, as purified Ded1p did not inhibit translation from IRES constructs (data not shown). These results suggest RCK/p54 and Dhh1p mediate translational repression by acting directly on the mRNA itself or by impairing 40S ribosome function. More importantly, these results demonstrate that RCK/p54 and Dhh1p are general translational repressors.
Figure 5. General Translational Inhibition by RCK/p54.

LUC activity of various IRES-containing mRNAs in rabbit reticulocyte lysate with 1 μM, 2 μM, or no recombinant RCK/p54. Activity is normalized to translation in absence of RCK/p54. Identity of each construct is indicated below graph, and corresponds to (A).
Translational Initiation Is Required for Dhh1p Effects In Vivo
If Dhh1p is a translational repressor, then we predicted that blocking translation by a strong stem-loop in the 5′UTR would bypass Dhh1p function in vivo (Muhlrad et al., 1995). We observed that the PGK1 mRNA bearing a stem-loop in the 5′UTR (SL-PGK1) decays rapidly in wild-type cells (t1/2 = 5.4′; Figure 6A). Importantly, decay of the SL-PGK1 was not affected in dhh1Δ strains (t1/2 = 4.8′; Figure 6D). Similar results were seen with other nontranslating substrates such as the MFA2 gene with a 5′UTR stem-loop (data not shown). Since inhibition of translation in cis abrogates need for Dhh1p in turnover, this provides an additional line of evidence that Dhh1p functions as a translational repressor and is not simply required for assembly of the decapping complex per se. In contrast, we observed decreased decay rates of the SL-PGK1 mRNA in lsm1Δ or pat1Δ (t1/2 = 15′ and 12′; Figures 6B and 6C). These data demonstrate that Dhh1p and Pat1p function in distinct manners with Pat1p having an additional role in mRNA decay separate from inhibition of translation. Because Dhh1p and Pat1p show numerous physical interactions with the decapping enzyme (Coller et al., 2001; Tharun and Parker, 2001; Bonnerot et al., 2000), Pat1p’s second role in mRNA turnover may be in helping to recruit the decapping enzyme to mRNA. For Dhh1p, however, recruitment of decapping enzyme by physical interactions cannot be rate limiting for decapping to occur, as WT decapping rates are seen for SL-PGK1 in dhh1Δ mutants.
Figure 6. Dhh1p Function Requires Translational Initiation In Vivo.
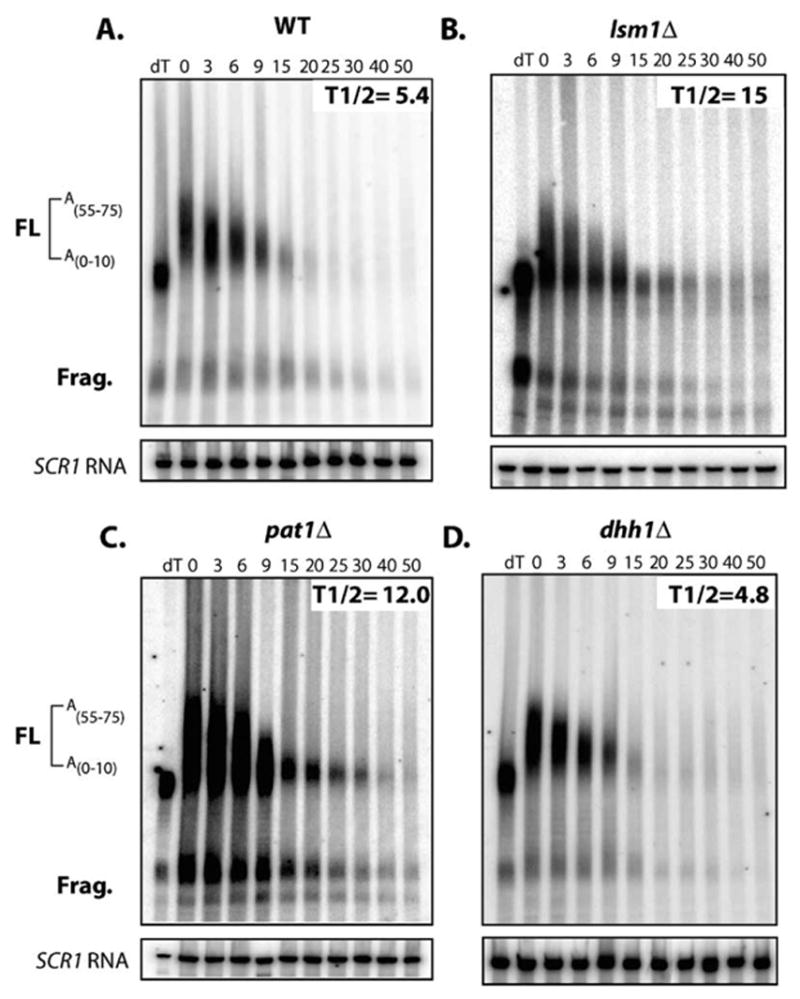
Decay of SL-PGK1 reporter in (A) WT, (B) lsm1Δ, (C) pat1Δ, and (D) dhh1Δ strains. Time points are indicated above each lane. Half-lives are indicated in minutes. Diagram indicates position of the full-length mRNA and polyguanosine decay intermediate. Poly(A) tail lengths are determined by comparing mRNA size to a oligo dT/RNaseH treated control (dT).
Discussion
Regulators of mRNA Decapping Are Translational Repressors
Previous work has documented a reciprocal relationship between mRNA translation and accumulation of transcripts within P bodies, their decapping, and 5′ to 3′ degradation. For example, mutations decreasing translational initiation cause both an increase in decapping rate and the production of P bodies (Schwartz and Parker, 1999, 2000; Teixeira et al., 2005). Conversely, mutations or conditions stabilizing mRNAs within the polysome pool leads to a decrease in mRNA decapping and P body accumulation (Sheth and Parker, 2003; Teixeira et al., 2005). An unresolved issue has been to determine which proteins are involved in facilitating movement of mRNAs from polysomes into a translationally repressed mRNP that is capable of decapping and/or accumulation in P bodies.
Several observations now indicate that Dhh1p and Pat1p are critical proteins that function in repressing translation of mRNAs and facilitating formation of P bodies. First, while strains lacking either Dhh1p or Pat1p show reductions in decapping rates and the ability to repress translation in response to glucose deprivation (Bonnerot et al., 2000; Coller et al., 2001; Fischer and Weis, 2002; Holmes et al., 2004), strains lacking both proteins are severely blocked for decapping, P body formation, and the ability to repress translation (Figures 1 and 2). Second, overexpression of either Pat1p or Dhh1p inhibits growth, decreases polysomes, and drives P body accumulation (Figures 3A, 3B, and 3D). Third, Dhh1p and its human homolog RCK/p54 function as translation repressors in vitro (Figure 4D). Fourth, inhibiting translational initiation in cis bypasses Dhh1p function in vivo (Figure 6). These results document that Dhh1p and Pat1p, which had been characterized as activators of the mRNA decapping reaction, function at least in part to repress translation and facilitate the transition in mRNP organization that leads to mRNAs being subject to decapping. One anticipates that other activators of decapping will also function by inhibiting translation. For example, mRNAs targeted for decapping by nonsense-mediated decay are also translationally repressed in a manner dependent upon Upf1p (Muhlrad and Parker, 1999), which suggests that Upf1p will act as a repressor of translation in some manner.
There are two possible mechanisms by which Dhh1p might inhibit translation. In the simplest model, Dhh1p might promote the assembly of a translation repression complex, which sequesters the mRNA into an mRNP that is unable to be accessed by translation initiation factors. Thus, translation repression occurs by sequestration of the mRNA away from the translation machinery. Consistent with this hypothesis. Dhh1p accumulates within P bodies in conjunction with translationally repressed mRNAs (Teixeira et al., 2005). Because Dhh1p is a member of the DEAD box family of “RNA-helicases,” Dhh1p could utilizes the energy of ATP hydrolysis, coupled with RNA binding, to create an irreversible step in assembly that commits an mRNA to the translationally repressed state. Such an mRNP rearrangement may facilitate further association and aggregation of the repression complex, thereby creating a cascade of events that sequester the mRNA into a quiescent mRNP particle, such as a P body. Alternatively, Dhh1p may interact with and inhibit the function of a required translation factor. However, if Dhh1p works in this manner, its inhibition of 48S complex formation on capped mRNAs and repression of translation from the CrPV IRES, which bypasses the need for all initiation factors, implies that it targets the 40S ribosomal subunit directly rather than a specific initiation factor per se.
Several observations suggest the role of Dhh1p and Pat1p in translational repression will be conserved in eukaryotes cells, including mammalian. First, Dhh1p is highly conserved protein and is 66% identical and 81% similar between yeast and humans. Second, the human homolog RCK/p54 represses translation in vitro similar to Dhh1p (Figure 4D). Third, homologs of Dhh1p are required for translation repression events in Drosophila, Xenopus, and C. elegans (Ladomery et al., 1997, Minshall et al., 2001; Nakamura et al., 2001). Similarly, Pat1p homologs in metazoans include the Xenopus protein, P100 (Figure S6), which is also likely to participate in the storage of maternal mRNAs (Murray et al., 1991; Rother et al., 1992). Fourth, RCK/p54 accumulates within P bodies in mammalian cells (Cougot et al., 2004), and RCK/p54 knockdowns with siRNA reduce P body formation (Andrei et al., 2005). Interestingly, inappropriate regulation of RCK/p54 expression has been implicated in many types of tumors and in precancerous conditions, such as hepatitis C infection (Nakagawa et al., 1999; Hashimoto et al., 2001; Miyaji et al., 2003). A clear implication of this conservation of function is that these alterations in mammalian cell physiology associated with RCK/p54 are related to its role in controlling translation and mRNA decay.
A General and Active Machinery for Translational Repression
Several observations indicate that translational repression promoted by Dhh1p and Pat1p targets the majority of cytoplasmic mRNA and functions in translational control independent of mRNA degradation, both in yeast and in other eukaryotes. First, decapping is the predominant mRNA decay pathway in yeast, and all normal mRNAs examined to date are affected by Dhh1p and Pat1p (for review, see Coller and Parker, 2004). Second, Dhh1p and/or Pat1p are required for global translational repression in response to glucose deprivation or amino acid starvation, wherein the vast majority of mRNAs undergo translational repression (Figures 2B and 2E). Third, overexpression of Dhh1p and Pat1p leads to a dramatic decrease in global translation (Figures 2A–2C). Fourth, RCK/p54 represses translation of all mRNAs tested in vitro indicating the effects are not specific to the cap, poly(A) tail, or a specific initiation step (Figures 4D and 5B). Thus, these results identify a general mechanism of translational repression that can actively inhibit the translation of the majority of cytoplasmic mRNAs. Importantly, because Dhh1p and Pat1p are required for the ongoing process of mRNA turnover in normally growing cells, this repression mechanism is constantly functioning and in competition with translation.
The presence of this active repression machinery suggests that the status of translation, both globally and on specific mRNAs, is due to the constant competition between the translational machinery and the repression machinery (Figure 7). For example, in the absence of Dhh1p and Pat1p, global translation is only marginally impaired by glucose deprivation or amino acid starvation. This argues that the decrease in translation caused by these stresses is only sufficient to repress translation when there is a competing mechanism to sequester the mRNAs in a repressed state (Figures 1B and 1E). This suggests that the translation and repression mechanisms are finely balanced, and relatively subtle alterations in function of either may have profound consequences for the distribution of mRNAs between translation and repression.
Figure 7. A General Active Repression Machinery Exists that Is in Competition with Translation.

This competition creates a finely balanced system setting the relative translation rate for an mRNA. mRNAs can be driven into either translation or repression by tipping the balance of this competition via any number of events.
The generality of Dhh1p and Pat1p translational repression suggests that translation and decay are fundamentally coupled because the mode of translational repression that accesses decapping is a major mode of general translational repression. This provides an explanation for the observation that numerous cis-acting sequences controlling mRNA decay also affect translational repression. For example, the AU-rich destabilizing elements, referred to as AREs, often found in unstable metazoan mRNAs, can serve as translational repression elements in some biological contexts (Otero et al., 2001). Similarly, cis-acting elements in mRNAs important for Drosophila development can modulate both decay and translation (Sonoda and Wharton, 1999). A simple model is that some of these control elements recruit translational repressors, possibly Dhh1p or Pat1p, which then promote mRNAs exiting translation. Because there are clearly additional steps in the decay pathway before decapping actually occurs, the context of the translational repression could lead either to translational repression and mRNA storage or to mRNA degradation.
The competition between assembly of a translation complex and repression suggests the translation rate of individual mRNAs can be consequence of its relative ability to either assemble translation factors or interact with various components of the repression machinery. Thus, an mRNP that effectively recruits the repression complex may strongly shift an mRNA into the repressed state. In addition, several observations suggest mRNA-specific repression complexes might feed into the general repression system we have described. For example, in Drosophila, the Oskar mRNA assembles a tripartite complex wherein eIF-4E is bound to the cap, but prevented from interaction with eIF-4G by the eIF-4E binding protein Cup, which is delivered to the mRNA by an interaction with the sequence specific binding of Bruno to the 3′ UTR (Nakamura et al., 2004). Despite this repression complex, efficient translational repression of the Oskar mRNA during early development requires the Drosophila homolog of Dhh1p, Me31b (Nakamura et al., 2001). Similarly, translation of Xenopus cyclin B1 mRNA is repressed by a tripartite complex of eIF-4E, maskin, which binds eIF-4E, and CPEB, which binds to both the mRNA 3′ UTR and to maskin (Groisman et al., 2000). Yet, CPEB interacts with Xp54, the Xenopus homolog of Dhh1p, which might reinforce this repression (Minshall et al., 2001; Nakahata et al., 2001). Consistent with that view, overexpression of CPEB in mammalian cells increases P bodies, and CPEB accumulates within the P body foci (Wilczynska et al., 2005). More recently, the demonstration that mRNAs translationally repressed by miRNAs accumulate in P bodies suggests that miRNA based translational repression might also interact with this general translation repression machinery (Liu et al., 2005). These interactions suggest that mRNA-specific translational repression mechanisms might create an initial translationally repressed mRNP, which then either directly or indirectly interacts with the general translational repression machinery to create a multilayered and robust system of translational control.
Conclusion
The general system of translational repression described here provides a mechanistic understanding into the interwoven nature of translation and mRNA decay. Since movement of an mRNA into translational repression stimulates assembly of the decapping complex, shifting the balance toward assembly of the translational repression complex will, in many cases, facilitate removal of the mRNA from the cytoplasmic pool by decapping and thus maintain a linear movement of mRNA from translation to repression to decay. In cases where decapping is inhibited, either globally or in a mRNA-specific manner, movement between translation and repression might occur interchangeably and presumably in a regulated fashion to control protein production.
Importantly, the presence of a general active mechanism of translational repression implies the translation status of a cytoplasmic mRNA is the consequence of a constant competition between the translational apparatus and the repression apparatus. This is analogous to regulation of gene expression by alterations in chromatin states, where shifting DNA from chromatin to heterochromatin impacts transcription by driving and maintaining active and inactive states. Analogously, the general translational repression machinery we describe creates an inactive mRNP state, with formation of a P body being the ultimate manifestation of this event. The key switch that stimulates the shift from translation to repression could be any number of events, including attenuation of the translational machinery, deadenylation, activation of the repression machinery, or specific recruitment of the repression complex. While the repression machinery may be ultimately what tips the scale toward inactivation, it may be sequestration of mRNA into a P body that, much like sequestration of DNA into heterochromatin, maintains the repression event, thereby limiting reentry of mRNA into translation.
Experimental Procedures
Yeast Strains, Plasmids, and Oligonucleotides
See Table S1.
RNA Analysis
Performed as in Coller et al. (2001). Cells with pRP485 (Figure S1) or pRP543 (Figure 6), were grown in 2% gal, 1% suc media, harvested, and resuspended in 2% dex media. Following transcriptional repression, aliquots were taken and RNA analyzed by PAGE and Northern. For Figures 1C and 2C, RNA was analyzed by Northern blotting following polysome separation.
Polysome Analysis
Performed as described in Teixeira et al. (2005). Cyclohexamide was omitted as it alters decapping and P bodies (Sheth and Parker, 2003). In Figures 1B and 1E, cells were grown in 2% dex media, harvested, and resuspended in either media plus or minus dex (Figure 2B), or plus or minus amino acids (Figure 2E) and incubated for 10 min. In Figures 2A, 3B, and 3C, cells were grown in 2% suc media, harvested, and resuspended in 2% suc or 2% gal media. Cultures were incubated for 120 min. and then harvested. Figure 4F cells were grown in 2% dex media, and polysomes were analyzed using 7%–47% sucrose gradients. In Figure 4F, a 5%–25% gradient was used.
In Vivo Labeling
[35S]methionine incorporation was performed as described in Ashe et al. (2000).
Microscopy
For Figures 1A, 2A, 3B, 3C, and S3, P bodies were visualized with pRP1175 (DCP2-GFP) or pRP1176 (LSM1-GFP). In Figure 1A, cells were grown in 2% dex media until OD600 = 0.3. Cells were harvested in media lacking glu and visualized by microscopy. In Figures 2A, 3B, and 3C, cells were grown to OD600 = 0.3 in 2% suc media and resuspended in either 2% suc or 2% gal media and visualized after 120 min.
Western Blot Analysis
Protein analysis was performed as described in Sheth and Parker (2003), using anti-GFP antibody (Covance).
In Vitro Translation Assays
Performed as described in Ostareck et al. (2001). Purified GST-Dhh1p or BSA (in 20 mM MES [pH 6.0], 200mM NaCl, 2mM DTT, and 10% glycerol) was added to nuclease treated yeast extracts made as in Iizuka and Sarnow (1997) or rabbit reticulocyte lysate (Promega). In Figures 4A–4D, 250 ng of capped, poly(A)− LUC mRNA was added to each reaction. Extracts were programmed with 250 ng of capped or uncapped LUC mRNA (Figure 4E) or 250ng of IRES mRNA (Figure 5B). In Figure 4F, 1 μg of radiolabeled, capped, poly(A)− mRNA was used plus 0.5mM GMPPNP. Reactions were assembled and incubated 1 hr at 30°C for retic or 22°C yeast extract experiments. In Figures 4A–4E and 5B, LUC expression was monitored using a LUC enzymatic assay (Promega). In Figure 4F, translation was monitored by sucrose gradient, using a 5%–25% gradient followed by Scintillation counting after fractionation.
Supplementary Material
Acknowledgments
We thank Drs. Eckhard Jankowsky, Quansheng Yang, Yukihiro Akao, Peter Sarnow, and Sunnie Thompson for generous gifts of reagents and Drs. Kristian Baker, Daniela Texeira, Nancy Standart, and Carolyn Decker for thoughtful advice and discussion. This work was supported by Howard Hughes Medical Institute (HHMI) and a grant from National Institutes of Health (NIH) GM45443.
Footnotes
Supplemental Data
Supplemental Data include six figures and a table and can be found with this article online at http://www.cell.com/cgi/content/full/122/6/875/DC1/.
References
- Akao Y, Mizoguchi H, Ohishi N, Yagi K. Growth inhibition by overexpression of human DEAD box protein rck/p54 in cells of a guinea pig cell line. FEBS Lett. 1998;429:279–283. doi: 10.1016/s0014-5793(98)00605-x. [DOI] [PubMed] [Google Scholar]
- Andrei M, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Luhrmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe M, De Long S, Sachs A. Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirov V, Scherthan H, Solinger J, Buerstedde J, Heyer W. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnerot C, Boeck R, Lapeyre B. The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol Cell Biol. 2000;16:5939–5946. doi: 10.1128/mcb.20.16.5939-5946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret E, Rigaut G, Shevchenko A, Wilm M, Séraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Coller J, Parker R, Song H. Crystal structure of the DEAD box helicase, Dhh1p. RNA. 2005;8:1258–1270. doi: 10.1261/rna.2920905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- Coller J, Tucker M, Sheth U, Valencia M, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Weis K. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 2002;21:2788–2797. doi: 10.1093/emboj/21.11.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman I, Huang Y, Mendez R, Cao Q, Theurkauf W, Richter J. CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell. 2000;103:435–447. doi: 10.1016/s0092-8674(00)00135-5. [DOI] [PubMed] [Google Scholar]
- Gray N, Hentze M. Iron regulatory protein prevents binding of the 43S translation pre-initiation complex to ferritin and eALAS mRNA. EMBO J. 1994;13:3882–3891. doi: 10.1002/j.1460-2075.1994.tb06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Nakagawa Y, Morikawa H, Niki M, Egashira Y, Hirata I, Katsu K, Akao Y. Co-overexpression of DEAD box protein rck/p54 and c-myc protein in human colorectal adenomas and the relevance of their expression in cultured cell lines. Carcinogenesis. 2001;22:1965–1970. doi: 10.1093/carcin/22.12.1965. [DOI] [PubMed] [Google Scholar]
- Holmes L, Campbell S, De Long S, Sachs A, Ashe M. Loss of translational control in yeast compromised for the major mRNA decay pathway. Mol Cell Biol. 2004;24:2998–3010. doi: 10.1128/MCB.24.7.2998-3010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka N, Sarnow P. Translation-competent extracts from Saccharomyces cerevisiae, effects of L-A RNA, 5′ cap, and 3′ poly(A) tail on translational efficiency of mRNAs. Methods. 1997;11:353–360. doi: 10.1006/meth.1996.0433. [DOI] [PubMed] [Google Scholar]
- Ingelfinger D, Arndt-Jovin D, Luhrmann R, Achsel T. The human LSm1–7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Ladomery M, Wade E, Sommerville J. Xp54, the Xenopus homologue of human RNA helicase p54, is an integral component of stored mRNP particles in oocytes. Nucleic Acids Res. 1997;25:965–973. doi: 10.1093/nar/25.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez M, Hannon G, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall N, Thom G, Standart N. A conserved role of a DEAD box helicase in mRNA masking. RNA. 2001;12:1728–1742. doi: 10.1017/s135583820101158x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaji K, Nakagawa Y, Matsumoto K, Yoshida H, Morikawa H, Hongou Y, Arisaka Y, Kojima H, Inoue T, Hirata I, et al. Overexpression of a DEAD box/RNA helicase protein, rck/p54, in human hepatocytes from patients with hepatitis C virus-related chronic hepatitis and its implication in hepatocellular carcinogenesis. J Viral Hepat. 2003;10:241–248. doi: 10.1046/j.1365-2893.2003.00447.x. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. Recognition of yeast mRNAs as “nonsense containing” leads to both inhibition of mRNA translation and mRNA degradation: implications for the control of mRNA decapping. Mol Biol Cell. 1999;10:3971–3978. doi: 10.1091/mbc.10.11.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Decker C, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Decker C, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M, Krohne G, Franke W. Different forms of soluble cytoplasmic mRNA binding proteins and particles in Xenopus laevis oocytes and embryos. J Cell Biol. 1991;112:1–11. doi: 10.1083/jcb.112.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa Y, Morikawa H, Hirata I, Shiozaki M, Matsumoto A, Maemura K, Nishikawa T, Niki M, Tanigawa N, Ikegami M, et al. Overexpression of rck/p54, a DEAD box protein, in human colorectal tumours. Br J Cancer. 1999;80:914–917. doi: 10.1038/sj.bjc.6690441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata S, Katsu Y, Mita K, Inoue K, Nagahama Y, Yamashita M. Biochemical identification of Xenopus Pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a Nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element-binding protein. J Biol Chem. 2001;276:20945–20953. doi: 10.1074/jbc.M010528200. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Amikura R, Hanyu K, Kobayashi S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development. 2001;128:3233–3242. doi: 10.1242/dev.128.17.3233. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila Cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- Otero L, Devaux A, Standart N. A 250-nucleotide UA-rich element in the 3′ untranslated region of Xenopus laevis Vg1 mRNA represses translation both in vivo and in vitro. RNA. 2001;7:1753–1767. [PMC free article] [PubMed] [Google Scholar]
- Ostareck D, Ostareck-Lederer A, Shatsky I, Hentze M. Lipoxygenase mRNA silencing in erythroid differentiation: The 3′UTR regulatory complex controls 60S ribosomal subunit joining. Cell. 2001;104:281–290. doi: 10.1016/s0092-8674(01)00212-4. [DOI] [PubMed] [Google Scholar]
- Rother R, Frank M, Thomas P. Purification, primary structure, bacterial expression and subcellular distribution of an oocyte-specific protein in Xenopus. Eur J Biochem. 1992;206:673–683. doi: 10.1111/j.1432-1033.1992.tb16973.x. [DOI] [PubMed] [Google Scholar]
- Schwartz D, Parker R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D, Parker R. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol Cell Biol. 2000;20:7933–7942. doi: 10.1128/mcb.20.21.7933-7942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Wharton R. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 1999;13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain non-translating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S, He W, Mayes A, Lennertz P, Beggs J, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- Tharun S, Parker R. Targeting an mRNA for decapping, displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- Tseng-Rogenski S, Chong J, Thomas C, Enomoto S, Berman J, Chang T. Functional conservation of Dhh1p, a cytoplasmic DExD/H-box protein present in large complexes. Nucleic Acids Res. 2003;31:4995–5002. doi: 10.1093/nar/gkg712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Séraphin B. Human Dcp2, a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland T, Olson J, Saito W, Huper G, Marks J, Bennett C. Dhh1 regulates the G1/S-checkpoint following DNA damage or BRCA1 expression in yeast. J Surg Res. 2003;113:62–73. doi: 10.1016/s0022-4804(03)00155-0. [DOI] [PubMed] [Google Scholar]
- Wilczynska A, Aigueperse C, Kress M, Dautry F, Weil D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J Cell Sci. 2005;118:981–992. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- Wilson J, Pestova T, Hellen C, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- Wyers F, Minet M, Dufour M, Vo L, Lacroute F. Deletion of the PAT1 gene affects translation initiation and suppresses a PAB1 gene deletion in yeast. Mol Cell Biol. 2000;20:3538–3549. doi: 10.1128/mcb.20.10.3538-3549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


