Abstract
Background
Although alterations in nonspecific (or global) DNA methylation (GDM) in specific cells are known to be involved in the process of lung carcinogenesis, similar associations have not been evaluated in other smoking-related cancers of the head and neck.
Methods
We evaluated the status of GDM by using monoclonal antibodies specific for 5-methylcytosine (5-mc) in oral squamous cell carcinoma (SCC) specimens of 48 cigarette smokers who had SCC develop and in 93 age-, race-, and sex-matched smokers who did not.
Results
Percentages of cells positive for 5-mc immunostaining of DNA of SCC and dysplastic lesions were significantly higher than those of normal oral epithelial cells from cancer subjects and from noncancer subjects. The degree of DNA methylation was unrelated to DNA content.
Conclusions
The pattern of GDM in oral SCCs is different from that of lung SCCs. The differences in nutrient risk factor profiles that are related to GDM and differential activity of DNA methyltranferases between oral and lung SCCs may explain these observations.
Keywords: DNA, methylation, oral cancer
Despite abundant evidence for the mutational basis of cancer, epigenetic changes, which are events that alter the way genes are expressed without affecting genetic coding, also may contribute to carcinogenesis. Methylation of DNA, which occurs at the cytosine residues of cytosine guanine dinucleotide (CpG) dinucleotides by an enzymatic reaction that produces 5-methylcytosine (5-mc), is an extensively characterized mechanism for epigenetic gene regulation. Neoplastic cells may simultaneously harbor widespread (global) genomic hypomethylation, regional areas of hypermethylation, and increased DNA-methyltransferase (Dnmt) activity. Although the precise roles of these components are unclear, each component of this “methylation imbalance” may fundamentally contribute to tumor progression. One of the first alterations of DNA methylation to be recognized in neoplastic cells was a decrease in nonspecific (or global) DNA methylation (GDM). Recent attention has been directed toward investigating the role of regional hypermethylation of tumor suppressor genes.1, 2 Despite the frequently observed cancer-associated increases of regional hypermethylation, the prevalence of nonspecific (or global) DNA hypomethylation in many types of human cancer3-6 suggests that such hypomethylation plays a significant and fundamental role in tumorigenesis. The most likely mechanisms through which nonspecific (or global) DNA hypomethylation may induce neoplastic transformation (ie, inducing genomic instability,7, 8 causing abnormal chromosomal structures,9 and activating oncogenes10-14) are biologically plausible. We believe that analysis of GDM can provide insights that may be missed in studies limited to specific genes whose state of methylation may not mirror most of the methylation changes in the genome.
The methods commonly used to study GDM rely either on quantitative measurement of 5-mc by high-performance liquid chromatography or on quantification of in vitro transfer of radiolabeled methyl groups from S-adenosyl methionine (SAM) to sites in DNA that were not methylated in vivo. Besides being cumbersome, these techniques may have limited value in the evaluation of GDM in the process of carcinogenesis, because they do not permit an evaluation of DNA methylation in intact or specific types of cells.
In a previous study, using a monoclonal antibody against 5-mc, we demonstrated that alterations in GDM play an important role in the development of squamous cell carcinomas (SCCs) of the lung. The 5-mc monoclonal antibody used in this study was produced by one of the coauthors (A.N.). This antibody has been well characterized15 and used with success in several laboratories.16-20 As previously described, these antibodies do not bind to nonmethylated DNA,21 and the specificity of these antibodies for methylated DNA has been well documented.22 The validity of using 5-mc antibodies in GDM assessment is supported by studies that compared and documented similar results of GDM by the radiolabeled methyl incorporation assay23 and by the high-performance capillary electrophoresis assay.24 The purpose of this study was to assess GDM status using the same antibody and to evaluate whether such methylation plays a similar role in the process of oral carcinogenesis, a cancer that shares common risk factors such as cigarette smoking.
MATERIALS AND METHODS
Patients and Tissue Specimens
Among patients diagnosed with oral lesions between 1984 and 2000 at the Veterans Administration Medical Center in Birmingham, Alabama, 57 and 115 subjects had oral SCCs and normal oral epithelium, respectively. Among the subjects with SCC, 48 subjects had adequate tissue specimens (formalinfixed and paraffin-embedded sections). Within these, we identified malignant lesions adequate for evaluation in 39 specimens and uninvolved oral epithelium (histopathologically normal appearing) and dysplastic lesions in 44 and 42 specimens, respectively. All of the normal subjects had adequate normal oral epithelium for evaluation. All 39 subjects with SCCs were included in this study. The control group (n = 93) consisted of age-, race-, and sex-matched subjects (two each for 45 cases of cancer and one each for three cases of cancer) with normal oral epithelium. The specimens (gingiva) from the control group were removed during routine periodontal surgery. The exposure to cigarette smoke was similar between subjects with and without SCCs; all subjects reported long-term smoking habits.
One tissue block from each control subject was selected to provide a section that contained adequate normal and uninflamed oral epithelium. One or more tissue blocks from each subject with SCC were selected to provide sections that contained samples of uninvolved oral epithelium and dysplasia, in addition to invasive carcinomas.
Histologic Criteria and Classification of Lesions
Oral epithelium was classified histologically as normal (in noncancer specimens), uninvolved (SCC-associated, but histopathologically normal), or dysplastic by one of the pathologists (W.C.B.) involved in the study.
Immunohistochemical Analysis
Our methods of immunohistochemical analysis with various antibodies (with/without various antigen retrieval techniques) have been reported previously.25 A comparison of results with and without different antigen retrieval protocols for anti-5-mc antibody revealed that the following antigen retrieval protocol gave the optimal results: slides containing 4-μm thick tissue sections were placed in 0.01 M citric acid, pH 6.0, in a microwave oven set at full potency for 10 minutes. After the antigen retrieval, the slides were immersed in 3.5 N HCl for 15 minutes at room temperature to expose the CpGs. The sections then were treated with 3.0% H2O2 for 4 minutes to quench endogenous peroxidase activity. Sections were incubated with preimmune goat serum (1%) for 20 minutes at room temperature to suppress nonspecific staining and then subsequently incubated with a hybridoma supernatant containing anti-5-mc monoclonal antibody at a concentration of 5 μg/mL for 1 hour at room temperature. Companion matching slides processed identically and stained without primary antibody served as controls (deletes). The remainder of the staining procedure was performed using a biotin-streptavidin detection system (Signet, Dedham, MA). The substrate diaminobenzidine tetrahydrochloride was used for visualization of the antigen-antibody complex, and sections were counterstained lightly with hematoxylin.
Immunostaining was performed without knowledge or grouping of the specimens by diagnosis (SCC/noncancer). To determine the reproducibility of the assay, five sections each from two tissue blocks were stained on 5 different days. In addition, sections from 10 randomly selected tissue blocks were stained on 2 different days, and the mean immunostaining score was calculated and compared for each group.
Assessment of Immunostaining
The immunostaining for 5-mc was localized in the nuclei of the cells. Two observers (C.J.P. and W.C.B.) scored the 5-mc immunostaining. Each observer graded the intensity of immunostaining independently on a scale of 0 (no staining) to 4+ (intense staining) in normal and uninvolved oral epithelial cells, dysplastic cells, and SCC. In addition, the percentage of cells at each intensity was estimated and multiplied by the appropriate intensity score to obtain a weighted average of the immunostaining score. The final score reported is the average of the two observers. A lower 5-mc score reflects a lower degree of GDM or hypomethylation.
To test whether the higher GDM in cancer cells compared with normal cells is due to higher DNA content in cancer cells. Two serial 4-μmthick tissue sections from formalin-fixed paraffin-embedded oral tissue blocks from a subject diagnosed with oral cancer were stained with the Feulgen stain for DNA content (Bacus Laboratories, Inc., Lombard, IL) and for DNA methylation using the 5-mc antibody. Slides were scanned using the BLISS imaging system. Nuclear images from areas of cancer and normal (approximately 2000 cells from each group of cells) cells were extracted using Cell Finder software, and optical density (OD) measurements were performed using Nuclear Grade Software (Bacus Laboratories, Inc.). The raw OD measurements were exported for statistical analysis.
Statistical Analysis
Descriptive statistics such as mean, median, standard deviation (SD) of the mean, and range were calculated for the percentage of cells positive for 5-mc and the weighted average of the 5-mc score for each type of tissue. Either the Wilcoxon rank sum or the Wilcoxon sign rank test was used to test for differential staining between different types of tissues. The association between DNA content and DNA methylation was examined by classifying ODs of DNA content and methylation into categories of low, medium, and high levels using the 33rd and 66th percentile values and by calculating the kappa statistic. The association between DNA content and methylation was also determined by calculating the Spearman rank correlation coefficient between direct ODs of DNA content and methylation.
RESULTS
The 5-mc scores were reproducible for individual cases and for groups of tissue specimens. The average coefficient of variation of the assay was 4%. The mean ± SD for 5-mc scores of the 10 tissue sections stained and evaluated on 2 different days was 1.17 ± 0.3 and 1.16 ± 0.2. Figure 1 shows the representative pattern of immunohistochemical staining for 5-mc in the normal oral epithelium of noncancer (A), uninvolved oral epithelium (B), and dysplasia (C) of an SCC specimen and SCC (D). The 5-mc immunostaining results of different types of tissue are shown in Table 1. There was no significant difference in the percentages of cells positive for 5-mc or 5-mc scores between uninvolved normal oral epithelium of patients with cancer and normal oral epithelium of noncancer subjects (p = .41 and p = .44, respectively). The percentages of cells positive for 5-mc and 5-mc scores were significantly higher in dysplastic lesions (p = .01, .02, respectively) or cancer cells (p = .03, .03, respectively) compared with normal oral epithelium of subjects without cancer. Among subjects with cancer, the percentages of cells positive for 5-mc and 5-mc scores were significantly higher in dysplastic lesions (p = .003, .007, respectively) and in cancer tissues (p = .003, .003, respectively) compared with uninvolved oral epithelium. When results were compared taking matching into account, the results remained the same: there were no differences in the percent of cells positive for 5-mc or 5-mc scores between uninvolved normal oral epithelium of patients with cancer and normal oral epithelium of subjects without cancer (p = .32 and p = .27 respectively), but the percentages of cells positive for 5-mc and 5-mc scores were significantly higher in dysplastic lesions (p = .01, .04, respectively) and cancer cells (p = .005, .005, respectively) than in normal oral epithelial cells from subjects without cancer.
FIGURE 1.
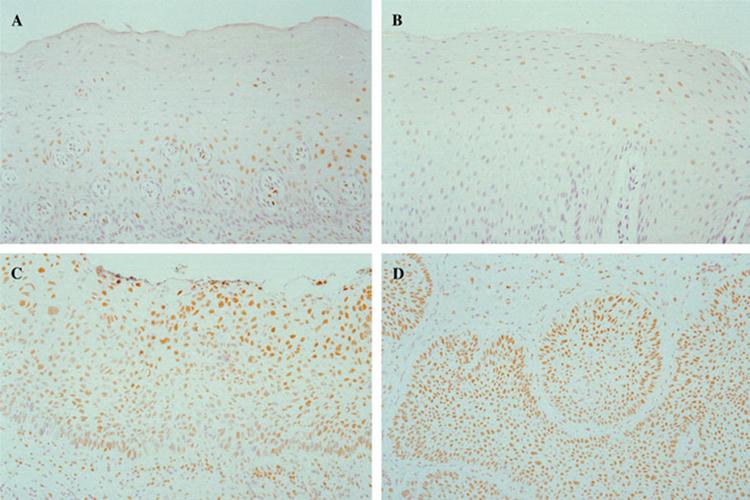
The immunohistochemical staining for 5-methylcytosine in normal oral epithelium of noncancer (A), uninvolved oral epithelium (B), and dysplasia (C) of a squamous cell carcinoma (SCC) specimen and SCC (D). [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Table 1.
Percentages of cells positive for 5-mc and weighted averages of immunostaining scores for 5-mc in normal and uninvolved oral epithelium, dysplasia, and squamous cell carcinoma.
| Tissue type | % of cells positive for 5-mc, mean (median) | Weighted average of 5-mc score, mean (median) |
|---|---|---|
| Normal oral epithelium of noncancer (n = 93) | 37 (35)a | 0.53 (0.47)b |
| Uninvolved oral epithelium of cancer (n = 44) | 34 (33)c | 0.48 (0.39)d |
| Dysplasia of cancer (n = 42) | 48 (50)e | 0.72 (0.69)f |
| Cancer (n = 39) | 51 (55)g | 0.77 (0.80)h |
Abbreviation: 5-mc, 5-methylcytosine.
p values; a, c, .41; b, d, .44; a, e, .01; b, f, .02; a, g, .03; b, h, .03; c, e,.003; d, f, .007; c, g, .003; d, h, .003.
The results of the experiment that assessed DNA content and methylation by ODs showed that the DNA content was significantly higher in cancer cells than normal cells. Global methylation status was also significantly higher in cancer cells than normal cells, similar to the overall methylation results observed. As shown in Table 2, no association was found between DNA content and DNA methylation in cancer or normal tissues, when low, medium, and high categories were compared by Kappa statistic or when direct ODs were compared by Spearman rank correlation. This experiment was repeated with five additional cases of oral cancer, and the results were similar. These observations indicate that the higher GDM in cancer cells is unlikely to be due to higher DNA content in these cells.
Table 2.
Associations between DNA methylation and DNA content in cancer and normal cells.
| Type of cells | Categories of low, medium, and high DNA content and methylation | Direct ODs of DNA content and methylation |
|---|---|---|
| Cancer cells | Kappa = -10; p = .34 | Spearman rank correlation coefficient = -0.05; p = .55 |
| Normal cells | Kappa = 0.002; p = .54 | Spearman rank correlation coefficient = -0.033; p = .66 |
Abbreviation: OD, optical density.
DISCUSSION
Although several primary cancers have been reported to be globally hypomethylated compared with normal tissues, as summarized by Feinberg et al,26 the degrees of GDM in primary tumors compared with normal tissues have been inconsistent. Some investigators have reported no differences or even increased methylation in tumors. We have previously reported that SCCs of the lung are hypomethylated compared with normal bronchial epithelium, epithelial hyperplastic lesions, and dysplastic lesions.27 We now report that dysplastic lesions and SCCs of the oral cavity are significantly hypermethylated compared with uninvolved oral epithelium of the same subjects and to normal oral epithelium of subjects without cancer. We ruled out the possibility that higher methylation in cancer cells may be due to higher DNA content in these cells. These observations demonstrate the site-specific differences in methylation patterns in primary cancers of the head and neck. Such differences challenge the scientific view of the effects of GDM on carcinogenesis.
We considered the possibility that the differences observed in methylation patterns in tumors of the lung and tumors of the oral cavity could be due to differing embryologic origin. The epithelium of the lung is entirely of endodermal origin, whereas the epithelium of the oropharynx has anatomic sites of both ectodermal and endodermal origin. The oral cavity mucosa and mucosa of the anterior two thirds of the tongue arise from the primitive stomadeum and are of ectodermal origin. The pharyngeal mucosa and posterior tongue arise from the foregut and are endodermally derived.28 To evaluate for possible differences in methylation from embryologic origin, the oral lesions were divided by anatomic site into endodermal and ectodermal groups. No significant differences in methylation patterns were observed between these two groups (data not shown).
We entertain the possibility that differences in vitamin profiles that influence GDM in the head and neck area may partially explain these observations. Folate and vitamin B12 are of particular interest because of their inactivation by exposure to cigarette smoke, a common risk factor for head and neck cancer. Both folate and vitamin B12 are essential cofactors in the synthesis of adenosyl-methionine (SAM), the body’s primary donor of methyl groups for methylation of DNA. We have previously reported that both folate and vitamin B12 are deficient in SCCs of the lung, and lower levels of both vitamins are associated with global DNA hypomethylation in these tissues.29 Multiple regression models of that study, however, suggested that folate is the limiting vitamin for DNA methylation in both cancerous tissues and at-risk tissues of the lung. A preliminary study conducted with SCCs of the oral cavity and matched normal oral tissues demonstrated that folate concentrations are not significantly different between these tissues, but vitamin B12 levels are significantly lower in oral SCCs than normal oral tissues.30 Although these observations suggest that folate is likely to be the limiting vitamin for GDM in the oral cavity as well, significantly higher methylation in oral SCCs compared with normal oral epithelium is not explained by these vitamin profiles. As discussed later, the role of vitamins in the process of carcinogenesis may vary, depending on the site of cancers in the head and neck.
Although a large number of prospective and retrospective studies have documented a consistent relationship between reduced risk of some cancers (eg, lung) and high levels of vegetable and/or fruit consumption,31 this association is weak for certain head and neck cancers such as those of the oral cavity. A diet deficient in foods of animal origin seems to be a more significant riskfactor for oral premalignancy than is a diet deficient in fruits and vegetables that contain folate and other antioxidants.32 Dietary intake of folate also showed no consistent relationship to risk of oral and pharyngeal cancer.33 Although these observations suggest that vitamins that are important in lung SCCs may not be beneficial in prevention of oral SCCs, the roles of other vitamins and nutrients in relation to oral carcinogenesis need to be investigated.
Most commonly, cancer cells are characterized by global DNA hypomethylation, regional hypermethylation of CpG islands, and an increase in Dnmt.34 Global DNA hypomethylation in these cancers, despite increased Dnmt activity, is thought to be because Dnmt may be incapable of methylating double-stranded unmethylated DNA present in these tissues. Increased methylation of specific CpG islands under similar conditions may be due to the establishment of a cancer-specific DNA methylation profile in these tissues. Our results, however, suggest that Dnmt may be capable of methylating DNA globally and in specific CpG islands, because promoter hypermethylation profiles of several tumor-associated genes (p16, p15, hMLH1, MGMT, and E-cadherin) have been reported in oral SCCs.35, 36 Aberrant methylation of multiple genes (p14ARF, DCC, DAP kinase, MINT1, and MINT31) have also been correlated with clinicopathologic features of oral SCCs.37 Hypermethylation of specific genes (p16) have also been reported in oral epithelial dysplasia.38 If a significant association between nonspecific global DNA hypermethylation and gene-specific methylation can be established, alterations in GDM may serve as a marker for methylation changes in specific genes in these tissues. Further studies are needed to evaluate these associations. However, because the GDM status of normal oral epithelium of at-risk subjects (smokers without cancer) is not significantly different from that of uninvolved oral epithelium of subjects with cancer, it is unlikely that evaluation of GDM in normal tissues of at-risk subjects will help identify subjects who are at imminent risk of oral cancer developing. However, it is possible that alterations in GDM could have been unidentified in at-risk tissues used in this study, because only one site of the mucosa of these subjects was examined rather than multiple sites. Also, this study does not rule out the possibility of alterations in specific genes. Further studies are warranted to evaluate whether altered methylation in specific genes in preneoplastic lesions of the oral cavity may predict the development of oral cancer.
Footnotes
Contract grant sponsor: Supported by Grant R03 CA 91273 from the National Cancer Institute and a pilot grant from the NYU Oral Cancer RAAHP Center.
REFERENCES
- 1.Herman JG, Merlo A, Mao L, et al. Inactivation of theCDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 2.Graff JR, Herman JG, Lapidus RG, et al. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 1995;55:5195–5199. [PubMed] [Google Scholar]
- 3.Gama-Sosa MA, Slagel VA, Trewyn RW, et al. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983;11:6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 1988;48:1159–1161. [PubMed] [Google Scholar]
- 5.Kliasheva RI. DNA methylation in human lung tumors[abstract] Voprosy Onkologii. 1990;36:186–189. [PubMed] [Google Scholar]
- 6.Cheng P, Schmutte C, Cofer KF, Felix JC, Yu MC, Dubeau L. Alterations in DNA methylation are early, but not initial, events in ovarian tumorigenesis. Br J Cancer. 1997;75:396–402. doi: 10.1038/bjc.1997.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laird PW, Jaenisch R. The role of DNA methylation in cancer genetic and epigenetics. Annu Rev Genet. 1996;30:411–464. doi: 10.1146/annurev.genet.30.1.441. [DOI] [PubMed] [Google Scholar]
- 8.Chen RZ, Pettersson U, Beard C, Jackson-Grushby L, Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature. 1998;395:89–92. doi: 10.1038/25779. [DOI] [PubMed] [Google Scholar]
- 9.Lewis J, Bird A. DNA methylation and chromatin structure. FEBS Lett. 1991;285:155–159. doi: 10.1016/0014-5793(91)80795-5. [DOI] [PubMed] [Google Scholar]
- 10.Feinberg AP, Vogelstein B. Hypomethylation of ras oncogenes in primary human cancers. Biochem Biophys Res Commun. 1983;111:47–54. doi: 10.1016/s0006-291x(83)80115-6. [DOI] [PubMed] [Google Scholar]
- 11.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 12.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 13.Razin A, Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991;55:451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weissbach A, Ward C, Bolden A. Eukaryotic DNA methylation and gene expression. Curr Topics Cell Regul. 1989;30:1–21. doi: 10.1016/b978-0-12-152830-0.50003-7. [DOI] [PubMed] [Google Scholar]
- 15.Reynaud C, Bruno C, Boullanger P, Grange J, Barbesti S, Niveleau A. Monitoring of urinary excretion of modified nucleosides in cancer patients using a set of six monoclonal antibodies. Cancer Lett. 1992;61:255–262. doi: 10.1016/0304-3835(92)90296-8. [DOI] [PubMed] [Google Scholar]
- 16.Miniou P, Jeanpierre M, Blanquet V, et al. Abnormal methylation pattern in constitutive and facultative (X inactive chromosome) heterochromatin of ICF patients. Hum Mol Genet. 1994;3:2093–2102. doi: 10.1093/hmg/3.12.2093. [DOI] [PubMed] [Google Scholar]
- 17.Bensaada M, Kiefer H, Tachdjian G, et al. Altered patterns of DNA methylation on chromosomes from leukemia cell lines: identification of 5-methylcytosines by indirect immunodetection. Cancer Genet Cytogenet. 1998;103:101–109. doi: 10.1016/s0165-4608(97)00409-3. [DOI] [PubMed] [Google Scholar]
- 18.De Capoa A, Di Leandro M, Grappelli C, et al. Computer-assisted analysis of methylation status of individual interphase nuclei in human cultured cells. Cytometry. 1998;31:85–92. doi: 10.1002/(sici)1097-0320(19980201)31:2<85::aid-cyto3>3.3.co;2-8. [DOI] [PubMed] [Google Scholar]
- 19.Habib M, Fares F, Bourgeois CA, et al. DNA global hypomethylation in EBV-transformed interphase nuclei. Exp Cell Res. 1999;249:46–53. doi: 10.1006/excr.1999.4434. [DOI] [PubMed] [Google Scholar]
- 20.De Capoa A, Febbo FR, Giovannelli F, et al. Reduced levels of poly(ADP-ribosyl)ation result in chromatincompaction and hypermethylation as shown by cell-by-cell computer-assisted quantitative analysis. FASEB J. 1999;13:89–93. doi: 10.1096/fasebj.13.1.89. [DOI] [PubMed] [Google Scholar]
- 21.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 22.Hernandez-Blazquez FJ, Habib M, Dumollard JM, et al. Evaluation of global DNA hypomethylation in human colon cancer tissues by immunohistochemistry and image analysis. Gut. 2000;47:689–693. doi: 10.1136/gut.47.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piyathilake CJ, Johanning GL, Frost AR, et al. Immunohistochemical evaluation of global DNA methylation: comparison with in vitro radiolabeled methyl incorporation assay. Biotech Histochem. 2000;75:251–258. doi: 10.3109/10520290009085128. [DOI] [PubMed] [Google Scholar]
- 24.Fraga MF, Herranz M, Espada J, et al. A mouse skin multistage carcinogenesis model reflects the aberrant DNA methylation patterns of human tumors. Cancer Res. 2004;64:5527–5534. doi: 10.1158/0008-5472.CAN-03-4061. [DOI] [PubMed] [Google Scholar]
- 25.Grizzle WE, Myers RB, Manne U, Srivastava S. Immunohistochemical evaluation of biomarkers in prostatic and colorectal neoplasia. In: Hanausek M, Walaszek Z, editors. John Walker’s Methods in molecular medicine — tumor marker protocols. Humana Press Inc.; Totowa, NJ: 1998. pp. 143–160. [Google Scholar]
- 26.Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 1988;48:1159–1161. [PubMed] [Google Scholar]
- 27.Piyathilake CJ, Frost AR, Bell WC, et al. Altered global methylation of DNA: an epigenetic difference in susceptibility for lung cancer is associated with its progression. Hum Pathol. 2001;32:856–862. doi: 10.1053/hupa.2001.26471. [DOI] [PubMed] [Google Scholar]
- 28.Moore KL, Persand TVN. The developing human. 5th ed. WB Saunders Company; Philadelphia: 1993. pp. 186–225. [Google Scholar]
- 29.Piyathilake CJ, Johanning GL, Macaluso M, Whiteside MA, Heimburger DC, Grizzle WE. Localized deficiencies of folate and vitamin B12 in lung tissues are associated with global DNA methylation. Nutr Cancer. 2000;37:99–107. doi: 10.1207/S15327914NC3701_13. [DOI] [PubMed] [Google Scholar]
- 30.Piyathilake CJ, Oelschlager D, Bell WC, et al. Variations of vitamin concentrations and DNA methylation in squamous cell cancers of the oral cavity, larynx and lung. FASEB J. 2001;15:A619–A619. [Google Scholar]
- 31.Ziegler RG, Mayne ST, Swanson CA. Nutrition and lung cancer. Cancer Causes Control. 1996;7:157–177. doi: 10.1007/BF00115646. [DOI] [PubMed] [Google Scholar]
- 32.Carley KW, Puttaiah R, Alvarez JO, Heimburger DC, Anantha N. Diet and oral premalignancy in female south Indian tobacco and betel chewers: a case-control study. Nutr Cancer. 1994;22:73–84. doi: 10.1080/01635589409514333. [DOI] [PubMed] [Google Scholar]
- 33.McLaughlin JK, Gridley G, Block G, et al. Dietary factors in oral and pharyngeal cancer. J Natl Cancer Inst. 1988;80:1237–1243. doi: 10.1093/jnci/80.15.1237. [DOI] [PubMed] [Google Scholar]
- 34.Jones PA, Laird PW. Cancer epigenetics comes of age. NatGenet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 35.Viswanathan M, Tsuchida N, Shanmugam G. Promoter hypermethylation profile of tumor-associated genes p16, p15, hMLH1, MGMT and E-cadherin in oral squamous cell carcinoma. Int J Cancer. 2003;105:41–46. doi: 10.1002/ijc.11028. [DOI] [PubMed] [Google Scholar]
- 36.Yeh KT, Chang JG, Lin TH, et al. Epigenetic changes of tumor suppressor genes, p15, p16, VHL and p53 in oral cancer. Oncol Rep. 2003;10:659–663. [PubMed] [Google Scholar]
- 37.Ogi K, Toyota M, Ohe-Toyota M, et al. Aberrant methylation of multiple genes and clinicopathological features in oral squamous cell carcinoma. Clin Cancer Res. 2002;8:3164–3171. [PubMed] [Google Scholar]
- 38.Kresty LA, Mallery SR, Knobloch TJ, et al. Alterations of p16 (INK4a) and p14 (ARF) in patients with severe oral epithelial dysplasia. Cancer Res. 2002;62:5295–5300. [PubMed] [Google Scholar]


