Abstract
Anopheles funestus, together with Anopheles gambiae, is responsible for most malaria transmission in sub-Saharan Africa, but little is known about molecular aspects of its biology. To investigate the salivary repertoire of this mosquito, we randomly sequenced 916 clones from a salivary-gland cDNA library from adult female F1 offspring of field-caught An. funestus. Thirty-three protein sequences, mostly full-length transcripts, are predicted to be secreted salivary proteins. We additionally describe 25 full-length housekeeping-associated transcripts. In accumulating mosquito sialotranscriptome information—which includes An. gambiae, Anopheles stephensi, Anopheles darlingi, Aedes aegypti, Aedes albopictus, Culex pipiens quinquefasciatus, and now An. funestus—a pattern is emerging. First, ubiquitous protein families are recruited for a salivary role, such as members of the antigen-5 family and enzymes of nucleotide and carbohydrate catabolism. Second, a group of protein families exclusive to blood-feeding Nematocera includes the abundantly expressed D7 proteins also found in sand flies and Culicoides. A third group of proteins, only found in Culicidae, includes the 30-kDa allergen family and several mucins. Finally, ten protein and peptide families, five of them multigenic, are exclusive to anophelines. Among these proteins may reside good epidemiological markers to measure human exposure to anopheline species such as An. funestus and An. gambiae.
Keywords: Malaria, Hematophagy, Salivary glands, Vector, Saliva
1. Introduction
A highly anthropophagic mosquito, Anopheles funestus, together with Anopheles gambiae, accounts for most malaria transmission in sub-Saharan Africa. While An. gambiae larval habitats are transient bodies of water created by rain, An. funestus larvæ develop in permanent water pools; for this reason, An. funestus is the vector primarily responsible for malaria transmission in the dry season. Both mosquito species belong to the subgenus Cellia and are thus closely related (Coetzee and Fontenille, 2004).
The salivary glands of the adult female mosquito serve a dual function, assisting both blood and sugar feeding. Sugary solutions are delivered to the crop, a cuticle-lined, inert organ from which food is slowly delivered to the anterior midgut, while blood meals are delivered directly to the midgut. Only adult females take blood meals, and this is reflected in their much enlarged salivary glands, which have distinct regions where glycosidases and lysozyme (which prevent bacterial growth in the sugar meal) reside along with anticlotting and other products assisting the blood meal (Rossignol and Lueders, 1986; Marinotti et al., 1990; Rodriguez and Hernandez-Hernandez, 2004).
In their adaptation to blood feeding, mosquitoes and other blood sucking animals have evolved a salivary cocktail of pharmacologically active molecules that disarm host hemostasis (platelet aggregation, blood clotting, and vasoconstriction) and several arms of the inflammatory response (Ribeiro, 1995; Ribeiro and Francischetti, 2003). Within the anophelines, this salivary cocktail has been exposed through transcriptome analysis of the adult female salivary glands. In An. gambiae, where a more extensive sialotranscriptome analysis has been completed (Arcà et al., 2005), a catalogue of ~70 possibly secreted salivary proteins has been identified assisting both sugar and blood meals. Sialotranscriptome analysis of An. stephensi (Valenzuela et al., 2003) and An. darlingi (Calvo et al., 2004) has been conducted before. In this work, we describe the results from the analysis of 916 expressed sequence tags (EST) generated from random sequencing of a salivary gland cDNA library made from adult An. funestus originating from Mali.
2. Materials and methods
2.1. Mosquitoes
Adult female An. funestus were caught in Niono (irrigated area of Mali, 1415N 00600W) and sent to the Malaria Research and Training Center of the University of Bamako. The captured females were maintained in an insectary under standard conditions for oviposition and the F1 progeny used for salivary glands extraction.
Salivary glands (50 pair) were dissected and placed into a solution of 75% RNA-Later (Ambion Inc.), 25% 1× PBS (RNAse free) and stored in 100% RNA-Later at −20°C for isolating polyA+ RNA at the Laboratory of Malaria and Vector Biology (NIAID/NIH).
2.2. Library construction
An. funestus salivary gland mRNA was isolated from 50 salivary-gland pairs from adult females using the Micro-FastTrack mRNA isolation kit (Invitrogen). The polymerase chain reaction (PCR)-based cDNA library was made following the instructions for the SMART cDNA library construction kit (Clontech). Salivary gland polyA+ RNA was used for reverse transcription to cDNA using PowerScript reverse transcriptase (Clontech), the SMART IV oligonucleotide, and the CDS III/3′ primer (Clontech). The reaction was carried out at 42°C for 1 h. Second-strand synthesis was performed by a long-distance (LD), PCR-based protocol using the 5′ PCR primer and the CDS III/3′ primer as sense and anti-sense primers, respectively. These two primers also create Sfi1 A and B restriction enzyme sites at the end of nascent cDNA. Advantage™ Taq polymerase mix (Clontech) was used to carry out the LD PCR reaction on a GeneAmp® PCR System 9700 (Perkin Elmer Corp.). The PCR conditions were: 95°C for 20 s; 24 cycles of 95°C for 5 s; 68°C for 6 min. A small portion of the cDNA was analyzed on a 1.1% agarose/EtBr (0.1 μg/ml) gel to check for the quality and the range of the cDNA synthesized. Double-stranded cDNA was immediately treated with proteinase K (0.8 μg/ml) at 45°C for 20 min. Proteinase K was removed using a Microcon YM-100 mini-column (100,000 MWCO; Millipore) following the manufacturer’s recommendations.
The clean, double-stranded cDNA was then digested with SfiI restriction enzyme at 50°C for 2 h, followed by size fractionation on a ChromaSpin–400 drip column (Clontech). The profiles of the fractions were checked on a 1.1% agarose/EtBr (0.1 μg/ml), and fractions containing cDNA of more than 400 bp were pooled and concentrated by mini-column as described above. The cDNAs were then ligated into a λ TriplEx2 vector (Clontech), and the resulting ligation mixture was packaged using GigaPack® III Plus packaging extract (Stratagene) according to the manufacturer’s instructions. The packaged library was plated by infecting log-phase XL1-Blue E. coli cells (Clontech). The percentage of recombinant clones was determined by performing a blue-white selection screening on LB/MgSO4 plates containing X-gal/IPTG. Recombinants were also determined by PCR, using vector primers (5′ λ TriplEx2 and 3′ λ TriplEx2 sequencing primers) flanking the inserted cDNA and visualizing the products on a 1.1% agarose/EtBr gel.
2.3. Sequencing of the An. funestus cDNA library
The An. funestus salivary gland cDNA library was plated on LB/MgSO4 plates containing X-gal/IPTG to an average of 250 plaques per 150-mm Petri dish. Recombinant (white) plaques were randomly selected and transferred to 96-well Microtest™ U-bottom plates (BD BioSciences) containing 100 μl of SM buffer (0.1 M NaCl, 0.01 M MgSO4.7 H2O, 0.035 M Tris-HCl [pH 7.5], 0.01% gelatin) per well. The plates were covered and placed on a gyrating shaker for 30 min at room temperature. The phage suspension was either immediately used for PCR or stored at 4°C for future use.
To amplify the cDNA using PCR, 4 μl of the phage sample was used as a template. The primers were sequences from the λ TriplEx2 vector and named pTEx2 5seq (5′-TCC GAG ATC TGG ACG AGC-3′) and pTEx2 3LD (5′-ATA CGA CTC ACT ATA GGG CGA ATT GGC-3′), positioned at the 5′ and 3′ end of the cDNA insert, respectively. The reaction was carried out in 96-well flexible PCR plates (Fisher Scientific) using Platinum SuperMix (Invitrogen) on a GeneAmp® PCR system 9700. The PCR conditions were: 1 hold at 95°C for 3 min; 25 cycles of 95°C for 1 min, 61°C for 30 sec; 72°C for 2 min. Amplified products were analyzed on a 1.5% agarose/EtBr gel. cDNA library clones (1100 clones) were PCR amplified; those showing a single band were selected for sequencing. Approximately 200–250 ng of each PCR product was used for DNA sequencing. cDNA sequencing was carried out using a BigDye® Terminator v3.1 cycle sequencing kit (Applied Biosystems), and reaction products were analyzed on an ABI 3730xl DNA analyzer (Applied Biosystems). A total of 1,017 cDNA library clones were sequenced, of which 916 were used in this work.
2.4. Bioinformatic tools and procedures used
EST were trimmed of primer and vector sequences, clusterized, and compared with other databases as previously described (Valenzuela et al., 2003). For functional annotation of the transcripts, we used the program blastx (Altschul et al., 1997) to compare nucleotide sequences to the nonredundant (NR) protein database of the National Center for Biotechnology Information (NCBI) and to the gene ontology database (Ashburner et al., 2000). The tool rpsblast (Schaffer et al., 2001) was used to search for conserved protein domains in the Pfam (Bateman et al., 2000), Smart (Letunic et al., 2002), Kog (Tatusov et al., 2003) and conserved domains databases (Marchler-Bauer et al., 2002). We also compared the transcripts with other subsets of mitochondrial and rRNA nucleotide sequences downloaded from NCBI, and to several organism proteomes downloaded from the NCBI (yeast), Flybase (Drosophila melanogaster), or ENSEMBL (An. gambiae). All blast comparisons were done with the complexity filter off, but segments of polymonucleotides of 20 bases were masked. All six frame translations were used in the case of blastx or rpsblast. To identify possible transcripts coding for secreted proteins, segments of the three-frame translations of all EST’s (because the libraries are unidirectional, we did not use six-frame translations) starting with a methionine found in the first 100 predicted amino acids (aa), or the predicted protein translation in the case of complete coding sequences, were submitted to the SignalP server (Nielsen et al., 1997) to help identify translation products that could be secreted. O-glycosylation sites on the proteins were predicted with the program NetOGlyc (http://www.cbs.dtu.dk/services/NetOGlyc/) (Hansen et al., 1998). Functional annotation of the transcripts was based on all the comparisons above. Following inspection of all these results, transcripts were classified as either Secretory (S), Housekeeping (H), or of Unknown (U) function, with further subdivisions based on function and/or protein families. Sequence alignments were performed with the ClustalX (Thompson et al., 1997) software package. Phylogenetic analysis and statistical neighbor-joining bootstrap tests of the phylogenies were carried out with the Mega package (Kumar et al., 2004). The programs hmmbuild and hmmcalibrate of the hmmer package (version 2.0) (Eddy, 1998) were used to make a hidden Markov model from clustal alignments, and the program hmmsearch was used to search the NR protein database of the NCBI for matches.
3. Results and Discussion
3.1. cDNA library characteristics
A total of 1,017 clones were sequenced from which 916 (having less than 5% N and larger than 100 bp) were used to assemble a clusterized database (Supplemental Table S1), yielding 390 clusters of related sequences, 319 of which contained only one EST. The consensus sequence of each cluster is named either a contig (deriving from two or more sequences) or a singleton (deriving from a single sequence); in this paper, for simplicity sake, we will use the denomination cluster to address sequences deriving both from consensus sequences and from singletons. The 390 clusters were compared by blastx, blastn, or rpsblast (Altschul et al., 1997) to several databases and to the SignalP server. The EST assembly, BLAST, and signal peptide results were transferred into an Excel spreadsheet for manual annotation.
Four categories of expressed genes derived from the manual annotation of the contigs (Table 1). The S category contained 13.1% of the clusters and 54.4% of the sequences, with an average number of 9.8 sequences per cluster. The H category had 28.7% and 19.1% of the clusters and sequences, respectively, and an average of 1.6 sequences per cluster. Fifty-seven percent of the clusters, containing 26% of all sequences, were classified U because no assignment for their function could be made; they had an average of 1.0 sequence per cluster. A good proportion of these transcripts could have derived from truncated 3′ or 5′ untranslated regions of genes of the above two categories, as was recently indicated for a sialotranscriptome of An. gambiae (Arcà et al., 2005). Indeed, the average length of the U class was 344 bp, while the average length for the S and H class were 710 and 630, respectively. Probable transposable elements originated six singletons representing either active transposition or, more likely, expression of transposable element regulatory transcripts in An. funestus. Notably, three of the transcripts code for antisense transcripts of reverse polymerase enzymes. To the extent that our transcripts are unidirectional, these transcripts may indicate that transposition suppression may occur by antisense elimination of transposase messages. Transposable element transcripts have been a regular finding in most sialotranscriptomes to date.
Table 1.
Types of transcripts found in adult female Anopheles funestus salivary glands
| Types of transcripts | Clusters | Sequences | Sequences/cluster | % of clusters | % of sequences |
|---|---|---|---|---|---|
| Secreted (S) | 49 | 496 | 10.1 | 12.6 | 54.1 |
| Housekeeping (H) | 113 | 176 | 1.6 | 29.0 | 19.2 |
| Unknown (U) | 222 | 238 | 1.1 | 56.9 | 26.0 |
| Transposable elements (TE) | 6 | 6 | 1.0 | 1.5 | 0.7 |
| Total | 390 | 916 |
3.2. H genes
The 117 clusters (comprising 188 EST) attributed to H genes expressed in the salivary glands of An. funestus were further characterized into 14 subgroups according to function (Table 2). As observed in previous sialotranscriptomes (Francischetti et al., 2002; Ribeiro et al., 2004a,b), the two larger sets were associated with protein synthesis machinery (76 EST in 31 clusters) and with energy metabolism (18 clusters containing 30 EST). We have also included in the H category a group of 16 singleton EST that code for conserved proteins of unknown function presumably associated with cellular metabolism. This group includes a protein previously identified in the sialotranscriptome of An. gambiae, possibly acquired in mosquito genomes by lateral transfer from a prokaryote, and identified as a salivary membrane protein associated with Plasmodium invasion of the gland (Arcà et al., 2005; Korochkina et al., 2006). ESTs coding for proteins associated with signal transduction, protein modification, and protein export machineries were also abundant. Transporters were also identified, including those coding for two subunits of the V-ATPase complex, which was found necessary for mosquito salivation (Novak and Rowley, 1994; Novak et al., 1995). Transcripts coding for V-ATPases are a common finding in mosquito sialomes (Francischetti et al., 2002; Valenzuela et al., 2002b, 2003; Calvo et al., 2004; Ribeiro et al., 2004b). Additional inspection of each cluster for further information can be done online with Supplemental Table S1.
Table 2.
Functional classification of the housekeeping genes expressed in adult female Anopheles funestus salivary glands
| Types of transcripts | Clusters | Sequences | Sequences/cluster | % of all clusters | % of all sequences |
|---|---|---|---|---|---|
| Protein synthesis machinery | 31 | 76 | 2.5 | 27.4 | 43.2 |
| Metabolism, energy | 20 | 32 | 1.6 | 17.7 | 18.2 |
| Conserved | 14 | 14 | 1.0 | 12.4 | 8.0 |
| Signal transduction | 10 | 12 | 1.2 | 8.8 | 6.8 |
| Transporters | 8 | 9 | 1.1 | 7.1 | 5.1 |
| Protein modification | 6 | 7 | 1.2 | 5.3 | 4.0 |
| Protein export machinery | 6 | 7 | 1.2 | 5.3 | 4.0 |
| Cytoskeletal | 5 | 5 | 1.0 | 4.4 | 2.8 |
| Proteasome machinery | 4 | 5 | 1.3 | 3.5 | 2.8 |
| Transcription factors | 2 | 2 | 1.0 | 1.8 | 1.1 |
| Metabolism, carbohydrate | 2 | 2 | 1.0 | 1.8 | 1.1 |
| Nuclear regulation | 2 | 2 | 1.0 | 1.8 | 1.1 |
| Transcription machinery | 1 | 1 | 1.0 | 0.9 | 0.6 |
| Metabolism, oxidant | 1 | 1 | 1.0 | 0.9 | 0.6 |
| Metabolism, amino acid | 1 | 1 | 1.0 | 0.9 | 0.6 |
| Total | 113 | 176 |
3.3. S proteins and peptides
Inspection of Supplemental Table S1 indicates the presence of several gene families previously described in the salivary glands of mosquitoes, including the relatively abundantly expressed D7, gSG6, gSG2, 30-kDa allergen, antigen 5 (AG5), and SG1 families, as well as enzymes associated with sugar (maltase) and blood (apyrase, adenosine deaminase) feeding. A summary of these transcripts organized by their abundance of protein family is shown in Table 3.
Table 3.
Classification of transcripts coding for putative secreted proteins in adult female Anopheles funestus salivary glands
| Types of transcripts | Number of clusters | Number of sequences | % of all S-type sequences |
|---|---|---|---|
| D7 | 6 | 72 | 14.5 |
| gSG6 family | 1 | 59 | 11.9 |
| Others | 8 | 53 | 10.7 |
| gSG2 family | 3 | 53 | 10.7 |
| 30-kDa allergen | 2 | 48 | 9.7 |
| gSG7 family | 2 | 43 | 8.7 |
| Mucins | 5 | 41 | 8.3 |
| Anophelin | 1 | 34 | 6.9 |
| Antigen 5 | 1 | 31 | 6.3 |
| SG1/Trio family | 9 | 20 | 4.0 |
| Maltase | 1 | 17 | 3.4 |
| Apyrase | 2 | 10 | 2.0 |
| Adenosine deaminase | 1 | 5 | 1.0 |
| Immunity related | 3 | 3 | 0.6 |
| 37.3-kDa family | 1 | 3 | 0.6 |
| Phospholipase A2 | 1 | 2 | 0.4 |
| Peroxidase | 1 | 1 | 0.2 |
| Serpin | 1 | 1 | 0.2 |
| Total | 49 | 496 |
3.4. Analysis of the adult female An. funestus sialotranscriptome
Several clusters of sequences coding for H and putative S polypeptides indicated in Supplemental Table S1 are abundant and complete enough to extract consensus sequences of novel sequences. Additionally, we have performed primer extension studies in several clones to obtain full- or near full-length sequences of products of interest. A total of 58 novel sequences, 33 of which code for S proteins, are grouped together in Supplemental Table S2.
A detailed description of the transcripts found in the salivary glands of adult An. funestus follows.
3.4.1. Secreted proteins belonging to ubiquitous protein families
AG5 family
This is a family of secreted proteins that belong to the CAP family (cysteine-rich secretory proteins; AG5 proteins of insects; pathogenesis-related protein 1 of plants) (Megraw et al., 1998). The CAP family is related to venom allergens in social wasps and ants (Hoffman, 1993; King and Spangfort, 2000) and to antifungal proteins in plants (Stintzi et al., 1993; Szyperski et al., 1998). Members of this protein family are found in the salivary glands of many blood-sucking insects and ticks (Li et al., 2001; Francischetti et al., 2002; Valenzuela et al., 2002b). In An. gambiae, four such proteins were identified in sialotranscriptomes, but only one (putative gVAG protein precursor) had coding transcripts enriched in the adult female salivary glands (Arcà et al., 2005). The An. funestus orthologue of the gVAG protein precursor (84% sequence identity) was represented with 31 EST in the sialotranscriptome. The An. stephensi homologue was 85% identical. The function of any AG5 protein in the saliva of any blood-sucking arthropod is still unknown.
Enzymes
Supplemental Table S2 presents partial coding sequences (truncated in the 5′ region) for the enzymes maltase, apyrase, 5′ nucleotidase, and adenosine deaminase. These enzymes are ubiquitously found in the salivary gland of mosquitoes, where they assist in sugar feeding (maltase) or in degradation of purinergic mediators of platelet aggregation and inflammation.
3.4.2. Secreted protein families found exclusively in Diptera
D7 protein family
The D7 proteins belong to the superfamily of odorant-binding proteins (Hekmat-Scafe et al., 2000), but are peculiar to the salivary glands of blood-sucking Nematocera, including mosquitoes, sand flies, and Culicoides (Arcà et al., 2002; Valenzuela et al., 2002a; Campbell et al., 2005). Short (~17 kDa) and long (~30 kDa) forms are recognized, short forms being found only in mosquitoes. Some of these proteins have been associated with binding of biogenic amines such as serotonin, histamine, and norepinephrine (Calvo et al., 2006). Additionally, one short D7 protein from An. stephensi, named hamadarin, was shown to prevent kallikrein activation by Factor XIIa (Isawa et al., 2002). In An. gambiae, five short and three long D7 proteins were shown to occur as tandem duplications in chromosome 3R. In An. funestus, we presently describe one long and four short D7 proteins (Supplemental Table S2). Overall, the An. funestus D7 proteins vary between 64% and 75% identity with their An. gambiae closest match. Phylogenetic analysis of the known anopheline D7 sequences shows strong bootstrap support for the separation between long and short proteins (Fig. 1). Notice also that the short D7 proteins fall into three robust clades, those containing the An. gambiae D7-r1 and D7-r4, D7-r2 and D7-r3, and D7r5. D7r5 is poorly transcribed in An. gambiae (Arcà et al., 2005), and homologues are not described in any other sialotranscriptome, probably because of its transcript rarity. This cladogram indicates that D7r1 and D7r4 probably arose from a single gene duplication event, as did D7r2 and D7r3. The four short An. funestus proteins co-cluster with the orthologous An. gambiae proteins, something that is not observed for the three short forms of An. stephensi proteins included in the cladogram. The inner branches of the long D7 clade, however, do not give strong bootstrap support for the observed clusters. The D7 family of genes is highly represented in the sialotranscriptome of An. funestus, with a total of 72 transcripts or 14.5% of the EST of the S class (Table 3).
Fig. 1.
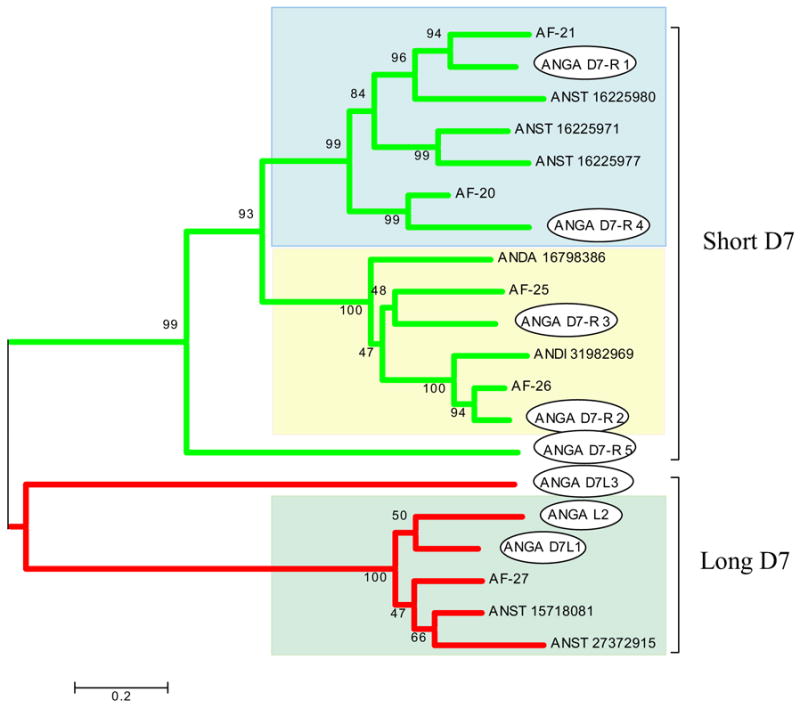
Phylogram of the salivary D7 proteins of anopheline mosquitoes showing the short and long D7 clades. An. gambiae proteins are marked with an oval box. An. gambiae sequences (starting with ANGA) originate from the annotation given by Arcà et al. (2005) and can be accessed online here. An. funestus sequences from this work can be recognized by the AF prefix. An. stephensi, An. dirus, and An. darlingi sequences can be recognized by the four letters originating from the genus and species name and by NCBI accession numbers. The numbers in the phylogram nodes indicate percent bootstrap support for the phylogeny. The bar at the bottom indicates 20% amino acid divergence in the sequences. The three different background shades indicate the three robust clades in the family.
Other Diptera-specific families
The full-length sequence of a peptide containing Gly-Gly-Tyr repeats is described. AFC-383 is similar to a previously reported salivary Ae. aegypti peptide and one An. gambiae sequence and less so to D. melanogaster predicted peptides. It is rich in repeats previously found in worm antimicrobial peptides and may assist the antimicrobial function of saliva. AFC-151 is 96% identical to a previously reported peptide in the salivary glands of An. gambiae containing Drosophila retinin domain. Its function is unknown.
3.4.3. Secreted protein families found exclusively in mosquitoes
30-kDa antigen family
Transcripts coding for members of this acidic protein family, first identified as the 30-kDa Aedes allergen (Simons and Peng, 2001) and also named GE-rich protein (Valenzuela et al., 2003), were found in all previously described transcriptomes of both culicine and anopheline mosquitoes. Only one gene is known in An. gambiae, the expression of which is enriched in salivary glands of adult females. The An. funestus homologue is also abundantly expressed in the sialotranscriptome, with 48 transcripts (9.6% of the S-class EST). It shares 63% identity with the An. gambiae orthologue. The function of this protein family is still unknown.
Mucins
Supplemental Table S2 presents two full-length and one partial coding sequences for three proteins containing >20–58 Ser/Thr galactosylation sites. These proteins have 10.5% to 35% Ser+Thr in their composition and present high similarity to previously described salivary proteins of mosquitoes.
Salivary peptide similar to that in Aedes
AF-41 codes for a mature peptide of 7.3 kDa that is similar to an An. darlingi salivary peptide and weakly similar to salivary peptides of Ae. aegypti and Ae. albopictus. No significant similarities to An. gambiae proteins can be found when comparing AF-41 to ENSEMBL or the NCBI NR databases. However, comparison of AF-41 using tblastn to previously reported salivary gland ESTs from An. gambiae (Arcà et al., 2005) yielded 63% identity to Ag-contig_222, which has not been identified before as coding for a putative secreted protein (the An. gambiae peptide has now been deposited in GenBank). This family of peptides appear to be unique to mosquitoes.
3.4.4. Secreted protein families found exclusively in Anophelines
SG1 protein family
The SG1 (or gSG1) family is thought to be found uniquely in anopheline mosquitoes (Arcà et al., 2005). Six genes of this family are known in An. gambiae, five of which reside in chromosome X (four of them in a tandem configuration), while the gene coding for the TRIO protein is in the 2R chromosome arm. All mature proteins have molecular weight near 41 kDa. Their transcripts are found uniquely or enriched in the salivary glands of adult females, suggesting a function in blood feeding (Arcà et al., 2005). They do not yield significant similarities by blastp to other proteins in the NCBI database except for other anopheline proteins. In An. funestus, we presently report full-length sequences for two of the six An. gambiae orthologues, plus two truncated sequences of two others. We additionally provide one allele of gSG1b (Supplemental Table S2). Overall, these five sequences are only 52% to 61% identical to the An. gambiae orthologues. Members of this protein family may be good immunological markers of human anopheline exposure.
gSG2 family
In An. gambiae, two genes code for the glycine- and proline-rich proteins SG2 and SG2A, which have mature molecular weights of 9.5 and 15.5 kDa, respectively. Their genes reside close to each other in chromosome 2L and may be similarly regulated. Their transcripts are found in female salivary glands and in whole males but not in female carcasses deprived of the salivary glands, indicating expression in both male and female salivary glands, where they may assist sugar feeding, possibly as an antimicrobial, or some other unique glandular function. Their glycine-rich composition is reminiscent of some antimicrobial peptides (Otvos, 2000). In An. funestus, 53 transcripts were found coding for these twp family members, accounting for 10.7% of the S-class transcripts. The An. funestus homologues are quite divergent, having 54% and 62% identity with the An. gambiae orthologues. Two orthologues were also found in the An. stephensi sialotranscriptome (Valenzuela et al., 2003). The New World species An. darlingi appears to have at least three members of the family. Alignment of the nine protein sequences shows that the SG2a of An. gambiae acquired a 30-aa insertion (Fig. 2A). The phylogram separates well the members of the Cellia subgenus from the New World Nyssorhynchus. Notice also that the two subgenus sequences diverged to the point that no strong bootstrap support is found to join the Old World and New World protein sequences, indicating the fast evolutionary pace for this protein family. A hidden Markov model was built from the alignment (excluding the first 20 aa in the signal peptide region) and used to search the NR database in an unsuccessful attempt to find other family relatives (results not shown).
Fig. 2.
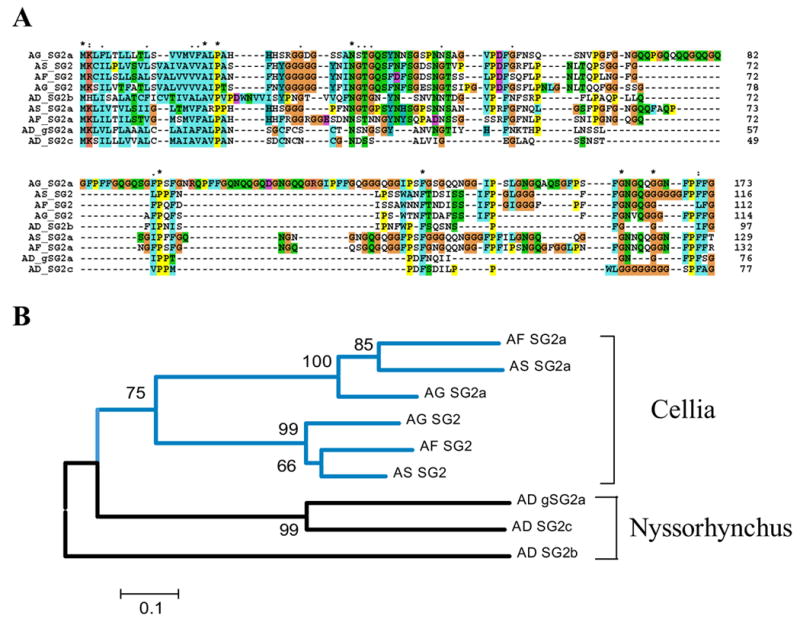
(A) Clustal alignment of the unique SG2 family of anopheline salivary peptides. The sequences shown are from An. darlingi (AD), An. stephensi (AS), An. funestus (AF), and An. gambiae (AG). (B) Neighbor-joining phylogram. The numbers in the phylogram nodes indicate percent bootstrap support for the phylogeny. The vertical bars show the Cellia and Nyssorhynchus subgroups. The bar at the bottom indicates 10% amino acid divergence in the sequences.
gSG6 peptide
The gSG6 peptide was first described in An. gambiae (Lanfrancotti et al., 2002) and found to be a unique protein sequence coding for a mature peptide of ~10 kDa with ten cysteine residues making probably five disulphide bonds (Fig. 3). A homologue was later found in the sialotranscriptome of An. stephensi. We here describe the An. funestus orthologue, AF-1, having 81% and 76% identities with An. stephensi and An. gambiae polypeptides, respectively. Fifty-nine EST were found in the An. funestus sialotranscriptome coding for AF-1, accounting for 12% of all S-class transcripts.. The spacing of the ten cysteines is unique to this protein family; when the pattern was searched against >3.5 million sequences on the NR database, only gSG6 proteins resulted with the ten perfectly spaced residues (not shown).
Fig. 3.

Clustal alignment of the unique SG6 family of anopheline salivary peptides. The ten cysteines are shown in black background. Identities are marked with ‘*’, strong amino acid conservations with ‘:’, and other conserved amino acids with ‘.’. The sequences shown are from An. stephensi (AS), An. funestus (AF), and An. gambiae (AG).
In An. gambiae, the transcript coding for gSG6 was found specifically in adult female salivary glands by RT-PCR, a result also supported by the Affymetrix chip array indicating the gSG6 gene expression to be 16 times larger in whole females than in males (Arcà et al., 2005). The function of this peptide is not known but is possibly associated with the blood-feeding function.
gSG7 family
Two genes for this uniquely anopheline family are known to exist in An. gambiae. Both gene transcription products were shown enriched in adult female salivary glands. The An. funestus sequences are 69% and 67% identical to the An. gambiae orthologues, and one of them is 70% identical to one previously described An. stephensi salivary protein. The New World species An. darlingi also has two members of the family, 49% and 47% identical to the An. funestus orthologues. Alignment of the seven known protein sequences of this superfamily (Fig. 4A) shows a conserved framework of four cysteines and also that a subset of three sequences has an extra odd cysteine found only in the protein sequences of the Old World SG7_2 subfamily (Fig. 4A). Similarly to the SG2 family, the phylogram (Fig. 4B) indicates the ancestral mosquito originating these four species had both members of the gene family, because the interspecies and not intraspecies sequences form a cluster with strong bootstrap support.
Fig. 4.
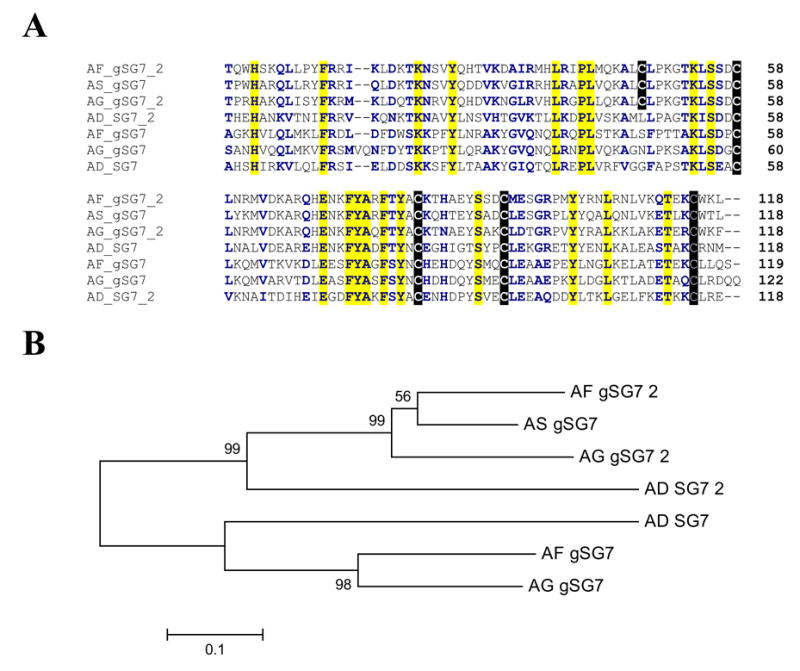
(A) Clustal alignment of the unique SG7 family of anopheline salivary peptides. The sequences shown are from An. darlingi (AD), An. stephensi (AS), An. funestus (AF), and An. gambiae (AG). The signal peptide region is not shown. Cysteines have black background, conserved amino acids (aa) yellow background. Positions of similar aa are shown with bold characters on white background. (B) Neighbor-joining phylogram. The numbers in the phylogram nodes indicate percent bootstrap support for the phylogeny. The bar at the bottom indicates 10% aa divergence in the sequences.
cE5/Anophelin family
Thirty-four EST from An. funestus sialotranscriptome matched uniquely anopheline peptides similar to the previously described antithrombin of An. albimanus named anophelin (Francischetti et al., 1999; Valenzuela et al., 1999). An. funestus anophelin is only 59% identical to the An. gambiae orthologue. Notice that overall there is less than 50% identity among five known family members, including two New World species that have shorter carboxyterminal regions when compared with Old World species (Fig. 5). The alignment shows three distinct regions: the conserved aminoterminus Ala-Pro-Gln-Tyr, which may be important for forming the pyroglutamic acid in the aminoterminus (Abraham and Podell, 1981; Valenzuela et al., 1999), and an acidic region that might be important for interaction of the peptide with the anion-binding exosite of thrombin. It is tempting to speculate that the conserved Asp-Pro-Gly-Lys may be the residue that locks into the enzyme active site.
Fig. 5.

Clustal alignment of the anopheline family of antithrombin peptides. The sequences shown are from An. gambiae (AG), An. funestus (AF), An. stephensi (AS), An. darlingi (AD), and An. albimanus (AA). The signal peptide region is not shown. Acidic amino acids (aa) are red on yellow background. Identical aa are marked in yellow background. Other symbols are as in Figs. 3 and 4.
8.2-kDa family
Sixteen EST from the sialotranscriptome of An. funestus coded for a peptide having ~42% identity to the 8.2-kDa salivary peptide of An. stephensi and similar proteins from An. gambiae and An. darlingi. Peptides of this family have a high composition of serine and threonine aa (10% in AF-9, while the average on all proteins of Supplemental Table S2 is 2.7, including the mucins) and no cysteine residues in the mature peptide (Fig. 6). There are nine predicted N-acetyl-galactosylation sites. In An. gambiae, this peptide was found enriched in adult female salivary glands, suggesting a role in blood feeding.
Fig. 6.

Clustal alignment of the 8.2-kDa family of peptides. The sequences shown are from An. gambiae (AG), An. funestus (AF), An. stephensi (AS), and An. darlingi (AD). The signal peptide region is not shown. Serine and threonine amino acids (aa) are marked in red on yellow background. Identical aa are marked in yellow background. Other symbols are as in Figs. 3 and 4.
6.2-kDa family
The first member of this peptide family was described in a sialotranscriptome of An. gambiae (Arcà et al., 2005), where it was found enriched in adult female salivary glands compared with other tissues. The An. funestus member of this family is 61% identical to the An. gambiae homologue, and 53% identical to an An. darlingi peptide. Alignment of the three peptides (Fig. 7) shows a remarkable conservation on the middle and carboxyterminal regions. The aminoterminal region of the mature peptide has three conserved prolines probably making two loops with variable lengths, with the An. darlingi sequencebeing the minimalist in this region, lacking the third conserved proline.
Fig. 7.

Clustal alignment of the 6.2-kDa family of peptides. The sequences shown are from An. gambiae (AG), An. funestus (AF), An. stephensi (AS), and An. darlingi (AD). Symbols above sequences are explained in Figs. 3 and 4.
Hypothetical family 13
AFC-202 has 46% identity to a polypeptide previously identified in a sialotranscriptome of An. gambiae annotated as hypothetical protein 13 (Francischetti et al., 2002), which codes for a mature peptide of 3.6 kDa of unknown function. This peptide was found ubiquitously expressed in An. gambiae tissues, and it is possible that it plays a housekeeping or antimicrobial role.
Hypothetical family 15/17
AF-24 is 56% identical to an An. gambiae salivary peptide previously annotated as hypothetical salivary protein 15 (Francischetti et al., 2002) and is also 40% identical to the salivary hypothetical protein 17 of the same mosquito. In An. gambiae, the two genes coding for this protein family are found as a tandem repeat in chromosome X. Both gene transcripts are enriched in adult female salivary glands, indicating a role in blood feeding for this peptide family. An. stephensi and An. darlingi also have protein family members as deducted from previous sialotranscriptomes. Alignment of all known sequences of this family (Fig. 8) shows only 11 absolutely conserved residues in the mature peptide region. No cysteines are found in the mature peptides.
Fig. 8.

Clustal alignment of hypothetical family 15/17 of salivary peptides. The sequences shown are from An. gambiae (AG), An. funestus (AF), and An. darlingi (AD). Symbols above sequences are explained in Figs. 3 and 4.
Hypothetical 10/12 family
Previous anopheline sialotranscriptomes (Francischetti et al., 2002; Valenzuela et al., 2003) identified a family of related peptides coded by two similar genes and their products named hypothetical 10 and hypothetical 12 salivary proteins. The genes coding for the An. gambiae protein lie as a tandem repeat on chromosome arm 3R. Mature peptides have predicted mass of 7.5–8 kDa. Both gene transcripts were overrepresented in female salivary glands and in males compared with female carcasses deprived of salivary glands, suggesting a salivary role common to male and females mosquitoes, possibly antimicrobial (Arcà et al., 2005). An. funestus putative salivary protein AF-29 is 46% and 43% identical to An. gambiae proteins 12 and 10, respectively, and 55% and 34% identical to the homologous proteins of An. stephensi. This protein family has four conserved cysteines in the mature peptide (Fig. 9) and only eight positionally conserved aa; however, within each family, a higher conservation is observed (29 and 19 residues for families 10 and 12, respectively).
Fig. 9.
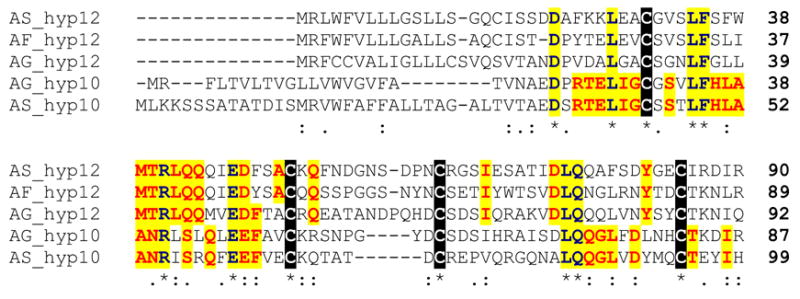
Clustal alignment of hypothetical family 10/12 of salivary secreted peptides. The sequences shown are from An. gambiae (AG), An. funestus (AF), and An. stephensi (AS). Cysteines are marked in black background. Conserved amino acids (aa) are shown in yellow background. Conserved aa in either the 10 or 12 amily are shown in red over yellow background. Symbols below sequences are explained in Figs. 3 and 4.
3.4.5. H proteins
Supplemental Table S2 presents 25 additional proteins that are classified as H, including in this group five hypothetical conserved proteins, nine ribosomal proteins, two cytoskeletal proteins, five enzymes involved in energy metabolism, and proteins related to the protein modification and protein export machineries. This group of 25 proteins is 95.9% ± 0.84 (mean ± SE) identical to their closest anopheline match in the NR protein database, as opposed to 66.7% ± 1.9 identity observed for the group of salivary proteins, a highly significant difference similar to a previous comparison made between the An. stephensi and An. gambiae proteins that indicated a fast pace of evolution for salivary proteins of mosquitoes (Valenzuela et al., 2003).
3.4.6. Concluding remarks
The accumulating information on mosquito sialotranscriptomes, including An. gambiae, An. stephensi, An. darlingi, Ae. aegypti, Ae. albopictus , C. p. quinquefasciatus, and now An. funestus, is allowing a pattern to emerge in the complex salivary composition of mosquitoes. First, ubiquitous protein families are found recruited for a salivary role, such as members of the AG5 family and enzymes of nucleotide and carbohydrate catabolism. Second, a group of protein families exclusive to Diptera includes the abundantly expressed D7 proteins, also found in sand flies and Culicoides. A third group of proteins is found only in Culicidae, including the 30-kDa allergen family and several mucins. Ten protein and peptide families, five of which are multigenic, are exclusive to anophelines. Among these proteins may reside good epidemiological markers to measure human exposure to anophelines, and even between anopheline species such as An. funestus and An. gambiae. Finally, among the 33 proteins presented in Supplemental Table S2 as possibly secreted, we can ascribe at least one function for only 10 of them, including the enzymes, the D7 proteins, and anophelin, based on previous studies with homologous proteins. The addition of An. funestus salivary proteins to this protein pool of unknown function represents a natural ‘mutagenesis’ experiment that might help future studies attempting to solve the function of these proteins involved in blood or sugar feeding or antimicrobial activity.
Supplementary Material
Acknowledgments
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. We thank NIAID intramural editor Brenda Rae Marshall for assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraham GN, Podell DN. Pyroglutamic acid. Non-metabolic formation, function in proteins and peptides, and characteristics of the enzymes effecting its removal. Molecular and Cellular Biochemistry. 1981;38:181–190. doi: 10.1007/BF00235695. Spec No. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W. Local alignment statistics. Methods in Enzymology. 1996;266:460–480. doi: 10.1016/s0076-6879(96)66029-7. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcà B, Lombardo F, Lanfrancotti A, Spanos L, Veneri M, Louis C, Coluzzi M. A cluster of four D7-related genes is expressed in the salivary glands of the African malaria vector Anopheles gambiae. Insect Moleuclar Biology. 2002;11:47–55. doi: 10.1046/j.0962-1075.2001.00309.x. [DOI] [PubMed] [Google Scholar]
- Arcà B, Lombardo F, Valenzuela JG, Francischetti IM, Marinotti O, Coluzzi M, Ribeiro JMC. An updated catalogue of salivary gland transcripts in the adult female mosquito, Anopheles gambiae. Journal of Experimental Biology. 2005;208:3971–3986. doi: 10.1242/jeb.01849. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. NatureGenetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Research. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, Andersen J, Francischetti IM, deL Capurro M, deBianchi AG, James AA, Ribeiro JMC, Marinotti O. The transcriptome of adult female Anopheles darlingi salivary glands. Insect Molecular Biology. 2004;13:73–88. doi: 10.1111/j.1365-2583.2004.00463.x. [DOI] [PubMed] [Google Scholar]
- Calvo E, Mans BJ, Andersen JF, Ribeiro JMC. Function and evolution of a mosquito salivary protein family. Journal of Biological Chemistry. 2006;281:1935–1942. doi: 10.1074/jbc.M510359200. [DOI] [PubMed] [Google Scholar]
- Campbell CL, Vandyke KA, Letchworth GJ, Drolet BS, Hanekamp T, Wilson WC. Midgut and salivary gland transcriptomes of the arbovirus vector Culicoides sonorensis (Diptera: Ceratopogonidae) Insect Molecular Biology. 2005;14:121–136. doi: 10.1111/j.1365-2583.2004.00537.x. [DOI] [PubMed] [Google Scholar]
- Coetzee M, Fontenille D. Advances in the study of Anopheles funestus, a major vector of malaria in Africa. Insect Biochemistry and Molecular Biology. 2004;34:599–605. doi: 10.1016/j.ibmb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Francischetti IM, Valenzuela JG, Pham VM, Garfield MK, Ribeiro JMC. Toward a catalog for the transcripts and proteins (sialome) from the salivary gland of the malaria vector Anopheles gambiae. Journal of Experimental Biology. 2002;205:2429–2451. doi: 10.1242/jeb.205.16.2429. [DOI] [PubMed] [Google Scholar]
- Francischetti IM, Valenzuela JG, Ribeiro JMC. Anophelin: kinetics and mechanism of thrombin inhibition. Biochemistry. 1999;38:16678–16685. doi: 10.1021/bi991231p. [DOI] [PubMed] [Google Scholar]
- Hansen JE, Lund O, Tolstrup N, Gooley AA, Williams KL, Brunak S. NetOglyc: prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconjugate Journal. 1998;15:115–130. doi: 10.1023/a:1006960004440. [DOI] [PubMed] [Google Scholar]
- Hekmat-Scafe DS, Dorit RL, Carlson JR. Molecular evolution of odorant-binding protein genes OS-E and OS-F in Drosophila. Genetics. 2000;155:117–127. doi: 10.1093/genetics/155.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman DR. Allergens in Hymenoptera venom. XXV: The amino acid sequences of antigen 5 molecules and the structural basis of antigenic cross-reactivity. Journal of Allergy and Clinical Immunology. 1993;92:707–716. doi: 10.1016/0091-6749(93)90014-7. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Research. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isawa H, Yuda M, Orito Y, Chinzei Y. A mosquito salivary protein inhibits activation of the plasma contact system by binding to factor XII and high molecular weight kininogen. Journal of Biological Chemistry. 2002;13:13. doi: 10.1074/jbc.M203505200. [DOI] [PubMed] [Google Scholar]
- King TP, Spangfort MD. Structure and biology of stinging insect venom allergens. International Archives of Allergy and Immunology. 2000;123:99–106. doi: 10.1159/000024440. [DOI] [PubMed] [Google Scholar]
- Korochkina S, Barreau C, Pradel G, Jeffery E, Li J, Natarajan R, Shabanowitz J, Hunt D, Frevert U, Vernick KD. A mosquito-specific protein family includes candidate receptors for malaria sporozoite invasion of salivary glands. Cellular Microbiology. 2006;8:163–175. doi: 10.1111/j.1462-5822.2005.00611.x. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lanfrancotti A, Lombardo F, Santolamazza F, Veneri M, Castrignano T, Coluzzi M, Arcà B. Novel cDNAs encoding salivary proteins from the malaria vector Anopheles gambiae. Federation of European Biochemical Societies Letters. 2002;517:67–71. doi: 10.1016/s0014-5793(02)02578-4. [DOI] [PubMed] [Google Scholar]
- Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley RR, Ponting CP, Bork P. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Research. 2002;30:242–244. doi: 10.1093/nar/30.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Kwon J, Aksoy S. Characterization of genes expressed in the salivary glands of the tsetse fly, Glossina morsitans morsitans. Insect Molecular Biology. 2001;10:69–76. doi: 10.1046/j.1365-2583.2001.00240.x. [DOI] [PubMed] [Google Scholar]
- Lynn AM, Jain CK, Kosalai K, Barman P, Thakur N, Batra H, Bhattacharya A. An automated annotation tool for genomic DNA sequences using GeneScan and BLAST. Journal of Genetics. 2001;80:9–16. doi: 10.1007/BF02811413. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Research. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinotti O, James A, Ribeiro JMC. Diet and salivation in female Aedes aegypti mosquitoes. Journal of Insect Physiology. 1990;36:545–548. [Google Scholar]
- Megraw T, Kaufman TC, Kovalick GE. Sequence and expression of Drosophila antigen 5-related 2, a new member of the CAP gene family. Gene. 1998;222:297–304. doi: 10.1016/s0378-1119(98)00489-2. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Engineering. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Novak MG, Ribeiro JMC, Hildebrand JG. 5-Hydroxytriptamine in the salivaryt glands of adult female Aedes aegypti and its role in regulation of salivation. Journal of Experimental Biology. 1995;198:167–174. doi: 10.1242/jeb.198.1.167. [DOI] [PubMed] [Google Scholar]
- Novak MG, Rowley WA. Serotonin depletion affects blood-feeding but not host-seeking in Aedes triseriatus (Diptera: Culicidae) Journal of Medical Entomology. 1994;31:600–606. doi: 10.1093/jmedent/31.4.600. [DOI] [PubMed] [Google Scholar]
- Otvos L., Jr Antibacterial peptides isolated from insects. Journal of Peptide Science. 2000;6:497–511. doi: 10.1002/1099-1387(200010)6:10<497::AID-PSC277>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC. Blood-feeding arthropods: lLive syringes or invertebrate pharmacologists? Infectious Agents and Disease. 1995;4:143–152. [PubMed] [Google Scholar]
- Ribeiro JMC, Andersen J, Silva-Neto MA, Pham VM, Garfield MK, Valenzuela JG. Exploring the sialome of the blood-sucking bug Rhodnius prolixus. Insect Biochemistry and Molecular Biology. 2004a;34:61–79. doi: 10.1016/j.ibmb.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC, Charlab R, Pham VM, Garfield M, Valenzuela JG. An insight into the salivary transcriptome and proteome of the adult female mosquito Culex pipiens quinquefasciatus. Insect Biochemistry and Molecular Biology. 2004b;34:543–563. doi: 10.1016/j.ibmb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC, Francischetti IM. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annual Reviews in Entomology. 2003;48:73–88. doi: 10.1146/annurev.ento.48.060402.102812. [DOI] [PubMed] [Google Scholar]
- Rodriguez MH, Hernandez-Hernandez F. Insect-malaria parasites interactions: the salivary gland. Insect Biochemistry and Molecular Biology. 2004;34:615–624. doi: 10.1016/j.ibmb.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Rossignol PA, Lueders AM. Bacteriolytic factor in the salivary glands of Aedes aegypti. Comparative Biochemistry and Physiology. 1986;83B:819–822. doi: 10.1016/0305-0491(86)90153-7. [DOI] [PubMed] [Google Scholar]
- Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Research. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons FE, Peng Z. Mosquito allergy: recombinant mosquito salivary antigens for new diagnostic tests. International Archives of Allergy and Immunology. 2001;124:403–405. doi: 10.1159/000053771. [DOI] [PubMed] [Google Scholar]
- Stintzi A, Heitz T, Prasad V, Wiedemann-Merdinoglu S, Kauffmann S, Geoffroy P, Legrand M, Fritig B. Plant ‘pathogenesis-related’ proteins and their role in defense against pathogens. Biochimie. 1993;75:687–706. doi: 10.1016/0300-9084(93)90100-7. [DOI] [PubMed] [Google Scholar]
- Szyperski T, Fernandez C, Mumenthaler C, Wuthrich K. Structure comparison of human glioma pathogenesis-related protein GliPR and the plant pathogenesis-related protein P14a indicates a functional link between the human immune system and a plant defense system. Proceedings of the National. Academy of Science USA. 1998;95:2262–2266. doi: 10.1073/pnas.95.5.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. The COG database: an updated version includes eukaryotes. BioMed Central Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela JG, Charlab R, Gonzalez EC, Miranda-Santos IKF, Marinotti O, Francischetti IM, Ribeiro JMC. The D7 family of salivary proteins in blood sucking Diptera. Insect Molecular Biology. 2002a;11:149–155. doi: 10.1046/j.1365-2583.2002.00319.x. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IM, Pham VM, Garfield MK, Ribeiro JMC. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochemistry and Molecular Biology. 2003;33:717–732. doi: 10.1016/s0965-1748(03)00067-5. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IM, Ribeiro JMC. Purification, cloning, and synthesis of a novel salivary anti-thrombin from the mosquito Anopheles albimanus. Biochemistry. 1999;38:11209–11215. doi: 10.1021/bi990761i. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Pham VM, Garfield MK, Francischetti IM, Ribeiro JMC. Toward a description of the sialome of the adult female mosquito Aedes aegypti. Insect Biochemistry and Molecular Biology. 2002b;32:1101–1122. doi: 10.1016/s0965-1748(02)00047-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


