Abstract
We have investigated intragenic recombination in Block 2 of the merozoite surface protein-1 (MSP-1), where three allele-specific families: K1, Mad20, and RO33 were previously known. Using parasites from western Kenya, we have found a fourth Block 2 allele type, which is a recombinant between Mad20 and RO33 alleles. These recombinant alleles, which we have termed MR, contain sequence from the 5′ region of Mad20 and the 3′ region of RO33. The results of this study provide new data on the complexity of the MSP-1 antigen gene, which is a candidate vaccine antigen, and further support the importance of intragenic recombination in generating genetic variability in Plasmodium falciparum parasites in nature.
Keywords: Merozoite protein, Genetic diversity, Vaccine, Recombination, Plasmodium, Allele family
1. Introduction
With 300–500 million clinical cases per year resulting in over 1 million deaths, malaria continues to be one of the leading causes of childhood morbidity and mortality in sub-Saharan Africa [1]. Malaria infection during pregnancy also poses a serious health problem for women of child-bearing age and is associated with adverse effects during fetal development. Four known species of Plasmodium cause disease in humans, but almost all malaria-attributable mortality is due to Plasmodium falciparum. The malaria parasite’s life cycle alternates between the human host and the mosquito vector; it is complex, and the parasite stages exhibit extensive genetic and antigenic diversity.
Genetic diversity of the parasites contributes to the failure of anti-malaria parasite control measures. For instance, the use of antimalarial drugs has led to the emergence and dispersal of drug resistant parasites that render the drugs ineffective. Similarly, antigenic diversity allows the parasite to evade natural immune responses and may jeopardize the effectiveness of vaccines.
Over the past several years, parallel to vaccine development and testing efforts, several molecular epidemiologic studies have been conducted to provide information on the nature and extent of genetic diversity of promising candidate vaccine antigen genes [2–4]. Two major factors have been considered to explain the observed genetic diversity in a specific gene: natural selection and intragenic recombination [5–7].
Given that the malaria parasite’s life cycle includes a sexual stage, intragenic recombination during meiosis has been proposed as an important mechanism for the generation of novel genetic variants in malarial antigens [8–11]. However, finding direct evidence for intragenic recombination is hampered by the slow accumulation of synonymous substitutions that makes it difficult to separate recombination from convergence due to positive natural selection [7]. The importance of intragenic recombination in the generation of new alleles has remained an elusive issue in the context of malarial vaccine antigens [4].
The merozoite surface protein-1 (MSP-1) is a promising candidate vaccine antigen. It is the most abundant surface protein on the merozoite of P. falciparum, and it is thought to play a role in erythrocyte invasion. The gene consists of both conserved and polymorphic regions, the most polymorphic of which is the 100–400 base pair (bp) Block 2 repetitive region.
Given the fact that MSP-1 Block 2 has been shown to play a role in the development of immunity [13], it is an important region of the protein for studying diversity and intragenic recombination. MSP1 Block 2 has been divided into three allele-specific families: K1, Mad20, and RO33. Alleles in K1 and Mad20 contain different numbers of unique, tripeptide repeats. K1 and Mad20 are the most diverse families in terms of the number of alleles identified using the fragment length polymorphism of Block 2. RO33 is monomorphic, lacking the tripeptide repeats observed in the other two families. Several studies have used MSP-1 Block 2-based PCR-genotyping to estimate the number of genetically distinct genotypes within an infection [14–16]. One study suggested the presence of alleles in Block 2 that are recombinants between two different main families, but only PCR-based typing methods were used [17]. Another study that used parasite specimens from Mali also indicated an apparent Mad20 and RO33 recombinant using PCR amplification of MSP-1 Block 2 with Mad20 and RO33 primers [18]. One of the limitations of PCR-based genetic analysis is that it does not allow detection of novel sequences that may exist in natural populations. Even more important in the context of studies of intragenic recombination, PCR-based methods can detect false recombinants due to nonspecific amplification.
Direct evidence of intragenic recombination in the MSP-1 gene of malaria parasites was provided by laboratory cross studies [12]. Studies of recombination in MSP-1 have largely focused on regions other than Block 2 [9,12,19–22]. Four studies have included MSP-1 Block 2 in the regions analyzed [14,23–25]. Studies on the pattern of linkage disequilibrium among polymorphic sites suggested a high recombination rate in MSP-1 [23,24]. However, this kind of analysis does not address the issue of convergent mutations due to selection versus intragenic recombination in malarial vaccine antigens [7].
We are exploring the role of intragenic recombination in the generation of new alleles in P. falciparum. In this study, we have investigated genetic diversity of the Block 2 region of the MSP-1 gene using parasites from a birth cohort in western Kenya. We used allele-specific and mismatched amplifying primers in PCR assays and sequence-characterized the amplified fragments to determine the nature of the Block 2 region. We have found a novel fourth allele family of Block 2. This new allele family is a recombinant between the Mad20 and RO33 allele families and is found in more than 25% of the Kenyan field samples studied, showing that intragenic recombination is a factor in the generation of diversity in the MSP-1 antigen gene.
2. Materials and methods
2.1. Sampling and DNA extraction
The samples used in this study came from participants of a birth cohort study (Asembo Bay Cohort Project) which took place in an area of high malaria transmission in western Kenya [26]. We extracted DNA from 173 parasitized blood samples taken from 37 different infants between 1992 and 1994. All infants were parasitemia positive by microscopy. DNA was extracted from approximately 300 ul of infected pRBC’s using the PureGene extraction method (Gentra Systems).
2.2. Nested PCR to detect Block 2 alleles
We used a nested PCR method where the first PCR was done with the external 5′ (AAGCTTTGAAGATG-CAGTATTGAC) and 3′ (ATTCATTAATTTCTTCA-TATCCATC) primers, which amplify Block 2 plus some of the flanking regions. The second PCR was done with allele specific primers (K1; 5′GAAATTACTAC AAAAGGTGCAAGTG and 3′AGATGAAGTATT TGAACGAGGTAAAGTG: M20; 5′GCTGTTAC AACTAGTACACC and 3′TGAATTATCTGAAT TTGTACGTCTTGA: and RO33; 5′GCAAATACT CAAGTTGTTGCAAAGC and 3′AGGATTTGCAG CACCTGGAGATCT) [16] in three different non-recombinant primer combinations (K1 5′and 3′, M20 5′ and 3′, and RO33 5′and 3′) and six different recombinant primer combinations (K1 5′ and M20 3′, M20 5′ and K1 3′, K1 5′ and RO33 3′, RO33 5′ and K1 3′, M20 5′ and RO33 3′, and RO33 5′ and M20 3′). For the PCR amplification mixture, 5 μl of each of the appropriate 5′ and 3′ primers (40 ng μl−1) were added to 12 μl of dNTP mixture (Promega, 107 μmol per reaction), 3 μl of MgCl2 (25 mM), 10 μl of PCR 10× buffer containing 15 mM MgCl2 (Perkin – Elmer), 0.5 μl Taq polymerase, and 59.5 μl double distilled water. For the first PCR, 5 μl of DNA (adjusted to a concentration of 150 ng μl−1) was added to the 95 μl reaction mixture, and amplified for 25 cycles at a 55 °C annealing temperature. For each internal PCR, 5 μl of the first (external) PCR product was added to the 95 μl reaction mixture and amplified for 30 cycles at a 64 °C annealing temperature. Each internal PCR product was electrophoresed on a 3% Agarose-1000 gel (Gibco) to determine the alleles present. We could consistently detect length differences of 10 bp.
2.3. Sequencing of Block 2 alleles
We sequenced products obtained from the recombinant PCR representing a range of sizes and band intensities to confirm whether or not the products were actual recombinants or PCR artifacts. Products were cut from 3% Agarose-1000 gels using a clean sterile scalpel, and the DNA was purified using the Qiaquick Gel Extraction Kit (Qiagen). The purified products were sequenced on an ABI automated sequencer.
3. Results
3.1. Amplification of non-recombinant MSP-1 Block 2 alleles
One hundred and seventy three samples taken from 37 parasitemic infants were genotyped using Block 2 family specific primers for K1, Mad20, and RO33. Of the 173 samples genotyped 97, 76, and 85% contained one or more K1, Mad20, or RO33 alleles, respectively. Each sample contained an average of 2.35 K1 alleles (S.E. = 0.09) and 1.31 Mad20 alleles (S.E. = 0.08). Since RO33 is monomorphic, it was either present or absent in the samples tested. The frequency of the K1, Mad20, and RO33 alleles were similar to that observed in other samples in this area of western Kenya [14]. We sequenced products representing a range of sizes and band intensities, and most of the non-recombinant products were ‘real’ (i.e. not PCR artifact).
3.2. Amplification of recombinant MSP-1 Block 2 alleles
The same 173 samples that were genotyped using the non-recombinant primer combinations were also genotyped using the six recombinant primer combinations (K1-Mad20, Mad20-K1, K1-RO33, RO33-K1, Mad20-RO33, and RO33-Mad20). Most of the products we observed using this method were faint spurious bands that upon sequence analysis appeared to be PCR artifacts rather than recombinant alleles. There was one recombinant type, made up of sequence from the 5′ end of Mad20 and the 3′ end of RO33, which we have termed ‘MR’. The most common MR allele detected was 140 bp in length, containing 72 bp of Mad20 sequence followed by 69 bp of RO33 sequence. Six additional sizes of MR alleles, ranging in size from approximately 130 to 220 bp, were also detected, and these differed only by the number of times the sequence GGTTCAGGT was repeated in the Mad20 region of the recombinant (Fig. 1A). The 200 bp MR allele also contains an extra 9 bp in the RO33 region. The amino acid sequences for the recombinants are shown in Fig. 1B, and the amino acid sequences for one representative of each of the non-recombinant families (K1, Mad20, and RO33) are shown in Fig. 1C. Comparing Fig. 1B and C demonstrates that the 5′ end of MR is the same as Mad20, and the 3′ end of MR is the same as RO33, while K1 is different from all the other sequences. Excluding artifacts, verified by sequencing of the PCR products, 48 (28%) of the 173 samples contained one or more MR alleles. Of the 37 infants tested, 21 (57%) had at least one infection containing a MR allele. To examine the effect of recrudescence on the MR frequency we examined 81 samples (of the original 173) that were preceded by aparasitemia either due to successful treatment or self-clearance of parasites. Of these 81 samples, 21 (26%) contained one or more MR alleles. These results suggest that the MR frequency estimate is not strongly affected by recrudescence.
Fig. 1.
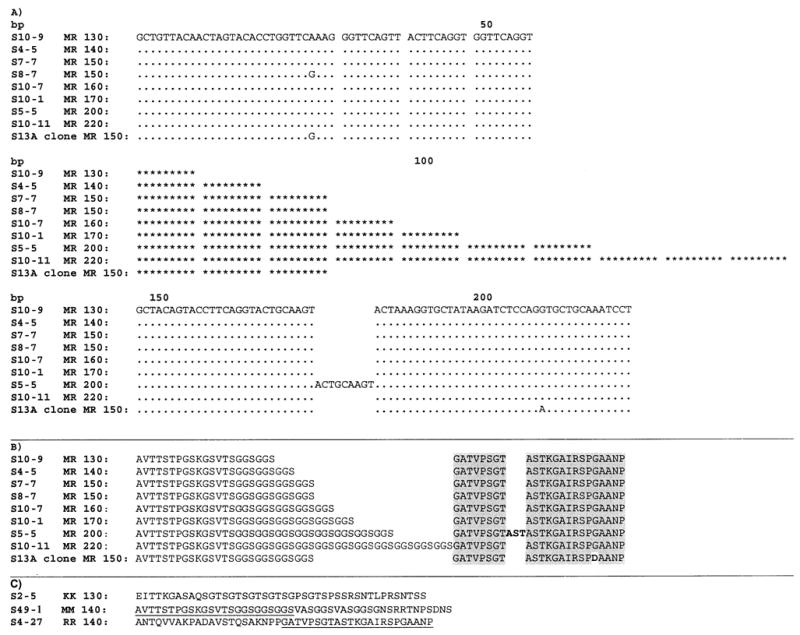
Alignment of MR sequences. One representative of each different type is shown. Alleles range in size from 130 to 220 bp, and primarily differ by the number of times the sequence GGTTCAGGT is repeated in the Mad20 region. Dots indicate identity with the S10-9 sequence listed first in the alignment. The repeat GGTTCAGGT has been replaced with *********.
3.3. Addressing template switching
Though these MR alleles appear to be genuine recombinant alleles based on their DNA sequence, false recombinant alleles can be generated during PCR as a result of template switching [27–32]. Saiki et al. [27] suggest that PCR-mediated recombinants can occur as a result of incompletely extended products acting as primers by annealing to other allelic templates. Odelberg et al. [32] suggest that such recombinants can be generated when either the polymerase or the nascent strand switches from the original template to a secondary template during DNA synthesis. Regardless of the mechanism, these PCR-mediated recombinants would have to contain sequence from templates present in the reaction. Consequently, in order for the observed MR alleles to be a result of template switching, both Mad20 and RO33 DNA would need to be present in the PCR reaction. Therefore, if a MR recombinant is detected in a sample that lacks Mad20 or RO33 alleles, one would not expect this product to be a result of in vitro PCR-mediated recombination.
In order to determine whether the observed MR alleles were the result of template switching, we did nested PCR using the Mad20 5′ primer and the RO33 3′ primer on 37 additional samples lacking both Mad20 and RO33 alleles. These samples were obtained from another study conducted in the same region of western Kenya [14]. These additional samples were used to rule out template switching as an explanation for the presence of MR alleles and were not included in the MR frequency estimate. We were able to detect 140 bp recombinant alleles in two samples lacking all three main families of non-recombinant alleles (i.e. K1, Mad20, and RO33 alleles were not present). These 140 bp recombinants were identical in sequence to the 140 bp recombinants observed previously.
To further test the possibility of template switching, we made experimental mixtures of Mad20 and RO33 DNA from whole MSP-1 clones, generated for another study, from five infected Kenyan children. These clones contain the entire MSP-1 gene sequence amplified using primers that were outside of the MSP-1 Block 2 region. In the other study, 15 clones were identified as having different MSP-1 Block 2 genotypes. These clones were used to make the experimental mixtures of Mad20 and RO33 DNA to determine if we would get MR-positive PCR-genotyping. The Mad20 block 2 alleles used in the experimental mixtures were the same size as the alleles present in the majority of the samples containing MR recombinant alleles. Each clone contained only one insert, and only one type of Block 2 allele. We made a 2:1 mix of Mad20 DNA to RO33 DNA, a 1:1 mix, and a 1:2 mix. We then performed nested PCR using the Mad20 5′ primer and the RO33 3′ primer with the experimental mixtures as the template. Because we knew that there were no recombinant alleles present in the original template, any recombinant alleles detected in this PCR would have resulted from template switching. The PCR was repeated ten times for each of the experimental mixtures. PCR-genotyping of one of the experimental mixtures yielded a MR PCR product. Therefore, we suspected that this product was a PCR-artifact. However, upon sequencing the MSP-1 clones that made up the mixture, we found the MSP-1 clone (Clone S13A) thought to contain a Mad20 Block 2 actually contained a MR Block 2 region. Sequencing of this entire MSP-1 clone showed further proof of this fourth type of Block 2. No other recombinant products were generated from PCR-genotyping of the experimental mixtures of DNA, which provides further evidence to rule out template switching.
3.4. Origins of MR
We next investigated the origin of the MR recombinant. Based on genetic comparison of the sequences, a recombination event between Mad20 and RO33 appears to be the most likely reason for the emergence of the MR allele. The parent alleles of this recombinant would need to have a sequence similar to the 5′ end (Mad20 progenitor) and 3′ end (RO33 progenitor) of MR. All of the MR alleles are identical in the RO33 region, with the exception of the 200 bp MR allele (extra 9 bp in RO33 region) and the S13A clone (Fig. 1). The S13A clone has a single nucleotide substitution in the region spanned by the 3′ RO33 primer. Because this substitution is in the primer region, we cannot determine whether this substitution is present in the alleles amplified with this primer. Only one out of 15 full length Mad20 MSP-1 sequences in Genbank contains this same nucleotide substitution (data not shown). Since almost all of the RO33 sequences obtained from this study and recorded in Genbank are identical to the 3′ end of the MR alleles detected, any one of these could be the RO33 progenitor of MR. The Mad20 region of MR differs only by the number of times the sequence GGTTCAGGT is repeated. The S13A clone and one other sample also have a single A–G nucleotide substitution (Fig. 1).
We examined the available sequences from Genbank and this study to look for a Mad20 allele that resembles the Mad20 portion of MR (Table 1). This sequence would have the sequence CAAAG preceding the MR repeat region and at least two GGTTCAGGT to explain the length polymorphism. In addition, the sequence would need to be similar in length to the RO33 allele for successful recombination. Considering these criteria, 45 Mad20 sequences could be divided into 21 Types (Table 1). Of these, the MSP-1 clone from Kenya (Clone S49-3) with the 150 bp Mad20 Block 2 allele, sequenced in this study, was consistent with the Mad20 portion of MR.
Table 1.
Comparison of MSP-1 Mad20 sequences with the sequence detected in the Mad20 region of MR, showing the S49-3 M20-150 clone to be the closest match
| Kn: MR consensus sequence | (0) ...CAAAG-GGTTCAGTT-ACTTCAGGT-GGTTCAGGT-GGTTCAGGT ...| 0 | |
| 0 substitutions in repeat region shown, 0 substitutions in the upstream Block 1 sequence, same Block 2 length | |
| Kn: S49-3 M20-150 clone | (0) ...*****-*********-*********-*********-********* ...| 0 | |
| 2 substitutions in repeat region shown | |
| Kn: M20-190 | (nd) ...***GT-*********-*********-*********-********* ...| 0 | |
| 3 substitutions in repeat region shown | |
| Tz: HN3 | (0) ...*****-*******G*-*********-*********-**C****T*...| 1 | |
| Tz: HN2 | (0) ...*****-*******G*-G********-*C*******-********* ...| 3 | |
| Ti: 828 | (0) ...*****-*********-*********-*******T*-AC******* ...| 2 | |
| Sg: Bandia | (3) ...*****-*********-G********-*********-**C****T* ...| 2 | |
| Pfant195, 195A, Hd: Gp185, Ti : 837, 822b, 827, 808 | (0) ...*****-*********-G********-*********-**C****T* ...| 2 | |
| Vt: 195RR, Ti 806 | (1) ...*****-*********-G********-*********-**C****T* ...| 3 | |
| PfANT1, AF218248, Tz: HN627, Ti: MSA1aJ | (0) ...***GT-*********-*********-*********-*******T* ...| 0 | |
| Kn: M20-190 | (nd) ...***GT-*********-*********-*********-*******T* ...| 3 | |
| Tz: IFA9-10 | (3) ...***GT-*********-*********-*********-*******T* ...| 4 | |
| Tz: IFA10 | (5) ...***GT-*********-*********-*********-*******T* ...| 5 | |
| 4 substitutions in repeat region shown | |
| Hd: HB3a | (0) ...*****-*********-G********-********T-**C****T* ...| 1 | |
| So: PaloAlto | (3) ...****A-*********-G********-*********-**C****T* ...| 1 | |
| Ti: 842, Tz: 2/M1 | (0) ...***GT-*********-********-*********-**C****T* ...| 1 | |
| Ti: 822a | (0) ...***GT-*********-*********-*********-**C****T* ...| 2 | |
| Kn: M20-180 | (nd) ...***GT-*********-*********-*********-**C****T* ...| 2 | |
| 5 substitutions in repeat region shown | |
| Tz: HN624 | (0) ...***GT-*********-*********-*******T*-AC******* ...| 0 | |
| Ti: 834a | (1) ...***GT-*********-*********-*******T*-AC******* ...| 1 | |
| Ti: 947, 836, 807, 815, 835b, PNG: FC-27 | (0) ...***GT-*********-*********-*******T*-AC******* ...| 1 | |
| Pfp190G1 | (0) ...***GT-*********-*********-*******T*-******GT* ...| 1 | |
| Kn: M20-200 | (nd) ...****n-*******G*-GG*****T*-*C*******-********* ...| 6 | |
| > 5 substitutions in repeat region shown | |
| D43960 | (0) ...C*GGT-*C*****G*-*********-**C****T*-*C******* ...| 1 | |
| Tz: IFA11 | (0) ...C*GGT-*********-G********-**C****T*-*C******* ...| 5 | |
| Kn: M20-170 | (nd) ...C*GGT-*C*****G*-GG*****T*-*C*******-********* ...| 3 | |
| Tz: IFA125 | (1) ...C*GGT-*C*****G*-GG*****T*-*C*******-********* ...| 3 | |
| Tz: IFA12 | (0) ...C*GGT-*C*****G*-GG*****T*-*C*******-********* ...| 3 | |
| Kn: M20-180 | (nd) ...C*GGT-*C*****G-GG*****T*-*C*******-********* ...| 4 | |
| 3 substitutions and one deleted repeat in the repeat region shown | |
| Ti: 843a | (0) ...*****-*********-G*****A**-*******T*---------- ...| 4 | |
Forty-five available Mad20 (M20) sequences were compared with the consensus for the MR sequences (see Fig. 1). Only the S49-3 M20-150 clone was an exact match to these characteristics. The other sequences were grouped by the number of substitutions in the repeat region involved in the recombination (sequence shown). The other sequences had at least two substitutions in the repeat region shown. Subtypes were defined by the count of substitutions in the MSP-1 5′ Block1 region when compared with the MR-150 S13A clone (shown in the 5′ parentheses). The value in bars shows the difference in Block 2 repeat unit copy number vs. the MR-150 S13A clone, indicating differences in length. The sequences are identified by geographic origin when available (Ti, Thai; Kn, Kenya; PNG, Papua New Guinea; Tz, Tanzania; Sg, Senegal; So, South America; Hd, Hondoras; Vt, Vietnam) and by key characters in their Genbank accession number or identification number used for sequences reported in this study. Not enough data, nd. Unassigned nucleotide, n.
Alignment of RO33 with a hypothetical Mad20 progenitor showed how the MR 130 bp MR might have arisen (Fig. 2a). The S49-3 and a RO33 sequence also showed the potential for recombination to yield a 130 bp MR allele (Fig. 2b). Alignment of both the hypothetical progenitor and RO33 and the Kenya Mad20-150 Clone S49-3 showed that the predicted recombination site was CAGGTG (Fig. 2a and b). This recombination site is similar to the recombination hotspot, Chi (GCTGGTGG), documented in bacteria [33].
Fig. 2.

Alignment of Mad20 and RO33 showing the predicted recombination site (underlined). (a) Hypothetical Mad20 aligned with existing RO33 yielding a 130 bp MR. (b) Existing Mad20 aligned with existing RO33 yielding a 130 bp MR. Also shown is the sequence of the S13A clone containing the 150 bp MR Block 2 region. (c) Existing K1 aligned with existing RO33. K1 lacks the recombination site observed in Mad20 and RO33.
4. Discussion
The first evidence of intragenic recombination in malaria parasites was provided by laboratory cross studies [12]. Since then several studies have suggested intragenic recombination as an important factor in the generation of new alleles in malarial parasites [8–11]. As we have shown in this study, recombinants reported solely on the basis of PCR fragment analysis could simply be amplification artifacts. We have investigated recombination in the Block 2 region of the MSP-1, and provide evidence of a novel, fourth allele family, resulting from intragenic recombination between the Mad20 and RO33 alleles.
This Block 2 allele, which we have termed MR, is comprised of Mad20 sequence at the 5′ end and RO33 sequence at the 3′ end. The information on the diversity of the Block 2 region along with the results of recent studies of diversity of the C-terminal region of the protein [3,9], provide a better picture of the extent of genetic diversity in this important vaccine antigen gene (Fig. 3).
Fig. 3.
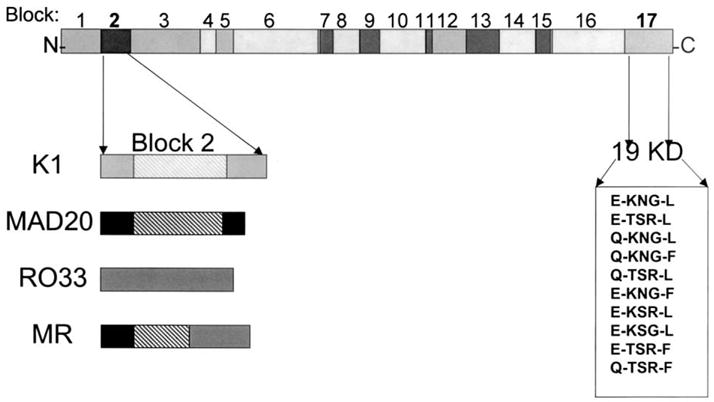
Schematic of MSP-1 showing known diversity in Block 2 and Block 17 (19KD fragment). The hatched areas in K1, Mad20, and MR indicate tripeptide repeats.
Two lines of evidence support the conclusion that the MR is a recombinant. First, the MR allele could not be explained by the occurrence of template switching during PCR. Second, we obtained a full-length MSP-1 clone containing a 150 bp MR Block 2 region from a parasite sample from western Kenya.
With very few exceptions noted in the results, these alleles differ only by the number of times the sequence GGTTCAGGT is repeated in the Mad20 region of the recombinant. This result suggests that these alleles arose from a single recombination event and diversified through expansion or contraction of repeats, either through slipped-strand mismatch repair or through recombination between MR alleles. We suggest that this Mad20-RO33 recombination was a single event, given that a crossing over takes place in the P. falciparum genome on an average every 1.67–106 bp [34]. Although it is possible that recombination either between two MR alleles or between a MR and Mad20 might have later contributed to the MR repeat copy number diversity, slipped strand mismatch repair is a more parsimonious explanation. This suggestion is based on two lines of evidence. First, considering the frequency of MR alleles detected and the P. falciparum crossover rate [34], the probability of two MR alleles being present in an infection and recombining in a mosquito would be less than 10−5 per meiosis. This is a significantly lower rate than the estimated rate of slipped strand mismatch repair: 10−3 per meiosis or mitosis. Second, we did not detect any diversity in MR alleles indicative of recombination between Mad20 and MR alleles. Therefore, we suggest that an initial recombination between Mad20 and RO33 gave rise to MR and then MR repeat copy number diversity arose from slipped strand mismatch repair.
The age of the MR allele in relation to other alleles cannot be established at this time; however, it does not appear to be a ‘recent’ event in relation to the P. falciparum population expansion. The reason for this speculation is that the MR allele is also found in Asia and the Americas (S. Takala et al., unpublished), in addition to its presence in Kenya and Mali.
The predicted recombination site between Mad20 and RO33 occurs at the sequence CAGGTG underlined in Fig. 2. The absence of this site in the K1 allelic form of the gene may explain the lack of recombination observed between K1 and the other allele families. The predicted recombination site is similar to the Chi sequence (GCTGGTGG), which locally increases recombination in Escherichia coli. Similar crossover hot spot instigator sequences occur in other bacteria. In addition to the presence of this putative recombination site in the MSP-1 gene, this site is also present in several other P. falciparum protein genes that are distributed throughout chromosome 2 and 3 (Genbank). Interestingly, these chromosomes contain variable genes (e.g. var and rifin genes), which contain Chi and/or Chi-like sequences.
Though less prevalent than K1, Mad20, and RO33, MR is still detected in more than one-fourth of the samples we have genotyped. Of the 45 sequences available in Genbank or determined in this study, only one, Kenya Mad20-150 S49-3, had a sequence and length consistent with the hypothetical Mad20 progenitor of the MR allele family. If the Mad20 allelic diversity detected at this time is representative of the Mad20 allelic diversity at the time MR arose, then the recombination between the RO33 and Mad20 progenitor was followed by expansion of this MR recombinant into the population. It has been proposed that the allele families in P. falciparum MSP-1 are maintained by balancing selection [35], so it may be possible that the high frequency of this novel allele family in the sample under study may be due to some selective advantage that has allowed it to survive and amplify in the population. Further investigations will be needed to determine the worldwide frequency of this new allele family and if these alleles are under positive selection, and if so, what factors drive that selection such as differential recognition by the host immune system.
In conclusion, we have provided evidence that intragenic recombination has generated a novel, fourth allelic form of the Block 2 region of the MSP-1 antigen of P. falciparum. We have also identified a putative recombination spot in the MSP-1 gene that bears homology to the prokaryotic recombination hot spot. This new information on the genetic diversity of MSP-1 gene would be useful for vaccine development and testing efforts as well as in molecular epidemiologic investigations. This study also provides another example of intragenic recombination as an important factor for generating genetic variability in P. falciparum parasites in nature.
Acknowledgments
We thank the study participants for their willingness to participate in the study, the CDC/KEMRI ABCP staff for their support, and A. Barskey for his technical support. We thank one of the reviewers for bringing up several important points for inclusion in the manuscript. We also thank the Director of the Kenya Medical Research Institute (KEMRI) for approving the publication of this research. Shannon Takala was supported by the Emerging Infectious Diseases Fellowship Program administered through APHL and CDC. Ananias A. Escalante is supported by the grant NIH R01 GM60740.
References
- 1.WHO. Malaria Fact Sheet. 1998 http://www.who.int/inf-fs/en/fact094.html.
- 2.Binks RH, Baum J, Oduola AM, et al. Population genetic analysis of the Plasmodium falciparum erythrocyte binding antigen-175 (eba-175) gene. Mol Biochem Parasitol. 2001;114:63–70. doi: 10.1016/s0166-6851(01)00240-7. [DOI] [PubMed] [Google Scholar]
- 3.Da Silveira LA, Ribeiro WL, Kirchgatter K, et al. Sequence diversity and linkage disequilibrium within the merozoite surface protein-1 (Msp-1) locus of Plasmodium falciparum: a longitudinal study in Brazil. J Eukaryot Microbiol. 2001;48:433–9. doi: 10.1111/j.1550-7408.2001.tb00176.x. [DOI] [PubMed] [Google Scholar]
- 4.Escalante AA, Grebert HM, Chaiyaroj SC, et al. Polymorphism in the gene encoding the apical membrane antigen-1 (AMA-1) of Plasmodium falciparum X. Asembo Bay Cohort Project Mol Biochem Parasitol. 2001;113:279–87. doi: 10.1016/s0166-6851(01)00229-8. [DOI] [PubMed] [Google Scholar]
- 5.Hughes MK, Hughes AL. Natural selection on Plasmodium surface proteins. Mol Biochem Parasitol. 1995;71:99–113. doi: 10.1016/0166-6851(95)00037-2. [DOI] [PubMed] [Google Scholar]
- 6.Conway D. Natural selection on polymorphic malaria antigens and the search for a vaccine. Parasitol Today. 1997;13:26–9. doi: 10.1016/s0169-4758(96)10077-6. [DOI] [PubMed] [Google Scholar]
- 7.Escalante AA, Lal AA, Ayala FJ. Genetic polymorphism and natural selection in the malaria parasite Plasmodium falciparum. Genetics. 1998;149:189–202. doi: 10.1093/genetics/149.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller LH, Roberts T, Shahabuddin M, McCutchan TF. Analysis of sequence diversity in the Plasmodium falciparum merozoite surface protein-1 (MSP-1) Mol Biochem Parasitol. 1993;59:1–14. doi: 10.1016/0166-6851(93)90002-f. [DOI] [PubMed] [Google Scholar]
- 9.Qari SH, Shi YP, Goldman IF, Nahlen BL, Tibayrenc M, Lal AA. Predicted and observed alleles of Plasmodium falciparum merozoite surface protein-1 (MSP-1), a potential malaria vaccine antigen. Mol Biochem Parasitol. 1998;92:241–52. doi: 10.1016/s0166-6851(98)00010-3. [DOI] [PubMed] [Google Scholar]
- 10.Eisen DP, Marshall VM, Billman-Jacobe H, Coppel RL. A Plasmodium falciparum apical membrane antigen-1 (AMA-1) gene apparently generated by intragenic recombination. Mol Biochem Parasitol. 1999;100:243–6. doi: 10.1016/s0166-6851(99)00054-7. [DOI] [PubMed] [Google Scholar]
- 11.Taylor HM, Kyes SA, Newbold CI. Var gene diversity in Plasmodium falciparum is generated by frequent recombination events. Mol Biochem Parasitol. 2000;110:391–7. doi: 10.1016/s0166-6851(00)00286-3. [DOI] [PubMed] [Google Scholar]
- 12.Kerr PJ, Ranford-Cartwright LC, Walliker D. Proof of intragenic recombination in Plasmodium falciparum. Mol Biochem Parasitol. 1994;66:241–8. doi: 10.1016/0166-6851(94)90151-1. [DOI] [PubMed] [Google Scholar]
- 13.Conway DJ, Cavanagh DR, Tanabe K, et al. A principal target of human immunity to malaria identified by molecular population genetic and immunological analyses. Nat Med. 2000;6:689–92. doi: 10.1038/76272. [DOI] [PubMed] [Google Scholar]
- 14.Branch OH, Takala S, Kariuki S, et al. Plasmodium falciparum genotypes, low complexity of infection, and resistance to subsequent malaria in participants in the Asembo Bay Cohort Project. Infect Immun. 2001;69:7783–92. doi: 10.1128/IAI.69.12.7783-7792.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brockman A, Paul RE, Anderson TJ, et al. Application of genetic markers to the identification of recrudescent Plasmodium falciparum infections on the northwestern border of Thailand. Am J Trop Med Hyg. 1999;60:14–21. doi: 10.4269/ajtmh.1999.60.14. [DOI] [PubMed] [Google Scholar]
- 16.Robert F, Ntoumi F, Angel G, et al. Extensive genetic diversity of Plasmodium falciparum isolates collected from patients with severe malaria in Dakar, Senegal. Trans R Soc Trop Med Hyg. 1996;90:704–11. doi: 10.1016/s0035-9203(96)90446-0. [DOI] [PubMed] [Google Scholar]
- 17.Contamin H, Fandeur T, Rogier C, et al. Different genetic characteristics of Plasmodium falciparum isolates collected during successive clinical malaria episodes in Senegalese children. Am J Trop Med Hyg. 1996;54:632–43. doi: 10.4269/ajtmh.1996.54.632. [DOI] [PubMed] [Google Scholar]
- 18.Ouattara A, Koita O, Doumbo O, Diakite M, Proudiougo B, Diawara S, Thera MA, Traore O, Dolo A, Toure O, Djimbe A, Krogstad D. Application of molecular markers as an epidemiological tool in a malaria endemic area of Mali. Suppl Am J Trop Med Hyg. 2000;62:391–2. [Google Scholar]
- 19.Jongwutiwes S, Tanabe K, Nakazawa S, Yanagi T, Kanbara H. Sequence variation in the tripeptide repeats and T cell epitopes in P190 (MSA-1) of Plasmodium falciparum from field isolates. Mol Biochem Parasitol. 1992;51:81–9. doi: 10.1016/0166-6851(92)90203-v. [DOI] [PubMed] [Google Scholar]
- 20.Tanabe K, Mackay M, Goman M, Scaife JG. Allelic dimorphism in a surface antigen gene of the malaria parasite Plasmodium falciparum. J Mol Biol. 1987;195:273–87. doi: 10.1016/0022-2836(87)90649-8. [DOI] [PubMed] [Google Scholar]
- 21.Ranford-Cartwright L, Walliker D. Intragenic recombinants of Plasmodium falciparum identified by in situ polymerase chain reaction. Mol Biochem Parasitol. 1999;102:13–20. doi: 10.1016/s0166-6851(99)00069-9. [DOI] [PubMed] [Google Scholar]
- 22.Conway DJ, Rosario V, Oduola AM, Salako LA, Greenwood BM, McBride JS. Plasmodium falciparum: intragenic recombination and nonrandom associations between polymorphic domains of the precursor to the major merozoite surface antigens. Exp Parasitol. 1991;73:469–80. doi: 10.1016/0014-4894(91)90071-4. [DOI] [PubMed] [Google Scholar]
- 23.Conway DJ, Roper C, Oduola AM, et al. High recombination rate in natural populations of Plasmodium falciparum. Proc Natl Acad Sci USA. 1999;96:4506–11. doi: 10.1073/pnas.96.8.4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakihama N, Kimura M, Hirayama K, et al. Allelic recombination and linkage disequilibrium within Msp-1 of Plasmodium falciparum, the malignant human malaria parasite. Gene. 1999;230:47–54. doi: 10.1016/s0378-1119(99)00069-4. [DOI] [PubMed] [Google Scholar]
- 25.Li M, Xie Y, Li Y, Ren D. Cloning and sequence analysis of the gene encoding the second region of MSA1 from three Chinese isolates of Plasmodium falciparum. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 1996;18:161–5. [PubMed] [Google Scholar]
- 26.Bloland PB, Boriga DA, Ruebush TK, et al. Longitudinal cohort study of the epidemiology of malaria infections in an area of intense malaria transmission II. Descriptive epidemiology of malaria infection and disease among children. Am J Trop Med Hyg. 1999;60:641–8. doi: 10.4269/ajtmh.1999.60.641. [DOI] [PubMed] [Google Scholar]
- 27.Saiki RK, Gelfand DH, Stoffel S, et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–91. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 28.Paabo S, Irwin DM, Wilson AC. DNA damage promotes jumping between templates during enzymatic amplification. J Biol Chem. 1990;265:4718–21. [PubMed] [Google Scholar]
- 29.Jansen R, Ledley FD. Disruption of phase during PCR amplification and cloning of heterozygous target sequences. Nucleic Acids Res. 1990;18:5153–6. doi: 10.1093/nar/18.17.5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyerhans A, Vartanian JP, Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990;18:1687–91. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marton A, Delbecchi L, Bourgaux P. DNA nicking favors PCR recombination. Nucleic Acids Res. 1991;19:2423–6. doi: 10.1093/nar/19.9.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Odelberg SJ, Weiss RB, Hata A, White R. Template-switching during DNA synthesis by Thermus aquaticus DNA polymerase I. Nucleic Acids Res. 1995;23:2049–57. doi: 10.1093/nar/23.11.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lam ST, Stahl MM, McMilin KD, Stahl FW. Rec-mediated recombinational hot spot activity in bacteriophage lambda. II. A mutation which causes hot spot activity. Genetics. 1974;77:425–33. doi: 10.1093/genetics/77.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su X, Ferdig MT, Huang Y, et al. A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science. 1999;286:1351–3. doi: 10.1126/science.286.5443.1351. [DOI] [PubMed] [Google Scholar]
- 35.Hughes AL. Positive selection and interallelic recombination at the merozoite surface antigen-1 (MSA-1) locus of Plasmodium falciparum. Mol Biol Evol. 1992;9:381–93. doi: 10.1093/oxfordjournals.molbev.a040730. [DOI] [PubMed] [Google Scholar]


