Abstract
Eukaryotic chromosome segregation depends on the mitotic spindle apparatus, a bipolar array of microtubules nucleated from centrosomes. Centrosomal microtubule nucleation requires attachment of γ-tubulin ring complexes to a salt-insoluble centrosomal core, but the factor(s) underlying this attachment remains unknown. In budding yeast, this attachment is provided by the coiled-coil protein Spc110p, which links the yeast γ-tubulin complex to the core of the yeast centrosome. Here, we show that the large coiled-coil protein kendrin is a human orthologue of Spc110p. We identified kendrin by its C-terminal calmodulin-binding site, which shares homology with the Spc110p calmodulin-binding site. Kendrin localizes specifically to centrosomes throughout the cell cycle. N-terminal regions of kendrin share significant sequence homology with pericentrin, a previously identified murine centrosome component known to interact with γ-tubulin. In mitotic human breast carcinoma cells containing abundant centrosome-like structures, kendrin is found only at centrosomes associated with spindle microtubules.
Eukaryotic cell division requires assembly of a mitotic spindle to faithfully segregate replicated chromosomes. A key step in spindle assembly is the nucleation of stable microtubules from the centrosome. One protein involved in nucleation is γ-tubulin, which forms a ring complex with approximately seven other proteins (1, 2). Immunoelectron microscopic tomography of purified centrosomes indicates that γ-tubulin ring complexes (γ-TURCs) localize to centrosomal pericentriolar material and associate with the minus ends of microtubules during in vitro nucleation from purified centrosomes (3). Fluorescence microscopy of stably integrated γ-tubulin-green fluorescent protein (GFP) fusions has demonstrated that γ-tubulin is rapidly recruited to the centrosome during mitosis (4), suggesting that recruitment and attachment of soluble γ-TURCs to the centrosome are essential precursors to microtubule nucleation. Recruitment of γ-TURCs to the centrosome is likely facilitated by pericentrin, a centrosomal protein that interacts with both γ-tubulin (5) and the motor protein dynein (6). The centrosomal proteins that subsequently anchor γ-TURCs to the centrosome have not yet been identified, but in vitro fractionation of purified centrosomes indicates that one or more salt-insoluble centrosomal proteins of the centrosomal core, or “centromatrix,” are essential for nucleation. These centromatrix proteins may serve as anchors for γ-TURCs during nucleation (7, 8).
In budding yeast, the coiled-coil protein Spc110p anchors the yeast γ-tubulin complex to the yeast centrosome, or spindle pole body (SPB) (9). The N-terminal region of Spc110p binds directly to the yeast γ-tubulin complex (9, 10), whereas the Spc110p C-terminal region, when bound to calmodulin, binds to the core of the SPB (11–13). The 70-kDa coiled-coil central region of Spc110p acts as a spacer between the spindle microtubules and the core of the SPB (14). The conservation of both calmodulin (15–17) and Spc110p epitopes (18) in mammalian centrosomes suggested the existence of a calmodulin-binding Spc110p orthologue, but its identity has remained elusive. Moreover, the striking structural differences between the yeast spindle pole body and the centrosome cast doubt that the protein or proteins conferring anchoring activity in higher eukaryotic cells would bear any resemblance to Spc110p. However, using knowledge of calmodulin-binding site sequences, we have identified a calmodulin-binding centrosomal protein in human cells whose molecular properties and subcellular distribution indicate a functional relatedness to yeast Spc110p. Like Spc110p in budding yeast, this protein may anchor γ-TURCs to the centrosomal core in human cells.
Materials and Methods
Identification and Analysis of Kendrin cDNA.
Murine and human expressed sequence tag (EST) clones with potential calmodulin-binding sites were identified by using Advanced blast (http://www.ncbi.nlm.nih.gov/blast/blast.cgi) using tblastn with “expect” set at 1000.¶ A human breast carcinoma STRETCH cDNA library (CLONTECH) was screened with the 32P-labeled human EST cDNA using Hybond-N filters (Amersham Pharmacia) according to the manufacturer's recommendations. We screened approximately 1 × 106 plaques and obtained 18 positive clones. PCR on plaque-purified positive clones using Expand polymerase (Roche Molecular Biochemicals) identified a clone with an insert of 5.8 kb for further analysis. A full-length human cDNA, named kendrin, was subsequently deposited in the database by Li and Joshi.¶
Coiled-coil secondary structure was predicted by using paircoil (19) (http://nightingale.lcs.mit.edu/cgi-bin/score) with default settings. All numbering of nucleotides and amino acids refers to kendrin cDNA and protein sequences, respectively. The kendrin database entry now includes the designation PCNT, for Homo sapiens pericentrin.¶ However, as discussed below, evidence suggests that kendrin is not the human form of pericentrin. Therefore, we use the designation kendrin, rather than pericentrin, to refer to the human protein.
Northern Blotting.
Multiple tissue and cancer line Northern blots (CLONTECH) were probed with 32P-labeled DNA fragments according to the manufacturer's recommendations. Hybridization signals were detected on Hyperfilm-MP (Amersham Pharmacia) or by using a Storm PhosphorImager (Molecular Dynamics).
Protein A-Calmodulin Overlay Blotting.
Protein A-calmodulin overlay blotting was done as previously described (20) with the following modifications. Protein A and a protein A-vertebrate calmodulin fusion were expressed in Escherichia coli by using plasmid pRIT-2T (Amersham Pharmacia). Fusions of glutathione S-transferase (GST) to kendrin fragments (amino acids 2628–2969 and 3158–3321) and GST alone were expressed in E. coli by using plasmid pGEX-2T (Amersham Pharmacia). A mutagenized GST-kendrin fusion (3158–3321) containing five alanine substitution mutations in the calmodulin-binding site (Fig. 1 a) was also expressed. Bacterial lysates were prepared in 0.01 M sodium phosphate, pH 7.2, 1% β-mercaptoethanol, 1% SDS, and 6 M urea. Protein A alone stains only the upper light background band seen in all lanes of the calmodulin overlay blots shown in Fig. 2 a and b.
Figure 1.
Kendrin, a large coiled-coil protein, contains a C-terminal calmodulin-binding site and shares homology with murine pericentrin. (a) Alignment of the Saccharomyces cerevisiae Spc110p calmodulin-binding site with related protein sequences detected in the fission yeast S. pombe (M.F. and T.D., unpublished data), the filamentous fungus A. nidulans (J. Joseph and A. Means, unpublished data), and humans (kendrin). Green, unanimously conserved residues at one position; yellow, three or four conserved residues in one position; purple, additional similar residues. Kendrin calmodulin-binding site residues changed to alanine by mutagenesis are underlined. (b) Schematic drawing of the kendrin protein showing locations of coiled-coil regions (blue lines and numbers), calmodulin-binding site (CaM), and region used for antibody generation (hatched area). Gray lines, human kendrin EST and two Northern blotting probes encoding corresponding kendrin segments. (c) Analysis of the N-terminal 2250 amino acids that share high overall homology with murine pericentrin. Schematic drawn to scale indicates segments of kendrin that share >80% identity with pericentrin (red boxes), >80% identity or conservative replacement with pericentrin (purple boxes), <80% identity or conservative replacement with pericentrin (gray boxes), or that are not found in pericentrin (white boxes). The C-terminal region of kendrin beyond residue 2250 is not shown here as it is unique and is not found in pericentrin. (d) Full-length kendrin is encoded by a 10-kb transcript, which shares homology with a 7.5-kb transcript, as shown by Northern blotting. Probe 1 (5.8 kb cDNA library clone insert) and Probe 2 (cDNA including kendrin nucleotides 9008–9976), the relative positions of which are shown in b, were hybridized to both a multiple tissue Northern blot (CLONTECH) and a cancer line blot (CLONTECH). Arrows indicate transcripts at ≈10 kb and ≈7.5 kb (Top) or at 10 kb (Bottom). Human tissues are as indicated. Cancer cell lines are SW480, colorectal adenocarcinoma; S3, HeLa cell.
Figure 2.
The predicted calmodulin-binding site in the kendrin C-terminal region physically binds calmodulin as shown by calmodulin overlay blotting and mutation analysis. (a) (Left) Coomassie blue-stained acrylamide gel of protein extracts from E. coli cells expressing either fusions of GST to indicated residues of kendrin or GST alone. (Right) Calmodulin overlay blot showing specific binding of protein A-tagged calmodulin to the GST-kendrin (3158–3321) fusion protein. Arrows indicate the position of the full-length GST-kendrin fusion protein (3158–3321), which binds calmodulin. A fusion between GST and a coiled-coil region of kendrin does not bind calmodulin, nor does GST alone. (b) Same as in a, but comparing binding of calmodulin to GST-kendrin (3158–3321) fusion protein (CTL, control fragment) and a GST-kendrin (3158–3321) fusion protein (MUT, mutagenized fragment) containing five alanine substitutions in residues predicted to be crucial for calmodulin binding. Positions of alanine substitution mutations shown in Fig. 1a.
Antibody Generation.
GST-kendrin (amino acids 2628–2969) fusion protein was expressed in bacteria using plasmid pGEX-2T (Amersham Pharmacia), solubilized in 150 mM Tris, pH 8, 500 mM NaCl, 1% Triton X-100, and purified using glutathione Sepharose according to the manufacturer (Amersham Pharmacia). Antibodies against this fusion were prepared in hens, and the IgY fraction was purified from egg white (Aves Laboratories, Tigard, OR). The purified IgY fraction recognized a ≈370-kDa band on immunoblots of whole cell extracts prepared from cultured human cells (not shown).
Cell Culture.
Primary human diploid fibroblasts and HS578T human breast carcinoma cells were grown on glass coverslips in high glucose (4.5 g/liter) DMEM supplemented with 10% FCS (HyClone) and, for HS578T cells, 10 μg/ml bovine insulin. HCC1937 human colon carcinoma cells were grown in RPMI 1640 medium supplemented with 5% FCS, 1 mM sodium pyruvate and buffered with 25 mM Hepes, 25 mM sodium bicarbonate. All culture media contained 100 units/ml penicillin G sulfate and 100 μg/ml streptomycin sulfate. Cultures were grown in a humidified, 5% CO2, 37°C incubator.
Immunofluorescence Microscopy.
Coverslips with attached cells were washed once in PBS and then fixed at −20° in 70% acetone/30% methanol for ≥20 min or methanol for 6 min, dried at room temperature for 30 min, and either used immediately or stored at −20°. All incubations were <1 h at room temperature. Coverslips were rehydrated in blocking solution (PBS supplemented with 3% BSA and 0.04% sodium azide). Primary antibodies were diluted in blocking solution as follows: chicken IgY anti-kendrin 1618 at 1:800, rabbit anti-γ-tubulin at 1:3000 (Sigma), mouse ascites anti-centrin 20H5 at 1:500 (21), rabbit anti-pericentrin M1-100-2 at 1:800 (generous gift from S. Doxsey, University of Massachusetts Medical Center, Worcester, MA) and rat anti-α-tubulin at 1:250 (Harlan, Haslett, MI). After five washes in blocking solution, bound antibody was detected with secondary antibody diluted in blocking solution: goat anti-chicken IgY Alexa 568 at 1:1000 (Molecular Probes) and either goat anti-rabbit FITC at 1:1000 (Molecular Probes), goat anti-mouse FITC at 1:50 (Roche Molecular Biochemicals), or goat anti-rat Oregon Green at 1:500 (Molecular Probes). After five washes in blocking solution, coverslips were briefly incubated in PBS containing 100 ng/ml 4′,6-diamidino-2-phenylindole (Sigma) and then mounted in Citifluor (Ted Pella, Redding, CA).
For control experiments, anti-kendrin IgY antibodies were incubated with nitrocellulose strips containing immobilized GST, and unbound antibodies were collected. Antibodies not binding to GST strips were subsequently incubated with strips containing the GST-kendrin (amino acids 2628–2969) fusion protein, and the resulting flow-through collected. Unbound antibodies collected after initial incubation with GST strips and flow-through after subsequent incubation with GST-kendrin strips were used to stain human diploid fibroblasts in parallel at a final dilution of 1:800.
For standard immunofluorescence microscopy, cells were imaged by using a Zeiss Axioplan microscope with 40× and 100× objectives and an Optivar set at 1.25. Images were captured by using Imagepoint or Quantix cooled charge-coupled device (CCD) video cameras (Photometrics, Tucson, AZ). For deconvolution immunofluorescence microscopy (Fig. 2k), cells prepared as described above were imaged by using a Zeiss Axiovert microscope with a 63× objective. The images were captured by using a Quantix-LC cooled CCD video camera (Photometrics) and analyzed by using deltavision software (Applied Precision, Issaquah, WA). Fig. 3k shows a projection of the seven sections containing kendrin and centrin (centriolar) staining.
Figure 3.
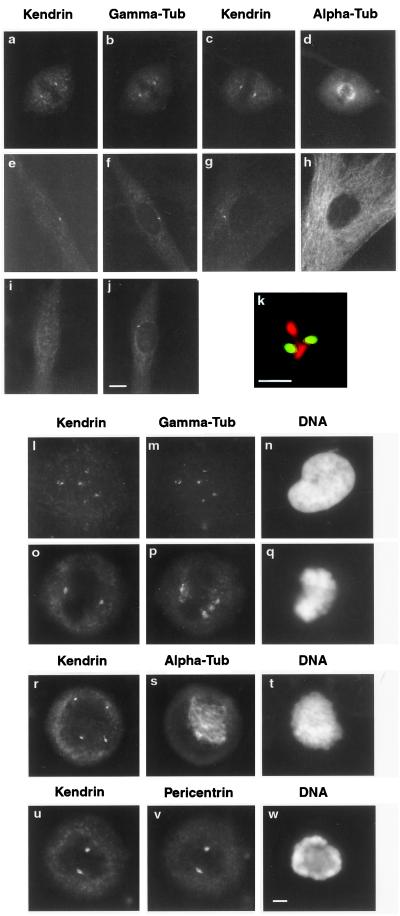
Kendrin localizes to centrosomes throughout the cell cycle and is restricted to spindle poles in mitotic cancer cells. (a and b) Colocalization of kendrin and γ-tubulin at centrosomes in a human diploid fibroblast transiting mitosis. (c and d) Kendrin labels the poles of the spindle apparatus in a mitotic fibroblast. (e and f) Colocalization of kendrin and γ-tubulin at an interphase fibroblast centrosome. (g and h) Localization of kendrin at an interphase fibroblast centrosome near the rim of the nucleus, from which cytoplasmic microtubules are excluded. (i) Depletion of anti-kendrin antibodies eliminates staining of the centrosome, which has been labeled in the same cell by anti-γ-tubulin antibodies (j). (k) Kendrin (red) localizes in close proximity to paired centrioles (green), labeled with anti-centrin antibodies (21). (l–n) Colocalization of kendrin and γ-tubulin to multiple centrosomal structures in an interphase HS578T breast cancer cell with uncondensed DNA. (o–q) A mitotic HS578T cancer cell containing condensed chromatin in which kendrin is restricted to spindle poles, whereas γ-tubulin labels both spindle poles and extra centrosomal structures. (r–t) Kendrin at all poles of a tetrapolar spindle in a HS578T cancer cell attempting mitosis; the kendrin focus at the lower right in r was visible in a separate focal plane and was digitally merged with the focal plane containing the other three kendrin foci. (u–w) Colocalization of kendrin and pericentrin in a mitotic HS578T cancer cell containing condensed DNA. (Size bar in j, 10 μm, applies to a–j; size bar in k, 1 μm; size bar in w, 5 μm, applies to l–w.)
Results and Discussion
To identify vertebrate proteins similar to yeast Spc110p, we aligned the calmodulin-binding site of budding yeast Spc110p with the calmodulin-binding sites of two Spc110p homologues identified in the filamentous fungus Aspergillus nidulans and the fission yeast Schizosaccharomyces pombe (Fig. 1a). These calmodulin site sequences share homology with one another (Fig. 1a) but are distinct from the IQ-type calmodulin-binding site in myosins [consensus (I/V/L)Qxxx(R/K)Gxxx(R/K), (22)]. EST database searches using these calmodulin-binding site sequences identified two murine EST clones and a related human EST clone. We used the human EST clone to probe a cDNA library and identified a large cDNA predicted to encode a protein named kendrin.
Kendrin contains two long central regions predicted to form coiled-coils (amino acid residues 256-1874 and 2559–3075) flanked by noncoiled regions (Fig. 1b). The noncoiled N-terminal domain and the first coiled-coil region of kendrin (amino acid residues 1–2250) share striking homology with murine pericentrin, a centrosome component that binds γ-tubulin (5) (global alignment score = 5954, 61% identity; Fig. 1c). The C-terminal 1071 amino acids of kendrin containing the calmodulin-binding site show no significant homology to pericentrin (global alignment score = −757). Northern blotting revealed that full-length kendrin is encoded by a 10-kb transcript. A related 7.5-kb transcript encoding the portion of kendrin homologous to pericentrin was also detected (Fig. 1d).
Although early reports labeled human kendrin as human pericentrin (23, 24), three lines of evidence indicate that kendrin and pericentrin are different proteins encoded by different genes. First, both mouse and human kendrin ESTs¶ contain sequences that are not present in the murine pericentrin cDNA (25). Conversely, mouse and human pericentrin ESTs (defined as those corresponding to a unique 3′ untranslated region in murine pericentrin)¶ contain sequences that are not present in kendrin cDNA. Second, human pericentrin is not encoded at the human kendrin locus. Kendrin has been mapped to chromosome 21 (21q22.3), and genomic sequence from this region includes 247 kb immediately downstream of the kendrin locus.¶ This region appears to contain two additional genes (S100 and an unidentified ORF); however, the sequence for the human pericentrin ESTs is not present. A final line of evidence comes from a comparison of the predicted kendrin and pericentrin proteins. If kendrin were simply a version of pericentrin with a long extension, we would predict that the shared N-terminal 2250 amino acids would closely resemble or be identical to one another. In contrast, kendrin shows a mosaic of regions sharing high homology with murine pericentrin (>80% identity), interspersed with multiple segments that share little or no sequence similarity (e.g., 186 residue segment with <22% conservative replacements; Fig. 1c). Thus, mouse and human appear to contain two different genes, at different chromosomal loci, one encoding pericentrin and the other encoding kendrin.
The unique C-terminal region of kendrin directly binds calmodulin, as shown by protein A-calmodulin overlay blotting (Fig. 2a). Alanine mutagenesis of five residues in the predicted calmodulin-binding site (Fig. 1a) dramatically decreased calmodulin binding (Fig. 2b), confirming that calmodulin-binding activity maps to the residues predicted by sequence analysis. The five mutated residues were chosen on the basis of their conservation (Fig. 1a) and their importance predicted from previously characterized families of calmodulin-binding sites catalogued by Dr. M. Ikura and coworkers (http://calcium.oci.utoronto.ca/ctdb/). Thus, calmodulin binding to kendrin is disrupted by mutations in five conserved residues of the predicted calmodulin-binding site.
To better understand the in vivo function of kendrin, we examined the subcellular localization of kendrin throughout the cell cycle in primary human diploid fibroblasts and in human cancer cell lines that demonstrate abnormal centrosomal structures (26). In diploid fibroblasts, antibodies raised against a unique fragment of the kendrin C terminus (amino acids 2628–2969; Fig. 1b) recognized centrosomes, but not microtubules, in both mitotic (Fig. 3 a–d) and interphase (Fig. 3 e–h) cells costained for γ- or α-tubulin. In control experiments, depletion of anti-kendrin antibodies eliminated staining of centrosomes (Fig. 3 i vs. j), whereas selective removal of anti-GST antibodies did not diminish centrosomal staining (not shown). Also, in diploid fibroblasts, immunofluorescence microscopy in combination with optical sectioning and deconvolution localized kendrin in close proximity to the distal portion of the centrioles stained by anti-centrin antibodies (Fig. 3k).
In interphase HS578T breast carcinoma cells, kendrin colocalized with γ-tubulin to multiple aberrant centrosomal structures (226/237 foci or 95%; Fig. 3 l and m). Colocalization of kendrin and γ-tubulin to more than two structures was observed in 90% of cells (52/58 cells; Fig. 3 l and m). In striking contrast, a majority of mitotic HS578T breast cancer cells demonstrated a restriction of kendrin staining to two dots at the mitotic spindle poles (52/67 cells or 78%; Fig. 3o). Costaining for kendrin and α-tubulin revealed that mitotic HS578T cells containing two dots of kendrin staining formed normal bipolar mitotic spindles (52/52 cells). Although γ-tubulin localized to all structures in mitotic cells that contain kendrin (151/151 foci), the converse was not true: a majority of mitotic cells exhibited additional structures containing γ-tubulin but not kendrin (43/67 cells or 64%; Fig. 3p). A smaller fraction of mitotic cells had one, three, or greater than three structures containing kendrin (15/59 cells or 25%), and all of these cells formed aberrant, multipolar spindles (15/15 cells, Fig. 3 r–t). In mitotic breast cancer cells costained for kendrin and α-tubulin, every focus of kendrin staining was associated with microtubules (59/59 cells, 127/127 foci). Finally, in mitotic breast cancer cells, every focus of kendrin contained pericentrin (131/131 foci, Fig. 3 u–w), even in cells containing abnormal numbers of kendrin foci. Thus, our localization data indicate that kendrin and pericentrin, but not γ-tubulin, are restricted to centrosomes at mitotic spindle ends in mitotic breast cancer cells. We have observed similar localization patterns for kendrin in mitotic colon cancer cells (not shown), indicating that kendrin localization is consistent in different types of human cancer cells.
In summary, several lines of evidence indicate that kendrin is a human orthologue of budding yeast Spc110p. Spc110p and kendrin share similar predicted protein structures and localize to analogous structures, the SPB (27) and centrosome, respectively. The N-terminal region of Spc110p binds the γ-tubulin complex (9, 10), and the N-terminal region of kendrin is highly homologous to murine pericentrin, which binds γ-tubulin as shown by coimmunoprecipitation, copurification, colocalization, and fluorescence resonance energy transfer experiments (5). The C-terminal region of Spc110p binds core SPB components (28, 29), and kendrin localizes near centrioles at the vertebrate centrosomal core. Finally, the C-terminal regions of Spc110p (27) and kendrin each contain a distinct calmodulin-binding site that targets Spc110p to the SPB (11–13) and that may target kendrin to spindle poles.
The distinct subcellular localization pattern of kendrin in interphase and mitotic cancer cells is consistent with kendrin playing an essential role in regulating the formation of spindle microtubules. Kendrin is tightly restricted during mitosis to the poles of the mitotic spindle in cancer cells containing extra centrosomal material, and all mitotic cancer cells containing tri- and tetrapolar spindles contained three or four kendrin foci, respectively, at the abnormal spindle poles. Kendrin may regulate spindle formation by recruiting pericentrin and γ-tubulin to nascent spindle poles.
Recently, a similar role has been suggested for the Drosophila melanogaster abnormal spindle protein (Asp). Asp, a centrosomal protein containing potential calmodulin-binding sites (30), appears to regulate the mitotic spindle apparatus by tethering γ-TURCs together (31). Despite the similarities between Asp and kendrin, the functions of these two proteins are likely distinct. Kendrin and Asp share no homology with one another, whereas kendrin is clearly related to pericentrin, which interacts with γ-tubulin (5). The predicted structure of kendrin, like that of Spc110p, contains long central coiled-coil domains flanked by noncoiled ends, whereas the secondary structure of Asp is predicted to be primarily α-helical with short stretches of coiled-coil near its C terminus (30). Additionally, Asp is predicted to contain an actin-binding domain (30), a feature found in neither kendrin nor Spc110p. The calmodulin-binding site of kendrin is similar to that of S. cerevisiae Spc110p and of the Spc110p homologues we identified in A. nidulans and S. pombe, whereas the IQ-type calmodulin-binding site of Asp is more similar to those found in myosins. Finally, Asp localizes to both the centrosome and the spindle and was initially purified as a microtubule-associated protein (30), whereas kendrin is restricted to the centrosome, as is Spc110p. These differences indicate that the activities of kendrin may be more similar to those of Spc110p than to those of Asp. Further analysis of the functional relationships among kendrin, pericentrin, γ-tubulin, and Asp will shed light on the mechanisms controlling the complex process of mitotic spindle formation and should aid in the understanding of centrosomal abnormalities that accompany cancerous growth.
Acknowledgments
We thank James Joseph and Anthony Means for providing data before publication, Steve Doxsey for antibodies to pericentrin, and Shannon Payne for human breast carcinoma lines. We would also like to thank Susan Francis, Eric Muller, Breck Byers, and David Morris for helpful comments on the manuscript. This work was supported by National Institutes of Health Grant R01 GM40506 (to T.N.D.), Training Grant T32 GM-07270-22 (to M.R.F.), and Grants R01 CA48022 and P01 AG01751 (to R.J.M. and M.J.M.).
Abbreviations
- SPB
spindle pole body
- γ-TURC
γ-tubulin ring complex
- GST
glutathione S-transferase
- EST
expressed sequence tag
Footnotes
Accession numbers: Murine EST clones homologous to the C-terminal calmodulin-binding domain of three Spc110p-related fungal proteins, AA600395 and AA000932; human EST clone, AA333809; kendrin cDNA, U52962; PCNT, NM_006031; representative human EST clone homologous to the 3′ untranslated region of murine pericentrin but not homologous to any sequenced regions of human chromosome 21 (as of 1/25/2000), AI970199; human chromosome 21 contigs containing the kendrin genomic locus, NT_002306 and NT_002102.
References
- 1.Zheng Y, Wong M L, Alberts B, Mitchison T. Nature (London) 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
- 2.Oegema K, Wiese C, Martin O C, Milligan R A, Iwamatsu A, Mitchison T J, Zheng Y. J Cell Biol. 1999;144:721–733. doi: 10.1083/jcb.144.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moritz M, Braunfeld M B, Sedat J W, Alberts B, Agard D A. Nature (London) 1995;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- 4.Khodjakov A, Rieder C L. J Cell Biol. 1999;146:585–596. doi: 10.1083/jcb.146.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dictenberg J B, Zimmerman W, Sparks C A, Young A, Vidair C, Zheng Y, Carrington W, Fay F S, Doxsey S J. J Cell Biol. 1998;141:163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Purohit A, Tynan S H, Vallee R, Doxsey S J. J Cell Biol. 1999;147:481–492. doi: 10.1083/jcb.147.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moritz M, Zheng Y, Alberts B M, Oegema K. J Cell Biol. 1998;142:775–786. doi: 10.1083/jcb.142.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schnackenberg B J, Khodjakov A, Rieder C L, Palazzo R E. Proc Natl Acad Sci USA. 1998;95:9295–9300. doi: 10.1073/pnas.95.16.9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knop M, Schiebel E. EMBO J. 1997;16:6985–6995. doi: 10.1093/emboj/16.23.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen T, Vinh D B N, Crawford D K, Davis T N. Mol Biol Cell. 1998;9:2201–2216. doi: 10.1091/mbc.9.8.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sundberg H A, Goetsch L, Byers B, Davis T N. J Cell Biol. 1996;133:111–124. doi: 10.1083/jcb.133.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kilmartin J V, Goh P Y. EMBO J. 1996;15:4592–4602. [PMC free article] [PubMed] [Google Scholar]
- 13.Stirling D A, Rayner T F, Prescott A R, Stark M J. J Cell Sci. 1996;109:1297–1310. doi: 10.1242/jcs.109.6.1297. [DOI] [PubMed] [Google Scholar]
- 14.Kilmartin J V, Dyos S L, Kershaw D, Finch J T. J Cell Biol. 1993;123:1175–1184. doi: 10.1083/jcb.123.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willingham M C, Wehland J, Klee C B, Richert N D, Rutherford A V, Pastan I H. J Histochem Cytochem. 1983;31:445–461. doi: 10.1177/31.4.6338107. [DOI] [PubMed] [Google Scholar]
- 16.Zavortink M, Welsh M J, McIntosh J R. Exp Cell Res. 1983;149:375–385. doi: 10.1016/0014-4827(83)90350-6. [DOI] [PubMed] [Google Scholar]
- 17.Li C J, Heim R, Lu P, Pu Y, Tsien R Y, Chang D C. J Cell Sci. 1999;112:1567–1577. doi: 10.1242/jcs.112.10.1567. [DOI] [PubMed] [Google Scholar]
- 18.Tassin A M, Celati C, Paintrand M, Bornens M. J Cell Sci. 1997;110:2533–2545. doi: 10.1242/jcs.110.20.2533. [DOI] [PubMed] [Google Scholar]
- 19.Berger B, Wilson D B, Wolf E, Tonchev T, Milla M, Kim P S. Proc Natl Acad Sci USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stirling D A, Petrie A, Pulford D J, Paterson D T, Stark M J. Mol Microbiol. 1992;6:703–713. doi: 10.1111/j.1365-2958.1992.tb01519.x. [DOI] [PubMed] [Google Scholar]
- 21.Salisbury J L, Baron A T, Sanders M A. J Cell Biol. 1988;107:635–641. doi: 10.1083/jcb.107.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhoads A R, Friedberg F. FASEB J. 1997;11:331–340. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- 23.Chen H, Gos A, Morris M A, Antonarakis S E. Genomics. 1996;35:620–624. doi: 10.1006/geno.1996.0411. [DOI] [PubMed] [Google Scholar]
- 24.Lapenta V, Sossi V, Gosset P, Vayssettes C, Vitali T, Rabatel N, Tassone F, Blouin J L, Scott H S, Antonarakis S E, et al. Genomics. 1998;49:1–13. doi: 10.1006/geno.1997.5185. [DOI] [PubMed] [Google Scholar]
- 25.Doxsey S J, Stein P, Evans L, Calarco P D, Kirschner M. Cell. 1994;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- 26.Pihan G A, Purohit A, Wallace J, Knecht H, Woda B, Quesenberry P, Doxsey S J. Cancer Res. 1998;58:3974–3985. [PubMed] [Google Scholar]
- 27.Geiser J R, Sundberg H A, Chang B H, Muller E G, Davis T N. Mol Cell Biol. 1993;13:7913–7924. doi: 10.1128/mcb.13.12.7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams I R, Kilmartin J V. J Cell Biol. 1999;145:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elliott S, Knop M, Schlenstedt G, Schiebel E. Proc Natl Acad Sci USA. 1999;96:6205–6210. doi: 10.1073/pnas.96.11.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saunders R D, Avides M C, Howard T, Gonzalez C, Glover D M. J Cell Biol. 1997;137:881–890. doi: 10.1083/jcb.137.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.do Carmo Avides M, Glover D M. Science. 1999;283:1733–1735. doi: 10.1126/science.283.5408.1733. [DOI] [PubMed] [Google Scholar]