Abstract
Papillary renal-cell carcinoma (RCC) is a renal carcinoma variant with distinct gross, microscopic, and cytogenetic features. Recently, a type 1 (pale cytoplasm, small-cell) and a type 2 (eosinophilic cytoplasm, large-cell) subtype of papillary RCC have been described. Chromosomal alterations associated with these tumor types were examined in 25 papillary RCCs by comparative genomic hybridization. Relative copy number gains were frequently detected at chromosomes 7p (56%), 7q (44%), 12q (28%), 16q (32%), 17p (56%), 17q (76%), and 20q (32%). Chromosomal regions that were most often lost included 1p (24%), 4q (36%), 6q (40%), 9p (36%), 13q (36%), Xp (28%), Xq (36%), and Y (73%). There were clinical and genetic differences between the subtypes of papillary RCC. Type 2 tumors were of higher nuclear grade (P = 0.0012) and higher stage (P = 0.01) and had a worse prognosis (P = 0.03) than type 1 tumors. The number of DNA gains per tumor, especially gains of 7p and 17p, was significantly higher in type 1 than in type 2 tumors (P < 0.01). These data suggest the existence of two distinct morphological and genetic subgroups of papillary RCC. Losses of chromosome Xp were associated with short patient survival (P < 0.01). Despite the small number of cases, this finding suggests that a gene on chromosome Xp may contribute to papillary RCC progression.
Papillary renal-cell carcinomas (RCCs) have characteristic gross, histological, and cytogenetic features that separate them from other types of RCC, eg, clear-cell, chromophobe, and duct Bellini carcinomas. 1 Papillary RCCs constitute approximately 10% of renal epithelial tumors, when defined as tumors having at least 50% papillary structures. 2 In comparison with clear-cell (nonpapillary) RCC, papillary tumors are relatively overrepresented in end-stage renal disease and are more frequently multifocal than nonpapillary RCC.
Previous cytogenetic and molecular analyses have identified characteristic genetic alterations that distinguish different RCC subtypes. The most frequent alteration in clear-cell RCC is a deletion of the short arm of chromosome 3, 3-6 whereas papillary tumors are characterized cytogenetically by chromosomal trisomies/tetrasomies, most often including chromosomes 7 and 17, and losses of chromosome Y. 7,8 Only a few papillary RCCs have been included in loss of heterozygosity (LOH) studies by microsatellite or restriction fragment length polymorphism (RFLP) analysis. 9-11
Recently, Delahunt and Eble 12 proposed the existence of two different papillary tumor subtypes, type 1 with small cells and pale cytoplasm and type 2 with large cells and eosinophilic cytoplasm. Type 2 papillary RCC presented more often at higher stages and expressed cytokeratin 7 less frequently than type 1 tumors. Genetic differences between these papillary RCC subtypes have not been investigated yet.
In this study, comparative genomic hybridization (CGH) was applied to screen for genetic differences between type 1 and type 2 papillary RCC. CGH is based on an in situ hybridization of differentially labeled DNAs, one from the tumor and another from the normal reference to normal metaphase spreads, allowing a survey of all DNA copy number changes. 13
Materials and Methods
Patient Selection
All tumors were identified from the archives of the Institute of Pathology, University of Basel. Among 615 renal tumors that had been reviewed by one pathologist (H. Moch) there were 52 papillary RCCs having at least 75% of the tumor composed of true papillae with recognizable fibrovascular cores without clear-cell cytoplasm. The 75% criterion as defining feature was used according to recent recommendations by the UICC. 14 Twenty-five consecutive papillary RCCs (16 male and 9 female patients) were selected for this study. All tumors were diagnosed after 1985. Survival data were obtained by reviewing the hospital records, by direct communication with the attending physicians, and from the Cancer Registry of Basel. Patients were evaluated from the time of biopsy diagnosis to the last known follow-up. The 1996 cutoff date allowed for the possibility of at least 2 years of clinical follow-up.
Tumor material consisted of 14 fresh-frozen tumors and 11 formalin-fixed tumor blocks. One additional tumor was excluded because of insufficient DNA quality for CGH. Histological grading and tumor staging were done according to Fuhrman 15 and International Union Against Cancer (UICC). 16 There were one grade 1, six grade 2, twelve grade 3, and six grade 4 tumors. Ten tumors were stage pT1, seven were pT2, and eight were pT3. The tumor size ranged from 1.2 to 20 cm (median, 7 cm) in largest diameter. The tumors were divided into type 1 (pale cytoplasm) and type 2 (eosinophilic cytoplasm) papillary RCC as suggested by Delahunt and Eble. 12 Type 1 tumors consisted of papillae and tubular structures covered by epithelial cells with small oval nuclei, inconspicuous nucleoli, foamy macrophages in papillary cores, and frequent psammoma bodies (Figure 1A) ▶ . Type 2 tumors consisted of papillae covered by large cells with abundant eosinophilic cytoplasm and large nuclei with more prominent nucleoli (Figure 1B) ▶ . Nine tumors showed a type 1 and sixteen tumors a type 2 cell type. Type 1 tumors were significantly larger (P = 0.03) and had a higher tumor stage (P = 0.01) and a higher nuclear grade (P = 0.0012) more frequently than type 2 tumors (Table 1) ▶ .
Figure 1.
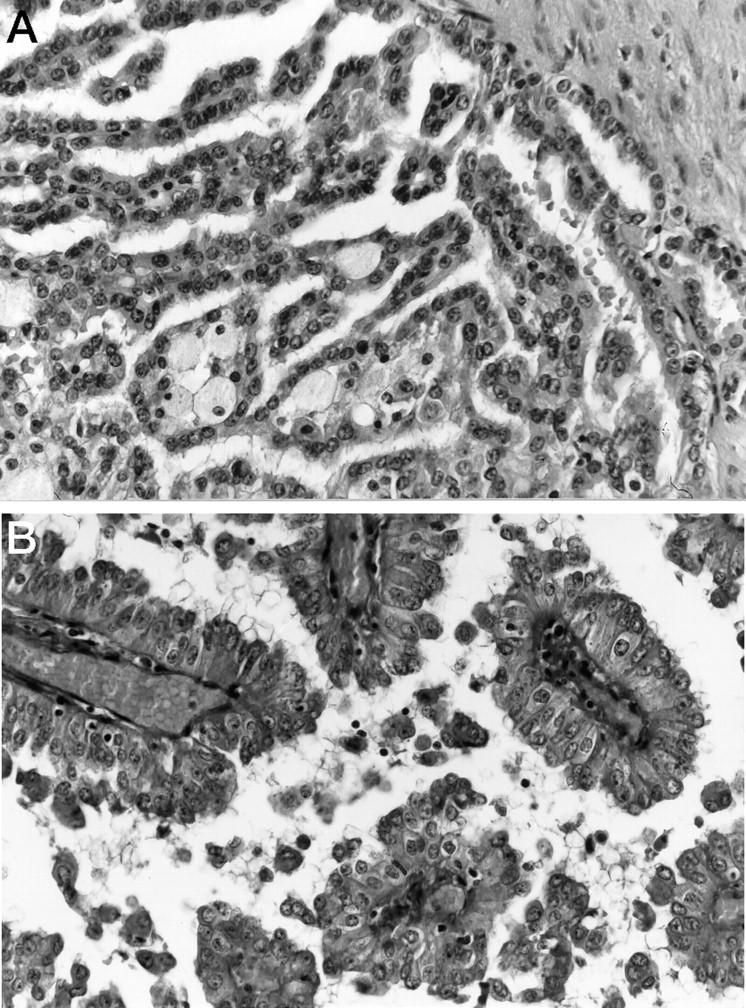
A: Type 1 papillary renal cell carcinoma. The papillae are covered by a single layer of small cells with small nuclei. Foamy macrophages are seen in the papillary cores. B: Type 2 papillary renal cell carcinoma. The papillae are covered by a pseudostratified layer of large cells. Nuclei with prominent nucleoli. H&E; magnification, ×280.
Table 1.
Morphological Findings in 25 Papillary RCCs
| Site, mean ± SD (cm) | Grade (%) | pT stage (%) | |||
|---|---|---|---|---|---|
| 1/2 | 3/4 | 1/2 | 3/4 | ||
| Type 1 (n = 9) | 4.7 ± 2.3 | 67 | 33 | 100 | 0 |
| Type 2 (n = 16) | 8.1 ± 4.1* | 6 | 94† | 50 | 50‡ |
*P = 0.03 (U test).
†P = 0.0012 (contingency table analysis).
‡P = 0.01 (contingency table analysis).
DNA Preparation
Specimens were trimmed to enrich for tumor by excising tumor tissue from the paraffin block. The excised tumor tissue was re-embedded in a paraffin block. Sections were cut from these tumor blocks and stained with hematoxylin and eosin (H&E) to ensure a minimum of 75% tumor cells in the sample. For frozen material, DNA was extracted from eight 50-μm sections. For paraffin material, 25 10-μm sections were taken for DNA extraction. DNA extraction and labeling were as follows. 17 One microgram of tumor DNA was nick translated by using a commercial kit (BioNick Kit, Life Technologies, Gaithersburg, MD) and Spectrum Green-dUTPs (Vysis, Downers Grove, IL) for direct labeling of tumor DNA. Spectrum Red-labeled normal reference DNA (Vysis) was used for co-hybridization.
CGH and Digital Image Analysis
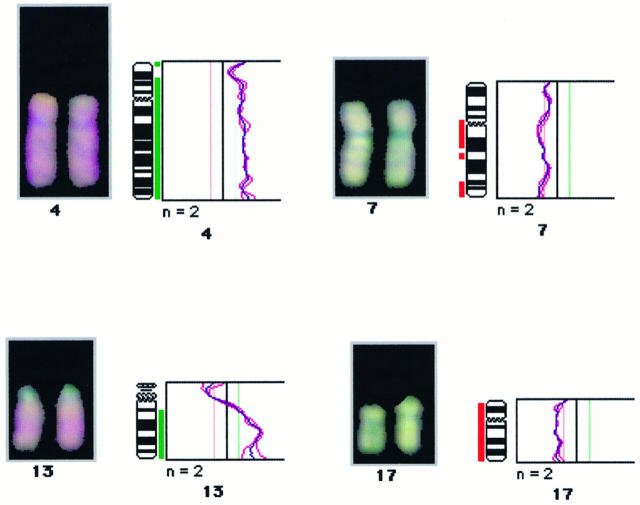
CGH and digital image analysis were carried out as described previously. 17-19 The hybridization mixture consisted of 200 ng of Spectrum Green-labeled tumor DNA, 200 ng of Spectrum Red-labeled normal reference DNA, and 20 μg of human Cot-1 DNA (GIBCO, Gaithersburg, MD) dissolved in 10 μg of hybridization buffer (50% formamide, 10% dextran sulfate, 2X SSC, pH 7.0). Hybridization was for 3 days at 37°C to normal metaphase spreads (Vysis). Post-hybridization washes were as described. 17 Digital images were collected from six to seven metaphases using a Photometrics cooled CCD camera (Microimager 1400, Xillix Technologies, Vancouver, British Columbia, Canada) and a Sun workstation. The Vysis software program was used to calculate average green-to-red ratio profiles for each chromosome. Four observations per autosome and at least two observations per sex chromosome were included in each analysis (Figure 2) ▶ .
Figure 2.
An example of a digital image of a CGH experiment illustrating chromosome 7 and 17 gains and chromosome 4 and 13q losses in a papillary renal cell carcinoma. DNA extracted from the tumor tissue (labeled in green) were hybridized simultaneously to a normal metaphase spread. Chromosomal regions that are overrepresented in the tumor are visualized in a predominantly greenish color, whereas regions that were lost are highlighted in a red color (relative lack of green). Also shown are the green-to-red ratio profiles generated by the image analysis program of all aberrant chromosomes of the same sample. Thin lines indicate normal range of the green-to-red ratios (0.8 to 1.2). Deletions are marked by bars to the right of the chromosome, and gains are marked by bars to the left of the chromosome.
Controls and Threshold Definition
CGH experiments included a tumor cell line (Spectrum Green-labeled MPE-600 DNA; Vysis) with known aberrations (positive control) and a hybridization of two differentially labeled sex-mismatched normal DNAs to each other (negative control). Sex-mismatched normal controls were also used to test the ability of each metaphase batch to allow for a linear relationship between fluorescence intensities and DNA sequence copy numbers. Metaphases were used only if the color ratio of sex-mismatched normal DNAs was ≤0.66 at the X chromosome. The thresholds used for definition of DNA sequence copy number gains and losses were based on the results of CGH analyses of formalin-fixed normal tissues. Gains of DNA sequences were defined as chromosomal regions where both the mean green-to-red fluorescence ratio and its SD were above 1.20, whereas losses were defined as regions where both the mean and its SD were below 0.80. Overrepresentations were considered amplifications when the fluorescence ratio values in a subregion of a chromosomal arm exceeded 1.5. In negative control hybridizations, the mean green-to-red ratio occasionally exceeded the fixed 1.2 cutoff level at the following chromosomal regions: 1p32-pter, 16p, 19, and 22. These known G-C-rich regions were therefore excluded from all analyses.
Statistics
Contingency table analysis was used to analyze the relationship between genomic alterations, grade, stage, and cell type. A U test was applied to compare the number of genomic alterations between different grades, stages, and histological subtypes. Overall survival rates were plotted by the Kaplan-Meier method using the median number of aberrations as a cutoff point for grouping between a low and high number of aberrations. The most common individual aberrations (occurring in more than 20% of tumors) were analyzed for their association with patient survival. Statistical differences between the groups were determined with the log-rank test. A Cox proportional hazards analysis was used to test for independent prognostic information.
Results
CGH Findings
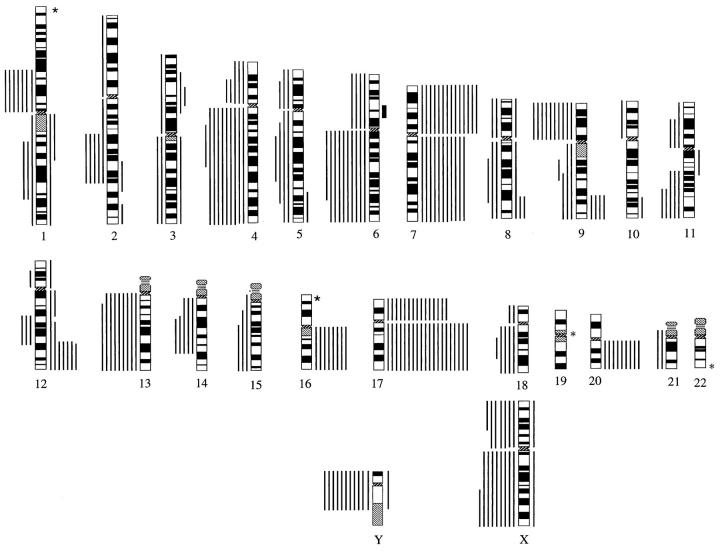
Twenty-five papillary RCCs were examined by CGH. A summary of all DNA sequence copy number aberrations is shown in Figure 3 ▶ . Twenty-two tumors showed DNA sequence copy number gains, and in all tumors losses at one or more chromosomal regions were present. There was a median of 7 aberrations per tumor (mean, 10.0; range, 2 to 29). The median number of gains was 5 (mean, 4.4; range, 0 to 10), and the median number of losses was 4 per tumor (mean, 5.6; range, 1 to 19).
Figure 3.
Summary of all relative DNA sequence copy number changes detected by CGH in 25 papillary renal cell carcinomas. The vertical lines on the right of the chromosome ideograms indicate gains; those on the left indicate losses. Solid bars indicate a gene amplification. *1p31-term, 16p, 19, and 22 were not analyzed.
Chromosomal regions that were most often lost included 1p (24%), 4p (20%), 4q (36%), 6q (40%), 9p, 13q (36% each), 14q, and 18q (20% each). There were frequent losses of sex chromosomes. Losses of Xp were seen in 28% and of Xq in 36% of patients. Chromosome X losses were more frequent in female than in male patients. Xp loss was seen in five (56%) and Xq loss in six (67%) female patients (n = 9). Xp loss was detected in two (12%), Xq loss in three (19%) male patients (n = 16). Chromosome Y loss was detected in 11 of 15 (73%) males.
Increased DNA sequence copy number was most often detected at chromosome 7p (56%), 7q (44%), 12q (28%), 16q (32%), 17p (56%), 17q (76%), and 20q (32%). The entire long arms of chromosomes 7 and 17 were often gained. One high-level amplification (green-to-red ratio > 1.5) was found in one tumor at the 6p12-6p21 region.
Histopathological Correlations
There was no association between tumor stage and the number of CGH aberrations (Table 2) ▶ . However, there was a strong tendency toward a higher number of DNA losses in tumors with high histological tumor grade (6.6 ± 5.4 per tumor) than in low-grade tumors (2.8 ± 3.0 per tumor). Even so, this difference did not reach statistical significance (P = 0.0893), nor was there a difference in the number of aberrations between low-grade and high-grade tumors.
Table 2.
Stage, Grade, Histological Subtype, and Number of CGH Aberrations
| n | Number of aberrations (mean ± SD) | |||
|---|---|---|---|---|
| All | Deletions | Gains | ||
| Stage pT1/2 | 17 | 10.3 ± 7.3 | 5.4 ± 5.1 | 4.9 ± 3.0 |
| Stage pT3/4 | 8 | 9.5 ± 7.0* | 5.9 ± 5.0† | 3.6 ± 2.8‡ |
| Grade 1/2 | 7 | 8.1 ± 4.0 | 2.8 ± 3.0 | 5.3 ± 1.5 |
| Grade 3/4 | 18 | 10.8 ± 8.0§ | 6.6 ± 5.2¶ | 4.2 ± 3.4∥ |
| Type 1 | 9 | 12.4 ± 7.9 | 6.0 ± 6.0 | 6.4 ± 2.0 |
| Type 2 | 16 | 8.8 ± 6.5** | 5.3 ± 4.4†† | 3.4 ± 2.9‡‡ |
P values for pT1/2 versus pT3/4; grade 1/2 versus grade 3/4 and type 1 versus type 2 subtype: *0.7861, †0.8323, ‡0.3143, §0.4069, ¶0.0893, ∥0.4359, **0.2192, ††0.747, ‡‡0.0123.
To screen for chromosomal alterations that might be linked to type 1 or type 2 papillary tumors we analyzed the association between the most frequent specific aberrations and the tumor subtype (Table 3) ▶ . There were no genetic differences between the two subtypes of papillary RCC with the exception of chromosome 7 and 17 gains. Gains of 7p and 17p were detected in all type 1 but in only 31% (7p) and 38% (17p) of type 2 tumors (P < 0.01). Accordingly, there was a slightly higher frequency of chromosome 7p and 17p gains in low-grade and low-stage tumors (P < 0.05; Table 3 ▶ ), as most type 1 RCCs were pT 1/2 stage and nuclear grade 1/2.
Table 3.
Most Frequent (>20%) DNA Sequence Copy Number Losses (−) and Gains (+) in Papillary RCC
| % of tumors | P Value | % of tumors | P value | % of tumors | P value | ||||
|---|---|---|---|---|---|---|---|---|---|
| Stage pT1/2 (n = 17) | Stage pT3/4 (n = 8) | Grade 1/2 (n = 7) | Grade 3/4 (n = 18) | Type 1 (n = 9) | Type 2 (n = 16) | ||||
| 7p+ | 64.7 | 37.5 | NS | 100 | 38.9 | 0.02 | 100 | 31.2 | 0.004 |
| 7q+ | 52.9 | 25 | NS | 71.4 | 33.3 | NS | 66.7 | 31.2 | NS |
| 12q+ | 29.4 | 25 | NS | 14.3 | 33.3 | NS | 22.2 | 31.2 | NS |
| 16q+ | 35.3 | 25 | NS | 41.8 | 27.8 | NS | 55.6 | 18.8 | NS |
| 17p+ | 76.5 | 25 | 0.04 | 85.7 | 50 | NS | 100 | 37.5 | 0.008 |
| 17q+ | 88.2 | 62.5 | NS | 100 | 72.2 | NS | 100 | 68.8 | NS |
| 20q+ | 41.2 | 25 | NS | 28.6 | 38.9 | NS | 55.6 | 25 | NS |
| 1p− | 29.4 | 25 | NS | 14.3 | 33.3 | NS | 33.3 | 25 | NS |
| 4q− | 52.9 | 12.5 | NS | 28.6 | 44.4 | NS | 55.6 | 31.2 | NS |
| 6q− | 41.2 | 37.5 | NS | 14.3 | 50 | NS | 44.4 | 37.5 | NS |
| 9p− | 52.9 | 12.5 | NS | 14.3 | 50 | NS | 44.4 | 37.5 | NS |
| 13q− | 41.2 | 25 | NS | 42.9 | 35.3 | NS | 66.7 | 18.8 | NS |
| Xp− | 17.6 | 50 | NS | 14.3 | 33.3 | NS | 22.2 | 31.2 | NS |
| Xq− | 17.6 | 75 | 0.02 | 14.3 | 47.1 | NS | 33.3 | 37.5 | NS |
P values are by χ2 test; all significant results (P ≤ 0.05) are shown. NS, not significant.
Clinical Outcome
Overall survival data were available for 24 patients. There was a median follow-up of 32 months (minimum, 1 month; maximum, 105 months). Nine patients had died, and fifteen patients were censored at the last clinical control. None of the patients with grade 1 and 2 tumors (n = 7) died of disease or developed metastases. Type 1 tumors had a significantly better prognosis (P = 0.03) than type 2 tumors. Clinical outcome of patients with stage pT1 and pT2 RCC was better than that of patients with stage pT3 (P = 0.08).
The number of aberrations (gains, losses, and total number) was not associated with patient prognosis when the median number of aberrations was used as a cutoff point. The evaluation of the most common individual aberrations for clinical significance revealed that Xp deletions were significantly associated with poor clinical outcome (P < 0.01; log-rank test; Figure 4A ▶ ). Five of seven patients with chromosome Xp loss died of disease during clinical follow-up. Proportional hazards analysis with the variables tumor stage and histological grade indicated that chromosome Xp loss was an independent predictor of prognosis, the relative risk being 5.4 (P = 0.03). Neither the tumor stage nor the histological grade provided additional prognostic information. Tumors with Xq loss had a worse prognosis than tumors without Xq loss (Figure 4B) ▶ , but this trend did not reach significance (P = 0.1). None of the other chromosomal alterations (losses of 1p, 4q, 9p, 13q, and 14q; gains of 7, 17, 12q, 16q, and 20q) was associated with patient prognosis.
Figure 4.
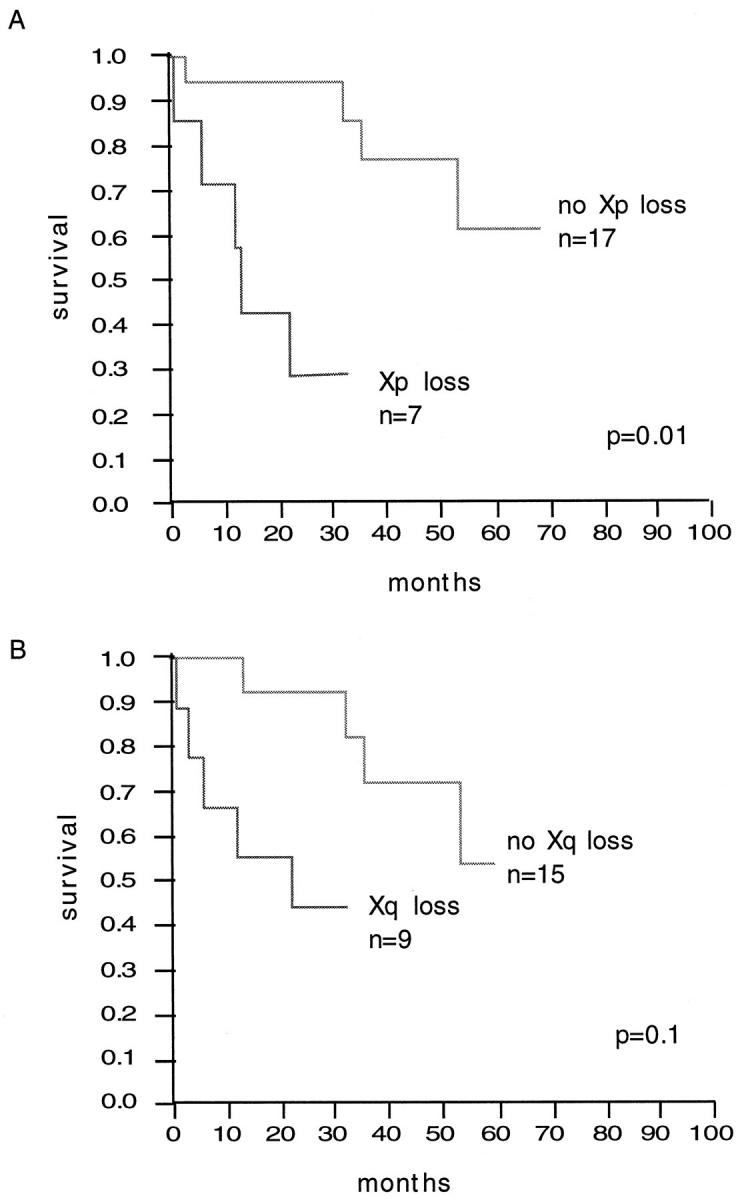
A: Overall survival analysis for patients with and without chromosome Xp losses (log-rank test). B: Overall survival analysis for patients with and without chromosome Xq losses (log-rank test).
Discussion
In this study, CGH was used to define genetic changes associated with papillary RCC. Stringent criteria were used for the definition of papillary tumors in this study to exclude nonpapillary (clear-cell) RCC, known to sometimes contain areas with papillary growth. Papillary RCCs were defined as tumors in which at least 75% of the tumor tissue was composed of true papillae and without clear-cell areas. A recent cytogenetic study 7 suggests that 75% is a better criterion for defining papillary RCC than the 50% criterion proposed by Mancilla-Jimenez. 2
A high prevalence of chromosome 7 and 17 gains was expected because both alterations have been described by cytogenetic and FISH analyses in most papillary RCCs. 20-22 Even in papillary renal cortical adenomas of <1 cm in largest diameter, gains of chromosomes 7 and 17 were found by cytogenetics 7 and CGH (J. Presti, unpublished observations). In this study, gains of chromosomes 7 and 17 were detected in almost all type 1 tumors but were considerably less frequent in type 2 papillary RCC. This lower percentage of chromosome 7 and 17 gains in a papillary RCC subtype has not been described in earlier cytogenetic analyses, possibly because papillary RCCs were not separated according to the cell type 20 or because this subtype was underrepresented in these studies. Only 2 of 11 papillary RCCs had an eosinophilic cell type in a cytogenetic analysis of Kovacs et al. 7 Interestingly, both tumors had no chromosome 7 gains. Lager et al 23 demonstrated trisomy 7 in 67% of low-grade but in only 43% of high-grade papillary RCCs. This is consistent with our results, because type 2 (large-cell) papillary RCCs typically show a higher histological grade than type 1 tumors.
The histogenetic relationship between type 1 and type 2 papillary RCC is currently unclear. Type 2 papillary tumors usually have a higher nuclear grade than type 1 tumors. They may therefore develop from type 1 papillary tumors with low nuclear grade. We feel that this is unlikely, because foam cells and psammoma bodies are more frequent in type 1 than in type 2 tumors. The existence of very small papillary adenomas showing large cells with eosinophilic cytoplasm, sometimes found at autopsy, might be a further argument for a de novo development of type 2 papillary RCC. Because of genetic differences between papillary tumors with different cell types, one might speculate that type 1 and type 2 papillary RCCs represent two different tumor entities.
It has been suggested that type 1 and type 2 tumors have a different clinical behavior. In this set of tumors, type 2 tumors had a 5-year survival rate of 36%, which is similar to clear-cell RCC, whereas none of nine patients with type 1 tumors died of disease. This observation is consistent with a report of Lager et al showing that none of 19 low-grade tumors with typical basophilic cells developed metastases. 23 However, we recently analyzed 208 renal tumors with a tumor diameter smaller than 3 cm at autopsy. We identified two type 1 papillary RCCs (tumor diameters of 2 and 2.8 cm, respectively) with metastases (unpublished observation), suggesting that small type 1 papillary RCCs are not always benign tumors.
Kovacs et al speculated that tumors with polysomy 7 and 17 as a sole aberration are papillary adenomas. 7 He suggested that accumulation of additional polysomies of chromosomes 12, 16, or 20 are linked to progression toward papillary carcinomas 6,7,9,21 with a more malignant behavior possibly because of activation of oncogenes. 21 However, neither the total number number of chromosomal gains detected by CGH nor specific gains, including gains of 12, 16, and 20 were associated with tumor stage or patient prognosis in this set of patients.
The clearly higher number of DNA losses in high-grade tumors (mean, 6.6) than in low-grade tumors (mean, 2.8) argues for a role of tumor suppressor genes in the progression of papillary RCC, because inactivation of tumor suppressor genes is usually accompanied by allelic losses. However, little is known about allelic losses in papillary RCC. Frequent sites of deletions included 4q, 6q, 9p, 13q, and X. These chromosomal loci are also subject to allelic losses in clear-cell RCC as determined by microsatellite or CGH analysis. Deletions at 6q have been reported in 18 to 22%, 10,18 at 9p in 16 to 33%, 24,25 at 13q in 50%, 26 and at Xq in 20% 18 of clear-cell carcinomas. Thrash-Bingham et al have recently shown 9p LOH in two of five papillary RCCs. 25 The finding that these relative DNA sequence copy number losses are found at similar frequencies in papillary and clear-cell RCC is consistent with a role of genes at these loci for progression of both tumor types. Interestingly, the only chromosomal aberration with a significant association to poor patient outcome was loss of X. A number of X-located genes, including STS (steroid sulfatase gene), XG (X-linked blood group gene), ZFX (zinc-finger protein, X-linked), KAL1 (Kallman syndrome gene), RPS4X (ribosomal protein S4 (X-linked), and XIST (X inactive specific transcript) are putative tumor suppressor genes. 27-29
X chromosome losses were found in approximately 60% of the papillary RCCs of female patients. The X chromosome has been rarely mentioned in cancer studies, 30 as LOH analyses cannot detect allelic imbalances in male patients. In contrast to LOH analyses, CGH allows detection of relative DNA sequence copy number changes of chromosome X in tumors of male patients. Chromosome X losses were detected more frequently in tumors of female (67%) than of male patients (19%). Interestingly, tumors of male patients have chromosome Y losses in more than 80% of papillary RCCs. 21 Recently, it has been suggested that tumor suppressor genes are located at pseudoautosomal regions (PARs) on chromosome X. The PAR is a zone at the end of the short arm of the sex chromosomes X and Y 28,31,32 showing marked genetic homology. It is tempting to speculate that one or more tumor suppressor genes at the PAR are also relevant for papillary RCC, as loss of the Y chromosome could uncover a mutant allele of a gene in the PAR region at the X chromosome in male patients. In contrast, inactivation of both alleles of a tumor suppressor gene at chromosome X might lead to papillary RCC progression in female patients. This mechanism might be less frequent, because there has been described a strong male predilection for papillary RCC in large surgical series (male-to-female ratio of 5:1). 12
Cytogenetic analyses were consistently negative for 3p deletions 6,7,21,33-35 in papillary RCC. Importantly, we have detected a 3p deletion in one unequivocal papillary RCC with 100% papillary differentiation. Our result is consistent with previous studies finding losses on 3p in 10 of 73 cases of papillary RCC (14%) examined to date 4,5,9,11,20,36-39 by molecular techniques. Also, chromosome 3p loss was detected in 1 of 11 papillary RCCs in another CGH study. 40 These results show that papillary RCC cannot be diagnosed on the basis of the absence of 3p deletion as was previously suggested. 10
There was only one papillary RCC with a gene amplification at chromosome 6p21. Potential candidate oncogenes on 6p21 may include the autosomal recessive polycystic kidney disease gene, 41 the metalloendopeptidase meprin gene MEP1A, encoding for kidney and intestinal proteases, 42 and the tumor necrosis factor-α gene. 43 The low frequency of amplifications in papillary RCC is in striking contrast to other solid tumors, eg, bladder and breast carcinomas. Recently, we found 12 amplifications at eight different loci in 56 noninvasive (pTa) and minimally invasive (pT1) bladder carcinomas by CGH 19 but no amplification in 41 locally advanced (pT3) clear-cell RCCs. 18
In summary, our data have shown that chromosome Xp losses are associated with poor patient prognosis in papillary RCC. Despite the small number of cases, this may suggest a role of genes on chromosome X for progression of papillary RCC. A high number of chromosomal gains was associated with type 1 papillary RCC, which is characterized by low nuclear grade and a significantly better prognosis. These data further suggest the existence of two distinct morphological subgroups of papillary RCC, which might be characterized by different genetic alterations.
Acknowledgments
We thank Dr. Lukas Rosenthaler for help with the computer systems. We thank C. Egenter, M. Storz, M. Mirlacher, H. Novotny, and the staff of the Pathology Department, University of Basel, for their technical support.
Footnotes
Address reprint requests to Dr. Holger Moch, Institut für Pathologie der Universität Basel, Schönbeinstrasse 40, CH-4003 Basel, Switzerland. E-mail: moch@ubaclu.unibas.ch.
Supported by the Swiss National Science Foundation (3200–043969.95/1 and 3100–050752.97/1), Schweizerische Krebsliga (291-2-1996), Krebsforschung Schweiz (367-9-1996), and Krebsliga beider Basel (12/95).
References
- 1.Amin M, Corless C, Renshaaw A, Tickoo S, Kubus J, Schultz D: Papillary (chromophil) renal cell carcinoma: histomorphologic characteristics and evaluation of conventional pathologic prognostic parameters in 62 cases. Am J Surg Pathol 1997, 21:621-635 [DOI] [PubMed] [Google Scholar]
- 2.Mancilla-Jimenez R, Stanley RJ, Blath RA: Papillary renal cell carcinoma. Cancer 1976, 38:2469-2480 [DOI] [PubMed] [Google Scholar]
- 3.Anglard P, Tory K, Brauch H, Weiss GH, Latif F, Merino MJ, Lerman MI, Zbar B, Linehan WM: Molecular analysis of genetic changes in the origin and development of renal cell carcinoma. Cancer Res 1991, 51:1071-1077 [PubMed] [Google Scholar]
- 4.Brooks JD, Bova GS, Marshall FF, Isaacs WB: Tumor suppressor gene allelic loss in human renal cancers. J Urol 1993, 150:1278-1283 [DOI] [PubMed] [Google Scholar]
- 5.el Naggar AK, Batsakis JG, Wang G, Lee MS: PCR-based RFLP screening of the commonly deleted 3p loci in renal cortical neoplasms. Diagn Mol Pathol 1993, 2:269-276 [PubMed] [Google Scholar]
- 6.Presti J, Rao H, Chen Q, Reuter V, Li F, Fair W, Jhanwar S: Histopathological, cytogenetic, and molecular characterization of renal cortical tumors. Cancer Res 1991, 51:1544-1552 [PubMed] [Google Scholar]
- 7.Kovacs G: Papillary renal cell carcinoma: a morphologic and cytogenetic study of 11 cases. Am J Pathol 1989, 134:27-34 [PMC free article] [PubMed] [Google Scholar]
- 8.Renshaw AA, Zhang H, Corless CL, Fletcher JA, Pins MR: Solid variants of papillary (chromophil) renal cell carcinoma: clinicopathologic and genetic features. Am J Surg Pathol 1997, 21:1203-1209 [DOI] [PubMed] [Google Scholar]
- 9.Presti J, Reuter V, Cordon-Cardo C, Mazumdar M, Fair W, Jhanwar S: Allelic deletions in renal tumors: histopathological correlations. Cancer Res 1993, 53:5780-5783 [PubMed] [Google Scholar]
- 10.Bugert P, Kovacs G: Molecular differential diagnosis of renal cell carcinomas by microsatellite analysis. Am J Pathol 1996, 149:2081-2088 [PMC free article] [PubMed] [Google Scholar]
- 11.Thrash-Bingham C, Salazar H, Freed J, Greenberg R, Tartof K: Genomic alterations and instabilities in renal cell carcinomas and their relationship to tumor pathology. Cancer Res 1995, 55:6189-6195 [PubMed] [Google Scholar]
- 12.Delahunt B, Eble J: Papillary renal cell carcinoma: a clinicopathologic and immunohistochemical study of 105 tumors. Mod Pathol 1997, 10:537-544 [PubMed] [Google Scholar]
- 13.Kallioniemi A, Kallioniemi O, Sudar D, Rutovitz D, Gray J, Waldman F, Pinkel D: Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 1992, 258:818-821 [DOI] [PubMed] [Google Scholar]
- 14.Bostwick D, Eble J, Denis L, Murphy G, von Eschenbach A: Diagnosis and prognosis of renal cell carcinoma. Cancer 1997, 80:973-10019307200 [Google Scholar]
- 15.Fuhrman S, Lasky L, Limas C: Prognostic significance of morphologic parameters in renal cell carcinoma. Am J Surg Pathol 1982, 6:655-663 [DOI] [PubMed] [Google Scholar]
- 16.International Union Against Cancer (UICC): TNM Classification of Malignant Tumours. Edited by Sobin L, Wittekind C. New York, Wiley-Liss, 1997
- 17.Jiang F, Moch H, Richter J, Gasser T, Gschwind R, Egenter C, Bubendorf L, Sauter G, Mihatsch M: Comparative genomic hybridization reveals frequent chromosome 13q and 4q losses in renal cell carcinomas with sarcomatoid transformation. J Pathol 1998, 185:382-388 [DOI] [PubMed] [Google Scholar]
- 18.Moch H, Presti JC, Jr, Sauter G, Buchholz N, Jordan P, Mihatsch MJ, Waldman FM: Genetic aberrations detected by comparative genomic hybridization are associated with clinical outcome in renal cell carcinoma. Cancer Res 1996, 56:27-30 [PubMed] [Google Scholar]
- 19.Richter J, Jiang F, Görög J, Sartorius G, Egenter C, Gasser T, Moch H, Mihatsch M, Sauter G: Marked genetic differences between stage pTa and stage pT1 papillary bladder cancer detected by comparative genomic hybridization. Cancer Res 1997, 57:2860-2864 [PubMed] [Google Scholar]
- 20.Corless C, Aburatani H, Fletcher J, Housman D, Amin M, Weinberg D: Papillary renal cell carcinoma: quantitation of chromosomes 7 and 17 by FISH, analysis of chromosome 3p for LOH, and DNA ploidy. Diagn Mol Pathol 1996, 5:53-64 [DOI] [PubMed] [Google Scholar]
- 21.Kovacs G, Fuzesi L, Emanual A, Kung HF: Cytogenetics of papillary renal cell tumors. Genes Chromosomes & Cancer 1991, 3:249-255 [DOI] [PubMed] [Google Scholar]
- 22.Moch H, Sauter G, Gasser T, Bubendorf L, Presti J, Mihatsch MJ, Waldman FM: EGF-r gene copy number gains detected by fluorescence in situ hybridization in renal cell carcinoma. J Pathol 1998, 184:424-429 [DOI] [PubMed] [Google Scholar]
- 23.Lager DJ, Huston BJ, Timmerman TG, Bonsib SM: Papillary renal tumors: morphologic, cytochemical, and genotypic features. Cancer 1995, 76:669-673 [DOI] [PubMed] [Google Scholar]
- 24.Schullerus D, Herbers J, Chudek J, Kanamura H, Kovacs G: Loss of heterozygosity at chromosomes 8p, 9p, and 14q is associated with stage and grade of non-papillary renal cell carcinomas. J Pathol 1997, 183:151-155 [DOI] [PubMed] [Google Scholar]
- 25.Thrash-Bingham CA, Greenberg RE, Howard S, Bruzel A, Bremer M, Goll A, Salazar H, Freed JJ, Tartof KD: Comprehensive allelotyping of human renal cell carcinomas using microsatellite DNA probes. Proc Natl Acad Sci USA 1995, 92:2854-2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai S, Bendict W, Silver S, El-Naggar A: Loss of retinoblastoma gene function and heterozygosity at the RB locus in renal cortical neoplasms. Hum Pathol 1997, 28:693-697 [DOI] [PubMed] [Google Scholar]
- 27.Ellis N, Goodfellow P: The mammalian pseudoautosomal region. Trends Genet 1989, 5:406-410 [DOI] [PubMed] [Google Scholar]
- 28.Brown C, Willard H: Localization of a gene that escapes inactivation to the X chromosome proximal short arm: implications for X inactivation. Am J Hum Genet 1990, 46:273-279 [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Passage M, Ellison J, Becker M, Yen P, Shapiro L, Mohandas T: Physical mapping of loci in the distal half of the short arm of the human X chromosome: implications for the spreading of the X-chromosome inactivation. Somat Cell Mol Genet 1992, 18:195-200 [DOI] [PubMed] [Google Scholar]
- 30.Seizinger B, Klinger H, Junien C, Nakamura Y, Le Beau M, Cavanee W, Emanuel B, Ponder B, Naylor S, Mitelman F, Louis D, Menon A, Newsham I, Decker J, Kaelbling M, Henry I, von Deimling A: Report of the committee on chromosome and gene loss in human neoplasia. Cytogenet Cell Genet 1991, 58:1080-1096 [Google Scholar]
- 31.Goodfellow P, Darling S, Thomas N, Goodfellow P: A pseudoautosomal gene in man. Science 1986, 234:740-743 [DOI] [PubMed] [Google Scholar]
- 32.Petit C, Levilliers J, Weissenbach J: Physical mapping of the human pseudo-autosomal region: comparison with genetic linkage map. EMBO J 1988, 7:236-2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emanuel A, Szucs S, Weier H, Kovacs G: Clonal aberrations of chromosomes X, Y, 7 and 10 in normal kidney tissue of patients with renal cell tumors. Genes Chromosomes & Cancer 1992, 4:75-77 [DOI] [PubMed] [Google Scholar]
- 34.Hughson M, Johnson L, Silva F, Kovacs G: Nonpapillary and papillary renal cell carcinoma: a cytogenetic and phenotypic study. Mod Pathol 1993, 6:449-456 [PubMed] [Google Scholar]
- 35.van den Berg E, van der Hout AH, Oosterhuis JW, Storkel S, Dijkhuizen T, Dam A, Zweers HM, Mensink HJ, Buys CH, de Jong B: Cytogenetic analysis of epithelial renal-cell tumors: relationship with a new histopathological classification. Int J Cancer 1993, 55:223-227 [DOI] [PubMed] [Google Scholar]
- 36.Hadaczek P, Podolski J, Toloczko A, Kurzawski G, Sikorski A, Rabbitts P, Huebner K, Lubinski J: Losses at 3p common deletion sites in subtypes of kidney tumors: histopathological correlations. Virchows Arch 1996, 429:37-42 [DOI] [PubMed] [Google Scholar]
- 37.Gnarra JR, Tory K, Weng Y, Schmidt L, Wei MH, Li H, Latif F, Liu S, Chen F, Duh FM, Lubensky J, Duan DR, Florence C, Pozzatti R, Walther MM, Bander NH, Grossmann HB, Brauch H, Pomer S, Brooks JD, Isaacs WB, Lerman MI, Zbar B, Linehan WM: Mutations of the VHL tumour suppressor gene in renal carcinoma. Nature Genet 1994, 7:85-90 [DOI] [PubMed] [Google Scholar]
- 38.van der Hout AH, van den Berg E, van der Vlies P, Dijkhuizen T, Storkel S, Oosterhuis JW, de Jong B, Buys CH: Loss of heterozygosity at the short arm of chromosome 3 in renal-cell cancer correlates with the cytological tumour type. Int J Cancer 1993, 53:353-357 [DOI] [PubMed] [Google Scholar]
- 39.Moch H, Schraml P, Bubendorf L, Richter J, Gasser T, Mihatsch M, Sauter G: Intratumoral heterogeneity of 3p deletions in renal cell carcinoma detected by fluorescence in situ hybridization. Cancer Res 1998, 58:2304-2309 [PubMed] [Google Scholar]
- 40.Bentz M, Bergerheim US, Li C, Joos S, Werner CA, Baudis M, Gnarra J, Merino MJ, Zbar B, Linehan WM, Lichter P: Chromosome imbalances in papillary renal cell carcinoma and first cytogenetic data of familial cases analyzed by comparative genomic hybridization. Cytogenet Cell Genet 1996, 75:17-21 [DOI] [PubMed] [Google Scholar]
- 41.Guay Woodford LM, Muecher G, Hopkins SD, Avner ED, Germino GG, Guillot AP, Herrin J, Holleman R, Irons DA, Primack W, Thomson PD, Waldo FB, Lunt PW, Zerres K: The severe perinatal form of autosomal recessive polycystic kidney disease maps to chromosome 6p21.1-p12: implications for genetic counseling. Am J Hum Genet 1995, 56:1101-1107 [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang W, Dewald G, Brundage E, Mucher G, Schildhaus HU, Zerres K, Bond JS: Fine mapping of MEP1A, the gene encoding the α subunit of the metalloendopeptidase meprin, to human chromosome 6P21. Biochem Biophys Res Commun 1995, 216:630-635 [DOI] [PubMed] [Google Scholar]
- 43.Honchel R, McDonnell S, Schaid DJ, Thibodeau SN: Tumor necrosis factor-α allelic frequency and chromosome 6 allelic imbalance in patients with colorectal cancer. Cancer Res 1996, 56:145-149 [PubMed] [Google Scholar]