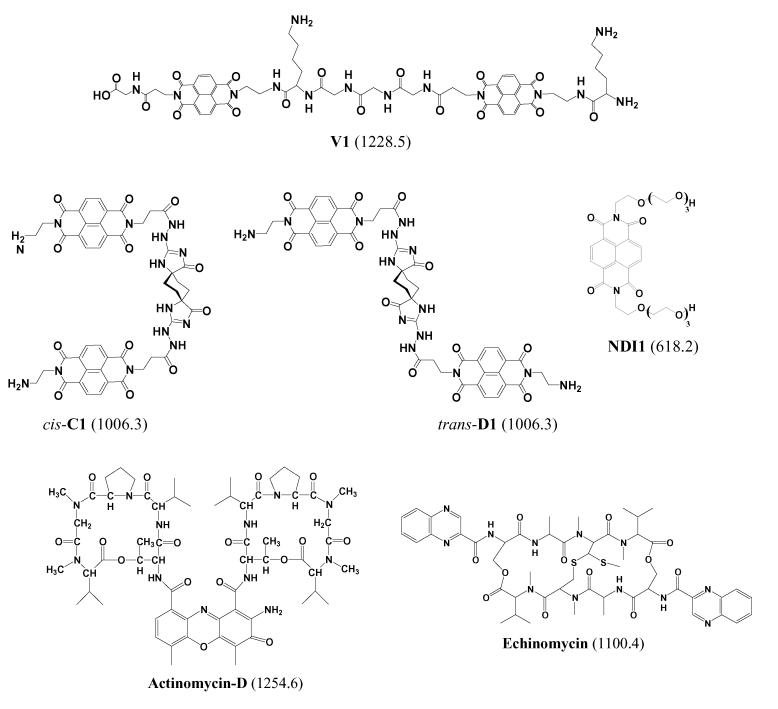
Abstract
The DNA binding of novel threading bis-intercalators V1, trans-D1, and cis-C1, which contain two naphthalene diimide (NDI) intercalation units connected by a scaffold, was evaluated using electrospray ionization mass spectrometry (ESI-MS) and DNAse footprinting techniques. ESI-MS experiments confirmed that V1, the ligand containing the –Gly3-Lys-peptide scaffold, binds to a DNA duplex containing the 5'-GGTACC-3' specific binding site identified in previous NMR-based studies. The ligand formed complexes with a ligand/DNA binding stoichiometry of 1:1, even when there was excess ligand in solution. Trans-D1 and cis-C1 are new ligands containing a rigid spiro-tricyclic scaffold in the trans- and cis- orientations, respectively. Preliminary DNAse footprinting experiments identified possible specific binding sites of 5'-CAGTGA-5' for trans-D1 and 5'-GGTACC-3' for cis-C1. ESI-MS experiments revealed that both ligands bound to DNA duplexes containing the respective specific binding sequences, with cis-C1 exhibiting the most extensive binding based on a higher fraction of bound DNA value. Cis-C1 formed complexes with a dominant 1:1 binding stoichiometry, whereas trans-D1 was able to form 2:1 complexes at ligand/DNA molar ratios ≥ 1 which is suggestive of non-specific binding. Collisional activated dissociation (CAD) experiments indicate that DNA complexes containing V1, trans-D1, and cis-C1 have a unique fragmentation pathway, which was also observed for complexes containing the commercially available bisintercalator echinomycin, as a result of similar binding interactions, marked by intercalation in addition to hydrogen bonding by the scaffold with the DNA major or minor groove.
INTRODUCTION
Many anticancer, antitumor, and antibacterial therapies are based on the interaction of small molecules with DNA [1, 2], fostering the need for sensitive and versatile analytical techniques that are both capable of characterizing the ligand/DNA interactions and compatible with library-based screening methods. Electrospray ionization mass spectrometry (ESI-MS) shows promise as a screening tool for the evaluation drug/DNA complexes due to its low sample consumption and rapid analysis time [3, 4]. During the electrospray process, non-covalent complexes are transferred to the gas-phase with minimal internal energy, allowing many of the binding interactions to be maintained. The preservation of these non-covalent complexes allows information about binding stoichiometry and selectivity to be elucidated from the mass spectra, while tandem mass spectrometry techniques, such as collisional activation dissociation (CAD), can be used to examine the binding mode.
One important class of DNA-interactive drugs are ligands that bind via intercalation of one or more aromatic groups between base pairs of duplex DNA [1, 2, 5]. There have been numerous ESI-MS studies that have examined the interaction between duplex DNA and well-studied, commercially available mono-intercalators such as the anthracyclines [6-11], porphyrins [12, 13], ruthenium compounds [11-15], ethidium bromide [16-18], actinomycin-D [12, 13], and aureolic acids [19]. Characteristics such as ligand binding stoichiometry [7, 8, 11-14, 16], sequence selectivity [7, 11, 13, 14, 16, 19], binding mode [12, 13, 15] and complex stability [9, 12, 16] have been examined with promising results correlating the binding trends observed in the mass spectra to known solution behavior. While the binding of many commercial monointercalators has been well-studied by ESI-MS, there has been only one study [20] that has focused on a bisintercalator, ditercalinium. Based on ESI-MS measurements of the complexation of ditercalinium to a series of DNA sequences, it was found that ditercalinum bound better to quadruplex structures than to duplexes [20].
The development of novel polyintercalating ligands is of interest because of the potential improvements in antitumor activity and sequence specificity. A novel class of DNA polyintercalators that shows great promise for binding to long stretches of DNA with sequence specificity and high affinity contain 1,4,5,8-tetracarboxylic naphthalene diimide (NDI) units connected in a head-to-tail arrangement by flexible scaffolds [21-24] (see Scheme 1). These compounds are known as threading polyintercalators because, upon intercalation, one of the functional groups attached to the diimide nitrogen resides in the DNA major groove, while the other is in the minor groove [21]. The potential advantages of developing a compound that binds to duplex DNA by threading polyintercalation include enhanced sequence specificity due to ligand interactions with the major and minor groove, the ability to bind to longer DNA sequences with a relatively low molecular weight compound, disruption of protein-ligand interactions that occur in both DNA grooves, and lower binding off-rates [22].
Scheme 1.
Structures of intercalator ligands. Molecular weights of compounds in Da are given in parenthesis.
A series of bis-intercalators, compounds that contain two NDI units, have been synthesized as precursors to longer compounds with more than two intercalation groups (Scheme 1). The primary objective of this study is to evaluate the binding behavior of trans-D1 and cis-C1 which are new ligands containing a rigid spiro-tricyclic scaffold in the trans- and cis-orientations, respectively. While few results have been reported for the newer ligands trans-D1 and cis-C1 [25], the binding behavior of V1, the ligand containing the peptide scaffold, has been examined in extensive footprinting and NMR-based studies [22, 23]. In the present study, the binding behavior of the well-characterized compound V1 will be examined to establish that the binding behavior observed by ESI-MS can be correlated to the results of traditional solution-based experiments. After developing a framework with V1, the binding of the new compounds, trans-D1 and cis-C1, will be reported with an emphasis on comparing differences in binding behavior of the compounds that result from the trans- versus cis-orientations of the scaffold. Here we examine binding stoichiometries, sequence selectivities, and concentration dependent binding of the bis-intercalators, evaluate the CAD fragmentation patterns of the observed DNA/drug complexes, and compare the results to ones obtained by conventional footprinting techniques. The binding of the bis-intercalator, echinomycin (Scheme 1) was also assessed by ESI-MS to serve as a comparison to the results of V1, cis-C1, and trans-D1. We also will compare the results of our study to other ESI-MS based studies involving mono-intercalators.
Another objective of this study is to demonstrate that mass spectrometry can be used as a screening tool for bis-intercalator ligands. To accomplish this aim, we demonstrate that the ESI-MS results mirror those established by traditional techniques such as NMR and DNAse I footprinting experiments to lend legitimacy to ESI-MS as an analysis tool for future studies involving novel polyintercalating compounds. It is not anticipated that ESI-MS will replace traditional techniques such as NMR and DNAse I footprinting, but instead that it be used as an initial screening tool. As drug discovery efforts shift to combinatorial synthesis of compound libraries, ESI-MS is suited to narrow down the pool of promising ligands that would then be examined in more detail using traditional methods.
EXPERIMENTAL
Chemicals
Single strand oligodeoxynucleotides (ODNs), custom synthesized as ammonium salts on the 1.0 μmole or 250 nmole scale with purification by HPLC, were obtained from Integrated DNA Technologies (Coralville, IA) and used without further purification. Stock solutions of each ODN were prepared at 2 mM concentration in deionized water. A portion of the stock solutions were diluted to 1 mM and set aside for experiments involving single strand ODNs. Duplex DNA was annealed by preparing solutions containing two complementary single strand ODNs, each at 0.7 mM concentration in 250 mM ammonium acetate. The annealing solutions were heated to 90 °C and then slowly cooled over a period of 4 hours. Table 1 shows the sequences used in this study. The synthesis of ligands V1 [26], trans-D1 [25], cis-C1 [25, and NDI1 [27] have been previously reported. Concentrations were determined spectroscopically using Beer's law. The extinction coefficients for the DNA strands were provided by the manufacturer and those of the ligands are 26 300 M−1 cm−1 (385 nm) for cis-C1, 36,000 M−1 cm−1 (383 nm) for trans-D1, and 26 300 M−1 cm−1 (385 nm) for V1 [25].
Table 1.
DNA sequences used in this study.
| Name | Sequence | Molecular Weight (Da) |
|---|---|---|
| ds1 | d(GGGCGGTACCGCGG/CCGCGGTACCGCCC) | 8531.5 |
| ds2 | d(GCGGGGATGGGGCG/CGCCCCATCCCCGC) | 8531.6 |
| ds3 | d(GCGGGAATTGGGCG/CGCCCAATTCCCGC) | 8529.6 |
| ds4 | d(GCGGAAATTTGGCG/CGCCAAATTTCCGC) | 8527.7 |
| ds5 | d(GGGACAGTGAGGGG/CCCCTCACTGTCCC) | 8529.6 |
| ds6 | d(GGTTGGGCCCAAGG/CCTTGGGCCCAACC) | 8529.6 |
| ds7 | d(GGGGTCGCCGGGGG/CCCCCGGCGACCCC) | 8532.6 |
DNAse I Footprinting
Plasmid pBR322 (New England BioLabs) was digested with NheI, dephosphorylated with CIAP, 5'-32P-end labeled with [γ-32P]-ATP and T4 kinase, digested with EcoRI (all enzymes were purchased from New England Biolabs) and purified by native polyacrylamide gel electrophoresis (PAGE), following standard protocols [28]. The 92 bp synthetic fragment (PAGE grade) was purchased from Midland Certified and labeled (32P) similarly. The DNAse I (Amersham) footprinting was carried out according to the procedure described previously [29]. The DNA fragments were separated on an 8% (231 bp) or 12% (92 bp) denaturing polyacrylamide gel. The gels were exposed on phosphor screen and analyzed with Quantity One 4.5 software from Bio-Rad.
Mass Spectrometry
Stock solutions of V1, trans-D1 and cis-C1 were prepared in deionized water at 1 mM. Analytical solutions containing duplex or single strand DNA and one ligand were prepared at equimolar 10 μM concentration (unless noted otherwise) in 50 mM ammonium acetate with 25% methanol to enhance the volatility of the solution. Lower concentrations of methanol result in similar binding trends, but the quality of the ESI-mass spectra is diminished. After allowing the solutions to equilibrate for 30 min., they were directly infused into a ThermoFinnigan LCQ Duo mass spectrometer (San Jose, CA) using a Harvard syringe pump (Holliston, MA) at 3 μL/min. Ions were generated in the negative ion mode with an electrospray voltage of 3.5 kV. The temperature of the heated capillary was set at 90 to 110 °C and nitrogen sheath and auxiliary gas flows of 10 and 40 arbitrary units, respectively, were used to aid in desolvation. The base pressure in the ion trap region was nominally ∼1 × 10−5 torr. Instrument conditions were optimized for each complex using the automatic tuning function of the Xcalibur software package (Finnigan, San Jose, CA). Spectra were acquired by summing 300 scans with an ion accumulation time of 100 ms.
Tandem mass spectrometry experiments were performed using collisional activated dissociation (CAD). The desired precursor ion was isolated in the trap using resonance ejection, followed by fragmentation induced by increasing the resonance voltage applied to the trap. An activation time of 30 ms was used for all experiments. The CAD energy was increased until the abundance of the precursor ion was reduced to ∼10% relative abundance. These experiments required CAD energies of ∼12 – 14%.
RESULTS and DISCUSSION
Complexes with V1
Evaluation of Binding Stoichiometry by ESI-MS
The DNA binding of ligand V1 has been previously examined by NMR and DNAse I footprinting techniques that identified the specific binding sites of this compound [23]. V1 was found to have a binding preference for d(GGTACC)2 sequences with NMR results confirming that the –Gly3-Lys- peptide scaffold was located in the major groove [23]. The V1-d(GGTACC)2 complex was formed with a dominant 1:1 binding stoichiometry.
The binding of V1 to a DNA duplex containing the preferred binding sequence was evaluated by ESI-MS to confirm that results revealed in the mass spectra can be correlated to the solution binding behavior. The 14-mer d(GGGCGGTACCGCGG/CCGCGGTACCGCCC) (ds1) was used for this study because it contains the specific binding sequence of V1 (GGTACC). All of the duplex DNA sequences selected for this study were non-self-complementary to allow the duplex and single strand ions to be unambiguously distinguished in the mass spectra. Duplexes with 14 base-pairs were selected for this study because previous ion mobility/molecular dynamics studies have reported that DNA duplexes greater than 12 base pairs better maintain the helical conformations in the gas-phase than smaller duplexes [37, 38]. Even larger duplexes were not chosen to ensure that there was only one high affinity binding site per duplex, allowing different binding sequences to be assessed individually. Each duplex was designed to contain the proposed high affinity ligand binding site in the center of the sequence. The terminal ends of the duplexes were selected to be G/C rich to enable good annealing of the sequences, and the sequences directly adjacent to the proposed specific binding site were selected based on the flanking sequences identified in DNAse I footprinting experiments to maintain consistency.
ESI mass spectra of solutions containing 10 μM of V1 and 10 μM of ds1 in an ammonium acetate/methanol buffer were evaluated first. Both the 5- and 6- charge states are prominent for the duplex/V1 complexes. For a typical solution containing V1 and ds1, the only complexes present in the spectrum possess a ligand/DNA binding stoichiometry of 1:1 with 2:1 complexes being at less than 5% of the relative abundance of the 1:1 complexes.
Concentration-dependent binding studies were undertaken to examine the extent of complexation changes as a result of varying the ligand/DNA ratios. Solutions containing ds1 at 10 μM and either 2.5, 5.0, 10, or 20 μM of V1 were analyzed by ESI-MS (spectra not shown). As the ligand/DNA molar ratio was increased, the relative ion abundance of the 1:1 complexes in the 5- and 6- charge states increased, while that of the free duplex decreased. However, no 2:1 complexes emerged, even with excess ligand in solution. While it is possible that there are multiple sites on the duplexes for which V1 could bind to form 2:1 complexes, binding of V1 at a second site is not anticipated to be as strong because the ligand bound at the high affinity site, GGTACC, would cover six base pairs and thus presumably hinder the bis-intercalative binding of a second ligand. These results are consistent with solution-based studies of V1 [23] and suggest ESI-MS is a promising tool for the analysis of DNA complexes containing bisintercalators.
Complexes with trans-D1 and cis-C1
To explore the effect of the scaffold on the binding specificity of the polyintercalators, two new bis-intercalators, trans-D1 and cis-C1, were synthesized containing a rigid spiro-tricyclic scaffold (Scheme 1). As shown in Scheme 1, trans-D1 is a trans-oriented ligand, while cis-C1 is cis-oriented. While the design and synthesis of these compounds has been previously reported [25], less is known about the DNA duplex binding of these compounds. We aimed to compare the binding of these compounds in a mass spectrometry based study.
Binding Selectivities
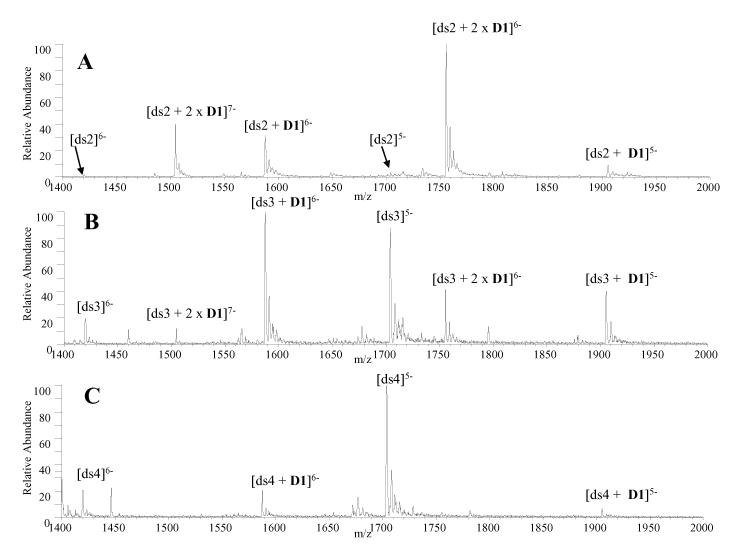
To begin an ESI-MS evaluation of the binding behavior of trans-D1 and cis-C1, the complexation of each ligand with DNA duplexes containing varying A-T and GC base pair composition was evaluated. The NDI intercalator unit has exhibited a preference for G-G steps [23], so it was of interest to assess the binding selectivities of the new compounds. ESI mass spectra were obtained for solutions containing trans-D1 or cis-C1 with a series of three duplexes with varying amounts of G/C and A/T base pair content: d(GCGGGGATGGGGCG/CGCCCCATCCCCGC) (ds2), d(GCGGGAATTGGGCG/CGCCCAATTCCCGC) (ds3), and d(GCGGAAATTTGGCG/CGCCAAATTTCCGC) (ds4).
While both ligands exhibited GC base pair selectivity, the preference was more pronounced for trans-D1. As shown in Figure 1A, trans-D1 readily forms abundant complexes with ds2, with ligand/DNA binding stoichiometries of 2:1 and 1:1, and little unbound DNA is present in the spectrum. The mass spectrum of trans-D1 with ds3 (Figure 1B) shows that the relative abundance of the 2:1 complexes is significantly decreased compared to Figure 1A, and the abundance of the unbound DNA has increased. The spectrum of trans-D1 with ds4, the duplex containing the most AT base pairs, reveals that only 1:1 complexes are present and with significantly lower abundances relative to the duplex ion present at m/z 1705 (Figure 1C). As the AT-content increases, both the binding stoichiometry and relative abundance of the complexes formed between trans-D1 and the duplex decrease dramatically.
Figure 1.
ESI mass spectra for complexes containing trans-D1 and equimolar (10 μM) amounts of (A) ds2, d(GCGGGGATGGGGCG/CGCCCCATCCCCGC) (B) ds3, d(GCGGGAATTGGGCG/CGCCCAATTCCCGC and (C) ds4, d(GCGGAAATTTGGCG/CGCCAAATTTCCGC).
The extent of complexation was calculated by expressing the sum of the abundances of ions from DNA/ligand complexes as a fraction of the total abundances of all ions from DNA as has been previously reported for ligand/DNA complexes [30]. Ions in the 5- and 6- charge states were used in this calculation. The binding results for trans-D1 (discussed above) and those for cis-C1 (spectra not show) with ds2, ds3, and ds4 are summarized in Table 2.
Table 2.
| ds1b,c | ds2 | ds3 | ds4 | ds5d | ds6c | ds7d | ss1 | ss5 | |
|---|---|---|---|---|---|---|---|---|---|
| V1 | 0.66 | n/ae | n/ae | n/ae | 0.52 | n/ae | n/ae | 0.00 | n/ae |
| cis-C1 | 0.86 | 0.52 | 0.39 | 0.32 | 0.38 | 0.51 | n/ae | 0.00 | n/ae |
| trans-D1 | 0.40 | 0.94 | 0.64 | 0.18 | 0.78 | n/ae | 0.85 | n/ae | 0.17 |
| NDI1 | 0.53 | n/ae | n/ae | n/ae | 0.41 | n/ae | n/ae | n/ae | n/ae |
All values +/− 0.05. This value was calculated to be the greatest standard deviation for the results of three experiments done with the samples.
Sequence contains proposed binding site of V1.
Sequence contains proposed binding site of cis-C1.
Sequence contains proposed binding site of trans-D1. dThe abundances for all of the sodium adducts ions associated with a complex were included in the relative abundance calculations.
“n/a” indicates data was not collected because the results were not relevant to the study.
Solutions contained equimolar (10 μM) concentrations of ligand and DNA.
Our results indicate that like trans-D1, cis-C1 also forms more abundant complexes with the GC-rich DNA duplexes, as demonstrated by a value of 0.52 for the fraction of bound DNA with ds2, compared to 0.39 with ds3 and 0.32 with ds4. These results are consistent with solution dissociation kinetics experiments in which cis-C1 and trans-D1 demonstrated a strong preference for binding to poly (dGdC) over poly (dAdT) sequences [25]. This is a trend that has been previously reported for NDI intercalation [21, 39]. The observed poly d(GdC) preference of the ligands may be the result of a variety of binding interactions. Increased hydrogen bonding between the intercalators and functional groups in the major and minor grooves of GC-rich sequences could account for this preference. The imide carbonyls on the NDI units may also undergo a favorable electrostatic interaction with the N2 amino group on G:C base pairs [39]. Steric and hydrophobic interactions could also play roles [21-23]. While the ligands demonstrate a general preference for GC-rich sequences, likely because of the NDI units, the functional linker imparts specific binding preferences.
DNAse I Footprinting
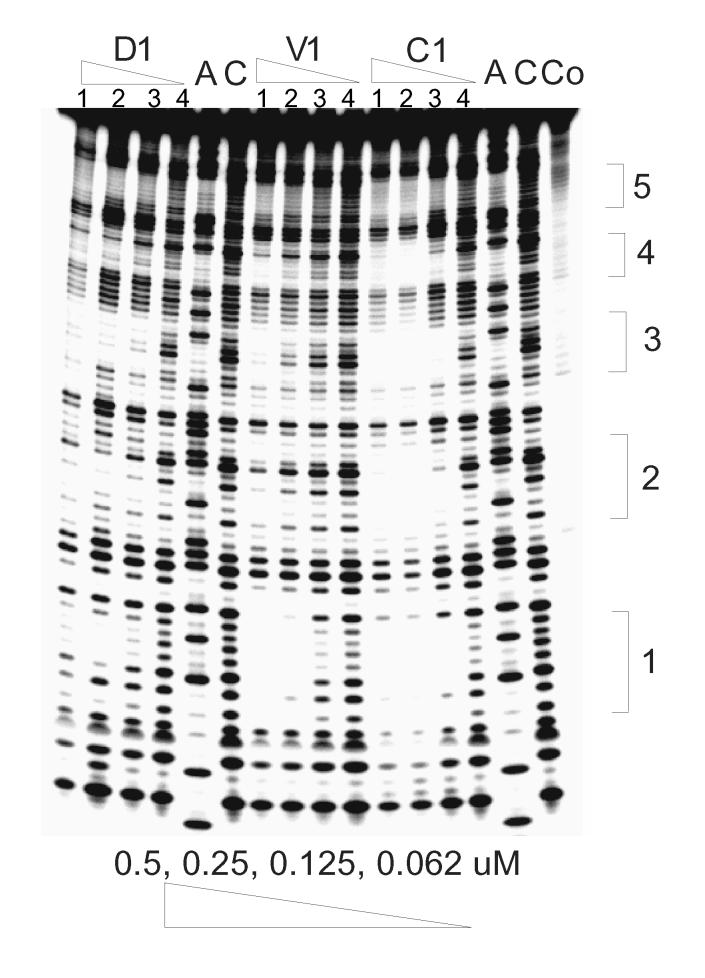
In addition to evaluating the GC versus AT sequence selectivity of trans-D1 and cis-C1, the binding of the compounds to duplexes containing potential specific binding sites was also evaluated by DNAse I footprinting and ESI-MS. It has been established that the naphthalenetetracarboxylic diimide based-intercalator units possess a preference for binding G-G steps [23]. In order to investigate the specificity of binding, DNAse I footprinting studies with trans-D1, cis-C1 and reference ligand V1 were carried out using a synthetic 92 bp DNA fragment containing 5 5'-GGNNCC-3' sites (Figure 2). As reported previously, V1 has a distinct binding site at 5'-GGTACC-3' with a Kd ∼100 nM [23].
Figure 2.
Footprinting of compounds cis-C1, trans-D1 and V1 on 92mer DNA with the (−) strand labeled on its 5'-end. Lane A represents Adenine-specific sequencing reaction. Lane Co contains DNA without DNAse I. For D1, V1, and C1, lanes 1-4 contain 0.5, 0.25, 0.125, and 0.062 μM ligand, respectively. Lane C contains DNA with DNAse but no compound. Sequences at 1: 5'-GGTACC; 2: 5'-GGATCC; 3: 5'-GGGCCC; 4: 5'-GGCGCC; 5: 5'-GGGGCC.
trans-D1 showed some non-specific binding behavior, from targeting less than six-base pairs to only binding at high concentration (> 0.25 μM) (Figure 2). cis-C1, however, shows very different binding characteristics from the others. It not only binds to GGTACC with an even higher affinity than V1, but it also binds to other sequences with good affinity, such as GGGCCC and GGATCC. The variety of binding sites with good binding affinity (Kd ∼100 nM) demonstrates the potential of spiro-tricyclic scaffold as a “universal” scaffold for polyintercalators.
To further investigate the binding specificity of trans-D1, a 231 bp EcoRI-NheI restriction fragment of plasmid pBR322 was chosen for a second round of footprinting experiments. This sequence was previously used to screen the binding specificity of NDI ligands [23]. V1 was used as a reference compound. As expected, one binding site for V1 was clearly seen with this DNA fragment at the GGTACC sequence (results not shown). For trans-D1, two potential binding sites were identified: CAGTGA and GGCGAC.
ESI-MS Evaluation of Ligand Binding Sequences
After identifying some possible specific binding sequences of cis-C1 and trans-D1 using DNAse I footprinting experiments, the binding of these ligands to DNA duplexes containing the most promising specific sequences were further evaluated by ESI-MS. Duplex ds1 contains the GGTACC sequence which is a potential specific binding site for cis-C1 and is the same sequence identified for V1, and duplex ds5 contains a possible specific binding sequence CAGTGA for trans-D1. The full sequences of these duplexes are shown in Table 1. Additional experiments were done with ds6, which contains a second possible binding site for cis-C1, GGGCCC, and ds7, containing the second potential site for trans-D1, GGCGAT.
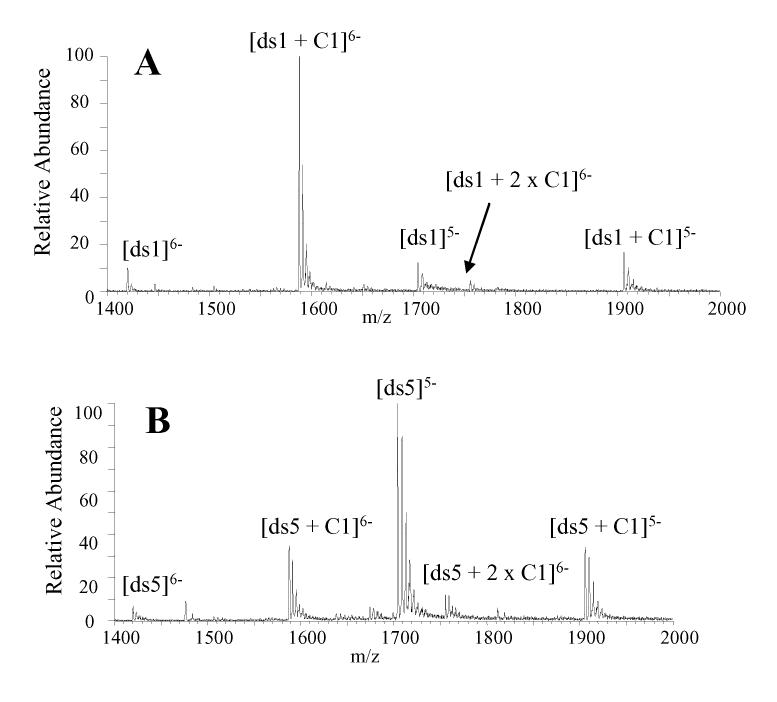
The mass spectrum acquired for a solution containing cis-C1 and ds1 at equimolar concentrations in 50 mM ammonium acetate with 25% methanol demonstrates that cis-C1 forms very abundant 1:1 complexes with this duplex (Figure 3A). The abundance of the unbound DNA ions are very low, suggesting cis-C1 undergoes extensive complexation with this duplex. In addition, there are no complexes with a 2:1 binding stoichiometry, which is expected since the duplex contains a specific binding site in the center of this sequence. Based on the abundances of the ions in Figure 3A, the fraction of bound DNA for the solution of ds1 with cis-C1 was calculated to be 0.86 (Table 2). These results suggest that the binding of cis-C1 with ds1 is more extensive than the binding of V1 with the same duplex, as the fraction of bound DNA for V1 with the duplex (shown in Figure 1B) was found to be 0.66 (Table 2). The higher binding affinity between cis-C1 and the GGTACC sequence was also demonstrated in the footprinting experiments discussed above. cis-C1 was also found to form complexes with ds6, which contained the GGGCCC binding site (spectra not shown). However, the fraction of bound DNA for cis-C1 and ds6 was 0.51, which suggests less extensive complexation between the cis-C1 and ds6 compared to ds1 and indicates a preference for the GGTACC binding sequence by the ligand.
Figure 3.
ESI mass spectra for complexes containing cis-C1 and equimolar (10 μM) amounts of (A) ds1 d(GGGCGGTACCGCGG/CCGCGGTACCGCCC) and (B) ds5, d(GGGACAGTGAGGGG/CCCCTCACTGTCCC).
Experiments aimed at evaluating the binding between cis-C1 and a DNA duplex that does not contain a target binding sequences were also undertaken. Duplex ds5 was selected for this experiment because the target binding sequence in the duplex, CAGTGA, was identified as a possible specific binding site for trans-D1 but not cis-C1. The ESI mass spectrum of a solution containing equimolar (10 μM) concentrations of cis-C1 and ds5 is shown in Figure 3B. The extent of complexation between cis-C1 and the duplex is lower than what was observed in the spectra of solutions containing cis-C1 with ds1 (Figure 3A) and ds6 (spectra not shown). In Figure 3B the abundances of the unbound DNA ions are considerably greater than the abundance of the 1:1 complexes. The fraction of bound DNA was calculated to be 0.38 which is significantly lower than the fraction of bound DNA for cis-C1 with ds1 (0.86) and moderately lower than that of cis-C1 with ds6 (0.51). These results indicate cis-C1 forms more abundant complexes with ds1 and ds6 which is consistent with the specific binding site identified by DNAse I footprinting experiments.
Similar experiments were undertaken involving trans-D1 and ds5 and ds7, each which contain a possible specific binding site of the ligand (CAGTGA and GGCGAC, respectively) and ds1, which was identified as a specific binding site for cis-C1 but not trans-D1. The results of these experiments (spectra not shown) are summarized in Table 2, and they suggest that while the structures of trans-D1 and cis-C1 are similar, they exhibit different binding behavior with the duplexes. The fraction of bound DNA in the spectrum of trans-D1 with ds5 was calculated to be 0.78, which is much greater than the fraction of bound DNA of cis-C1 with the same duplex (0.38). trans-D1 also formed abundant complexes with ds7, as indicated by a fraction of bound DNA of 0.85 for the ligand with the duplex. Conversely, the fraction bound for ds1 with trans-D1 was only 0.41 compared to 0.86 observed with cis-C1.
As a further comparison, the fraction of bound DNA of V1 with ds1 is also summarized in Table 2 since V1 and cis-C1 have the same possible specific binding sequence. The 0.66 value for fraction bound is not as great as that observed with cis-C1, suggesting cis-C1 might be an improvement over V1 in terms of forming abundant complexes with the GGTACC sequence. The fraction of bound DNA in a spectrum of V1 with ds5, a duplex that does not contain a specific binding site for V1, is 0.52 which is lower than that with ds1. However, the difference in the fraction of bound DNA for V1 with ds1 (containing the specific binding sequence) and ds5 (no specific binding sequence) is not as great as that observed with cis-C1. These results indicate that cis-C1 shows the most promising selectivity for binding to its target sequences over other sequences and demonstrate that the relative binding behavior of the bis-intercalators observed by ESI-MS correlates with DNAase I footprinting results.
Because NDI1 contains only one intercalating unit and lacks the scaffold designed to interact with the groove of duplex DNA, this compound functions as a mono-intercalator and was used as a reference ligand to compare its complexation with the same duplexes as used in the experiments above for cis-C1 and trans-D1. The ESI-mass spectra indicate the formation of 1:1 and 2:1 NDI1:duplex complexes, with the fractions bound for ds1 and ds5 summarized in Table 2. The greater abundances of the 2:1 NDI1:duplex complexes is consistent with a lower specificity of NDI1.
Concentration Dependent Binding
To further explore how the complexation of trans-D1 and cis-C1 changes with ligand/DNA molar ratios, concentration dependent binding was assessed. A series of solutions containing trans-D1 or cis-C1 with a duplex containing a proposed specific binding site (ds5 for trans-D1 and ds1 for cis-C1) were prepared with a DNA concentration of 10 μM and a variable ligand concentration of 2.5, 5.0, 10, or 20 μM. The mass spectra of trans-D1 with ds5 at these molar ratios demonstrate that when the ligand concentration is increased relative to the DNA concentration, changes in the extent of ligand complexation are observed in the mass spectra. At a trans-D1/ds5 molar ratio of 0.25, complexes with 1:1 binding stoichiometry are present, but with low relative abundances compared to the unbound DNA ions. As the molar ratio is increased to 0.5, the relative abundance of the 1:1 complexes increases, and low abundance 2:1 complexes emerge while the abundance of the free DNA decreases. When the molar ratio is increased to 1:1 or even 2:1, the abundances of the 2:1 complexes increase further, while the abundance of the unbound DNA ions diminish.
The appearance of the 2:1 complexes at higher trans-D1/ds5 molar ratios is notable since ds5 contains only one relatively high affinity binding site for trans-D1. The results imply that trans-D1 is able to bind to the DNA duplex in a non-specific manner when the ligand concentration is increased relative to the DNA. At this point, it is unknown how the 2:1 complexes are formed, but some possible scenarios include that the ligand may bind to the DNA via the intercalation of only one of the NDI units, two trans-D1 ligands could be aggregating in solution and then binding to the DNA duplex, the second ligand may non-specifically aggregate to the DNA, or there could be two binding sites on the duplex that are mutually exclusive. This last scenario is the least likely since the duplex contains one proposed specific binding site at the center of the sequence and the ligand bound at the higher affinity site would hinder the binding of the second molecule. The presence of D1 footprints that are less than six bases long in Figure 2 suggest the ligand is able to partially bind to the duplex via one NDI unit. Future NMR modeling work will shed light on the structure of these higher binding stoichiometry complexes.
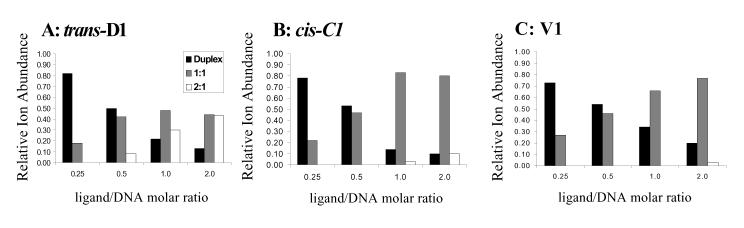
The results of the concentration dependent binding studies for trans-D1 with ds5, cis-C1 with ds1 (spectra not shown), and V1 with ds1 (discussed earlier) are summarized by the graphs in Figure 4, which reflect the relative ion abundance of the unbound duplex ions (black bars), 1:1 complexes (grey bars), and 2:1 complexes (light bars), grouped by ligand/DNA ratio. The results for cis-C1 with ds1 are significantly different from those of trans-D1 with ds5. As the ligand molar ratio is increased, the relative ion abundance of the free DNA duplex decreases and the abundances of the 1:1 complexes increase. However, when there is excess cis-C1 in solution, the 1:1 binding stoichiometry is dominant and only very low abundance 2:1 complexes are observed. The results of cis-C1 with ds1 are similar to V1 with ds1, which are also summarized in Figure 4C and suggest that like V1, cis-C1 is binding specifically to the DNA duplex that contains a target site.
Figure 4.
Summary plots of concentration dependent binding studies of (A) trans-D1 with ds5, (B) cis-C1 with ds1, and (C) V1 with ds1, indicating distribution of free duplexes, 1:1 ligand:duplex complexes, and 2:1 ligand:duplex complexes. Solutions contained the specified duplex DNA at 10 μM and ligand at, 2.5 μM, 5.0 μM, 10 μM, and 20 μM.
ESI-MS/MS Studies of Complexes Containing V1, trans-D1 and cis-C1
An additional goal of this study was to determine if a bis-intercalative binding mode could be distinguished from a mono-intercalative binding mode using CAD since full scan mass spectra provide little insight into binding interactions between ligands and DNA. Until now, no MS/MS studies have been done on complexes containing bis-intercalators. Our group [31] and others [12, 32] have used collisional activated dissociation experiments to examine the fragmentation patterns of intercalator/DNA complexes, including complexes of actinomycin-D [12, 32], daunomycin [31], and nogalamycin [31]. For complexes containing actinomycin-D, the predominant dissociation route is loss of the drug, but some strand separation (with retention of actinomycin D by one strand) and some nucleobase loss are also observed to a lesser extent [12, 32]. Our own CAD results for duplex/actinomycin-D complexes confirm that the primary fragmentation route is the disruption of the non-covalent interactions between actinomycin-D and the duplex, resulting in loss of actinomycin-D. Our earlier studies of complexes containing daunomycin or nogalamycin in a quadrupole ion trap mass spectrometer indicated that the drug/DNA complexes followed charge state dependent fragmentation patterns [31]. At lower charge states, the dominant fragmentation pathway was ejection of the drug leaving the intact duplex, while at higher charge states, separation of the individual single strand components of the duplex occurred, leaving the drug bound to one of the single strands. Dissociation by cleavage of a nucleobase was insignificant or not observed for complexes containing daunomycin or nogalamycin [31]. CAD of complexes containing reference ligand NDI1 [27], a mono-intercalator, resulted in fragmentation patterns consistent with those obtained for the daunomycin and nogalamycin complexes described above (spectra not shown). Complexes in the 5- charge state dissociated via loss of a neutral NDI1 ligand, while complexes in the 6-charge state produced fragment ions resulting from ligand ejection and predominant strand scission (spectra not shown).
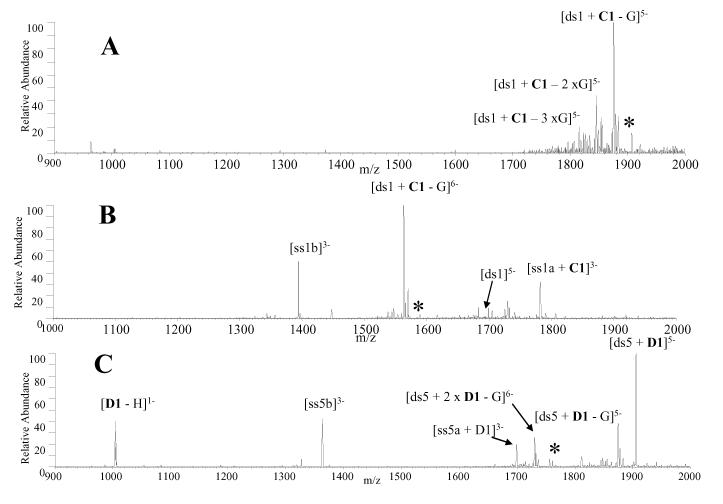
CAD experiments were undertaken in the present study to examine what, if any, differences exist in the fragmentation pathways of complexes containing V1, trans-D1, and cis-C1 compared to the mono-intercalators. The low charge state [ds + L]5- complexes, where L represents either V1, cis-C1, or trans-D1, produced the same fragmentation pattern, characterized by the guanine nucleobase loss (Figure 5A). This fragmentation pattern is different from what is commonly observed for the complexes containing commercial mono-intercalators, which predominantly dissociate via ejection of the ligand [12, 32]. For V1, trans-D1 and cis-C1, all 1:1 complexes in the 6- charge state dissociated via dominant guanine nucleobase loss in addition to strand scission, with the ligand remaining bound to a single strand as demonstrated by the CAD spectrum of [ds1 + C1]6- shown in Figure 5B. There were also very low abundance ions resulting from ejection of the negatively charged ligand, leaving the intact duplex. The predominance of nucleobase loss rather than ligand ejection of the 1:1 complexes in the 5- and 6-charge states is indicative of stronger binding interactions between the bis-intercalators and duplex DNA compared to traditional mono-intercalators, resulting from two intercalation sites and specific hydrogen bonding interactions between the scaffold and the DNA grooves. It is interesting that the strand scission pathway occurs in such a way that the ligand remains bound exclusively to only one of the ODNs (i.e. ss1a in Figure 5B, but not to ss1b). Although ss1a is more G-rich than ss1b, the underlying reason for this ODN selectivity is not clear.
Figure 5.
CAD mass spectra of (A) [ds1 + cis-C1]5-, (B) [ds1 + cis-C1]6- and (C) [ds5 + 2 × trans-D1]6-. The precursor ion is indicated by the asterisk. Solutions contained equimolar concentrations (10 μM) of the ligand and DNA duplex.
The 2:1 complexes in the 6- charge state produced a different CAD pattern than those described above for the 1:1 complexes. Trans-D1 was the only compound to form 2:1 complexes with sufficient abundance for CAD experiments, and the resulting CAD mass spectrum of [ds5 + 2 × D1]6- is shown in Figure 5C. The most abundant product ion results from ejection of the negatively charged ligand, leaving the [ds5 + D1]5- complex. Ions resulting from strand scission and guanine base loss ions from the precursor complex are present but with significantly lower abundances. This result suggests that the second molecule is bound differently (and likely more weakly) than the first ligand in the 2:1 complexes which is similar to what has been previously observed with complexes containing nogalamycin [31]. Furthermore, the duplex DNA used in these experiments contain only one specific binding site for the ligands, so the presence of the 2:1 complexes indicates some non-specific binding by the second ligand, which is expected to be a weaker binding interaction.
All complexes in the 7- charge state, regardless of binding stoichiometry, dissociated by strand scission leaving the ligands bound to a single strand (spectra not shown). This fragmentation pattern is consistent with past CAD studies involving intercalators [31] and is believed to result from coulombic repulsion of the more highly charged phosphate backbones.
The CAD fragmentation patterns of complexes containing echinomycin (Scheme 1) with duplex DNA were also evaluated in this study to serve as a comparison to the results of the new bis-intercalators. Echinomycin has been found to bind to duplex DNA via bis-intercalation, with the two quinoxaline rings preferably intercalating at CpG sites, and the bicyclic peptide scaffold oriented toward the DNA minor groove where hydrogen bonds are formed between the peptide and the nucleobases. In general, little ligand ejection was observed in the CAD spectra. Complexes in the 5- charge state undergo guanine base loss upon collisional activation. Guanine nucleobase loss is also observed in the CAD spectra of all complexes in the 6- charge state, in addition to ions resulting from strand scission. Complexes in the 7- charge state dissociated by strand separation. The CAD spectra of complexes containing enchinomycin are similar to those containing V1, trans-D1, or cis-C1, and are markedly different than those of the complexes containing mono-intercalators.
Single Strand Binding of V1, trans-D1 and cis-C1
The bis-intercalators evaluated in this study were designed to engage in two primary types of binding interactions with duplex DNA: intercalation interactions between the NDI units and the nucleobases, and hydrogen bonding interactions between the peptide scaffold and the minor or major groove of the DNA duplex. To determine if the ligands bind selectively to duplex DNA via these interactions over other DNA structures, ESI-MS was used to analyze solutions of trans-D1 or cis-C1 with single strand DNA. The single strand ODNs used for these experiments, d(GGGCGGTACCGCGG) (ss1) and d(GGGACAGTGAGGGG) (ss5), were one of the two complementary single strand ODNs used to anneal duplexes ds1 and ds5, respectively. Based on the results of the duplex DNA binding studies of the ligands discussed above, cis-C1 formed the most abundant complexes with ds1 so the binding of cis-C1 to ss1 was evaluated, while trans-D1 formed more abundant complexes with ds5, so ss5 was used. Using these single strand ODNs ensures that the same target binding sequences present in the duplexes are also found in the single strand sequences.
Solutions containing one single strand ODN and either V1, trans-D1 or cis-C1 at equimolar 10 μM concentration in ammonium acetate/methanol buffer were prepared and analyzed using the same instrument conditions used in experiments involving the duplex DNA discussed above. While cis-C1 and V1 did not form any complexes with ss1, trans-D1 formed low abundance ligand/DNA complexes with ss5 (spectra not shown). The results of the single strand binding study are summarized by the fraction of bound DNA values shown in Table 2. In general, the complexes that formed between the trans-D1 and the single strand DNA were relatively low in abundance compared to ligand/duplex DNA complexes (Table 2), suggesting that the ligand prefers binding to duplex DNA. It is also interesting to note that trans-D1 is less selective for duplex DNA compared to cis-C1, which echoes the earlier observation that trans-D1 exhibited considerable concentration dependent binding behavior with the duplex DNA and formed non-specific 2:1 complexes.
CONCLUSIONS
The utility of ESI-MS as a tool for screening non-covalent complexes formed between threading bis-intercalators and DNA is demonstrated in this study. Binding stoichiometries and ligand sequence selectivity can be quickly assessed and qualitatively compared using the ESI-mass spectra, while CAD experiments provide information about ligand binding interactions. Our results demonstrated that V1 forms abundant 1:1 complexes with ds1, the duplex containing its specific binding sequence 5'-GGTACC-3', and forms more abundant complexes with ds1 over ds5, which does not contain the specific binding sequence of the ligand. These results correlate well with previous DNAse I footprinting and NMR studies. Experiments involving trans-D1 indicate the ligand extensively binds to duplex ds5 containing the target sequence 5'-CAGTGA-3' and forms significantly less abundant complexes with ds1, which does not contain a specific binding site identified by footprinting experiments. However, at higher ligand/DNA molar ratios, trans-D1 forms 2:1 complexes which is indicative of non-specific binding by the ligand. Cis-C1 exhibited the most promising specific binding behavior to ds1 containing the 5'-GGTACC-3' target sequence as evidenced by the formation of significantly more abundant complexes with this duplex over ds5 which did not contain a specific binding site. Unlike trans-D1, cis-C1 did not form extensive 2:1 complexes at ligand/DNA molar ratios greater than one, conveying that the cis structure of C1 is favorable for more specific binding.
In general the CAD spectra of the bis-intercalators are characterized by dominant guanine base loss in the 5- and 6- charge states, with increasing degrees of strand scission as the charge state increases. The different CAD fragmentation patterns exhibited by the bisintercalator/duplex complexes compared to complexes containing known mono-intercalators mirror the shift in binding interaction of the ligands, characterized by intercalation at two sites and hydrogen bonding interactions with the DNA major or minor groove.
ACKNOWLEDGEMENTS
Funding from the Robert A. Welch Foundation (F1155 to JSB) and the National Institutes of Health (RO1 GM65956 to JSB and GM069647 to BLI) is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Goodman LS, Hardman JG, Limbird LE, Gilman AG. Goodman & Gilman's the Pharmacological Basis of Therapeutics. 10th ed. McGraw-Hill; New York: 2001. [Google Scholar]
- 2.Propst CL, Perun TJ. Nucleic Acid Targeted Drug Design. M. Dekker; New York: 1992. [Google Scholar]
- 3.Hofstadler SA, Griffey RH. Analysis of non-covalent complexes of DNA and RNA by mass spectrometry. Chem. Rev. 2001;101:377–390. doi: 10.1021/cr990105o. [DOI] [PubMed] [Google Scholar]
- 4.Beck JL, Colgrave ML, Ralph SF, Sheil MM. Electrospray ionization mass spectrometry of oligonucleotide complexes with drugs, metals, and proteins. Mass Spectrom. Rev. 2001;20:61–87. doi: 10.1002/mas.1003. [DOI] [PubMed] [Google Scholar]
- 5.Brana MF, Cacho M, Gradillas A, de Pascual-Teresa B, Ramos A. Intercalators as anticancer drugs. Curr. Pharm. Des. 2001;7:1745–1780. doi: 10.2174/1381612013397113. [DOI] [PubMed] [Google Scholar]
- 6.Triolo A, Arcamone FM, Raffaelli A, Salvadori P. Non-covalent complexes between DNA-binding drugs and doubly stranded deoxyoligonucleotides: A study by ionspray mass spectrometry. J. Mass Spectrom. 1997;32:1186–1194. doi: 10.1002/(SICI)1096-9888(199711)32:11<1186::AID-JMS575>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 7.Kapur A, Beck JL, Sheil MM. Observation of daunomycin and nogalamycin complexes with duplex DNA using electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 1999;13:2489–2497. doi: 10.1002/(SICI)1097-0231(19991230)13:24<2489::AID-RCM816>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 8.Gupta R, Kapur A, Beck JL, Sheil MM. Positive ion electrospray ionization mass spectrometry of double stranded DNA/drug complexes. Rapid Commun. Mass Spectrom. 2001;15:2472–2480. doi: 10.1002/rcm.524. [DOI] [PubMed] [Google Scholar]
- 9.Colgrave ML, Beck JL, Sheil MM, Searle MS. Electrospray ionization mass spectrometric detection of weak non-covalent interactions in nogalamycin-DNA complexes. Chem. Commun. 2002:556–557. doi: 10.1039/b200235n. [DOI] [PubMed] [Google Scholar]
- 10.Furlan RLA, Watt SJ, Garrido LM, Amarante-Mendes GP, Nur-EAlam M, Rohr J, Brana A, Mendez C, Salas JA, Sheil MM, Beck JL, Padilla G. DNA-binding properties of cosmomycin D, an anthracycline with two trisaccharide chains. J. Antibiot. 2004;57:647–654. doi: 10.7164/antibiotics.57.647. [DOI] [PubMed] [Google Scholar]
- 11.Urathamakul T, Beck JL, Sheil MM, Aldrich-Wright JR, Ralph SF. A mass spectrometric investigation of non-covalent interactions between ruthenium complexes and DNA. Dalton Trans. 2004:2683–2690. doi: 10.1039/B406889K. [DOI] [PubMed] [Google Scholar]
- 12.Wan KX, Gross ML, Shibue T. Gas-phase stability of double-stranded oligodeoxynucleotides and their noncovalent complexes with DNA-binding drugs as revealed by collisional activation in an ion trap. J. Am. Soc. Mass Spectrom. 2000;11:450–457. doi: 10.1016/S1044-0305(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 13.Wan KX, Shibue T, Gross ML. Non-covalent complexes between DNA-binding drugs and double stranded oligodeoxynucleotides: A study by ESI ion trap mass spectrometry. J. Am. Chem. Soc. 2000;122:300–307. doi: 10.1016/S1044-0305(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 14.Beck JL, Gupta R, Urathamakul T, Williamson NL, Sheil MM, Aldrich-Wright JR, Ralph SF. Probing DNA selectivity of ruthenium metallointercalators using ESI mass spectrometry. Chem. Commun. 2003:626–627. doi: 10.1039/b212132h. [DOI] [PubMed] [Google Scholar]
- 15.Gupta R, Beck JL, Ralph SF, Sheil MM, Aldrich-Wright JR. Comparison of the binding stoichiometries of positively charged DNA-binding drugs using positive and negative ion electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 2004;15:1382–1391. doi: 10.1016/j.jasms.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Gabelica V, De Pauw E, Rosu F. Interaction between antitumor drugs and a double stranded oligonucleotide studied by electrospray ionization mass spectrometry. J. Mass Spectrom. 1999;34:1328–1337. doi: 10.1002/(SICI)1096-9888(199912)34:12<1328::AID-JMS889>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 17.Greig MJ, Robinson JM. Detection of oligonucleotide-ligand complexes by ESI-MS (DOLCE-MS) as a component of high throughput screening. Journal of Biomolecular Screening. 2000;5:441–454. doi: 10.1177/108705710000500607. [DOI] [PubMed] [Google Scholar]
- 18.Rosu F, De Pauw E, Guittat L, Alberti P, Lacroix L, Mailliet P, Riou JF, Mergny JL. Selective interaction of ethidium derivatives with quadruplexes : An equilibrium dialysis and electrospray ionization mass spectrometry analysis. Biochemistry. 2003;42:10361–10371. doi: 10.1021/bi034531m. [DOI] [PubMed] [Google Scholar]
- 19.Reyzer ML, Brodbelt JS, Kerwin SM, Kumar D. Evaluation of complexatin of metal-mediated DNA-binding drugs to oligonucleotides via electrospray ionization mass spectrometry. Nucleic Acids Res. 2001;29:e103. doi: 10.1093/nar/29.21.e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carrasco C, Rosu F, Gabelica V, Houssier C, De Pauw E, GarbayJaureguiberry C, Roques B, Wilson WD, Chaires JB, Waring MJ, Bailly C. Tight binding of the anticancer drug ditercalinium to quadruplex DNA. Chembiochem. 2002;3:1235–1241. doi: 10.1002/1439-7633(20021202)3:12<1235::AID-CBIC1235>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 21.Lokey RS, Kwok Y, Guelev V, Pursell CJ, Hurley LH, Iverson BL. A new class of polyintercalating molecules. J. Am. Chem. Soc. 1997;119:7202–7210. [Google Scholar]
- 22.Lee J, Guelev V, Sorey S, Hoffman DW, Iverson BL. NMR structural analysis of a modular threading tetraintercalator bound to DNA. J. Am. Chem. Soc. 2004;126:14036–14042. doi: 10.1021/ja046335o. [DOI] [PubMed] [Google Scholar]
- 23.Guelev V, Lee J, Ward J, Sorey S, Hoffman DW, Iverson BL. Peptide bis-intercalator binds DNA via threading mode with sequence specific contacts in the major groove. Chem. Biol. 2001;8:415–425. doi: 10.1016/s1074-5521(01)00013-8. [DOI] [PubMed] [Google Scholar]
- 24.Guelev V, Sorey S, Hoffman DW, Iverson BL. Changing DNA grooves- A 1,4,5,8-naphthalene tetracarboxylic diimides bis-intercalator with the linker (β-Ala)3-Lys in the minor groove. J. Am. Chem Soc. 2002;124:2864–2865. doi: 10.1021/ja016834e. [DOI] [PubMed] [Google Scholar]
- 25.Chu Y, Lynch V, Iverson BL. Synthesis and DNA binding studies of bisintercalators with a novel spiro-cyclic linker. Tetrahedron. 2006;62:5536–5548. [Google Scholar]
- 26.Guelev VM, Cubberley MS, Murr MM, Lokey RS, Iverson BL. Design, synthesis, and characterization of polyintercalating ligands. Method. Enzymol. 2001;340:556–570. doi: 10.1016/s0076-6879(01)40442-3. [DOI] [PubMed] [Google Scholar]
- 27.Cubberley MS, Iverson BL. H-1 NMR investigation of solvent effects in aromatic stacking interactions. J. Am. Chem. Soc. 2001;123:7560–7563. doi: 10.1021/ja015817m. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Russell DW. Molecular cloning : a laboratory manual. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2001. [Google Scholar]
- 29.Guelev V. University of Texas at Austin; Austin, TX: 2002. Ph.D. Thesis. [Google Scholar]
- 30.Mazzitelli CL, Kern JT, Rodriguez M, Brodbelt JS, Kerwin SM. Evaluation of Binding of Perylene Diimide and Benzannulated Perylene Diimide Ligands to DNA by Electrospray Ionization Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2006;17:593–604. doi: 10.1016/j.jasms.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 31.Keller KM, Zhang JM, Oehlers L, Brodbelt JS. Influence of initial charge state on fragmentation patterns for noncovalent drug/DNA duplex complexes. J. Mass Spectrom. 2005;40:1362–1371. doi: 10.1002/jms.927. [DOI] [PubMed] [Google Scholar]
- 32.Gabelica V, De Pauw E. Comparison of the collision-induced dissociation of duplex DNA at different collision regimes: Evidence for a multistep dissociation mechanism. J. Am. So. Mass Spectrom. 2002;13:91–98. doi: 10.1016/s1044-0305(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 33.Yen SF, Gabbay EJ, Wilson WD. Interaction of aromatic imides with deoxyribonucleic-acid - Spectrophotometric and viscometric studies. Biochemistry. 1982;21:2070–2076. doi: 10.1021/bi00538a014. [DOI] [PubMed] [Google Scholar]
- 34.Low CML, Drew HR, Waring MJ. Sequence-specific binding of echinomycin to DNA - Evidence for conformational-changes affecting flanking sequences. Nucleic Acids Res. 1984;12:4865–4879. doi: 10.1093/nar/12.12.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vandyke MM, Dervan PB. Echinomycin binding-sites on DNA. Science. 1984;225:1122–1127. doi: 10.1126/science.6089341. [DOI] [PubMed] [Google Scholar]
- 36.Sobell HM, Jain SC, Sakore TD, Nordman CE. Stereochemistry of actinomycin-DNA binding. Nature. 1971;231:200–205. doi: 10.1038/newbio231200a0. [DOI] [PubMed] [Google Scholar]
- 37.Rueda M, Luque FJ, Orozco M. Nature of minor-groove binders - DNA complexes in the gas phase. J. Am. Chem. Soc. 2005;127:11690–11698. doi: 10.1021/ja0422110. [DOI] [PubMed] [Google Scholar]
- 38.Gidden J, Ferzoco A, Baker ES, Bowers MT. Duplex formation and the onset of helicity in poly d(CG)n oligonucleotides in a solvent-rree environment. J. Am. Chem. Soc. 2004;126:15132–15140. doi: 10.1021/ja046433+. [DOI] [PubMed] [Google Scholar]
- 39.Liu Z-R, Hecker KH, Rill RL. Selective DNA binding of (N-alkylamine)-substituted napthalene imides and diimides to G+C-rich DNA. J. Biomol. Struct. Dynam. 1996;14:331–339. doi: 10.1080/07391102.1996.10508128. [DOI] [PubMed] [Google Scholar]