Abstract
Despite the severe ecological and economic damage caused by introduced species, factors that allow invaders to become successful often remain elusive. Of invasive taxa, ants are among the most widespread and harmful. Highly invasive ants are often unicolonial, forming supercolonies in which workers and queens mix freely among physically separate nests. By reducing costs associated with territoriality, unicolonial species can attain high worker densities, allowing them to achieve interspecific dominance. Here we examine the behavior and population genetics of the invasive Argentine ant (Linepithema humile) in its native and introduced ranges, and we provide a mechanism to explain its success as an invader. Using microsatellite markers, we show that a population bottleneck has reduced the genetic diversity of introduced populations. This loss is associated with reduced intraspecific aggression among spatially separate nests, and leads to the formation of interspecifically dominant supercolonies. In contrast, native populations are more genetically variable and exhibit pronounced intraspecific aggression. Although reductions in genetic diversity are generally considered detrimental, these findings provide an example of how a genetic bottleneck can lead to widespread ecological success. In addition, these results provide insights into the origin and evolution of unicoloniality, which is often considered a challenge to kin selection theory.
Invasive species are recognized as a leading threat to biodiversity as well as an increasing economic concern (1–3). Despite these problems, the attributes responsible for the establishment and spread of specific invaders are often difficult to pinpoint or are unknown (4–6). In particular, studies that examine the biology of invasive species in both their native and introduced ranges are surprisingly rare, despite the potential insights that can be gained from such comparisons. In many cases, forces external to the biology of the invasive species, such as escape from predators, competitors, and parasites (4, 7) or characteristics of the invaded habitat (4, 6, 8, 9), are implicated as the primary causes of invasion success. When the characteristics of invasive species themselves are considered, it is usually in terms of general demographic or life history traits that typify a broad suite of taxa (4, 6). However, many insights may be gained by examining the specific attributes responsible for success of particular invasive species. Here we examine the behavior and population genetics of the highly invasive Argentine ant (Linepithema humile) in its native and introduced ranges, and provide a mechanism to explain its success as an invader.
The Argentine ant is a widespread and damaging invasive species (10). In the United States, it was first detected in New Orleans in 1891, and by 1907, was established in California (11), where it is now common in coastal habitats, riparian woodlands, and irrigated urban and agricultural areas (12, 13). Once established, the Argentine ant displaces most native ants (12–16) and detrimentally affects other non-ant arthropods (17). These direct effects reverberate through communities as other taxa are affected indirectly (18, 19). In contrast, Argentine ants in their native range coexist with other ants in species-rich communities (20, 21). This difference may correspond with variation in colony structure between native and introduced populations of Argentine ants (20). In their native range, Argentine ants appear to defend territories against conspecifics (20), whereas introduced populations are thought to be more unicolonial, forming expansive supercolonies (11, 22–24). Previous work has demonstrated that the absence of intraspecific aggression within supercolonies reduces costs associated with territoriality, and leads to increases in colony size (25). Because individual Argentine ant workers are small, they rely on numerical superiority in interference interactions with larger native species (26). The apparent differences between native and introduced populations in both colony structure and competitive dominance suggest that exploration of the causes of intraspecific aggression will illuminate factors responsible for the Argentine ant's success as an invader.
To elucidate the mechanisms responsible for the success of an invasive species, we compared behavioral and genetic characteristics of Linepithema humile at two spatial scales in both its native and introduced ranges. First, we quantified intraspecific aggression among nests at a local scale within seven introduced and eight native populations. Next, we examined the relationship between intraspecific aggression and genetic similarity among nests from sites along 1,000-km transects through California (introduced range) and northern Argentina (native range). Finally, to examine more closely the mechanisms that delineate colony structure, we investigated the relationship between genetic similarity and intraspecific aggression across supercolony boundaries.
Methods
Study Areas.
To examine the frequency and degree of intraspecific aggression among nests at a local spatial scale, behavioral data were collected in Argentina at five sites on the Rio Paraná [Reserva Otamendi (8 nests), Reserva Ecologíca Costanera Sur (9 nests), Parque Nacional PreDelta (9 nests), Porto Ocampo (9 nests), and Itá-Ibaté (9 nests)] and at three sites on the Rio Uruguay [Ibicuy (7 nests), Colón (6 nests), and Alvear (7 nests)]. In the introduced range, behavioral data were collected in Bermuda (6 nests) and Chile (12 nests) and from five sites in the United States [New Orleans, LA (6 nests); La Jolla, CA (12 nests); Encinitas, CA (10 nests); Temecula, CA (6 nests); and Palo Alto, CA (7 nests)] (Fig. 1).
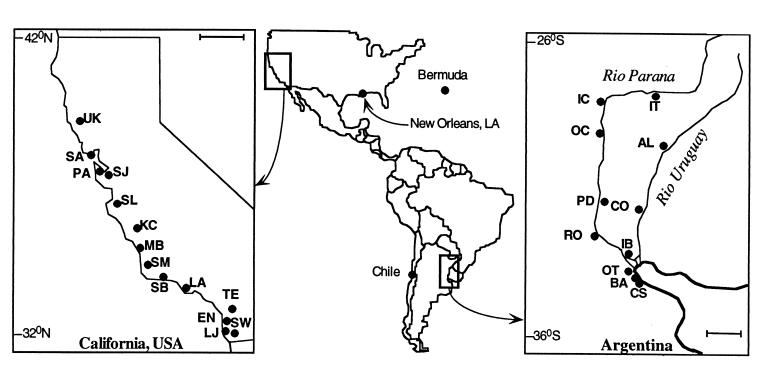
Figure 1.
Sites used in this study. Introduced populations of Argentine ants were sampled in Chile, Bermuda, and the United States. Sampling sites in the United States included New Orleans, Louisiana and the following sites in California: Ukiah (UK), Sausalito (SA), Palo Alto (PA), San Jose (SJ), Salinas (SL), King City (KC), Morro Bay (MB), Santa Maria (SM), Santa Barbara (SB), Los Angeles (LA), Temecula (TE), Encinitas (EN), La Jolla (LJ) and Sweetwater Reservoir (SW). Sites in San Diego County, California not shown: Solana Beach, Lake Hodges and Mission Trails Regional Park. Native populations in Argentina include: Itá-Ibaté (IT), Isla de las Cerritas (IC), Porto Ocampo (OC), Alvear (AL), Parque Nacional PreDelta (PD), Colón (CO), Rosario (RO), Ibicuy (IB), Reserva Nacional Ecológica Otamendi (OT), Buenos Aires (BA), and Reserva Ecológica Costanera Sur (CS). (Bars in the expanded maps represent 100 km.)
We collected workers for large-scale genetic and behavioral analyses from nests at six sites in Argentina: Buenos Aires, Rosario, Parque Nacional PreDelta, Porto Ocampo, Isla de las Cerritas, and Itá-Ibaté. In the United States, workers for these analyses were collected at nine sites within California: Los Angeles, Santa Barbara, Morro Bay, Santa Maria, King City, Salinas, San Jose, Sausalito, and Ukiah (Fig. 1).
To examine more closely the relationship between intraspecific aggression and genetic similarity, we also collected workers from different supercolonies. We defined a supercolony as a group of nests among which intraspecific aggression was absent. In Argentina, workers were collected from eight supercolonies at three sites: Reserva Otamendi (6 nests, 3 colonies), Reserva Ecológica Costanera Sur (9 nests, 4 colonies), and Buenos Aires (2 nests, 1 colony). Because intraspecific aggression is rare in the introduced range, extensive searching was necessary to find different supercolonies in California. In California, workers were collected from seven sites representing six supercolonies: Encinitas (9 nests, 2 supercolonies), La Jolla (8 nests, 1 supercolony), Temecula (4 nests, 2 supercolonies), Solana Beach (1 nest, 1 supercolony), Sweetwater Reservoir (2 nests, 1 supercolony), Lake Hodges (1 nest, 1 supercolony), and Mission Trails Regional Park (1 nest, 1 supercolony) (Fig. 1). One supercolony is represented by nests from multiple sites (Encinitas, La Jolla, Temecula, and Solana Beach).
Behavioral Assays.
We measured intraspecific aggression with a standard aggression assay (20, 25). We randomly selected a single worker from each of two nests and placed them together in a vial with Fluon-coated sides for 5–10 min. We scored the behavioral interactions that ensued in order of escalating aggression: touch = 1 (contacts that included prolonged antennation), avoid = 2 (contacts that resulted in one or both ants quickly retreating in opposite directions), aggression = 3 (lunging, biting, and pulling legs or antennae), or fight = 4 (prolonged aggression between individuals). We recorded the highest level of aggression for each trial and then used the mean of 5–10 trials for each nest pairing to calculate an average aggression score. This assay had a high repeatability (r = 0.881) (27) and, on average, variance among trials within nest pairings was low (s2 = 0.302).
Molecular Analysis.
Four microsatellite loci (Lihu C1.1, Lihu M1, Lihu S3, and Lihu T1) were cloned from a small-insert genomic library (20). Primers for three additional loci (Lhum-11/11B, Lhum-14, and Lhum-33) were designed on the basis of sequences present in GenBank (accession nos. AF093525, AF093520, and AF093517) (51). Ten to 15 individuals from each nest were genotyped at these seven microsatellite loci, and these data were used to calculate unbiased estimates of expected heterozygosity (28) and the percentage of alleles shared by each pair of nests. The percentage of alleles shared is a measure of genetic similarity and refers to the alleles shared at these focal loci, and not the alleles shared overall.
Statistical Analysis.
We used the percentage of alleles shared rather than relatedness coefficients (R) (29) as our measure of genetic similarity between nests. Estimates of percent alleles shared were calculated for each nest pair as the number of alleles shared by two nests divided by the sum of alleles possessed by both nests. We did not use R because it is a measure of genetic similarity calculated relative to a reference population (P*) (29). Because introduced populations are very genetically homogeneous, R will be low when calculated with only the introduced population as P*, regardless of the level of absolute genetic similarity. Alternatively, when P* includes both native and introduced populations, R values within introduced populations will be higher (30, 31). However, when two widely divergent groups are combined in this way, the allele frequencies of the reference population will fall between the values of each range and will not accurately represent either. We have therefore chosen to use a more absolute measure, the percentage of alleles shared, to determine levels of genetic similarity between groups.
To examine the relationship between genetic similarity and intraspecific aggression, we used linear regression. Each point in our regressions represents a unique pairing of nests, but some nests were used in more than one pairing. Consequently, these points may not be completely independent of one another. Therefore, we tested significance (one-tailed) by using a Mantel test and 10,000 permutations of the data. Nest pairing for behavioral assays was distributed across nests to prevent overrepresentation by a single or a few sites. In Argentina the mean number of trials per nest was 3.4 ± 0.3 (SE). In California each nest was used for 6.4 ± 0.5 trials. The Student t test was used to examine differences between the slopes and intercepts of the regressions.
Results
The Argentine ant passed through a genetic bottleneck during its introduction to California. Overall, the number of alleles in the introduced range is half that found in the native range, despite greater sampling in the introduced range (Table 1). Similarly, the average expected heterozygosity in California has decreased by 68% relative to that found in the native range (Table 1).
Table 1.
The expected heterozygosity (Hexp) and number of alleles at seven microsatellite loci for native (Argentina) and introduced (California) populations
| Locus | Argentina (n = 255)
|
California (n = 460)
|
||
|---|---|---|---|---|
| Hexp | No. of alleles | Hexp | No. of alleles | |
| Lhum-11/11B | 0.851 | 15 | 0.726 | 11 |
| Lhum-14 | 0.430 | 3 | 0.058 | 3 |
| Lhum-33 | 0.611 | 9 | 0.093 | 3 |
| Lihu C1.1 | 0.654 | 5 | 0.121 | 2 |
| Lihu M1 | 0.642 | 8 | 0.073 | 3 |
| Lihu S3 | 0.465 | 6 | 0.160 | 3 |
| Lihu T1 | 0.821 | 13 | 0.199 | 5 |
| Mean (SE) | 0.639 (0.060) | 59 | 0.204 (0.089) | 30 |
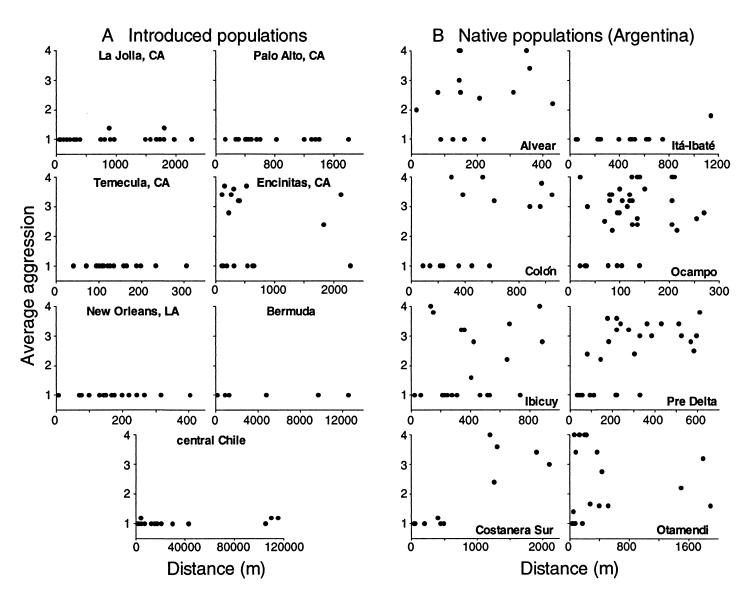
Within sites, patterns of intraspecific aggression differed between the native and introduced ranges. Intraspecific aggression was typically absent in introduced populations (Fig. 2A). In contrast, pronounced intraspecific aggression was common throughout the native range (Fig. 2B). These data illustrate the striking disparity in behavior between ants in the two geographic ranges.
Figure 2.
Relationship between degree of intraspecific aggression and distance between nest pairs at each of 15 sites within native (A) and introduced (B) ranges of the Argentine ant. Native populations typically exhibited pronounced intraspecific aggression over short distances. In contrast, intraspecific aggression was rare in introduced populations. Aggression between nests was measured by using a behavioral assay ranging from 1 (no aggression) to 4 (high aggression).
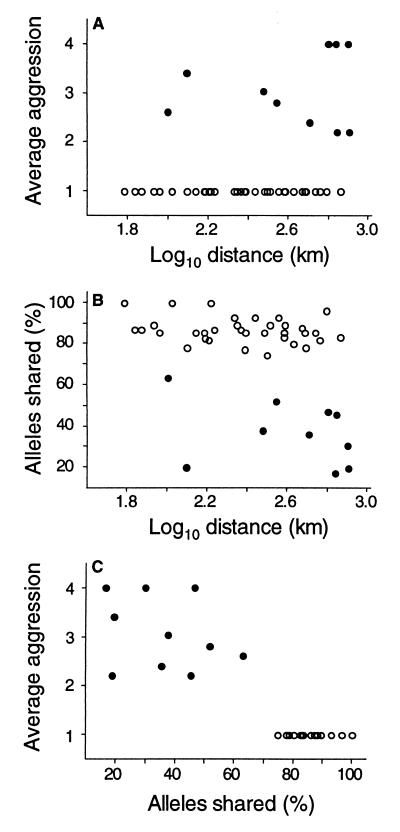
At large spatial scales, behavioral and genetic characteristics were also markedly different between the two ranges. In Argentina, distant nest pairs displayed high levels of aggression and low levels of genetic similarity (Fig. 3). In contrast, equally distant nest pairs in the introduced range were genetically similar, and aggression was not observed (Fig. 3). These data mirror the behavioral differences observed between the native and introduced populations at small spatial scales (Fig. 2).
Figure 3.
Relationships among intraspecific aggression, genetic similarity, and distance at large spatial scales (60–800 km) in native (Argentina, ●) and introduced (California, ○) populations. Aggression between nests was measured by using a behavioral assay ranging from 1 (no aggression) to 4 (high aggression). (A) Relationship of intraspecific aggression and distance between nests. Aggression scores did not overlap between native and introduced populations. All nest pairs in Argentina displayed high levels of aggression, whereas nest pairs in California did not. (B) Genetic similarity between nests versus distance. Nest pairs in California shared at least 75% of alleles. In contrast, 17–63% of alleles were shared between nest pairs in Argentina. (C) Relationship between aggression and genetic similarity of nests. In California, aggression was absent and nests were genetically similar, whereas in the native range the opposite pattern held.
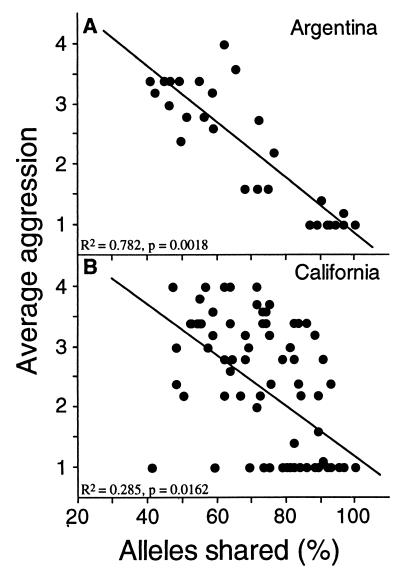
Once intercolony aggression was detected in the introduced range (e.g., Fig. 2, Encinitas), intensive sampling was initiated to locate other supercolonies and identify their boundaries. The results of these pairings are shown in Fig. 4. Despite the genetic and behavioral differences observed between native and introduced populations, the quantitative relationship between degree of intraspecific aggression and genetic similarity appears similar in both ranges. Specifically, when intraspecific aggression is detected in the introduced range, the degree of aggression among nests decreases with increasing genetic similarity, as in native populations (Fig. 4). In addition, the slopes and intercepts of these regressions did not differ (Fig. 4), suggesting that the mechanism underlying nestmate recognition has been retained in introduced populations. The greater variance about the regression in Fig. 4B is largely a consequence of reduced genetic variability of introduced populations. Because the total number of alleles in introduced populations is one-half that observed in native populations, the effect of sampling error (e.g., failure to detect an allele) will be twice as large in the introduced range. This sampling error disproportionately increases the variance around the regression in introduced populations. Additionally, the variation around the regression in both ranges will be increased because microsatellites are indicators of genetic similarity and presumably are not directly involved in the process of nestmate recognition. Therefore, the increased scatter around the regression in Fig. 4B is both expected and consistent with predictions based on the loss of genetic diversity experienced by introduced populations. Despite the influence of these factors, the relationship between genetic similarity and intraspecific aggression is highly significant in both ranges (Fig. 4).
Figure 4.
Relationship of aggression between nests and their genetic similarity. Aggression between nests was measured by using a behavioral assay ranging from 1 (no aggression) to 4 (high aggression). For this analysis, extensive sampling was necessary to locate nest pairs that exhibited intraspecific aggression within California. Average aggression decreased with increasing genetic similarity in both Argentina (A; 29 nest pairs, y = 5.48 − 0.046x) and California (B; 83 nest pairs, y = 5.37 − 0.042x). The slopes and intercepts of these regressions did not differ (t tests, P > 0.5). Additionally, adding the nest pairs from the long distance comparisons (Fig. 3) increases significance.
Discussion
Our findings demonstrate that, relative to native populations of Argentine ants, introduced populations are less diverse genetically and display intraspecific aggression less frequently. However, when introduced populations do exhibit intraspecific aggression, the relationship between genetic similarity and the degree of aggression resembles that of native populations. Therefore, the absence of intraspecific aggression in introduced populations has likely resulted from a loss of genetic variability associated with a population bottleneck. Because reduced intraspecific aggression leads to the higher population densities key to the ecological dominance of Argentine ants (25, 26), our results demonstrate how a relatively simple population genetic change can have dramatic ecological and economic consequences. The ecological success of this species is surprising, given that reductions in genetic diversity are generally believed to be harmful (32, 33).
Genetically based cues can be used to identify nestmates because colony members are often related in social insects (34–37). In such systems, selection should set a threshold of genetic similarity necessary for the acceptance of nestmates. In Argentine ants, this threshold will be set relative to the high genetic diversity in native populations. Although introduced populations still possess the ability to recognize genetically different individuals (Fig. 4), a consequence of their widespread genetic similarity is that there is rarely sufficient genetic differentiation to elicit intraspecific aggression. Thus, low levels of genetic differentiation could lead to the formation of expansive supercolonies as reported here (Figs. 2 and 3) and in other introduced populations (11, 23).
Unicoloniality is a recognized problem for kin selection theory because workers appear to bestow altruism upon unrelated individuals (38, 39). However, our findings suggest that unicoloniality may arise through the widespread loss of genetic diversity and the resulting subversion of kin recognition mechanisms. Additional support for this hypothesis comes from the widespread prevalence of unicoloniality among other invasive ant species (such as Wasmannia auropunctata, Pheidole megacephala, and Monomorium pharaonis) (24, 38, 40) many of which undoubtedly experienced a genetic bottleneck during introduction. In contrast, noninvasive unicolonial species, such as mound-building Formica, may have achieved unicoloniality via an alternative pathway involving the monopolization of stable resources and long-lived nest sites (38, 41).
One well-studied invasive species is the red imported fire ant (Solenopsis invicta), in which differences in colony structure exist between native and introduced populations (42–44). In its introduced range, the more ecologically destructive multiple-queened (polygyne) form of S. invicta (45) exhibits reduced intraspecific aggression relative to the single-queened (monogyne) form (46). Introduced populations of S. invicta have also passed through a population bottleneck that has reduced genetic variation (43) and affected the sex-determining system (47). It will be of interest to determine whether the occurrence of intraspecific aggression differs between native and introduced populations of other invasive species (such as S. invicta) and, if so, to determine whether similar underlying mechanisms are involved. In addition, an examination of native populations of Argentine ants that lack intraspecific aggression over hundreds of meters (e.g., Itá-Ibaté, Fig. 2B) will provide further insights into the transition from multicoloniality to unicoloniality.
Although our results suggest that genetic factors play an important role in nestmate recognition in Argentine ants, environmental cues are likely also involved, as they are in other social insects (34, 48). When nestmates share food or nesting material, odors from these sources can be used to assess group membership (34, 48). Three lines of evidence suggest that environmental factors cannot solely explain the occurrence of intraspecific aggression in this system. First, in California, intraspecific aggression was absent across an environmentally heterogeneous 1,000-km transect. Second, in a previous study, intraspecifically aggressive nests of Argentine ants reared on identical diets and under similar lab conditions maintained high levels of aggression over a 3-month period (25). Finally, mate preference studies in the Argentine ant have shown that queens avoid inbreeding by preferentially mating with unrelated males, apparently by using genetically based cues (49).
Despite the apparent short-term success of unicolonial species such as the Argentine ant in its introduced range, there are several reasons why unicoloniality may not persist over evolutionary time. Because many unrelated queens each contribute to the worker caste in unicolonial species, relatedness among nestmates (30, 38, 50), and therefore the heritability of worker behaviors (39), can approach zero. Under these circumstances, adaptive changes in worker behavior could not evolve (39). Alternatively, genetic differentiation among spatially distant populations could lead to increased nestmate recognition and the breakdown of widespread unicoloniality. Additionally, mutations that promote nepotism should be favored and could lead to greater multicoloniality (38). Evidence that unicoloniality may be evolutionarily unstable is provided by its patchy phylogenetic distribution across ant taxa (34, 38).
Our results also suggest a possible control strategy for the Argentine ant. Given the association between genetic variability and intraspecific aggression, the introduction of new alleles into introduced populations could increase genetic differentiation to a level sufficient to trigger intraspecific aggression. An increase in intraspecific competition within introduced populations should decrease the density of Argentine ants, allowing native ant species to compete more effectively, thereby facilitating the recovery of invaded ecosystems. However, a possible risk of this approach is that increasing genetic diversity may undermine future control strategies designed to exploit the genetic homogeneity of introduced populations.
Acknowledgments
We thank B. Epstein, K. Haight, K. Howard, J. LaBonte, K. Pease, and J. Shanahan for help with aggression assays. Work in Argentina was made possible by P. Cichero, F. Menvielle, and L. Raffo from the Administracion de Parques Nacionales Argentinas, and S. Rocio at R. E. Costanera Sur. For logistical support in Argentina we also thank R. (Mima) Venguet and I. C. Quilmes. The manuscript benefited from comments by J. D. Evans, T. Price, D. C. Queller, J. W. Terborgh, and two anonymous reviewers. DNA sequencing was performed in part by the Molecular Pathology Shared Resource, University of California at San Diego Cancer Center, which is funded in part by National Cancer Institute Cancer Center Support Grant 5P0CA23100-16. This work was supported by the Canon National Parks Science Scholars Program (A.V.S.), a U.S. Department of Agriculture National Research Initiative Competitive Grants Program Postdoctoral Fellowship (D.A.H.), and the National Science Foundation (T.J.C.).
Footnotes
Data deposition: The sequences of the microsatellite loci reported in this paper have been deposited in the GenBank database (accession nos. AF173162, AF173163, AF173164, and AF254979).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100110397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100110397
References
- 1.Wilcove D S, Rothstein D, Dubow J, Phillips A, Losos E. Bioscience. 1998;48:607–615. [Google Scholar]
- 2.Pimentel D, Lach L, Zuniga R, Morrison D. Bioscience. 2000;50:53–65. [Google Scholar]
- 3.Vitousek P M, D'Antonio C M, Loope L L, Westbrooks R. Am Sci. 1996;84:468–477. [Google Scholar]
- 4.Lodge D M. Trends Ecol Evol. 1993;8:133–137. doi: 10.1016/0169-5347(93)90025-K. [DOI] [PubMed] [Google Scholar]
- 5.Kareiva P. Ecology. 1996;77:1651–1697. [Google Scholar]
- 6.Simberloff D. In: Biological Invasions: A Global Perspective. Drake J A, Mooney H A, DiCastri F, Groves R H, Kruger F J, Rejmanek M, Williamson M, editors. New York: Wiley; 1989. pp. 61–75. [Google Scholar]
- 7.Porter S D, Williams D F, Patterson R S, Fowler H G. Env Entomol. 1997;26:373–384. [Google Scholar]
- 8.Orians G H. In: Ecology of Biological Invasions of North America and Hawaii. Mooney H A, Drake J A, editors. New York: Springer; 1986. pp. 133–148. [Google Scholar]
- 9.Levine J M, D'Antonio C M. Oikos. 1999;87:15–26. [Google Scholar]
- 10.Williams D F, editor. Exotic Ants: Biology, Impact and Control of Introduced Species. Boulder, CO: Westview; 1994. [Google Scholar]
- 11.Newell W, Barber T C. USDA Bur Entomol Bull. 1913;122:1–98. [Google Scholar]
- 12.Ward P S. Hilgardia. 1987;55:1–16. [Google Scholar]
- 13.Suarez A V, Bolger D T, Case T J. Ecology. 1998;79:2041–2056. [Google Scholar]
- 14.Erickson J M. Psyche. 1971;78:257–266. [Google Scholar]
- 15.Human K, Gordon D M. Oecologia. 1996;105:405–412. doi: 10.1007/BF00328744. [DOI] [PubMed] [Google Scholar]
- 16.Holway D A. Oecologia. 1998;116:252–258. doi: 10.1007/s004420050586. [DOI] [PubMed] [Google Scholar]
- 17.Cole F R, Medeiros A C, Loope L L, Zuehlke W W. Ecology. 1992;73:1313–1322. [Google Scholar]
- 18.Bond W, Slingsby P. Ecology. 1984;65:1031–1037. [Google Scholar]
- 19.Suarez, A. V., Richmond, J. Q. & Case, T. J. (2000) Ecol. Appl.10, in press.
- 20.Suarez A V, Tsutsui N D, Holway D A, Case T J. Biol Invasions. 1999;1:43–53. [Google Scholar]
- 21.Majer J D. In: Exotic Ants: Biology, Impact and Control of Introduced Species. Williams D F, editor. Boulder, CO: Westview; 1994. pp. 163–173. [Google Scholar]
- 22.Markin G P. J Kans Entomol Soc. 1968;41:511–516. [Google Scholar]
- 23.Way M J, Cammell M E, Paiva M R, Collingwood C A. Insectes Soc. 1997;44:415–433. [Google Scholar]
- 24.Hölldobler B, Wilson E O. Naturwissenschaften. 1977;64:8–15. [Google Scholar]
- 25.Holway D A, Suarez A V, Case T J. Science. 1998;282:949–952. doi: 10.1126/science.282.5390.949. [DOI] [PubMed] [Google Scholar]
- 26.Holway D A. Ecology. 1999;80:238–251. [Google Scholar]
- 27.Lessells C M, Boag P T. Auk. 1987;104:116–121. [Google Scholar]
- 28.Nei M. Molecular Evolutionary Genetics. New York: Columbia Univ. Press; 1987. [Google Scholar]
- 29.Queller D C, Goodnight K F. Evolution. 1989;43:258–275. doi: 10.1111/j.1558-5646.1989.tb04226.x. [DOI] [PubMed] [Google Scholar]
- 30.Crozier R H, Pamilo P. Evolution of Insect Colonies: Sex Allocation and Kin Selection. New York: Oxford Univ. Press; 1996. [Google Scholar]
- 31.Pamilo P. Trends Ecol Evol. 1989;4:353–355. doi: 10.1016/0169-5347(89)90091-8. [DOI] [PubMed] [Google Scholar]
- 32.Soulé M E. Conservation Biology: The Science of Scarcity and Diversity. Sunderland, MA: Sinauer; 1986. [Google Scholar]
- 33.Saccheri I, Kuussaari M, Kankare M, Vikman P, Fortelius W, Hanski I. Nature (London) 1998;392:491–494. [Google Scholar]
- 34.Hölldobler B, Wilson E O. The Ants. Cambridge, MA: Harvard Univ. Press; 1990. [Google Scholar]
- 35.Hölldobler B, Michener C D. In: Evolution of Social Behaviour: Hypotheses and Empirical Tests. Markl H, editor. Weinheim, Germany: Verlag Chemie; 1980. pp. 35–58. [Google Scholar]
- 36.Buckle G R, Greenberg L. Anim Behav. 1981;29:802–809. [Google Scholar]
- 37.Beye M, Neumann P, Chapuisat M, Pamilo P, Moritz R F A. Behav Ecol Sociobiol. 1998;43:67–72. [Google Scholar]
- 38.Bourke A F G, Franks N R. Social Evolution in Ants. Princeton, NJ: Princeton Univ. Press; 1995. [Google Scholar]
- 39.Queller D C, Strassman J E. Bioscience. 1998;48:165–175. [Google Scholar]
- 40.Passera L. In: Exotic Ants: Biology, Impact and Control of Introduced Species. Williams D F, editor. Boulder, CO: Westview; 1994. pp. 23–43. [Google Scholar]
- 41.Chapuisat M, Keller L. Behav Ecol Sociobiol. 1999;46:405–412. [Google Scholar]
- 42.Ross K G, Keller L. Annu Rev Ecol Syst. 1995;26:631–656. [Google Scholar]
- 43.Ross K G, Vargo E L, Keller L. Proc Natl Acad Sci USA. 1996;93:3021–3025. doi: 10.1073/pnas.93.7.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tschinkel W R. Bioscience. 1998;48:593–605. [Google Scholar]
- 45.Porter S D, Savignano D S. Ecology. 1990;71:2095–2106. [Google Scholar]
- 46.Vander Meer R K, Obin M S, Morel L. In: Applied Myrmecology: A World Perspective. Vander Meer R K, Jaffe K, Cedeno A, editors. Boulder, CO: Westview; 1990. pp. 322–328. [Google Scholar]
- 47.Ross K G, Vargo E L, Keller L, Trager J C. Genetics. 1993;135:843–854. doi: 10.1093/genetics/135.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carlin N F. Neth J Zool. 1989;39:86–100. [Google Scholar]
- 49.Keller L, Passera L. Behav Ecol Sociobiol. 1993;33:191–199. [Google Scholar]
- 50.Keller L. Trends Ecol Evol. 1995;10:355–360. doi: 10.1016/s0169-5347(00)89133-8. [DOI] [PubMed] [Google Scholar]
- 51.Krieger M J B, Keller L. Mol Ecol. 1999;8:1075–1092. [Google Scholar]