Abstract
Hepatocellular carcinoma (HCC) is a common and highly malignant tumor that is prevalent in Southeast Asia. Although the etiological factors associated are now well recognized, the interactions between individual factors and the molecular mechanisms by which they lead to cancer remain unclear. Cytogenetic analysis on HCC has been limited because of poor hepatocyte growth in vitro. The recently developed technique of comparative genomic hybridization (CGH), however, permits screening of the entire genome without the need of cell culture. CGH was applied to the study of genomic aberrations in 67 surgically resected samples of HCC, 3 of adenomatous hyperplasia (AH), and 12 of nontumorous cirrhotic liver surrounding the tumors. All samples were from patients of a racially and etiologically homogeneous population in Southern China, where chronic hepatitis B virus infection is the main etiological factor. CGH analysis of the HCC samples revealed frequent copy number gain of 1q (48/67 cases, 72%), 8q (32/67 cases, 48%), 17q (20/67 cases, 30%), and 20q (25/67 cases, 37%) and common losses on 4q (29/67 cases, 43%), 8p (25/67 cases, 37%), 13q (25/67 cases, 37%), and 16q (20/67 cases, 30%). Our finding of a high incidence of 1q gain strongly suggested this aberration was associated with the development of HCC. Genomic abnormalities were detected in 1 of the 3 AH specimens but absent in all 12 cirrhotic tissues surrounding the tumor. Clinical staging classified 3/67 HCC cases as T1, 53 cases as T2, and 11 cases as T3. No significant difference in the pattern of genomic imbalances was detected between stages T2 and T3. A significant copy number loss of 4q11-q23 was, however, identified in those tumors larger than 3 cm in diameter. Of particular interest was the identification of 8q copy number gain in all 12 cases of HCC that arose in a noncirrhotic liver, compared with only 20/55 cases in HCC arising in a cirrhotic liver. We suggest that 8q over-representation is likely associated with a growth advantage and proliferative stimulation that have encouraged malignant changes in the noncirrhotic human liver.
Hepatocellular carcinoma (HCC) is responsible for at least 5% of all cancer deaths worldwide. 1 It is particularly common in Southeast Asia, China, and sub-Saharan Africa where the age-standardized annual incidence rate ranges from 20 to over 100 per 100,000. 2,3 The main etiological factor is chronic hepatitis B virus (HBV) infection, to which 50% to 80% of all cases are attributable. 4,5 Other etiological factors include chronic hepatitis C virus infection, exposure to aflatoxin, male gender, and chronic liver disease of any type. 5-7 Differences in exposure to these various risk factors account for the wide geographical variation in incidence. Surgical resection of the tumor offers the only hope of long-term survival, but because patients frequently present late in the natural history of the disease and often have poor underlying liver function, it is applicable to only a minority of patients. 8-10 The poor liver function is caused both by the tumor itself and by concomitant chronic liver disease, usually cirrhosis, which is present in 70% to 90% of cases. By the time of clinical presentation, intra- and extrahepatic metastases are common, which further limit the scope of surgical resection. 11,12
Molecular studies have demonstrated frequent loss of heterozygosity (LOH) on chromosomes 1p, 4q, 8p, 11p, 13q, 16q, and 17p 13-18 and multiplications of chromosome 8q 19 in HCC. Although cytogenetic analysis of HCC has received much less attention 20-23 due to the poor hepatocyte growth in vitro, structural aberrations of chromosome 1 were consistently reported. 20-23 Comparative genomic hybridization (CGH), first described by Kallioniemi in 1992, 24 is a powerful molecular cytogenetic tool that allows rapid screening for regions of DNA sequence gains and losses across the entire tumor genome without the need of cell culture. To date, there has been only a single report on the application of CGH to HCC 25 that described the recurring chromosomal changes of 4q loss (70%), 8p loss (65%), 8q gain (60%), and 1q gain (58%). However, the patients studied were from various, but unspecified, geographic locations, there was no clinicopathological data except all patients being HBV positive, and no attempts were made to correlate CGH findings with the clinical stage or the tumor size.
Here, we have applied CGH to the detection of changes in the genetic pattern of 67 cases of HCC and associated nonmalignant tissues (cirrhosis and adenomatous hyperplasia), in relation to clinical disease stage, tumor size, and the presence or absence of underlying liver cirrhosis. By comparing the pattern of genetic aberrations in these groups of tissues, we sought to identify those changes that may be associated with the development and progression of HCC.
Materials and Methods
Samples were collected from 70 ethnic Southern Chinese patients (aged 30 to 97 years, 81% male) undergoing hepatic resection of a liver tumor with curative intent. All but one case was tested for hepatitis B surface antigen (HBsAg) and serum α-fetoprotein (AFP), and 89% of cases were seropositive for HBsAg. Serum AFP levels ranged from less than 10 ng/ml (within the reference range) in 13 patients, to minimally elevated in 21 (10 to 50 ng/ml) and markedly elevated in the remaining 35 patients (50 to 40,000 ng/ml). The disease stage of the HCC cases was classified according to the tumor/node/metastasis (TNM) staging criteria. 26 Three cases (5%) were classified as stage I (T1N0M0), 53 (79%) as stage II (T2N0M0), and 11 (16%) as stage III (T3N0M0). The macro- and microscopic features of the resected specimens were reviewed by an experienced liver pathologist (C.-T. Liew) who confirmed the diagnosis of HCC, assessed the presence or absence of vascular invasion, and recorded the maximal diameter of the tumor. The presence or absence of cirrhosis in the nontumorous part of the resected specimen was also recorded. Cirrhosis was defined as the presence of complete fibrous septa separating regenerating nodules. 27 Within the nontumorous liver parenchyma, an increase in fibrous tissue alone was not classified as cirrhosis.
Sixty-seven samples of HCC tissue and three of adenomatous hyperplasia (AH) were received directly from the operating theater. They were snap-frozen in embedding medium (Tissue-Tek, Elkhart, IN) and stored at −80°C until analysis. Each resected specimen was evaluated for its tumor cell content on a hematoxylin and eosin (H&E)-stained section. For CGH analysis of HCC tissues and AH cases, only specimens with more than 80% tumor cell content were used. Twelve cirrhotic tissue samples, taken from the nontumorous part of the resected specimens, were also analyzed for CGH abnormalities.
Comparative Genomic Hybridization Analysis
High molecular weight DNA was extracted from the resected liver specimen and control DNA from blood lymphocytes of healthy volunteers by standard phenol/chloroform extraction procedures. Normal metaphase chromosomes were prepared from 72-hour phytohematoglutinine-stimulated peripheral blood lymphocyte cultures of healthy donors. The CGH protocol was carried out according to the method described in Chan et al. 28 Briefly, tumor and normal DNA labeled with biotin-16-dUTP (Boehringer Mannheim, Mannheim, Germany) and digoxigenin (dig)-11-dUTP (Boehringer Mannheim), respectively, by nick translation were co-precipitated in excess unlabeled Cot-1 DNA (Gibco Life Technologies, Gaithersburg, MD) and redissolved in hybridization solution containing 50% v/v formamide, 10% w/v dextran sulfate, and 2× SSC, pH 7. Slides containing normal metaphase chromosomes were denatured in 70% formamide/2× SSC (pH 7) at 70°C for 110 seconds. The hybridization was performed in a humid chamber at 37°C for 2 days. Biotin signals were detected through avidin-conjugated fluorescein isothiocyanate (FITC) antibodies (Sigma Chemical Co., St. Louis, MO), whereas dig-labeled DNA were visualized using antibodies conjugated with tetramethylrhodamine isothiocyanate (TRITC) (Sigma). The preparations were counterstained with 0.4 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) in anti-fade solution.
Hybridized metaphases were captured with a cooled CCD camera mounted on a Leitz DM RB (Leica, Wetzlar, Germany) fluorescence microscope. Three-band-pass filter (DAPI, FITC, and TRITC) sets arranged in an automated filter wheel were employed for image acquisition. CGH software version 3.1 on Cytovision (Applied Imaging, Sunderland, UK) was used for digital image analysis of fluorescence intensity. Chromosome identification was performed on the reverse DAPI banding images. The average ratio profiles were calculated based on the analysis of 5 to 10 selected metaphases. Thresholds for gains and losses were defined as the theoretical value of 1.25 and 0.75, respectively. High-level gain of a whole chromosome arm or amplification of a chromosomal region was considered to be present when ratios exceeded 1.5. Regions rich in heterochromatin (centromeres of chromosomes 1, 9, and 16, p-arm of acrocentromeric chromosomes, and Yq12) were excluded in the CGH analysis because the excess Cot-1 DNA present suppressed hybridization to these regions. 29
Statistical Analysis
Total DNA copy number aberrations, whether gains or losses, were compared between various groups by the two-tailed unpaired Student’s t-test. A difference was considered significant when the P value was less than 0.05. Individual chromosome copy number changes were compared by the nonparametric χ2 test and considered significant when the P value was less than 0.05.
Results
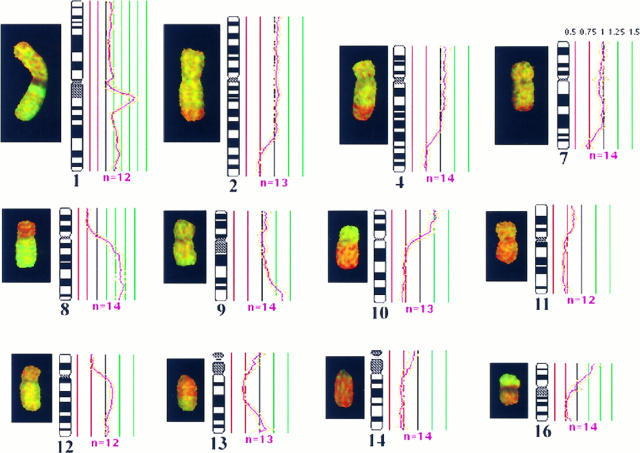
Of the 67 cases of HCC analyzed, frequent copy number gains were detected on chromosomes 1q (48/67 cases, 72%), 8q (32/67 cases, 48%), 17q (20/67 cases, 30%), and 20q (25/67 cases, 37%). Common losses were identified on 4q (29/67 cases, 43%), 8p (25/67 cases, 37%), 13q (25/67 cases, 37%), and 16q (20/67 cases, 30%). High-level gains of regional and/or the whole of 1q were also identified in 20/67 cases (30%), and a novel amplicon was mapped to 1q21-q22 (Figure 1) ▶ .
Figure 1.
CGH aberrations detected in case H2. A CGH image of hybridized chromosomes is shown with the corresponding fluorescence ratio profile plotted alongside the chromosome ideogram. Green regions represent gains, whereas red highlights the losses. The mean ratio profile of 12 analyzed chromosomes or more (n ≤ 12, pink line) is depicted with the 95% confidence interval (gold lines). Red and green lines represent thresholds for chromosomal losses (0.75) and gains (1.25). An amplicon on 1q21-22 and high-level gain of 8q can be readily identified by the intense green signals. The corresponding ratio profiles exceeded 1.5 (see Materials and Methods). The CGH profile of this specimen suggests high-level copy gains of 1q21-22 and 8q, low-level gains of 1q, 9q32-qter, 10p, and 16pter-p11.2, and losses of 2q32-qter, 4q28-qter, 7q32-qter, 8p, 10q, 11q, 12p, 13q12-q31, 14q, and 16q.
No sequence gains or losses were detected in any of the surrounding cirrhotic tissues. One of the three AH cases displayed a gain of 1q32-qter and 20. The remaining two cases had no detectable CGH abnormalities.
TNM Staging
There were no significant DNA sequence differences between the two major stage groups (T2 and T3) other than 8q over-representations, which were found mainly in stage T2 (P = 0.027; Table 1 ▶ ). A high incidence of 1q copy number gain was detected in both stage T2 (39/53 cases, 74%) and stage T3 (8/11 cases, 73%). Other common gains include 8q, 17q, and 20q and frequent losses on 4q, 8p, 13q, and 16q (Table 1) ▶ .
Table 1.
Comparison of Chromosomal Aberrations between Stages T2 and T3
| Stage T2 | Stage T3 | P value | |
|---|---|---|---|
| +1q | 39 /53 (74%) | 8 /11 (73%) | 0.953 |
| +8q | 29 /53 (55%) | 2 /11 (18%) | 0.027 |
| +17q | 17 /53 (32%) | 3 /11 (27%) | 0.755 |
| +20q | 19 /53 (36%) | 5 /11 (46%) | 0.549 |
| −4q | 22 /53 (42%) | 5 /11 (46%) | 0.810 |
| −8p | 20 /53 (38%) | 4 /11 (36%) | 0.932 |
| −13q | 20 /53 (38%) | 4 /11 (36%) | 0.932 |
| −16q | 17 /53 (32%) | 17 /53 (32%) | 0.359 |
Tumor Size
Twenty specimens fell into the category of small tumors (<3 cm) and forty-seven into the category of large tumors (>3 cm). Figure 2 ▶ summarizes the chromosomal aberrations detected in the 47 large tumors. No significant difference on the incidence of chromosomal gains and losses could be identified between the two groups, except diminution on 4q11-q23 was more profound in the larger HCC (P = 0.009; Table 2 ▶ ).
Figure 2.
Summary of gains and losses of DNA sequences identified by CGH in 47 HCCs >3 cm in diameter. Gains are shown on the right side of the chromosome ideogram and losses on the left. High-level gains are shown as thick lines. Each vertical line represents the affected chromosomal region seen in a single tumor specimen.
Table 2.
Comparison of Chromosomal Aberrations between Small (<3 cm) and Large (>3 cm) Tumors
| <3 cm | >3 cm | P value | |
|---|---|---|---|
| +1q | 12 /20 (60%) | 36 /47 (77%) | 0.168 |
| +8q | 9 /20 (45%) | 23 /47 (49%) | 0.768 |
| +17q | 6 /20 (30%) | 14 /47 (21%) | 0.986 |
| +20q | 5 /20 (25%) | 20 /47 (43%) | 0.174 |
| −4q | 7 /20 (35%) | 22 /47 (47%) | 0.372 |
| −4q11-q21 | 3 /20 (15%) | 20 /47 (43%) | 0.009 |
| −8p | 9 /20 (45%) | 16 /47 (40%) | 0.396 |
| −13q | 6 /20 (30%) | 19 /47 (40%) | 0.419 |
| −16q | 4 /20 (20%) | 16 /47 (34%) | 0.250 |
Cirrhotic and Noncirrhotic HCC
Twelve of the sixty-seven HCC specimens arose in a noncirrhotic liver; the remaining fifty-five cases had associated liver cirrhosis. All 12 tumors that had no underlying liver cirrhosis exhibited an 8q copy number gain. This incidence was significantly lower in the cirrhotic HCC cases (20/55 cases; P = 0.0001). Other significant gains include 20q, which was found in 9/12 of those without cirrhosis but in only 16/55 cases with cirrhosis (P = 0.003). Losses on 4q were also marked in the noncirrhotic cases (9/12 cases) in comparison with the cirrhotic cases (20/55 cases; P = 0.014; Table 3 ▶ ). The mean number (±SD) of DNA sequence copy changes per tumor in the cirrhotic and noncirrhotic groups were 7.4 ± 5.3 and 12.8 ± 5.0, respectively (P = 0.002). On subdivision of the total aberrations into gains (including amplifications) and losses, a mean of 4.1 ± 2.7 gains was found in the cirrhotic group compared with 7.5 ± 2.8 in the noncirrhotic group (P = 0.0002). The mean copy number losses were 3.3 ± 3.3 in the cirrhotic group and 5.3 ± 2.9 in those without underlying cirrhosis (P = 0.046).
Table 3.
Comparison of Chromosomal Aberrations between HCC with and without Underlying Liver Cirrhosis
| Cirrhotic HCC | Non-cirrhotic HCC | P value | |
|---|---|---|---|
| +1q | 38 /55 (69%) | 10 /12 (83%) | 0.321 |
| +8q | 20 /55 (36%) | 12 /12 (100%) | 0.0001 |
| +17q | 15 /55 (27%) | 5 /12 (42%) | 0.324 |
| +20q | 16 /55 (29%) | 9 /12 (75%) | 0.003 |
| −4q | 20 /55 (36%) | 9 /12 (75%) | 0.014 |
| −8p | 18 /55 (33%) | 7 /12 (58%) | 0.097 |
| −13q | 18 /55 (33%) | 7 /12 (58%) | 0.097 |
| −16q | 15 /55 (27%) | 5 /12 (42%) | 0.324 |
Discussion
The present study represents the first genome-wide investigation on the genetic imbalances in HCC in relation to TNM staging, tumor size, and underlying cirrhosis. Our series has the advantage of being particularly homogeneous, all samples coming from Southern Chinese patients of whom the great majority were chronic carriers of HBV. As noted previously, this series is typical of the majority of cases in Southeast Asia, although there may still be additional etiological factors even among HBV-related cases, such as aflatoxin exposure and HCV co-infection. 4,7,30 The two most striking features of our analysis were the finding of 8q copy number gain in all HCC cases without underlying liver cirrhosis and the very high incidence of 1q gain (48/67 cases, 72%). These observations strongly suggest that 8q plays an important role in the malignant changes in the noncirrhotic liver and that the gain of 1q is an early aberrant event in HCC development.
High-level gain of regional and/or whole q-arm of chromosome 1 was also observed in 20 of the 48 HCC cases (42%) that exhibited a 1q copy number gain. This observation, together with the finding of an amplicon on 1q21-22, indicates the likelihood of important proto-oncogenes residing in this region. According to the Genome Data Base, 1q21 harbors the gene that encodes the human mRNA for hepatoma-derived growth factor. The enhanced expression of this gene could be associated with the paracrine and/or autocrine activity that supports tumor growth. CGH studies on soft tissue sarcomas, osteosarcoma, and the Ewing family of tumors have also reported the presence of a recurring 1q21-q22 amplicon. 31-33 Amplification of the FLG and SPRR3 genes, also located on 1q21, have been identified in several sarcoma cell lines. 34 An increased expression of CACY and CAPL of the S-100 family calcium-binding proteins have been mapped to the same region and implicated in tumor progression and metastasis. 35 We are currently examining the significance of 1q21-q22 amplifications using interphase fluorescence in situ hybridization analysis on a large series of archival paraffin-embedded material and examining the expression pattern of candidate genes described in this region. Although a previous CGH study on HCC also noted a raised incidence of 1q gain (58%), the amplicon 1q21-q22 was not detected. 25 Sequence amplifications detected in the latter study were defined within one band and mapped to 11q12, 12p11, and 14q12 (1/50 cases each) and 19q31.1 (2/50 cases). 25 In our present study, the amplicons identified by Marchio et al 25 were not detected. Rather, frequent high-level gains of regional and/or whole chromosome arms were seen.
In nearly 90% of our cases the HCC arose in a liver that was cirrhotic. This figure is typical of our area and similar to that reported from most other countries. 6,36 Cirrhosis may be considered as the final common pathway of several chronic liver insults, including viral hepatitis types B and C, excessive alcohol consumption, and various metabolic diseases, and indeed each of these is also strongly associated with the development of HCC. The extent to which it is cirrhosis per se or the factors responsible for the cirrhosis that cause the tumor to develop remains controversial although many authors have considered cirrhosis itself to be a premalignant condition. 6
Our present study reveals a significantly lower total number of aberrations in the cirrhosis-associated HCC compared with those without underlying liver cirrhosis. None of the surrounding cirrhotic livers displayed any CGH abnormalities. This suggests that, although liver cirrhosis itself is not associated with any gross genotypic changes, it may increase the susceptibility of the liver to HCC development so that fewer genetic aberrations are required for tumor development. Noncirrhotic HCC, on the other hand, exhibited significantly more genomic variants, in particular the gains of 8q (100%) and 20q (75%) and loss of 4q (75%). These aberrations may be essential for the transformation of noncirrhotic liver to HCC, providing the hepatocytes with proliferative stimulation and growth advantages. Gain of 8q material might be related to the CMYC proto-oncogene. Coexpression of CMYC and transforming growth factor (TGF)-α in a transgenic mouse liver have been reported to result in a major enhancement of HCC development. 37 The interaction between CMYC and TGF-α is believed to support the preneoplastic hepatocytes with growth advantages by disrupting the pRB/E2F pathway and TGF-α-mediated apoptosis. 38 The expression of CMYC and TGF-α in human HCC has yet to be confirmed. However, as we have shown that a gain of the whole chromosome 8 q-arm is often observed, the involvement of one or more other putative proto-oncogenes besides the CMYC gene (8q24) is also likely. An increase in 8q copy number gain has been reported in a number of malignant diseases. These include breast, pancreatic, head and neck, and lung cancers. 39-42 In this study, we observed a frequent loss of 8p accompanying the gain of 8q (50%, 16/32 cases with 8q gain). Deletion of one chromosome arm and multiplication of the other arm could be indicative of the respective iso-chromosomes formation as seen in many solid tumors. Allelic loss of 8p is well described in HCC and is also common in breast, bladder, and in particular recurrent prostate cancers. 39,43-44 The putative tumor suppressor gene (TSG) on 8p has been suggested to be PRLTS (platelet-derived growth factor receptor-β-like TSG). 18
Some authors have proposed that it is adenomatous hyperplasia (AH), rather than cirrhosis, that is the premalignant precursor of HCC. 45,46 In this series, we have examined three AH cases of which two had normal CGH profiles and one displayed a copy number gain of 1q32-qter and 20. The absence of CGH abnormalities in two cases of AH could be due to the insensitivity of CGH in detecting imbalances of small subchromosomal regions (<10 Mbp) and/or the inability to elucidate balanced translocations. 29 Frequent low-level gain of 20q and high-level amplification of 20q13 were also identified in the 67 HCC cases studied (37%). The finding of a chromosome 20 gain in a case of AH together with its frequent occurrence in HCC suggested that chromosome 20 has an important role in HCC development. Chromosome 20q over-representation has not been described previously in HCC but is common in pancreatic, bladder, colon, and breast cancers. 39,40,47,48 Studies on bladder cancer and transformed uroepithelial cells have shown that low-level 20q gain was associated with overcoming cellular senescence, which in turn enabled surviving cells to accumulate multiple genetic alterations, whereas 20q13 amplification was associated with chromosome instability. 48-50
The conventional TNM staging classification 51,52 is generally less widely used in HCC than other malignant tumors because the prognosis is related to the state of the underlying liver disease as much as the extent of the tumor itself. In this paper, we have studied exclusively surgical specimens, a subpopulation of HCC that represents only the early resectable tumors with neither lymph node (N0) involvement nor metastasis (M0). We compared the pattern of genetic alterations between 53 cases of T2 and 11 cases of T3, and found no significant difference except a higher incidence of 8q gain in stage T2. On further analysis, this raised incidence could be explained by the observation that all of the noncirrhotic HCC cases fell within the T2 stage. The absence of progressive genetic changes in relation to the clinical staging supports a recent report that suggested TNM classification in HCC has little prognostic implications among small tumors. 53 Rather, tumor size or the presence of vascular invasion in conjunction with measures of the underlying liver function may be better prognostic parameters for HCC. Tumors of less than 3 cm in diameter are often referred to as minute HCC, 54-58 and this forms the basis of our tumor classification into small and large sizes.
Frequent LOH on the long arm of chromosome 4 is well documented in HCC 13-17 with a putative TSG suggested to be HVBS6 (hepatitis B virus integration site 2). 18 Common deleted regions have been suggested in the vicinity of the albumin gene locus (4q11-q12), 13,59 4q12-q23, 60 4p11-q21, 61 and 4q35. 14 In the present investigation, we found no significant difference in the incidence of total number of deletions on 4q in relation to disease stage, tumor size, or underlying liver cirrhosis. However, when focusing on the distal q-arm, a significant increase in the incidence of 4q11-q23 copy number loss was found in tumors larger than 3 cm in diameter. Frequent interstitial deletions on 4q21-q22 have also been reported among the larger and more advanced HCC tumors. 17 Our present finding thus supports the previous study 17 and suggest a correlation between DNA sequence deletions on 4q11-q23 and tumor progression.
In the present study, we were unable to discern any obvious chromosomal difference in relation to AFP positivity. However, we have observed a marked difference in genomic alterations in relation to HCC with or without underlying liver cirrhosis and tumor size. A high frequency of 1q gain was also observed. In our future investigations, attempts to further define the gene(s) involved in the aberrant chromosomal regions identified here will be carried out. Also, a comparison of genomic aberrations between etiological factors, such as HBV- and HCV-related HCC, and correlation of genetic changes with clinical outcome will be undertaken.
Acknowledgments
We are indebted to Dr. Evelin Schröck of the National Institutes of Health, Cytogenetics Core, Bethesda, MD, for her helpful advice and criticisms during the preparation of this manuscript.
Footnotes
Address reprint requests to Prof. P. J. Johnson, Department of Clinical Oncology, The Chinese University of Hong Kong, Prince of Wales Hospital, Shatin, N.T., Hong Kong, China. E-mail: pjjohnson@cuhk.edu.hk.
Supported by a direct grant (2040613) from The Chinese University of Hong Kong, in part by a grant from the Research Grants Council of the Hong Kong Special Administrative Region, China (RGC Reference CUHK 4264/98M), and the Providence Foundation Limited, Hong Kong.
References
- 1.World Health Organization: Cancer Research for Cancer Control. International Agency for Cancer Research, Lyon (IARC Scientific Publication), 1997, p 5
- 2.Doll R, Muir C, Waterhouse J: Cancer Incidence in Five Continents, 1970, vol 2. Springer, Berlin
- 3.Waterhouse JAH, Muir CS, CoRrea P, Powell J: Cancer Incidence in Five Continents, vol 4. International Agency for Research on Cancer, Lyon (IARC Scientific Publications 42), 1982
- 4.Beasley RP: Hepatitis B virus: the major etiology of hepatocellular carcinoma. Cancer 1988, 61:1942-1946 [DOI] [PubMed] [Google Scholar]
- 5.Munoz N, Bosch X: Epidemiology of hepatocellular carcinoma. Neoplasms of the Liver. 1987, Ishak KG. London, Springer Verlag, Edited by Okuda K
- 6.Johnson PJ, Williams R: Cirrhosis and the aetiology of hepatocellular carcinoma. J Hepatol 1987, 4:140-147 [DOI] [PubMed] [Google Scholar]
- 7.Hasan F, Jeffers LJ, De Medina M, Reddy KR, Parker T, Schiff ER, Houghton M, Choo QL, Kuo G: Hepatitis C-associated hepatocellular carcinoma. Hepatology 1990, 12:589-591 [DOI] [PubMed] [Google Scholar]
- 8.Lim R, Bongard FS: Hepatocellular carcinoma: changing concepts in diagnosis and management. Arch Surg 1984, 119:637. [DOI] [PubMed] [Google Scholar]
- 9.Lin T-Y, Lee CS, Chen KM, Chen CC: Role of surgery in the treatment of primary carcinoma of the liver: a 31 year experience. Br J Surg 1987, 74:839-842 [DOI] [PubMed] [Google Scholar]
- 10.Fortner JG, MacLean BJ, Kim DK, Howland WS, Turnbull AD, Goldiner P, Carlon G, Beattie EJ, Jr: The seventies evolution in liver surgery for cancer. Cancer 1981, 47:2162-2166 [DOI] [PubMed] [Google Scholar]
- 11.Okuda K, Obata H, Nakajima Y, Ohtsuki T, Okazaki N, Ohnishi K: Prognosis of primary hepatocellular carcinoma. Hepatology 1984, 4:3-6 [DOI] [PubMed] [Google Scholar]
- 12.Sasaki Y, Imaoka S, Masutani S, Osashi I, Ishikawa O, Koyama H, Iwanaga T: Influence of coexisting cirrhosis on longer-term prognosis after surgery in patients with hepatocellular carcinoma. Surgery 1992, 112:515-521 [PubMed] [Google Scholar]
- 13.Buetow KH, Murray JC, Israel JL, London WT, Smith M, Kew M, Blanquet V, Brechot C, Redeker A, Govindarajah S: Loss of heterozygosity suggests tumour suppressor gene responsible for primary hepatocellular carcinoma. Proc Natl Acad Sci USA 1989, 86:8852-8856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagai H, Pineau P, Tiollais P, Buendia MA, Dejean A: Comprehensive allelotyping of human hepatocellular carcinoma. Oncogene 1997, 14:2927-2933 [DOI] [PubMed] [Google Scholar]
- 15.Fujimori M, Tokino T, Hino O, Kitagawa T, Imamura T, Okamoto E, Mitsunobu M, Ishikawa T, Nakagama H, Harada H, Yagura M, Matsubara K, Nakamura Y: Allelotype study of primary hepatocellular carcinoma. Cancer Res 1991, 51:89-93 [PubMed] [Google Scholar]
- 16.Fujimoto Y, Hampton LL, Wirth PJ, Wang NJ, Xie JP, Thorgeirsson SS: Alterations of tumor suppressor genes and allelic losses in human hepatocellular carcinomas in China. Cancer Res 1994, 54:281-285 [PubMed] [Google Scholar]
- 17.Kuroki T, Fujiwara Y, Tsuchiya E, Nakamori S, Imaoka S, Kanematsu T, Nakamura Y: Accumulation of genetic changes during development and progression of hepatocellular carcinoma: loss of heterozygosity on chromosome arm 1p occurs at an early stage of hepatocarcinogenesis. Genes Chromosomes & Cancer 1995, 13:163-167 [DOI] [PubMed] [Google Scholar]
- 18.Boige V, Laurent-Puig P, Fouchet P, Flejou JF, Monges G, Bedossa P, Bioulas S, Capron F, Schmitz A, Olschwang S, Thomas G: Concerted nonsyntenic allellic losses in hyperploid hepatocellular carcinoma as determined by a high-resolution allelotype. Cancer Res 1997, 57:1986-1990 [PubMed] [Google Scholar]
- 19.Fuijiwara Y, Monden M, Mori T, Nakamura Y, Emi M: Frequent multiplication of the long arm of chromosome 8 in hepatocellular carcinoma. Cancer Res 1993, 53:857-860 [PubMed] [Google Scholar]
- 20.Bardi G, Johansson B, Pandis N, Heim S, Mandahl N, Andren-Sandberg A, Hagerstrand I, Mitelman F: Cytogenetic findings in three primary hepatocellular carcinomas. Cancer Genet Cytogenet 1992, 58:191-195 [DOI] [PubMed] [Google Scholar]
- 21.Chen HL, Chen YC, Chen DS: Chromosome 1p aberrations are frequent in human primary hepatocellular carcinoma. Cancer Genet Cytogenet 1996, 86:102-106 [DOI] [PubMed] [Google Scholar]
- 22.Simon D, Munoz SJ, Maddrey WC, Knowles BB: Chromosomal rearrangements in a primary hepatocellular carcinoma. Cancer Genet Cytogenet 1990, 45:225-260 [DOI] [PubMed] [Google Scholar]
- 23.Werner M, Nolte M, Gergii M, Klempnauer J: Chromosome 1 abnormalities in hepatocellular carcinoma. Cancer Genet Cytogenet 1993, 66:130. [DOI] [PubMed] [Google Scholar]
- 24.Kallioniemi A, Kallioniemi O-P, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D: Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 1992, 258:818-821 [DOI] [PubMed] [Google Scholar]
- 25.Marchio A, Meddeb M, Pineau P, Danglot G, Tiollais P, Bernheim A, Dejean A: Recurrent chromosomal abnormalities in hepatocellular carcinoma detected by comparative genomic hybridization. Genes Chromosomes & Cancer 1997, 18:59-65 [PubMed] [Google Scholar]
- 26.Beahrs OH, Henson DE, Hutter RVP, Kennedy BJ (editors): Handbook for Staging of Cancer. Philadelphia, JB Lippincott, 1993
- 27.Antony PP, Ishak KG, Nayak NC, Poulsen HE, Scheuer PJ, Sobin LH: The morphology of cirrhosis: recommendations on definitions, nomenclature and classification by a working group sponsored by the World Health Organization. J Clin Pathol 1978, 31:395-414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan WY, Wong N, Chan ABW, Chow JHS, Lee JCK: Consistent copy number gain in chromosome 12 in primary diffuse large cell lymphomas of the stomach. Am J Pathol 1998, 152:11-16 [PMC free article] [PubMed] [Google Scholar]
- 29.Kallioniemi O-P, Kallioniemi A, Piper J, Isola J, Waldman FM, Gray JW, Pinkel D: Optimizing comparative genomic hybridization for the analysis of DNA sequence copy number changes in solid tumours. Genes Chromosomes & Cancer 1994, 10:231-243 [DOI] [PubMed] [Google Scholar]
- 30.Qian G-S, Ross RK, Yu MC, Yuan J-M, Gao Y-T, Henderson BE, Wogan GN, Groopman JD: A follow-up study of urinary markers of aflatoxin exposure and liver cancer risk in Shanghai, People’s Republic of China. Cancer Epidemiol Biomarkers Prevention 1994, 3:3-10 [PubMed] [Google Scholar]
- 31.Forus A, Weghuis DO, Smeets D, Fodstad Ø, Myklebost O, van Kessel AG: Comparative genomic hybridization analysis of human sarcomas. I. Occurrence of genomic imbalances and identification of a novel major amplicon at 1q21–q22 in soft tissue sarcomas. Genes Chromosomes & Cancer 1995, 14:8-14 [DOI] [PubMed] [Google Scholar]
- 32.Tarkkanen M, Karhu R, Kallioniemi A, Elomaa I, Kivioja AH, Nevalainen B, Karaharju E, Hyytinen E, Knuutila S, Kallioniemi OP: Gains and losses of DNA sequences in osteosarcomas by comparative genomic hybridization. Cancer Res 1995, 55:1334-1338 [PubMed] [Google Scholar]
- 33.Armengol G, Tarkkanen M, Virolainen M, Forus A, Valle J, Bohling T, Asko-Sel javaara S, Blomqvist C, Elomaa I, Karaharju E, Kivioja AH, Siimes MA, Tukiainen E, Caballin MR, Myklebost O, Knuutila S: Recurrent gains of 1q, 8 and 12 in Ewing family of tumors by comparative genomic hybridization. Br J Cancer 1997, 75:1403-1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forus A, Weterman MAJ, Van Kessel AG, Berner JM, Fodstad Ø, Myklebost O: Characterisation of 1q21–22 amplifications in human sarcomas by CGH and molecular analysis. Cytogenet Cell Genet 1996, 72:148 [Google Scholar]
- 35.Engelkamp D, Schafer BW, Mattei MG, Erne P, Heizmann CW: Six S100 genes are clustered on human chromosome 1q21: identification of two genes coding for previously unreported calcium-binding proteins S100D and S100E. Proc Natl Acad Sci USA 1993, 90:6547-6551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okuda K: Hepatocellular carcinoma: recent progress. Hepatology 1992, 15:948-963 [DOI] [PubMed] [Google Scholar]
- 37.Santoni-Rugiu E, Nagy P, Jensen MR, Factor VM, Thorgeirsson SS: Evolution of neoplastic development in the liver of transgenic mice co-expressing c-myc and transforming growth factor-α. Am J Pathol 1996, 149:407-428 [PMC free article] [PubMed] [Google Scholar]
- 38.Santoni-Rugiu E, Jensen MR, Thorgeirsson SS: Disruption of the pRb/E2F pathway and inhibition of apoptosis are major oncogenic events in liver constitutively expressing c-myc and transforming growth factor alpha. Cancer Res 1998, 58:123-134 [PubMed] [Google Scholar]
- 39.Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, Kallioniemi OP: Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosomes & Cancer 1998, 21:177-184 [PubMed] [Google Scholar]
- 40.Mahlamaki EH, Hoglund M, Gorunova L, Karhu R, Dawiskiba S, Andren-Sandberg A, Kallioniemi OP, Johansson B: Comparative genomic hybridization reveals frequent gains of 20q, 8q, 11q, 12p, and 17q, and losses of 18q, 9p, and 15q in pancreatic cancer. Genes Chromosomes & Cancer 1997, 20:383-391 [DOI] [PubMed] [Google Scholar]
- 41.Bockmuhl U, Wolf G, Schmidt S, Schwendel A, Jahnke V, Dietel M, Petersen I: Genomic alterations associated with malignancy in head and neck cancer. Head Neck 1998, 20:145-151 [DOI] [PubMed] [Google Scholar]
- 42.Bjorkqvist AM, Tammilehto L, Nordling S, Nurminen M, Anttila S, Mattson K, Knuutila S: Comparison of DNA copy number changes in malignant mesothelioma, adenocarcinoma and large-cell anaplastic carcinoma of the lung. Br J Cancer 1998, 77:260-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kallioniemi A, Kallioniemi OP, Citro G, Sauter G, DeVries S, Kerschmann R, Caroll P, Waldman F: Identification of gains and losses of DNA sequences in primary bladder cancer by comparative genomic hybridization. Genes Chromosomes & Cancer 1995, 12:213-219 [DOI] [PubMed] [Google Scholar]
- 44.Visakorpi T, Kallioniemi AH, Syvanen AC, Hyytinen ER, Karhu R, Tammela T, Isola JJ, Kallioniemi OP: Genetic changes in primary and recurrent prostate cancer by comparative genomic hybridization. Cancer Res 1995, 55:342-347 [PubMed] [Google Scholar]
- 45.Nakanuma Y, Terada T, Terasaki S, Ueda K, Nonomura A, Kawahara E, Matsui O: Atypical adenomatous hyperplasia in liver cirrhosis: low-grade hepatocellular carcinoma or borderline lesion? Histopathology 1990, 17:27-35 [DOI] [PubMed] [Google Scholar]
- 46.Lancioni R, Caramella D, Bartolozzi C, Di Coscio G: Long term follow-up study of adenomatous hyperplasia in liver cirrhosis. Ital J Gastroenterol 1994, 26:163-168 [PubMed] [Google Scholar]
- 47.Schlegel J, Stumm G, Scherthan H, Bocker T, Zirngibl H, Ruschoff J, Hofstadter F: Comparative genomic in situ hybridization of colon carcinomas with replication error. Cancer Res 1995, 55:6002-6005 [PubMed] [Google Scholar]
- 48.Reznikoff CA, Belair CD, Yeager TR, Savelieva E, Blelloch RH, Puthenveettil JA, Cuthill S: A molecular genetic model of human bladder cancer pathogenesis. Semin Oncol 1996, 23:571-584 [PubMed] [Google Scholar]
- 49.Yeager TR, De Vries S, Jarrard DF, Kao C, Nakada SY, Moon TD, Bruskewitz R, Stadler WM, Meisner LF, Gilchrist KW, Newton MA, Waldman FM, Reznikoff CA: Overcoming cellular senescence in human cancer pathogenesis. Genes Dev 1998, 12:163-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Savelieva E, Belair CD, Newton MA, De Vries S, Gray JW, Waldman F, Reznikoff CA: 20q gain associates with immortalization: 20q13.2 amplification correlates with genome instability in human papillomavirus 16 E7 transformed human uroepithelial cells. Oncogene 1997, 14:551-560 [DOI] [PubMed] [Google Scholar]
- 51.International Union Against Cancer: TNM classification of malignant tumor, ed. 4. Edited by Hermanek P, Sobin LH. Berlin, Springer-Verlag, 1987, pp 53–55
- 52.Yamamoto M, Sugahara K: Overview of the general rules for the clinical and pathological study of primary liver cancer in Japan. Tobe T eds. Primary Liver Cancer in Japan. 1992, :pp 385-401 Springer, Tokyo [Google Scholar]
- 53.Llovet JM, Jordi B, Fuster J, Castells A, Garcia-Valdecasas JC, Grande L, Franca A, Bru C, Navasa M, Ayuso MDC, Sole M, Real MI, Vilana R, Rimola A, Visa J, Rodes J: Liver transplantation for small hepatocellular carcinoma: the tumor-node-metastasis classification does not have prognostic power. Hepatology 1998, 27:1572-1577 [DOI] [PubMed] [Google Scholar]
- 54.Sasaki Y, Imaoka S, Ishiguro S, Nakano H, Kasugai H, Fujita M, Inoue E, Ishikawa O, Furukawa H, Nakamori S, Kuroda C, Iwanaga T: Clinical features of small hepatocellular carcinomas as assessed by histologic grades. Surgery 1996, 119:252-260 [DOI] [PubMed] [Google Scholar]
- 55.Kenmochi K, Sugihara S, Kojiro M: Relationship of histologic grade of hepatocellular carcinoma (HCC) to tumor size, and demonstration of tumor cells of multiple different grades in single small HCC. Liver 1987, 7:18-26 [DOI] [PubMed] [Google Scholar]
- 56.Ohnishi K, Yoshioka H, Ito S, Fujiwara K: Prospective randomized controlled trial comparing percutaneous acetic acid injection and percutaneous ethanol injection for small hepatocellular carcinoma. Hepatology 1998, 27:67-72 [DOI] [PubMed] [Google Scholar]
- 57.Tanaka K, Nakamura S, Numata K, Kondo M, Morita K, Kitamura T, Saito S, Kiba T, Okazaki H, Sekihara H: The long term efficacy of combined transcatheter arterial embolization and percutaneous ethanol injection in the treatment of patients with large hepatocellular carcinoma and cirrhosis. Cancer 1998, 82:78-85 [DOI] [PubMed] [Google Scholar]
- 58.Hiroshige K, Yokosuka O, Imazeki F, Saisho H, Omata M: Telomerase activity and telomere length in hepatocellular carcinoma and chronic liver disease. Gastroenterology 1997, 112:493-500 [DOI] [PubMed] [Google Scholar]
- 59.Urano Y, Watanabe K, Lin CC, Hino O, Tamaoki T: Interstitial chromosomal deletion within 4q11–q13 in human hepatoma cell line. Cancer Res 1991, 51:1460-1464 [PubMed] [Google Scholar]
- 60.Yeh SH, Chen PJ, Lai MY, Chen DS: Allelic loss on chromosomes 4q and 16q in hepatocellular carcinoma. Gasteroenterology 1996, 110:184-192 [DOI] [PubMed] [Google Scholar]
- 61.Zhang W, Hirohashi S, Tsuda H, Shimosato Y, Yokota J, Terada M, Sugimura T: Frequent loss of heterozygosity of chromosomes 16 and 4 in human hepatocellular carcinoma. Jpn J Cancer Res 1990, 81:108-111 [DOI] [PMC free article] [PubMed] [Google Scholar]