Abstract
Primary effusion lymphoma (PEL) is a recently described distinct subtype of non-Hodgkin’s lymphoma associated with infection by the Kaposi’s sarcoma-associated herpesvirus, also called human herpesvirus-8. Most cases of PEL are also associated with the Epstein-Barr virus (EBV). In order to better characterize the cellular origin of PEL, we investigated the immunoglobulin (Ig) heavy chain variable region (VH) genes expressed by tumor cells of the BC-1 and BC-3 cell lines derived from PELs and five original PEL specimens. In the six EBV-positive PELs examined, including the BC-1 cell line, the expressed VH gene sequences showed numerous point mutations relative to the putative germline VH gene sequences. In addition, the VH segment of one of these cases showed intraclonal sequence heterogeneity, indicating ongoing somatic mutation. In five cases, the distribution and type of mutations indicated that tumor cells had been selected by antigen. Because somatically mutated Ig genes are expressed by B cells that have reached a germinal center/post-germinal center stage of development, these findings suggest that the PEL cell of origin is a germinal center or post-germinal center B cell in most cases. In contrast, the VH gene segment expressed by tumor cells of the BC-3 cell line, which was originated from an EBV-negative PEL obtained from an HIV-negative patient, was unmutated, suggesting a pre-germinal center B cell origin for tumor cells of this particular PEL cell line. Taken together, these findings suggest that development of PELs may not be restricted to one stage of B cell differentiation and may represent transformation of B cells at different stages of ontogeny.
In 1989 Knowles et al, 1 and later other investigators, described an unusual type of aggressive non-Hodgkin’s lymphoma that grows in the pleural, pericardial, and abdominal cavities as a lymphomatous effusion, usually in the absence of a detectable tumor mass. 2-4 These so-called primary effusion lymphomas (PEL) have been recognized as a distinct clinicopathological and biological entity based on their unique constellation of clinical, morphological and immunophenotypic characteristics and consistent infection of the tumor cells by the Kaposi’s sarcoma-associated herpesvirus (KSHV)/human herpesvirus-8 (HHV-8). 5-9 PEL occurs predominantly but not exclusively among human immunodeficiency virus (HIV)-infected individuals. 5,10-13 These neoplasms exhibit cytomorphologic features that bridge large cell immunoblastic and anaplastic large cell lymphoma, usually lack surface immunoglobulin (Ig) and B-cell-associated antigens but express CD45 and antigens associated with late stages of B cell differentiation such as CD30, CD38, CD71, CD138, and epithelial membrane antigen (EMA). 5-7,13 Genotypic analysis of the tumor cells has revealed clonal Ig gene rearrangement in all cases and Epstein-Barr virus (EBV) infection in most cases. 6
Although the immunophenotypic and genotypic characteristics of PELs confirm their B cell lineage, the normal counterpart and stage of differentiation of the tumor cells remain controversial. The combination of the pleomorphic morphology, null cell immunophenotype, and CD30 antigen expression has led some authors to link PEL to anaplastic large cell lymphoma. 10 In contrast, other investigators have suggested that PEL cells represent a mature stage of B cell development closely associated with mature plasma cells based on the findings that the tumor cells coexpress CD30, CD38, CD138, and EMA. 6,9 Finally, some of the PELs display Ig heavy chain but not light chain gene rearrangement, causing some investigators to suggest that PELs may arise early in B cell development, following heavy chain gene but prior to light chain gene rearrangement. 4
Analysis of nucleotide sequences of Ig heavy chain variable region (VH) genes can provide insight into the stage of B cell development at which clonal expansion occurs in B cell tumors. It has been shown that pregerminal center B cells contain IgVH region genes with germ line sequence and postgerminal center B cells contain mutated Ig VH regions. Furthermore, antigen selected postgerminal center B cells display somatic mutations clustered in the complementary determining regions (CDR) sequences. Germinal center B cells are characterized by ongoing mutation, which is absent in postgerminal center B cells. 14,15
In order to determine the stage of B cell development at which clonal expansion of the lymphoma cells occurs in PEL, we analyzed nucleotide sequences of IgVH regions expressed by seven PELs. Our results suggest that the development of PELs may not be restricted to one stage of B cell differentiation.
Materials and Methods
Pathological Samples
A group of seven non-Hodgkin’s lymphomas containing KSHV sequences meeting the clinical, morphological, immunophenotypic, and molecular genetic criteria for PEL were included in this study. 5,6,10 Case 1 represents the KSHV-positive, EBV-negative BC-3 cell line established from a PEL occurring in an HIV-negative patient. 16 Case 2 represents the KSHV- and EBV-positive BC-1 cell line established from a PEL occurring in a patient with AIDS. 17 Cases 3–7 represent KSHV- and EBV-positive PELs collected from adult HIV-positive patients. Heparin-treated samples of the lymphomatous effusions were collected under sterile conditions during standard diagnostic procedures. Mononuclear cells were isolated and cryopreserved as described. 5
Immunophenotypic Analyses
The immunophenotype of the PEL tumor cells and cell lines was determined by direct and indirect immunofluorescent flow cytometry using the FACScan fluorescent activated cell sorter (Becton-Dickinson, Mountain View, CA) as previously described. 18 Monoclonal antibodies used include leukocyte common antigen (LCA, CD45), BerH2, Ki-1 (CD30), epithelial membrane antigen (EMA, Dako Corp, Santa Barbara, CA), B4 (CD19), B1 (CD20), B2 (CD21, Coulter Immunology, Hialeah, FL), Leu1 (CD5), Leu14 (CD22), Leu20 (CD23, Becton-Dickinson Immunocytometry Systems, Mountain View, CA), and T3 (CD3, United Biomedical, Hauppauge, NY). Antisera to total Ig, kappa (κ) and lambda (λ) Ig light chains were obtained from Dako.
Isolation of RNA and First-Strand cDNA Synthesis
Total RNA was extracted from cryopreserved mononuclear cell suspensions using guanidine isothiocyanate techniques. 19 Five mg of RNA was reverse transcribed into cDNA using M-MLV reverse transcriptase (Superscript RNase H Reverse Transcriptase, GIBCO-BRL, Grand Island, NY), in conjunction with a poly(dT)12–13 primer according to the manufacturer’s instructions.
PCR Amplification, Cloning, and Sequencing of the Expressed Ig VH-D-JH Genes
cDNAs were amplified by PCR using sense Ig VH gene family-specific (VH1, VH2, VH3, VH4, VH5, and VH6) leader primers in conjunction with an antisense consensus JH primer in independent reactions, as previously described. 20 After amplification, PCR products were electrophoresed in 2% agarose and visualized by ethidium bromide staining. Appropriately sized PCR products were isolated. PCR products were cloned in the pCR II vector using the TA cloning system (Invitrogen, San Diego, CA), following the manufacturer’s instructions. Plasmid DNA was isolated from overnight cultures of randomly selected white colonies using Wizard Mini-preps (Promega, Madison, WI). DNA sequencing was performed using the Sequenase version 2.0 (United States Biochemical, Cleveland, OH) system according to the manufacturer’s instructions. DNA sequences were analyzed using the MacVector version 4.5 (Eastman Kodak, New Haven, CT) software and the EMBL/GenBank data base.
Analysis of Mutations
We calculated the number of expected replacement (R) mutations in complementary determining regions (CDR) and framework regions (FR) for the Ig VH genes using the formula RCDR or RFR = n × CDR Rf or FR Rf × CDRf or FRf, where n is the total number of observed mutations; Rf is the replacement frequency intrinsic to each Ig VH gene, 21 and CDRf and FRf are the relative sizes of the CDRs and FRs. A binomial probability model was used to evaluate whether the observed R mutations in CDRs and the scarcity of silent (S) mutations in the FRs were due to chance alone: p = n!/k!(n − k)!qk(1 − q)n−k, where q = probability that an R mutation will localize to CDR or FR (q = CDRrel or FRrel × CDR Rf or FR Rf), and k = number of observed R mutations in the CDR or FR. 21
Results
Immunophenotypic Analysis
The immunophenotypic features of the PELs included in this study are summarized in Table 1 ▶ . All but one PEL (case 5) expressed CD45. However, only one case (case 3) expressed surface Ig. Also, all seven cases lacked B cell-associated antigens CD19 and CD20 and only case (case 6) expressed CD22. Five cases (cases 1–4 and 6) expressed CD30, three cases (cases 2, 6, 7) expressed EMA and two cases (cases 2 and 6) expressed HLA-DR. All seven cases lacked CD21 and CD23 and T cell-associated antigens.
Table 1.
Results of Immunophenotypic Analysis of Seven Cases of PEL
| Antigens | Case Number | ||||||
|---|---|---|---|---|---|---|---|
| 1 (BC-3) | 2 (BC-1) | 3 | 4 | 5 | 6 | 7 | |
| sIg | − | − | + | − | − | − | − |
| κ | − | − | − | − | − | − | − |
| λ | − | − | + | − | − | − | − |
| HLA-DR | − | + | − | − | − | + | − |
| CD3 | − | − | − | − | − | − | − |
| CD5 | − | − | − | − | − | − | − |
| CD19 | − | − | − | − | − | − | − |
| CD20 | − | − | − | − | − | − | − |
| CD21 | − | − | − | − | − | − | − |
| CD22 | − | − | − | − | − | + | − |
| CD23 | − | − | − | − | − | − | − |
| CD30 | + | + | + | + | − | + | − |
| CD45 | + | + | + | + | − | + | + |
| EMA | − | + | − | − | − | + | + |
+, positive; −, negative.
Sequence Analysis of the Expressed Ig VH-D-JH Genes
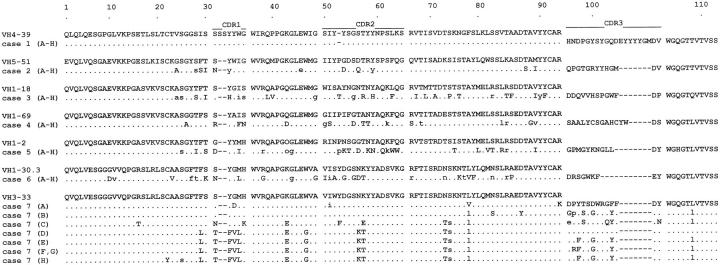
To analyze the Ig VH-D-JH genes expressed by the seven PELs, first-strand cDNAs were synthesized from total RNA of lymphoma cells and then PCR-amplified using the appropriate primers. The amplified DNAs were cloned and sequenced. Eight sequences from independent bacterial isolates were analyzed in each sample. The sequences obtained were compared with the corresponding germline sequences using EMBL/GenBank database to determine the respective homology of each gene with its closest germline counterpart. The closest germline VH genes and their degrees of similarity are shown in Table 1 ▶ . The deduced amino acid sequences of the VH-D-JH segments of the seven PELs and predicted amino acid changes when compared with the reported closest germline VH gene sequences are depicted in Figure 1 ▶ . All the VH-D-JH sequences were in-frame and no stop codons were found, suggesting that these cases expressed functional Ig. In cases 1–6, only identical and unique sequences were detected from analysis of the multiple isolates. Cases 3–5 used germline genes from the VH1 family and showed 88.4%, 93.5%, and 90.1% homology to their closest germline genes, respectively. Cases 1, 2, and 6 utilized germline genes from VH4, VH5, and VH3 families and displayed 100%, 94.9%, and 90.1% homology to their corresponding germline genes, respectively. In case 7, VH-D-JH gene sequences derived from eight independent isolates revealed seven unique but collinear sequences. Lymphoma cells of this case rearranged a member of VH3 family and sequences displayed 95.2% to 99.0% homology with the germline precursors (Table 2) ▶ . D segment sequences in cases 1 and 4 displayed the highest degree of identity to those of germline genes HD5-5 and HD2-15, respectively. The CDR3 sequences of the other PELs did not display any identity to those of known germline D genes. The HJ4 gene was used in five cases and the HJ6 gene in two cases.
Figure 1.
Deduced amino acid sequences of Ig VH-(D)-JH regions of the tumor-derived clones (A–H) from seven patients with PEL. Comparisons are made with the closest germline VH genes. The proportion of the CDR3 corresponding to the D and JH segments are not shown. The individual FR and CDR regions are indicated according to Kabat et al (1991). Identity with the VH germline sequences is shown by dots, replacement mutations by uppercase letters, and silent mutations by lowercase letters.
Table 2.
Analysis of Ig VH-D-JH Genes Expressed by the Neoplastic Cells in Seven Cases of PEL
| Case No. | Intraclonal Diversity | Clone | Ig VH Gene | D Gene | JH Gene | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Closest Germline Gene | Nucleotide Identity % | CDR1 and CDR2 | FR1, FR2, and FR3 | |||||||||||
| R | S | R:S | P | R | S | R:S | P | |||||||
| 1 (BC-3) | no | A–H | HV4-39 | 100 | 0 (0.00) | 0 | 0.00 | 0.00 | 0 (0.00) | 0 | 0.00 | 0.00 | HD5-5 | HJ6 |
| 2 (BC-1) | no | A–H | HV5-51 | 94.9 | 4 (9.03) | 3 | 1.33 | 7.08 × 10−3* | 5 (2.58) | 3 | 1.67 | 6.84 × 10−2 | ND | HJ6 |
| 3 | no | A–H | HV1-18 | 88.4 | 7 (20.56) | 4 | 1.75 | 0.20 × 10−5* | 15 (5.70) | 8 | 1.87 | 1.34 × 10−4* | ND | HJ4 |
| 4 | no | A–H | HV1-69 | 93.5 | 8 (11.49) | 2 | 4.00 | 4.95 × 10−2* | 5 (3.19) | 4 | 1.25 | 1.18 × 10−1 | HD2-15 | HJ4 |
| 5 | no | A–H | HV1-2 | 90.1 | 12 (18.28) | 2 | 6.00 | 9.18 × 10−3* | 8 (4.88) | 7 | 1.14 | 5.45 × 10−2 | ND | HJ4 |
| 6 | no | A–H | HV1-30.3 | 90.1 | 8 (17.89) | 2 | 4.00 | 1.64 × 10−4* | 10 (4.82) | 9 | 1.11 | 1.01 × 10−2* | ND | HJ4 |
| 7 | yes | A | HV3-33 | 98.6 | 1 (2.46) | 1 | 1.00 | 1.39 × 10−1 | 2 (0.7) | 0 | 2.00 | 1.15 × 10−1 | ND | HJ4 |
| B | HV3-33 | 99.0 | 0 (1.85) | 0 | 0.00 | 0.00 | 2 (0.50) | 1 | 2.00 | 6.91 × 10−2 | ND | HJ4 | ||
| C | HV3-33 | 96.3 | 5 (6.78) | 1 | 5.00 | 1.31 × 10−1 | 3 (1.8) | 2 | 1.50 | 1.76 × 10−1 | ND | HJ4 | ||
| D | HV3-33 | 96.0 | 6 (7.39) | 0 | 6.00 | 1.62 × 10−1 | 4 (2.0) | 2 | 2.00 | 8.83 × 10−2 | ND | HJ4 | ||
| E | HV3-33 | 96.0 | 6 (7.39) | 0 | 6.00 | 1.62 × 10−1 | 4 (2.0) | 2 | 2.00 | 8.83 × 10−2 | ND | HJ4 | ||
| F–G | HV3-33 | 96.0 | 6 (7.39) | 0 | 6.00 | 1.62 × 10−1 | 4 (2.0) | 2 | 2.00 | 8.83 × 10−2 | ND | HJ4 | ||
| H | HV3-33 | 95.2 | 6 (8.63) | 0 | 6.00 | 7.74 × 10−2 | 5 (2.3) | 3 | 1.67 | 4.95 × 10−2* | ND | HJ4 |
ND, not detected; R, number of detected and (expected) R mutations; S, number of detected S mutations; P, probability; *, statistically significant (P < 0.05).
Mutation Analysis
In six cases, the expressed VH genes showed numerous nucleotide differences from the closest germline gene sequences obtained from EMBL/GenBank database. Because germline VH genes were not identified in these patients, some of these nucleotide differences may represent individual polymorphism. However, sequence polymorphism among V genes is generally low 23 and in cases where we 24 and others 25,26 have analyzed germline genes of the patients, sequences corresponded exactly to those obtained from EMBL/GenBank database in most cases. Therefore, it has been concluded that polymorphism in VH genes is not sufficient to require analysis 27-29 and nucleotide differences in these cases have been classified as somatic mutation. Analysis of the distribution of somatic mutations in each sequence has been carried out by the method of Chang and Casali. 21 In this method, the binomial distribution model is used to calculate the probability that an excess in CDRs or scarcity in FRs of R mutations resulted from chance alone. The results of the analysis are summarized in Table 2 ▶ . The distribution of R and S mutations indicates a high R:S ratio in the CDRs of cases 4–7; however, this ratio did not exceed 2.9 for cases 2 and 3. Applying the binomial distribution model revealed more R mutations in the CDRs than expected in five sequences, with significant (P < 0.05) clustering. All mutated sequences had an R:S ratio equal to or less than 2 in their FR. Using the binomial distribution model, there were fewer R mutations in the FRs than expected due to chance in three sequences, with significant (P < 0.05) conservation of sequences.
Discussion
We have characterized the Ig VH gene configuration expressed by tumor cells of seven PELs in order to help determine their cellular origin. Both EBV-positive and EBV-negative cases were analyzed in order to cover the clinicopathological spectrum of the disease. The VH gene of the BC-3 cell line, which was the only EBV-negative PEL examined here and the only PEL obtained from an HIV-negative individual, showed complete nucleotide sequence identity with its putative germline precursor, suggesting a naive pregerminal center B cell origin of this PEL. In contrast, the six EBV-positive PELs, including the BC-1 cell line, displayed highly mutated VH genes. The average mutation frequency for the six cases analyzed was 7.7%, a value considerably higher than the 3–4% mutation frequency determined for nonmalignant germinal center and memory B cells, 30 suggesting that most PELs, and all of those containing EBV in this series, have reached a mature stage of B cell development.
The intraclonal heterogeneity of the expressed VH-D-JH gene sequences was variable among the six EBV-positive PELs. In five of these six cases, the expressed VH-D-JH sequences were extensively mutated but these sequences did not show intraclonal heterogeneity. These findings suggest that the tumor cells have traversed the germinal center but are no longer influenced by the somatic hypermutation mechanism. It could be concluded from this that the neoplastic event has occurred at a postgerminal center stage of B-cell maturation. The similar pattern of somatic mutation and antigen expression, including the absence of surface Ig in multiple myeloma and most of these PEL cases, suggests a plasma cell origin for some PEL cases. 31
In contrast to homogenous Ig gene sequences of PELs, considerable intraclonal heterogeneity, similar to the situation in follicle center cell lymphoma, was detected in one case. 32,33 The ongoing somatic mutation indicates that the tumor cells are still influenced by the mutation mechanism after malignant transformation. This finding is highly suggestive of a germinal center origin of this particular PEL. However, certain memory B cells may recirculate through the germinal center and undergo further rounds of somatic mutation and antigen selection that further improve and expand B-cell memory. 34
The germinal center/postgerminal center cellular origin of EBV-positive PELs analyzed is also suggested by the distribution of mutations in the VH genes of the tumor cells. Replacement mutations that affect CDRs can be positively or negatively selected by antigen, as these regions form the antigen binding sites. If a DNA segment displays more R mutations than would be expected by chance, it is likely that positive pressure was exerted on the gene product to select for these mutations. 21 In all but one EBV-positive PEL, the significant clustering of R mutations in the CDRs indicates that tumor cells have been selected by antigen or, in other words, that the tumor cells have undergone affinity maturation.
EBV-infected tumor cells of different lymphoid neoplasms frequently express mutated Ig VH genes. For example, it has been shown that the tumor cells of endemic Burkitt’s lymphomas, 35 posttransplantation lymphoproliferative disorders, 36 and Reed-Sternberg cells of Hodgkin’s disease 37 express mutated Ig VH genes. Our findings strengthen this observation by providing evidence that EBV-positive tumor cells of PEL also express mutated VH genes. However, the exact role of EBV infection in Ig gene somatic mutation of neoplastic cells is controversial. It has been demonstrated that EBV-infected normal B lymphocytes display VH genes with somatic hypermutation and evidence intraclonal heterogeneity. 38 Therefore, it appears that EBV infection plays a role in preneoplastic events by driving B cell proliferation, activation, and mutation of the Ig gene.
Several lines of evidence suggest that KSHV infection plays a central role in the development of PELs. KSHV carries different genes that may behave as oncogenes, including a gene homologous to bcl-2 (ORF/16), a gene homologous to the cellular D-type cyclins (ORF72/cyclin D), and a G-protein-coupled receptor displaying constitutive activation (ORF74/GPCR). 39,40 However, nonneoplastic lymphoid cells may be infected by KSHV. Ambroziak et al 41 have shown that the CD19-positive B cell population of HIV-infected and uninfected individuals may carry KSHV without clinical evidence of PEL or Kaposi’s sarcoma, and Luppi et al 42 found KSHV sequences in benign lymph nodes with florid germinal center hyperplasia. This finding suggests that latent infection of KSHV precedes malignant transformation of the cells and additional adequate stimuli may be necessary to initiate malignant transformation of the KSHV-infected cells, not unlike the EBV scenario. A possible explanation for the heterogeneous mutation profile of VH genes found in PELs is that malignant transformation is not restricted to specific stages of B cell development. Based on this hypothesis, B cells may carry KSHV in all stages of B cell maturation and events leading to the neoplastic transformation may occur independently of the stage of B cell development.
B cell tumors generally are thought to represent normal B cells that froze at a point in B cell differentiation. 43 Analysis of Ig genes expressed by B cell tumors shows that most of the non-Hodgkin’s lymphoma types represent a certain stage of B cell development at which clonal expansion occurred. Our results demonstrate that tumor cells of PEL may express unmutated, highly mutated, and intraclonal divergent Ig VH gene sequences suggesting that development of PELs is not restricted to one stage of B cell maturation. These results also suggest that different stages of B cell differentiation can be the targets of malignant transformation associated with KSHV infection.
Footnotes
Address reprint requests to Daniel M. Knowles, M.D., Department of Pathology, Weill Medical College of Cornell University, 525 East 68th Street, New York, NY 10021.
Supported by grants FKFP 0931/97, OTKA T25782, and ETT 365/96 from the Hungarian Ministry of Culture and Education (to AM) and NIH grants CA 68939 and CA 73531 (to EC).
References
- 1.Knowles DM, Inghirami G, Ubriaco A, Dalla-Favera R: Molecular genetic analysis of three AIDS-associated neoplasms of uncertain lineage demonstrates their B-cell derivation and the possible pathogenetic role of the Epstein-Barr virus. Blood 1989, 73:792-799 [PubMed] [Google Scholar]
- 2.Walts AE, Shintaku IP, Said JW: Diagnosis of malignant lymphoma in effusions from patients with AIDS by gene rearrangement. Am J Clin Pathol 1990, 94:170-175 [DOI] [PubMed] [Google Scholar]
- 3.Karcher DS, Dawkins F, Garrett CT, Schulof RS: Body cavity-based non-Hodgkin’s lymphoma (NHL) in HIV-infected patients, and genotypic features. Lab Invest 1992, 66:80 (abstr)
- 4.Green I, Espiritu E, Ladanyi M, Chaponda R, Wieczorek R, Gallo L, Feiner H: Primary lymphomatous effusions in AIDS: a morphological, immunophenotypic, and molecular study. Modern Pathol 1995, 8:39-45 [PubMed] [Google Scholar]
- 5.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM: Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med 1995, 332:1186-1191 [DOI] [PubMed] [Google Scholar]
- 6.Nador RG, Cesarman E, Chadburn A, Dawson DB, Ansari MQ, Said J, Knowles DM: Primary effusion lymphoma: a distinct clinicopathologic entity associated with the Kaposi’s sarcoma-associated herpes virus. Blood 1996, 88:645-656 [PubMed] [Google Scholar]
- 7.Said JW, Chien K, Takeuchi S, Tasaka T, Asou H, Cho SK, de Vos S, Cesarman E, Knowles DM, Koeffler HP: Kaposi’s sarcoma-associated herpesvirus (KSHV or HHV8) in primary effusion lymphoma: ultrastructural demonstration of herpesvirus in lymphoma cells. Blood 1996, 87:4937-4943 [PubMed] [Google Scholar]
- 8.Knowles DM, Cesarman E: The Kaposi’s sarcoma-associated herpesvirus (human herpesvirus-8) in Kaposi’s sarcoma, malignant lymphoma, and other diseases. Ann Oncol 1997, 8(suppl. 2):123-522 [PubMed] [Google Scholar]
- 9.Carbone A, Gaidano G: HHV-8-positive body-cavity-based lymphoma: a novel lymphoma entity. Br J Haematol 1997, 97:515-522 [DOI] [PubMed] [Google Scholar]
- 10.Ansari MQ, Dawson DB, Nador R, Rutherford C, Schneider NR, Latimer MJ, Picker L, Knowles DM, McKenna RW: Primary body cavity-based AIDS-related lymphomas. Am J Clin Pathol 1996, 105:221-229 [DOI] [PubMed] [Google Scholar]
- 11.Nador RG, Cesarman E, Knowles DM, Said JW: Herpes-like DNA sequences in a body-cavity-based lymphoma in an HIV-negative patient. N Engl J Med 1996, 333:943. [DOI] [PubMed] [Google Scholar]
- 12.Said JW, Tasaka T, Takeuchi S, Asou H, de Vos S, Cesarman E, Knowles DM, Koeffler HP: Primary effusion lymphoma in women: report of two cases of Kaposi’s sarcoma herpes virus-associated effusion-based lymphoma in human immunodeficiency virus-negative women. Blood 1996, 88:3124-3128 [PubMed] [Google Scholar]
- 13.Carbone A, Gloghini A, Vaccher E, Zagonel V, Pastore C, Palma PD, Branz F, Saglio G, Volpe R, Tirelli U, Gaidano G: Kaposi’s sarcoma-associated herpesvirus DNA sequences in AIDS-related and AIDS-unrelated lymphomatous effusions. Br J Haematol 1996, 94:533-543 [DOI] [PubMed] [Google Scholar]
- 14.Küppers R, Zhao M, Hansmann ML, Rajewsky K: Tracing B cell development in human germinal centres by molecular analysis of single cells picked from histological sections. EMBO J 1993, 12:4955-4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pascual V, Liu YJ, Magalski A, de Bouteiller O, Banchereau J, Capra JD: Analysis of somatic mutation in five B cell subsets of human tonsil. J Exp Med 1994, 180:329-339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arvanitakis L, Mesri EA, Nador RG, Said JW, Asch AS, Knowles DM, Cesarman E: Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood 1996, 88:2648-2654 [PubMed] [Google Scholar]
- 17.Cesarman E, Moore PS, Rao PH, Inghirami G, Knowles DM, Chang Y: In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi’s sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 1995, 86:2708-2714 [PubMed] [Google Scholar]
- 18.Inghirami G, Zhu BY, Chess L, Knowles DM: Flow cytometric and immunohistochemical characterization of the g/d T-lymphocyte population in normal human lymphoid tissue and peripheral blood. Am J Pathol 1990, 136:357-367 [PMC free article] [PubMed] [Google Scholar]
- 19.Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ: Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 1979, 18:5294-5299 [DOI] [PubMed] [Google Scholar]
- 20.Deane M, Norton JD: Immunoglobulin heavy chain variable region family usage is independent of tumor cell phenotype in human B lineage leukemias. Eur J Immunol 1990, 20:2209-2217 [DOI] [PubMed] [Google Scholar]
- 21.Chang B, Casali P: The CDR1 sequences of a major proportion of human germline Ig VH gene are inherently susceptible to amino acid replacement. Immunol Today 1994, 15:367-373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabat E, Wu TT, Reid-Miller M, Perry HM, K Gottesmann KS, Foeller C: Sequences of Proteins of Immunological Interest. Fifth Edition US Government Printing Office, Washington, DC, 1991
- 23.Cook GP, Tomlinson IM: The human immunoglobulin VH repertoire. Immunol Today 1995, 16:237-242 [DOI] [PubMed] [Google Scholar]
- 24.Matolcsy A, Casali P, Liu Y, Knowles DM: Molecular characterization of IgA- and/or IgG-switched chronic lymphocytic leukemia B cells. Blood 1997, 89:1732-1739 [PMC free article] [PubMed] [Google Scholar]
- 25.Shaota SS, Hamblin T, Oscier DG, Stevenson FK: Assessment of the role of clonogenic B lymphocytes in the pathogenesis of multiple myeloma. Leukemia 1994, 8:1285-1289 [PubMed] [Google Scholar]
- 26.Davi F, Maloum K, Michel A, Pritsch O, Magnac C, Macintyre E, Salomon-Nguyen F, Binet JL, Dighiero G, Merle-Béral H: High frequency of somatic mutations in the VH genes expressed in prolymphocytic leukemia. Blood 1996, 88:3953-3961 [PubMed] [Google Scholar]
- 27.Bahler DW, Miklos JA, Swerdlow SH: Ongoing Ig gene hypermutation in salivary gland mucosa-associated lymphoid tissue-type lymphomas. Blood 1997, 89:3335-3344 [PubMed] [Google Scholar]
- 28.Shaota SS, Leo R, Hamblin TJ, Stevenson FK: Myeloma VL and VH gene sequences reveal a complementary imprint of antigen selection in tumor cells. Blood 1997, 89:219-226 [PubMed] [Google Scholar]
- 29.Küppers R, Rajewsky K, Hansmann ML: Diffuse large cell lymphomas are derived from mature B cells carrying V region genes with a high load of somatic mutation and evidence of selection for antibody expression. Eur J Immunol 1997, 27:1398-1405 [DOI] [PubMed] [Google Scholar]
- 30.Klein U, Küppers R, Rajewsky K: Variable region gene analysis of B cell subsets derived from a 4-year-old child: somatically mutated memory B cells accumulate in the peripheral blood already at young age. J Exp Med 1994, 180:1383-1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakkus MHC, Heirman C, van Riet I, van Camp B, Thielemans K: Evidence that multiple myeloma Ig heavy-chain VDJ genes contain somatic mutations but show no intraclonal variation. Blood 1992, 80:2326-2332 [PubMed] [Google Scholar]
- 32.Bahler DW, Levy R: Clonal evolution of a follicular lymphoma: evidence for antigen selection. Proc Natl Acad Sci USA 1992, 89:6770-6774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zelenetz AD, Chen TT, Levy R: Clonal expansion in follicular lymphoma occurs subsequent to antigenic selection. J Exp Med 1992, 176:1137-1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manser T, Tumas-Brudage KM, Casson LP, Giusti AM, Hande S, Notidis E, Vora KA: The roles of antibody variable region hypermutation and selection in the development of the memory B-cell compartment. Immunol Rev 1998, 162:182-196 [DOI] [PubMed] [Google Scholar]
- 35.Chapman CJ, Mockridge CI, Rowe M, Rickinson AB, Stevenson FK: Analysis of VH genes used by neoplastic B cell in endemic Burkitt’s lymphoma shows somatic hypermutation and intraclonal heterogeneity. Blood 1995, 85:2176-2181 [PubMed] [Google Scholar]
- 36.Miklos JA, Locker J, Bahler DW: Evidence for antigen selection in posttransplant lymphoproliferative disorders. Lab Invest 1995, 72(suppl):116A [Google Scholar]
- 37.Kanzler H, Küppers R, Hansmann ML, Rajewsky K: Hodgkin and Reed-Sternberg cells in Hodgkin’s disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J Exp Med 1996, 184:1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chapman CJ, Spellerberg MB, Hamblin TJ, Stevenson FK: Pattern of usage of the VH4–21 gene by B lymphocytes in a patient with EBV infection indicates ongoing mutation and class switching. Mol Immunol 1995, 32:347-353 [DOI] [PubMed] [Google Scholar]
- 39.Cesarman E, Nador RG, Bai F, Bohenzky RA, Russo JJ, Moore PS, Chang Y, Knowles DM: Kaposi’s sarcoma associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi’s sarcoma and malignant lymphoma. J Virol 1996, 70:8218-8223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang Y, Moore PS, Talbot SJ, Boshoff CH, Zarkowska T, Godden-Kent D, Paterson H, Weiss RA, Mttnancht S: Cyclin encoded by KS herpesvirus. Nature 1996, 382:410. [DOI] [PubMed] [Google Scholar]
- 41.Ambroziak JA, Herndier BG, Glogau RG, Gullett JH, McDonald AR, Lennette ET, Levy JA: Herpes-like sequences in HIV-infected and uninfected Kaposi’s sarcoma patients. Science 1995, 268:582-583 [DOI] [PubMed] [Google Scholar]
- 42.Luppi M, Barozzi P, Maiorana A, Artusi T, Trovato R, Marasca R, Savarino M, Caccherini-Nelli L, Torelli G: Human herpesvirus-8 DNA sequences in human immunodeficiency virus-negative angioimmunoblastic lymphadenopathy and benign lymphadenopathy with giant germinal center hyperplasia and increased vascularity. Blood 1996, 87:3903-3909 [PubMed] [Google Scholar]
- 43.Salmon SE, Seligmann M: B-cell neoplasia in man. Lancet 1974, 2:1230-1233 [DOI] [PubMed] [Google Scholar]