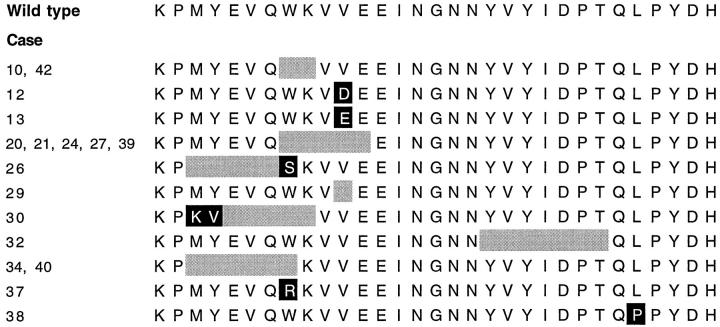Abstract
Gastrointestinal stromal tumors (GISTs) comprise the largest subset of mesenchymal tumors of the gastrointestinal tract. These neoplasms differ histologically and immunohistochemically from typical leiomyomas and leiomyosarcomas. Most GISTs express CD34 and CD117 (c-kit protein) but not desmin. Recently, gain-of-function mutations of c-kit proto-oncogene have been shown in five solitary GISTs and in tumors and leukocytes from a family with multiple GISTs. An in-frame deletion or a point mutation in exon 11 of c-kit was detected in these cases. Stable transfection of the mutant c-kit complementary DNA was also shown to induce malignant transformation of murine lymphoid cells, suggesting that the c-kit mutations contribute to tumor development. In this study, we evaluated 43 GISTs and 14 smooth muscle tumors for mutations in the exon 11 of c-kit by a PCR-assay. Half of the malignant GISTs (12/24) and only one benign GIST (1/19) revealed mutant bands. No mutant bands were found in 3 leiomyomas and 11 leiomyosarcomas. Sequence analysis confirmed the presence of an in-frame deletion of 3–21 bp in all 13 GISTs with mutant bands. Wild-type bands from 8 malignant and 11 benign GISTs and 7 smooth muscle tumors without mutant bands were cloned and sequenced. Additional mutations were found in 3 malignant and 2 benign GISTs. There were no mutations in 3 leiomyomas and 4 leiomyosarcomas. The mutation status of exon 11 did not correlate with immunohistochemically detectable expression of the CD117, as virtually all GISTs with or without such mutations showed CD117 immunoreactivity. The c-kit mutations occur preferentially in malignant GISTs and might be a clinically useful adjunct marker in the evaluation of GISTs. The conservation of the c-kit mutation pattern, observed in consecutive lesions from the same patients, suggests that these mutations might be useful tumor markers in monitoring recurrence or minimal residual disease.
Gastrointestinal stromal tumor (GIST) is the designation for a major subset of mesenchymal tumors of the gastrointestinal tract. These tumors differ from ordinary leiomyomas and leiomyosarcomas histologically, immunohistochemically and genetically. GISTs are typically more cellular than ordinary leiomyomas and are composed of spindle or epithelioid-appearing cells. Immunohistochemically they are typically positive for CD34, the hematopoietic progenitor cell antigen, but negative for desmin, in contrast to leiomyomas and leiomyosarcomas. 1-6
Recently, the expression of the c-kit proto-oncogene has been shown in GISTs. 7-10 The c-kit gene encodes for a receptor for a growth factor termed stem cell factor. The c-kit gene product (stem cell factor receptor) contains an internal tyrosine kinase component and regulates cell growth and survival. 11-14 Cell types in which c-kit expression plays a functionally important role include germ cells, melanocytes, hematopoietic stem cells, mast cells, and the interstitial cells of Cajal, the gastrointestinal pacemaker cells. 15-17
A specific mutation in the tyrosine kinase domain of c-kit has been found in mast cell neoplasms. 18 Mutations between the transmembrane and tyrosine kinase domains in exon 11 of c-kit have been shown in GISTs 7,19 and in a mast cell leukemia cell line. 20 The c-kit mutations in GISTs have been shown to lead to ligand-independent activation of the tyrosine kinase of c-kit and have a tumor promoting effect in vitro. 7
The c-kit mutation status of exon 11 have been studied in relatively few GISTs (five sporadic and three familial tumors) and several questions remain open. Unknown are the possible relationship of c-kit expression and the mutations, the relationship of the mutations and malignancy, and the possible specificity of the mutations for GISTs, as previously suggested. 7
In this study, we further examined the c-kit expression and mutation status of exon 11 of c-kit in a series of benign and malignant GISTs and smooth muscle tumors. The results showed a consistent c-kit expression in GISTs with or without detectable mutations. The mutations occur preferentially in malignant versus benign GISTs and do not occur in leiomyomas and leiomyosarcomas. These observation suggest that the mutations in the exon 11 of c-kit might represent useful molecular genetic markers for malignant GISTs.
Materials and Methods
Tissue Material
Nineteen benign and 24 malignant or potentially malignant gastrointestinal stromal tumors (GISTs) and three esophageal leiomyomas and 11 leiomyosarcomas from different sites were obtained from the files of the Haartman Institute of the University of Helsinki, Helsinki, Finland.
Immunohistochemistry
The tumors were immunohistochemically analyzed for CD34, CD117 (the c-kit proto-oncogene protein product), α-smooth muscle actin, desmin, and S100-protein. Immunohistochemistry was performed by using the avidin-biotin peroxidase complex system and diaminobenizidine as the chromogen, as previously described. 9 Negative and positive controls were included in each run. The primary antibodies, pretreatments, and dilutions are shown in Table 1 ▶ .
Table 1.
Monoclonal Antibodies and Antisera Used in this Study
| Molecule/antibody | Clone | Pretreatment, Antibody dilution | Source |
|---|---|---|---|
| CD117 (c-kit) | polyclonal | MW, 1:400 | Santa-Cruz Biotechnology, Santa Cruz, CA |
| CD34 | HPCA-1 | Pepsin, 1:50 | Becton-Dickinson, Mt View, CA |
| α-smooth muscle actin | 1A4 | No treatment, 1:1600 | Sigma, St. Louis, Mo |
| Desmin | D33 | No treatment, 1:50 | Dako Corp., Carpinteria, CA |
| S100-protein | polyclonal | No treatment, 1:1600 | Dako |
MW, microwave-based antigen retrieval with citrate buffer; Pepsin, pepsin digestion, 30 minutes at 37°C.
DNA Extraction and Evaluation of Template Quality
DNA for the PCR amplification was obtained from fresh tissue by standard phenol-chloroform-isoamylalcohol extraction and from FFPE tissues using a previously published procedure. 21 The quality of DNA templates was verified by PCR amplification of a 268-bp fragment of genomic DNA as previously described. 22
PCR Amplification of Exon 11 of c-kit
A PCR assay was developed to amplify exon 11 of c-kit. The forward primer CK10.4 (5′-CCA GAG TGC TCT AAT GAC TG-3′) and the reverse primer CK 11.2 (5′-ACT CAG CCT GTT TCT GGG AAA CTC-3′) were designed based on a previously published sequence of the human c-kit gene. 23 This PCR assay (30 cycles) yielded amplification products of 192 bp. The PCR reaction conditions were the standard ones recommended by Perkin Elmer (Norwalk, CT). The annealing temperature was 56°C. The PCR products were size fractionated on 5% polyacrylamide gels and stained with ethidium bromide.
Cloning and Sequencing of PCR Products
The PCR products were purified from the gels, treated with Klenow DNA polymerase (Boehringer Mannheim, Indianapolis, IN), blunt end-ligated into pBSK+ phagemid (Stratagene, La Jolla, CA), and transfected into DH5α-competent cells (Gibco-BRL, Gaithersburg, MD). White colonies obtained from the plating on IPTG/X-gal selective medium (Gibco-BRL) were picked and screened by PCR as described above to confirm the presence of the appropriate inserts. Double-stranded DNA templates were prepared from randomly selected recombinants using Qiagen plasmid kit (Qiagen Inc., Chatsworth, CA) and sequenced on a 373 DNA-sequencer (Applied Biosystems, Foster City, CA). Five to nine clones were sequenced per case using the M13 forward primer. The subsets of clones that revealed consistent nucleotide differences from the germline were sequenced using the reverse primers. To prevent PCR contamination, standard precautions were undertaken.
Evaluation of Taq-polymerase Error
To exclude Taq-polymerase error as the source of clonal variation, placental DNA was amplified, cloned, and sequenced as described above.
Sequence Analysis
Computer analysis of the DNA sequences was performed using the Lasergene software (DNASTAR, Madison, WI) in connection with the data of the GenBank 107/EMBL 54 database (July 1998 edition).
Results
Clinicopathological Features of GISTs
Demographic data and clinical and histological features of all GISTs analyzed in this study have been summarized in Tables 2 and 3 ▶ ▶ . There were 25 gastric, 12 small intestinal, and 2 rectal primary tumors. Five intraabdominal recurrences and 2 liver metastases were also analyzed from a total of 43 patients. Two different lesions from each of three patients were analyzed .
Table 2.
Clinical Data and c-kit Mutations of the Benign GISTs Analyzed in this Study
| Case | Age/Sex | Location | Cell type | Tumor size or maximum diameter (cm) | Status and follow-up in months | c-kit mutant band | Sequencing of exon 11 of c-kit |
|---|---|---|---|---|---|---|---|
| 1 | 68 /M | Stomach | Spindle | 2.0 | NED 27 | − | Not done |
| 2 | 66 /M | Stomach | Spindle | 3–4 | − | Not done | |
| 3 | 69 /M | Stomach | Epithelioid | 3–4 | − | Not done | |
| 4 | 63 /M | Stomach | Spindle | 3.5 | − | Not done | |
| 5 | 64 /F | Stomach | Spindle | 3.6 | NED 21 | − | Wild-type |
| 6 | 60 /F | Stomach | Spindle | 4× 4× 3 | − | Wild-type | |
| 7 | 57 /M | Stomach | Spindle | 4 | NED 56 | − | Not done |
| 8 | 70 /F | Stomach | Spindle* | 4.5× 4.5× 3.5 | NED 25 | − | Wild-type |
| 9 | 40 /M | Stomach | Epithelioid | 5 | NED 34 | − | Wild-type |
| 10 | 33 /M | Stomach | Spindle | 5× 3.5× 3.5 | NED 44 | + | Deletion |
| 11 | 61 /F | Stomach | Epithelioid | 5× 5 | NED 35 | − | Wild-type |
| 12 | 79 /F | Stomach | Spindle* | 8 | NED 58 | − | One point mutation |
| 13 | 45 /F | Stomach | Spindle | 9 | NED 24 | − | Two point mutations |
| 14 | 69 /M | Small bowel | Spindle | 2.5× 2.0× 2.0 | NED 8 | − | Not done |
| 15 | 35 /F | Small bowel | Spindle | 3–4 | − | Wild-type | |
| 16 | 57 /M | Small bowel | Epithelioid | 4 | NED 54 | − | Wild-type |
| 17 | 80 /M | Small bowel | Spindle | 4.5 | NED 31 | − | Not done |
| 18 | 60 /M | Small bowel | Spindle | 6× 6× 6 | − | Not done | |
| 19 | 52 /M | Small bowel | Spindle | 7 × 7× 6.5 | NED 27 | − | Not done |
NED, no evidence of disease.
*Case 8 showed 2 mitoses/10 HPF and case 12 showed 3 mitoses/10 HPF. All other cases showed ≤1 mitosis/10 HPF.
Table 3.
Clinical Data and c-kit Mutations of the Malignant and Potentially Malignant GISTs Analyzed in this Study
| Case | Age/Sex | Location | Cell type | Tumor size or maximum diameter (cm) | Mitoses/ 10 HPF | Clinical behavior, follow-up (months) | c-kit mutant band | Sequencing of exon 11 of c-kit |
|---|---|---|---|---|---|---|---|---|
| 20* | 62 /M | Stomach | Spindle | 15 | 2 | Died of lung cancer, 36 months. No evidence for IAR | + | Deletion |
| 21* | 80 /F | Stomach | Epithelioid | 5.5× 3.5× 1.7 | <1 | + | Deletion | |
| 22* | 72 /M | Stomach | Epithelioid | 11× 10× 3.5 | <1 | − | Wild type | |
| 23 | 26 /F | Stomach | Epithelioid | 6× 4.5× 2.5 | 18 | − | Wild type | |
| 24 | 47 /F | Stomach | Spindle | 10× 6× 5 | 11 | Omental seeding at present-ation. IAR at 15 months | + | Deletion |
| 25 | 57 /M | Stomach | Spindle | 9× 7.5× 7 | 12 | Mediastinal metastasis at presentation | − | Not done |
| 26 | 70 /M | Stomach | Spindle | 11× 5 | 18 | Omental seeding at presentation. IAR at 25 months | + | Deletion+ PM |
| 27 | 69 /F | Stomach | Spindle | 12 | 9 | NED 6 months | + | Deletion |
| 28 | 44 /M | Stomach | Epithelioid | 17 × 10× 5 | 9 | IAR at 25 months | − | Wild type |
| 29 | 44 /M | Stomach | Spindle | 17 | 6 | − | Deletion | |
| 30 | 63 /M | Stomach | Spindle | 19× 12× 7 | 8 | + | D+ 3 PM | |
| 31 | 41 /M | Stomach | 39× 18 | 30 | Alive with disease, 16 months | − | Not done | |
| 32 | 50 /M | SB | Spindle | 6.5× 4.5× 3.4 | 11 | Liver metastasis at presentation | + | Deletion |
| 33 | 68 /M | SB | Spindle | 7× 6 | 8 | IAR at 16 months | − | Not done |
| 34 | 60 /M | SB | Spindle | 7 | 6 | IAR at 12 months DOD 24 months | + | Deletion |
| 35 | 64 /F | SB | Spindle | 12–15 | >20 | DOD 15 months | − | Wild type |
| 36 | 54 /M | SB+ mesentery | Spindle | 15× 13× 6 | 15 | IAR 8 months | − | Wild type |
| 37 | 85 /M | SB | Spindle | 30× 30× 5 | 15 | DOD 16 months | − | PM |
| 38 | 63 /M | Rectum | Spindle | 7 | 13 | NED 20 months | − | PM |
| 39 | 73 /M | Rectum | Spindle | 10× 9× 8 | 10 | + | Deletion | |
| 40 | 62 /F | IAR (2 yrs) | Spindle | 2 kg, multiple tumors | IAR 2 and 3 years after a gastric GIST | + | Deletion | |
| 41 | 56 /F | IAR (5 yrs) | Spindle | >20 cm | IAR 4 and 5 years after a total gastrectomy for gastric malignant GIST | − | Wild type | |
| 42 | 59 /F | Liver metastasis | Spindle | 8 cm | Liver metastasis 4 years after malignant gastric GIST | + | Deletion | |
| 43 | 64 /M | Liver metastasis | Spindle | 6 cm | Liver metastasis 6 months after SB GIST | − | Not done |
*Cases classified as potentially malignant.
DOD, died of disease; NED, no evidence of disease; SB, small bowel; IAR, intra-abdominal recurrence; D, deletion; PM, point mutation.
Benign GISTs
Histological Features
Nineteen GISTs were classified as histologically benign based on low, if any, mitotic activity. All but two cases showed < 1 mitosis per 10 high power fields (HPF), one case showed 2 mitoses/10 HPF, and another 3 mitoses/10 HPF. The tumor size did not exceed 10 cm in the group of spindle cell tumors or 5 cm in the group of epithelioid tumors. Follow-up available in 13 cases did not show intra-abdominal recurrences or metastases in any of the cases.
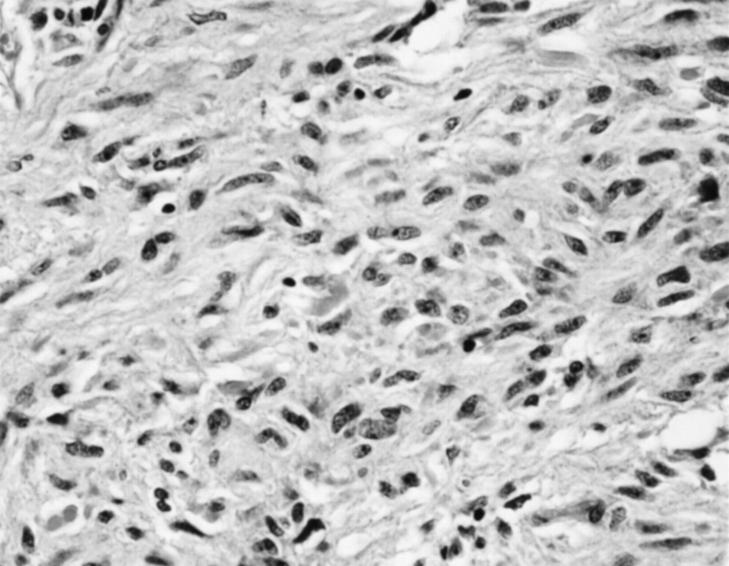
Histologically, 15 cases showed exclusively or predominantly a spindle cell pattern typically showing high cellularity, frequent perinuclear vacuoles, a rich capillary vascular pattern, and a general lack of cytoplasmic eosinophilia (Figure 1) ▶ . Epithelioid cytologic features with large cytoplasmic cells and distinct cell borders were seen exclusively or predominantly in four cases. All cases, irrespective of morphological subtype and location, were immunohistochemically positive for CD117 (Figure 2) ▶ . Most cases (74%) were positive for CD34, whereas a minority of the tumors (32%) expressed α-smooth muscle actin. All cases were negative for desmin except two epithelioid GISTs that showed focal desmin-reactivity seen in less than 10% of the tumor cells. All cases were negative for S100-protein.
Figure 1.
A benign small intestinal GIST of spindle cell type. Note the uniform appearance of bland spindle cells without mitotic activity. Hematoxylin and eosin, original magnification ×411.
Figure 2.
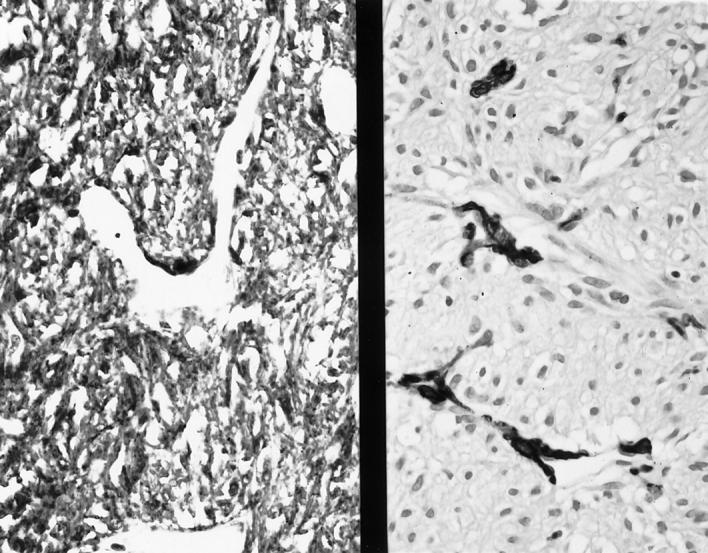
A benign spindle cell GIST (shown in Figure 1 ▶ ) is strongly positive for CD117 (left), but negative for α-smooth muscle actin; vessel walls are positive (right). ABC immunoperoxidase, original magnification ×300.
Evaluation of Mutations in Exon 11 of c-kit
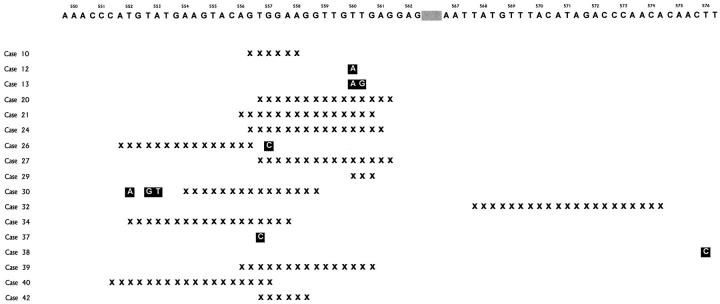
Analysis of the PCR amplification products of the critical region in exon 11 of c-kit revealed a mutant band in one of 19 benign GISTs (Case 10). Sequence analysis showed an in-frame deletion of 6 bp involving codons 556–558. The remaining 18 benign GISTs revealed only one wild-type band each. Wil-type bands from 11 cases were cloned and sequenced. In two benign GISTs (Case 12 and 13) sequencing showed mutations at codon 560 (Figure 3) ▶ . Predicted amino acid sequences of the mutant c-kit are shown in Figure 4 ▶ .
Figure 3.
Sequences of the exon 11 of c-kit from the mutant bands obtained from the GISTs. The deleted nucleotides are indicated with x, and point mutations with ▪. The fragments of germline sequence of exon 11 are shown according to Andre et al. 23 The codons are indicated above with numbers and the gray area indicates codons 563–566.
Figure 4.
Predicted amino acid sequences of the mutant c-kit. The sequence starts at codon 550 and ends at 580. The wild-type sequence is shown above. The shaded areas correspond to deletions. Point mutations are shown in ▪.
Malignant and Potentially Malignant GISTs
Histological Features
Twenty-one GISTs were classified as histologically malignant based on high mitotic activity (> 4 mitoses/10 HPF). The three tumors classified as potentially malignant showed a low mitotic activity but were large tumors and included a 15-cm spindle cell tumor from the stomach and two large epithelioid gastric tumors of 5.5 cm and 11 cm diameter.
Follow-up available in 15 cases of malignant or potentially malignant GISTs showed intraabdominal recurrences, metastases, or death from tumor in 13 of them.
Histologically, 19 tumors had a predominant spindle cell pattern (Figure 5) ▶ , and four were epithelioid. CD117-immunoreactivity was shown in all cases (Figure 6) ▶ , except in one (Case 26). The majority of cases (83%) were positive for CD34 and α-smooth muscle actin was present in 6 cases (26%), usually focally. All cases were negative for desmin and S100-protein.
Figure 5.
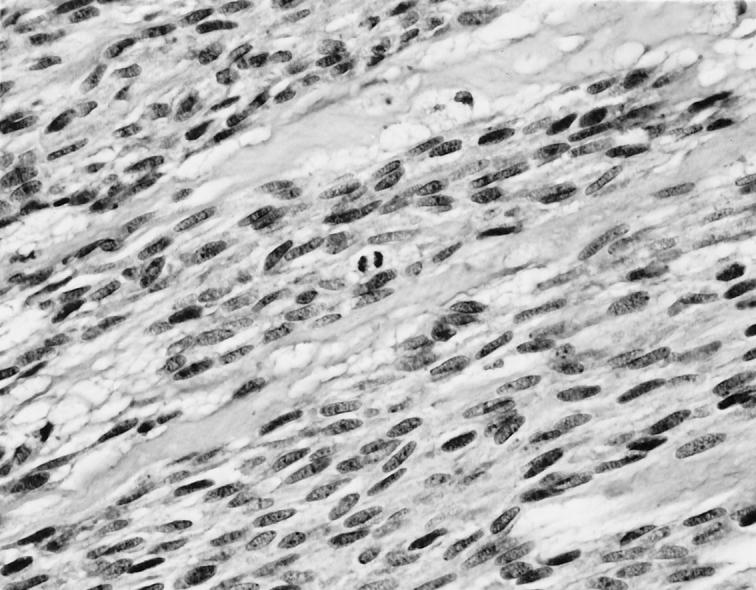
A malignant gastric GIST of spindle cell type shows plump spindle cells with conspicuous mitotic activity. Hematoxylin and eosin, original magnification ×482.
Figure 6.
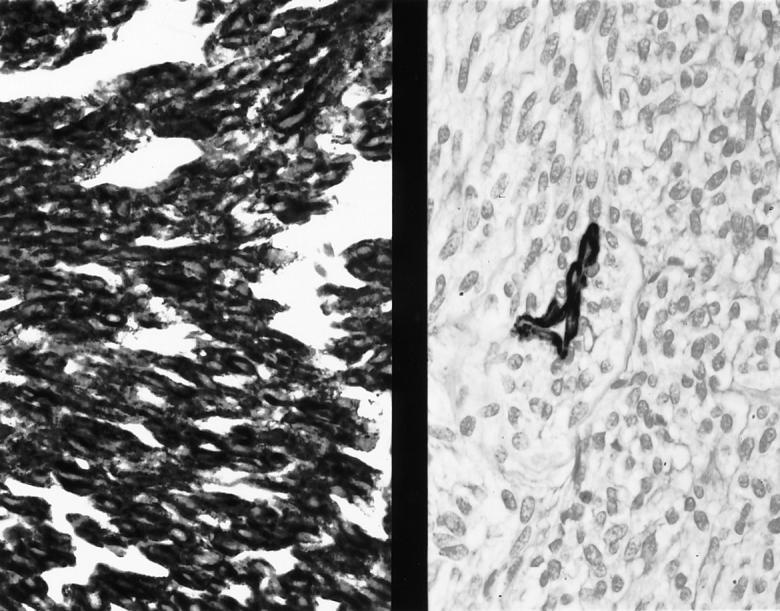
The malignant GIST shown in Figure 3 ▶ is strongly immunoreactive for CD117 (left), but negative for α-smooth muscle actin; a vessel wall is positive (right). ABC immunoperoxidase, original magnification ×411.
Evaluation of Mutations in Exon 11 of c-kit
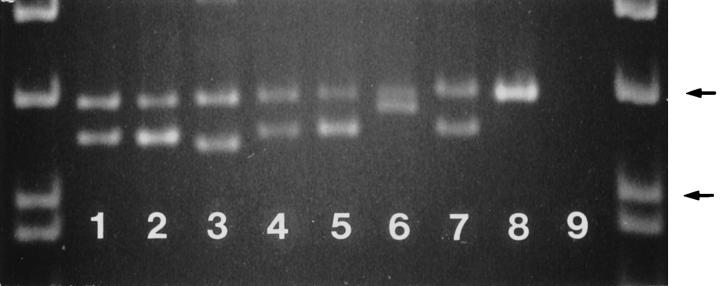
Analysis of the PCR amplification products of exon 11 of c-kit showed smaller mutant bands, in addition to the wild-type bands, in 12 of the 24 (50%) malignant GISTs (Figure 7) ▶ . Sequencing of the mutant bands revealed in-frame deletions of 6 to 21 bp (Figure 3) ▶ . Wild-type bands from these cases showed germline sequences of the c-kit. The remaining 12 malignant GISTs revealed only wild-type bands, and eight of them were sequenced. Three of the eight cases showed c-kit mutations. Two cases (Cases 37 and 38) showed point mutations, one at the codon 557 and another at the codon 576. The third case (Case 29) revealed a 3-bp in-frame deletion involving codons 560 and 561. Predicted amino acid sequences of the mutant c-kit are shown in Figure 4 ▶ . Among the malignant GISTs, c-kit mutations were seen in tumors of different sites including gastric, small intestinal, and rectal primary tumors.
Figure 7.
Analysis of the PCR amplification products of the exon 11 of c-kit in malignant GISTs. Note the different size mutant bands in different cases. Lane 1: Case 20. Lane 2: Case 34. Lane 3: Case 32. Lane 4: Case 21. Lane 5 : Case 27. Lane 6: Case 42. Lane 7: Case 30. Lane 8: Placenta (germline), Lane 9: Negative control. The samples are flanked by PhiX174 DNA/HinfI markers (Promega, Madison, WI). Arrows indicate 200- and 151-bp fragments. 5% polyacrylamide gel electrophoresis.
Analysis of Consecutive Lesions
In three cases, two consecutive lesions of the same patient were analyzed. They included primary tumor and an intraabdominal recurrence in Cases 24 and 34 and two intra-abdominal recurrences in Case 40. In all cases, the banding pattern and sequences of the mutant bands remained identical in primary and recurrent lesions.
Smooth Muscle Tumors
The two esophageal leiomyomas and one small intestinal leiomyoma were histologically spindle cell neoplasms with distinctly eosinophilic cytoplasm, moderate cellularity, and lack of mitotic activity. All cases were positive for α-smooth muscle actin and desmin and negative for CD34 and CD117.
The eleven leiomyosarcomas included six from the retroperitoneum, two from the thigh, one from the inferior vena cava and two metastatic uterine leiomyosarcomas, one from the buttock and another from the small intestine. Histologically these tumors showed intersecting fascicles of spindle cells with blunt-ended nuclei and variably eo- sinophilic cytoplasm. Significant mitotic activity was present in each case. All cases were positive for α-smooth muscle actin and 9 of 11 at least focally for desmin; all were negative CD117 and S100-protein. Two cases were focally positive for CD34; all other cases were negative.
Analysis of the PCR-amplification products of exon 11 of c-kit of three leiomyomas and 11 leiomyosarcomas showed wild-type bands similar to those seen in placental DNA. Sequencing revealed 100% homology with the c-kit germline.
Evaluation of Taq-polymerase Error
No nucleotide differences were identified in ten randomly selected PCR products from the amplification of exon 11 of c-kit from placental DNA. This gives a Taq-polymerase error rate of less than 1 nucleotide difference in 1920 bp and practically ruled out Taq-polymerase error as the source of the nucleotide variation seen in the exon 11 of c-kit in GISTs.
Discussion
In this study, we examined the sequences and expression of c-kit in the spectrum of gastrointestinal stromal tumors (GISTs), including benign and malignant variants from different sites. The sequences of the c-kit were also evaluated in typical smooth muscle tumors.
Previous molecular studies 7 have revealed mutations between the transmembrane and tyrosine kinase domains in five GISTs. Specifically, the mutations were located within codons 550 to 560 in exon 11, and no mutations were detectable in other domains of c-kit cDNA. Therefore, the exon 11 sequences coding amino acids 550–582 were analyzed in the current study.
Our series of 43 benign and malignant GISTs showed that mutations in exon 11 of c-kit occur predominantly in the malignant GISTs that were tumors with high mitotic activity. Furthermore, clinically malignant tumor behavior was evident in most cases by intra-abdominal recurrences, distant metastases, or both. Mutations in the exon 11 of c-kit were observed in 62% of these tumors. In contrast, only 3 benign GISTs (16%) showed mutations, suggesting that the mutations of exon 11 of c-kit may represent a genotypic marker with a correlation to malignancy. The c-kit mutations were seen in malignant GISTs of different sites including gastric, small intestinal, and rectal tumors, indicating that in respect to the c-kit mutation status, malignant GISTs appear molecularly similar.
Lack of c-kit mutations in 12/24 malignant and potentially malignant GISTs suggests that the mutations in the exon 11 of c-kit are not the only mechanisms related to tumorigenesis and malignancy and that other molecular mechanisms explaining the differences between benign and malignant GISTs must exist. Such mechanisms may include mutations in other regions of the c-kit. Involve- ment of other genes is suggested by the greater number of the DNA copy number changes in malignant versus benign GISTs as observed in comparative genomic hybridization studies. 5
Most c-kit mutations were found in the previously described hot spot region of exon 11 7 involving codons 550–560. Clustering of the c-kit mutations in the same region supports their specific biological significance.
However, in two cases, an additional region including codons 567–576 was found to be affected by both a point mutation and a deletion in two different cases. These findings suggest the possibility that mutations can involve other areas of the exon 11 of c-kit than previously described.
Analysis of the c-kit mutation pattern in consecutive lesions in 3 cases showed the persistence of the mutations that remained identical in the different recurrences. Therefore, the tumor-specific c-kit mutation pattern could be used as a potential marker of early recurrence or to trace minimal residual disease. Further studies are necessary to explore the tumor cell detection in such contexts.
The c-kit mutations were never observed in benign leiomyomas or well-documented leiomyosarcomas (actin-positive, usually desmin-positive, CD34-negative, CD117-negative). Therefore, the c-kit mutations may also represent a genotypic lineage marker for GISTs. In this respect, our findings confirm and expand on the earlier reports on the absence of c-kit mutations in smooth muscle tumors. 7 Other genetic changes found in GISTs but not in smooth muscle tumors include losses in the long arm of chromosome 14, as reported based on comparative genomic hybridization studies. 5,6
The presence of the mutations in exon 11 of c-kit did not correlate with c-kit expression as determined by immunohistochemistry. Whereas virtually all GISTs, including benign and malignant tumors, expressed c-kit, mutations occurred mostly in malignant cases. Therefore, explanations other than mutations in exon 11 have to be considered for the source of high c-kit expression. The interstitial cells of Cajal in the gastrointestinal nervous system show constitutional expression of c-kit and have recently been suggested as ancestor cells for GISTs. 7,8,10,16,17 Therefore, c-kit expression in GISTs could merely reflect the constitutional c-kit expression that is maintained in the neoplastic state of tumor cells, which may be related to the Cajal cells or show differentiation toward them. 8,10
In summary, we have evaluated a series of GISTs and typical smooth muscle tumors for the expression of c-kit protein and for the mutations in exon 11 of c-kit. The c-kit expression is consistent in both benign and malignant GISTs with and without the c-kit mutations. The c-kit mutations occur preferentially in malignant GISTs. The mutations seem to remain stable in recurrences, and could be useful tumor-specific markers. Leiomyomas and leiomyosarcomas show neither c-kit expression nor mutations, adding a new phenotypic and genotypic marker to distinguish these tumors from GISTs.
Footnotes
Address reprint requests to Markku Miettinen, M.D., Department of Soft Tissue Pathology, Armed Forces Institute of Pathology, 14th Street and Alaska Avenue, N.W., Washington, DC 20306-6000.
The opinions and assertions contained herein are the expressed views of the authors and are not to be construed as official or reflecting the views of the Departments of the Army or Defense.
References
- 1.Mikhael AI, Bacchi CE, Zarbo RJ, Ma CK, Gown AM: CD34 expression in stromal tumors of the gastrointestinal tract. Appl Immunohistochem 1994, 2:89-93 [Google Scholar]
- 2.van de Rijn M, Hendrickson MR, Rouse RV: The CD34 expression by gastrointestinal stromal tumors. Hum Pathol 1994, 25:766-771 [DOI] [PubMed] [Google Scholar]
- 3.Monihan JM, Carr NJ, Sobin LH: CD34 immunoexpression in stromal tumours of the gastrointestinal tract, and in mesenteric fibromatoses. Histopathology 1994, 25:469-473 [DOI] [PubMed] [Google Scholar]
- 4.Miettinen M, Virolainen M, Sarlomo-Rikala M: Gastrointestinal stromal tumors: value of CD34 antigen in their identification and separation from true leiomyomas and schwannomas. Am J Surg Pathol 1995, 19:207-216 [DOI] [PubMed] [Google Scholar]
- 5.El-Rifai W, Sarlomo-Rikala M, Miettinen M, Knuutila S, Andersson LC: DNA copy number losses in chromosome 14: an early change in gastrointestinal stromal tumors. Cancer Res 1996, 56:3230-3233 [PubMed] [Google Scholar]
- 6.Sarlomo-Rikala M, El-Rifai W, Andersson L, Miettinen M, Knuutila S: Different patterns of DNA copy number changes in gastrointestinal stromal tumors, leiomyomas and schwannomas. Hum Pathol 1998, 29:476-481 [DOI] [PubMed] [Google Scholar]
- 7.Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M, Tunio GM, Matsuzawa Y, Kanakura Y, Shinomura Y, Kitamura Y: Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science 1998, 279:577-580 [DOI] [PubMed] [Google Scholar]
- 8.Kindblom LG, Remotti HE, Aldenborg F, Meis-Kindblom JM: Gastrointestinal pacemaker cell tumor (GIPACT): gastrointestinal stromal tumors show phenotypic characteristics of the interstitial cells of Cajal. Am J Pathol 1998, 152:1259-1269 [PMC free article] [PubMed] [Google Scholar]
- 9.Sarlomo-Rikala M, Kovatich AJ, Barusevicius A, Miettinen M: CD117: A sensitive marker for gastrointestinal stromal tumors that is more specific than CD34. Mod Pathol 1998, 11:728-734 [PubMed] [Google Scholar]
- 10.Sircar K, Hewlett BR, Riddell RH: Most gastrointestinal stromal tumors arise from interstitial cells of Cajal (Abstract). Mod Pathol 1998, 11:71A [Google Scholar]
- 11.Vliagoftis H, Worobec AS, Metcalfe DD: The proto-oncogene c-kit and c-kit ligand in human disease. J Allerg Clin Immunol 1997, 100:435-440 [DOI] [PubMed] [Google Scholar]
- 12.Yarden Y, Kuang WS, Yang-Fend T, Coussens L, Munemitsas S, Dull TJ: Human proto-oncogene kit, a new cell surface-receptor tyrosine kinase for an unidentified ligand. EMBO J 1987, 6:3341-3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams DE, Eisenman J, Baird A, Rauch C, van Ness K, March CJ, Park LS, Martin U, Mochizuki DY, Boswell HS, Burgess GS, Cosman D, Lyman SD: Identification of a ligand for the c-kit proto-oncogene. Cell 1990, 63:167-174 [DOI] [PubMed] [Google Scholar]
- 14.Zsebo KM, Williams DA, Geissler EN, Broudy WC, Martin FH, Atkins HL, Hsu R-Y, Birkett NC, Okino KH, Murdock DC, Jacobsen FW, Langley KE, Smith KA, Takeishi T, Cattanach BM, Galli SJ, Suggs SV: Stem cell factor is encoded at the SI locus of the mouse and is ligand for the c-kit tyrosine kinase receptor. Cell 1990, 63:214-224 [DOI] [PubMed] [Google Scholar]
- 15.Tsuura Y, Hiraki H, Watanabe K, Igarashi S, Shimamura K, Fukuda T, Suzuki T, Seito T: Preferential localization of c-kit product in tissue mast cells, basal cells of skin, epithelial cells of breast, small cell lung carcinoma and seminoma/dysgerminoma in human: immunohistochemical study of formalin-fixed, paraffin-embedded tissues. Virchows Arch 1994, 424:135-141 [DOI] [PubMed] [Google Scholar]
- 16.Maeda H, Yamagata A, Nishikawa S, Yoshinaga K, Kobayshy S, Nishi K, Nishikawa S: Requirement of c-kit for development of intestinal pacemaker system. Development 1992, 116:369-375 [DOI] [PubMed] [Google Scholar]
- 17.Sanders KM: A case for interstitial cells of Cajal as pacemakers and mediators of neurotransmission in the gastrointestinal tract. Gastroenterology 1996, 111:492-515 [DOI] [PubMed] [Google Scholar]
- 18.Nagata H, Worobec AS, Oh CK, Chowdhury BA, Tannenbaum S, Suzuki Y, Metcalfe DD: Identification of a point mutation in the catalytic domain of the protooncogene c-kit in peripheral blood mononuclear cells of patients who have mastocytosis with an associated hematologic disorder. Proc Natl Acad Sci USA 1995, 92:10560-10564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishida T, Hirota S, Taniguchi M, Hashimoto K, Isozaki K, Nakamura H, Kanakura Y, Tanaka T, Takabayshi A, Matsuda H, Kitamura Y: Familial gastrointestinal stromal tumours with germline mutation of the KIT gene. Nat Genet 1998, 19:323-324 [DOI] [PubMed] [Google Scholar]
- 20.Furitsu T, Tsujimura T, Tono T, Ikeda H, Kitayama H, Koshimizu U, Sugahara H, Butterfielf JH, Ashman LK, Kanayama Y, Matsuzawa Y, Kitamura Y, Kanakura Y: Identification of mutations in the coding sequence of the proto-oncogene c-kit in a human mast cell leukemia cell line causing ligand-independent activation of c-kit product. J Clin Invest 1993, 92:1736-1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lasota J, Franssila K, Koo CH, Miettinen M: Molecular diagnosis of mantle cell lymphoma in paraffin-embedded tissue. Mod Pathol 1996, 9:361-366 [PubMed] [Google Scholar]
- 22.Greer CE, Peterson SL, Kiviat NB, Manos MM: PCR amplification from paraffin-embedded tissues: effects of fixative and fixation time. Am J Clin Pathol 1991, 95:117-124 [DOI] [PubMed] [Google Scholar]
- 23.Andre C, Hampe A, Lachaume P, Martin E, Wang XP, Manus V, Hu WX, Galibert F: Sequence analysis of two genomic regions containing the KIT and FMS receptor tyrosine kinase genes. Genomics 1997, 39:216-226 [DOI] [PubMed] [Google Scholar]