Abstract
A recombination system has been developed for efficient chromosome engineering in Escherichia coli by using electroporated linear DNA. A defective λ prophage supplies functions that protect and recombine an electroporated linear DNA substrate in the bacterial cell. The use of recombination eliminates the requirement for standard cloning as all novel joints are engineered by chemical synthesis in vitro and the linear DNA is efficiently recombined into place in vivo. The technology and manipulations required are simple and straightforward. A temperature-dependent repressor tightly controls prophage expression, and, thus, recombination functions can be transiently supplied by shifting cultures to 42°C for 15 min. The efficient prophage recombination system does not require host RecA function and depends primarily on Exo, Beta, and Gam functions expressed from the defective λ prophage. The defective prophage can be moved to other strains and can be easily removed from any strain. Gene disruptions and modifications of both the bacterial chromosome and bacterial plasmids are possible. This system will be especially useful for the engineering of large bacterial plasmids such as those from bacterial artificial chromosome libraries.
DNA engineering is conducted routinely in Escherichia coli, not only for genetic studies in bacteria, but also for constructing DNA molecules to be used in studies of other organisms. Most cloning methods use restriction endonuclease cleavage followed by DNA joining with DNA ligase. These in vitro cleavage and joining reactions are the basis for creating DNA recombinants and for cloning of DNA segments on plasmid vectors in which additional modification and amplification can take place. In the yeast Saccharomyces cerevisiae, strategies that generate novel DNA junctions exploit homologous recombination (1). The DNA double-strand break and repair recombination pathway is very efficient in yeast. Functions in this pathway recombine transformed linear DNA with homologous DNA in the yeast cell. Moreover, this recombination is proficient with short synthetic oligonucleotides, thereby permitting recombinant DNA to be generated in vivo without using DNA restriction and ligation (2–4). However, subsequent manipulation of recombinant DNA molecules in yeast, as opposed to E. coli, is laborious, especially if it is to be used for recombinant DNA work in other organisms.
Unlike yeast, E. coli is not readily transformed by linear DNA fragments due in part to the rapid degradation of the DNA by the intracellular RecBCD exonuclease (5). Mutant recBCD strains lacking the exonuclease do not rapidly degrade linear DNA; however, such strains are extremely poor growing, are defective for recombination, and do not support efficient replication of the many plasmids used in recombinant DNA work. In certain recA+ backgrounds, the recBCD defect for recombination is suppressed, allowing linear DNA to be taken up by the cell and to be recombined (6). Other homologous recombination strategies in E. coli recBC derivatives have been applied to generate recombinants between homologies on linear and/or circular plasmids (7, 8). Still, recombinants are rare, the recombination generally uses thousands of base pairs of homology, and plasmids are poorly maintained. Murphy (9) reported the highest frequencies of recombination with linear DNA containing long homologies by transforming in the presence of the bacteriophage λ recombination functions (Exo and Beta) in a bacterial recBCD mutant background. This λ recombination system, like yeast, uses double strand break repair (10, 11). In a recBC sbcA strain, short regions of homology can function in recombination of linear DNA; however, recombination is inefficient (12). In rec+ backgrounds, gene replacement on the chromosome has been accomplished by integration and excision of episomes carrying bacterial homology (13, 14).
To obtain efficient recombination of linear donor DNA in recA+ or recA− backgrounds, an E. coli strain was made that contains a λ prophage harboring the recombination genes exo, bet, and gam under control of a temperature-sensitive λ cI-repressor (Fig. 1). The exo, bet, and gam genes can be easily switched on at 42°C and off at 32°C. When λ functions are turned on for as short a time as 5 min, cells become more recombinogenic and take up linear DNA without its destruction. Gam inhibits the RecBCD nuclease from attacking linear DNA, and Exo and Beta generate recombination activity for that linear DNA. More importantly, this recombination is proficient with DNA homologies as short as 30–50 bp on the ends of linear DNA substrates.
Figure 1.
Description of the defective λ prophage on the E. coli chromosome. The defective prophage contains λ genes from cI to int. A deletion (dotted line) removes the right side of the prophage from cro through attR and including bioA (21). On the chromosome, the nadA and gal operons are to the left of the prophage, and the bio genes without bioA are to the right. Genes of the λ prophage are shown on the solid line, and genes of the host are shown on the broken line. pL and pR indicate the early left and right promoters of λ. attL and attR indicate the left and right attachment sites of λ. The λ genes and functions have been described (17).
Materials and Methods
Bacterial Strains.
Bacterial strains used in this work are listed in Table 1. Strain DY329 was constructed by transduction of ZH1141 with P1 phage grown on WJW23 (see Table 1), selecting for nadA∷Tn10 tetracycline resistance (TetR) at 32°C and then screening for the presence of a defective λ prophage, which causes temperature-sensitive cell growth at 42°C. Similar P1 transduction was used to create other strains described in Table 1 using standard media, methods, and selections (15).
Table 1.
Bacterial strains used in this work
| Strains | Genotype |
|---|---|
| WJW23 | his ilv rpsL ΔlacU169 nadA∷Tn10 gal490 λcI857 Δ(cro-bioA) |
| ZH1141 | W3110 ΔlacU169 gal490 |
| λN:lacZ Δ(N-int) cI857 Δ(cro-bioA) | |
| BR3677 | lacIq lacZ(M15) srl∷Tn10 ΔrecA |
| DY329 | W3110 ΔlacU169 nadA∷Tn10 gal490 λcI857 Δ(cro-bioA) |
| DY330 | W3110 ΔlacU169 gal490 λcI857 Δ(cro-bioA) |
| DY331 | W3110 ΔlacU169 srl∷Tn10 ΔrecA gal490 λcI857 Δ(cro-bioA) |
| DY378 | W3110 λcI857 Δ(cro-bioA) |
| HME5 | W3110 ΔlacU169 λcI857 Δ(cro-bioA) |
Induction of λ Recombination Functions and Preparation of Electroporation-Competent Cells.
Overnight cultures grown at 32°C from isolated colonies were diluted 50-fold in LB medium and were grown at 32°C with shaking to an OD600 = 0.4–0.6. Induction was performed on a 10-ml culture in a baffled conical flask (50 ml) by placing the flask in a water bath at 42°C with shaking (200 revolutions/min) for 15 min. Immediately after the 15-min induction, the flask was swirled in an ice water slurry to cool for 10 min. An uninduced control culture was also placed into the ice slurry. The cooled 10 ml cultures were centrifuged for 8 min at 5,500 × g at 4°C. Each cell pellet was suspended in 1 ml of ice-cold sterile water, was transferred to a 1.5-ml Eppendorf tube, and was spun for 20 sec at 4°C at maximum speed in a microfuge. After washing the cell pellets as described two more times, the cells were suspended in 100 μl of ice-cold sterile water. This volume of competent cells is sufficient for two standard electroporation reactions (≈108 cells per reaction). Larger cultures can be prepared for a greater number of reactions or for storage of electrocompetent cells at −80°C with 12% glycerol present. Fresh competent cells give highest efficiencies of recombination and were used here.
Preparation of Linear DNA Cassettes.
Standard PCR conditions were used to amplify linear DNA fragments with the Expand High Fidelity PCR system of Boehringer Mannheim. The chloramphenicol-resistant (CmR) cassette cat was amplified from pPCR-Script Cam (Stratagene) with primers 5′TGTGACGGAAGATCACTTCG and 5′ACCAGCAATAGACATAAGCG. The tetracycline-resistant (TetR) cassette tet was amplified from Tn10 with primers 5′CAAGAGGGTCATTATATTTCG and 5′ACTCGACATCTTGGTTACCG. The ampicillin-resistant (ApR) cassette amp was amplified from pBluescript SK(+) (Stratagene) with primers 5′CATTCAAATATGTATCCGCTC and 5′AGAGTTGGTAGCTCTTGATC. The kanamycin-resistant cassette kan was amplified from Tn5 with primers 5′TATGGACAGCAAGCGAACCG and 5′TCAGAAGAACTCGTCAAGAAG. PCR products were purified by using Qiagen (Chatsworth, CA) PCR purification kits and were concentrated if necessary by ethanol precipitation. The amplified linear DNAs were suspended in sterile water or TE buffer (10 mM Tris⋅Cl, pH7.5/1 mM EDTA) and were quantified by spectroscopy. DNA in water was stored at −20°C. We avoided PCR product purification schemes from gels in which the DNA is subject to UV irradiation.
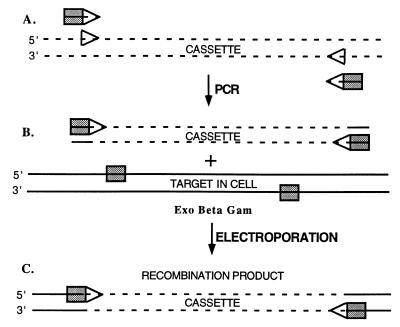
Fig. 2 illustrates the design of primers for amplification of a recombination cassette. The cassette is most often a drug marker but could be any DNA if the target sequence in the subsequent steps can be counterselected. The transcription of the marker cassette has been oriented arbitrarily in the same direction as the target region being replaced. The primers contain two parts: a 5′ end homologous to flanking regions of the target DNA, and a 3′ end that primes the cassette DNA for replication. The PCR using these primers and a DNA template containing the marker cassette generates a linear DNA product with the cassette flanked by target homology. Note that, if transformation with the template DNA will generate the selected phenotype (i.e., the template is a plasmid), the template must be eliminated. Plasmid template DNA can be destroyed by treatment with DpnI after the PCR; DpnI cuts methylated GATC template DNA, leaving the newly replicated unmethylated DNA intact. Once a linear cassette has been generated, it can be stored and used as the template for all subsequent PCRs.
Figure 2.
Strategy for generating recombinant DNA molecules and gene replacement. Three steps are outlined. (A) Recombinant oligonucleotides were chemically synthesized with the 5′ 30–50 nt (shaded rectangles) identical to sequences at the target and the 3′ 20 nt (arrowheads) homologous to the ends of the cassette to be introduced. A cassette is generated by PCR that is flanked by the 30- to 50-bp homologies present at the target. (B) Cells carrying the target DNA either on the chromosome or on a plasmid are induced for Exo, Beta, and Gam function. These cells are made competent for electroporation and mixed with the amplified cassette. (C) After electroporation, recombination occurs between the homologous sequences on the linear cassette and the target replacing the target segment with the cassette. The 50-nt galK homology segments (rectangles) used for the experiment described in Table 2 are 5′GTTTGCGCGCAGTCAGCGATATCCATTTTCGCGAATCCGGAGTGTAAGAA and 5′'TTCATATTGTTCAGCGACAGCTTGCTGTACGGCAGGCACCAGCTCTTCCG.
Cell Transformation.
Purified linear donor DNA (1–10 μl) was mixed with competent cells in a final volume of 50 μl on ice and then was pipetted into a precooled electroporation cuvette (0.1 cm). The amount of donor DNA used per reaction (usually 1–100 ng) is indicated for relevant experiments. Electroporation was performed by using a Bio-Rad Gene Pulser set at 1.8 kV, 25 μF with Pulse controller of 200 ohms. Two protocols have been used interchangeably to allow segregation of recombinant from parental chromosomes within the electroporated cells. In both protocols, the electroporated cells were immediately diluted with 1 ml of LB medium. In one, the cells were incubated for 1–2 h at 32°C before selecting for recombinants. In the other, the cells were immediately diluted and spread on sterile nitrocellulose filters (100 mm) on LB agar. After a 2-h incubation at 32°C, the filters were transferred to the appropriate agar plates required to select for recombinants. Aliquots were also directly spread on LB agar and were incubated at 32°C to determine total viable cells after electroporation. For drug resistant selection, each ml of LB medium contained 10 μg of chloramphenicol, 12.5 μg of tetracycline, 20 μg of kanamycin, or 15–30 μg of ampicillin.
Confirmation of Recombinants.
Although recombinants were verified by more than one method, the primary detection was for an altered phenotype caused by the modified target gene. Disruption or mutation of the galK gene was confirmed by the presence of white colonies on MacConkey galactose indicator agar, disruption of the rnc gene for the endoribonuclease RNaseIII was confirmed by the inability of lambdoid type phage to lysogenize (16), and deletion of gam, kil, and cIII in the pL operon was scored as an ability of the λ lysogen to survive growth at 42°C (17, 18). PCR analysis was used to confirm the altered structure caused by replacement of a gene (see text). Southern hybridization analyses of parental and recombinant DNAs confirmed structural changes, and, in critical experiments, DNA from the recombinant clones was amplified by PCR and sequenced.
Removing the Defective Prophage by Homologous Recombination.
Oligonucleotides 5′GAGGTACCAGCGCGGTTTGATC and 5′CTCCGGTCTTAATCGACAGCAAC were made and used as primers for PCR amplification of DNA from the attB bio region present in nonlysogens. The linear PCR product (≈3 kbp) was used to electroporate cells made competent for recombination. Recombinants were selected as bio+ on minimal media at 42°C. These recombinants have lost the defective prophage and replaced it with the wild-type attB bio region.
Results
The λ Recombination System.
Phage λ has a well characterized homologous recombination system. Double strand breaks in DNA are the initiation sites for this recombination (11). λ Exonuclease (Exo) degrades processively from the 5′ ends of these break sites (19), and λ Beta binds to the remaining 3′ single strand tail, protecting and preparing the recessed DNA for homologous strand invasion (20). Murphy (9) showed that the λ recombination system is very efficient at gene replacement using linear substrates with homologies of more than 1,000 bp. To test homologies less than 100 bp long as substrates for λ-mediated recombination, we modified a λ prophage to express high levels of phage recombination functions for a defined amount of time. In this prophage, the pL operon is intact and expressed under control of the temperature-sensitive λ cI-repressor (allele cI857). A deletion removes the right arm of the prophage from cro through the right attachment site (attR), and extends into the bacterial bioA gene (21). The absence of Cro-repressor allows pL operon expression to be fully derepressed when the cI-repressor is inactivated at 42°C. The cro to bioA deletion removes the lytic genes of the prophage. The functions encoded by these lytic genes are toxic to the cell and cause cell death within 7 min after a normal prophage induction (22). Functions present in the pL operon are also toxic but kill cells only after 60 min of continuous induction (18, 23). Thus, shifting cultures of cells containing the pL operon construct from repressed conditions at 32°C to induced conditions at 42°C allows pL operon expression; shifting the cells back from 42°C to 32°C within 60 min reestablishes repression and prevents cell death. We tested the recombination activity of these pL operon-induced cells when presented with a short linear DNA substrate that was homologous to a chromosome segment.
Homologous Recombination with Short Linear DNA Fragments.
Using our recombination system, we have created a single base pair change in the bacterial galK gene, which encodes a galactokinase that phosphorylates galactose and its derivatives. Nonmetabolized sugar-phosphate is inhibitory to cells, and feeding the nonmetabolite 2-deoxygalactose to galK+ cells causes bacteriostasis. Mutants defective in galK proliferate in the presence of 2-deoxygalactose (24). We synthesized and annealed complementary 70 base oligonucleotides that were homologous to an internal coding segment of the bacterial galK gene, except that a UAU codon (TYR-145) was changed to a UAG amber codon. This 70-bp mutant fragment was transferred by electroporation into galK+ cells (HME5) that had been induced for λ pL operon expression by growth at 42°C for 15 min. After electroporation with the mutant DNA (100 ng) or mock electroporation without DNA, the cells were spread on minimal 0.4% glycerol agar medium with 0.2% 2-deoxygalactose present. Spontaneous resistant mutants occurred frequently (10−4) in the absence of mutant DNA. Despite this, the addition of the mutant DNA enhanced the frequency of resistant mutants dramatically, generating one mutant per 500 electroporated cells. To determine that temperature induction was required, another batch of cells that had not been induced for recombination function was tested in the same way. In this treatment, no discernable effect of added mutant DNA was observed. This indicated that both induction of pL operon expression and mutant DNA addition were required for the enhanced survival. We assume that the expressed λ functions allowed for efficient recombination of this short linear mutant DNA with the chromosomal galK gene.
Colonies surviving 2-deoxygalactose treatment were screened for their Gal phenotype on indicator plates; all tested had the Gal− phenotype expected for a galK amber mutant. To test specifically whether the galK amber mutation was present, four independent Gal− colonies were tested by transducing cultures of each with a λimm21 phage that carries the tRNAtyr suppressor allele supF. The four mutants tested were suppressed to a Gal+ phenotype. Finally, we verified the presence of the amber mutation in galK by PCR amplification and sequence analysis of the galK gene segment from the chromosome.
The frequency of recombinants in this experiment, about 1 in 500, indicates that nonselective screens could be used to identify recombinants. DNA hybridization probes could be designed to detect point mutations or other modifications recombined into native DNA.
Gene Replacement by Targeted Homologous Recombination.
Having demonstrated that 70-bp linear DNA can direct mutations to a specific target, we tested 50-bp galK DNA segments flanking the cat (CmR) cassette for targeting a galK gene replacement by cat. The linear cat cassette with flanking galK DNA was made by PCR using chemically synthesized primers as described in Materials and Methods (Fig. 2). The cat cassette was transferred by electroporation into galK+ cells either induced (15 min) or not induced for pL operon expression. After electroporation, CmR recombinants were selected at 32°C. CmR colonies were only found in the induced culture (Table 2). All 50 CmR colonies tested had a Gal− phenotype on MacConkey galactose indicator agar, indicating the presence of the galK<>cat replacement. Note that we are using the symbol <> to indicate a replacement generated by homologous recombination techniques. In similar experiments using the same 50-bp homologous arms, we have exchanged galK for kan, amp, and tet cassettes by selecting for KmR, ApR, and TetR, creating galK<>kan, galK<>amp, and galK<>tet replacements, respectively (data not shown).
Table 2.
Targeting genes for replacement by the cat cassette
| Target Site* | 42°C, min | CmR Recombinants† |
|---|---|---|
| galK | 0 | <1 |
| galK | 15 | 2.5 × 104 |
| cIII kil gam | 0 | <1 |
| cIII kil gam | 15 | 5.0 × 104 |
DY330 competent cells were electroporated with 100 ng of the cat cassette targeted to replace either galK (galK<>cat) or prophage genes cIII kil gam (cIII kil gam<>cat).
† Total recombinants per electroporation are shown.
To test whether this approach also works at other positions on the bacterial chromosome, we created a linear cat cassette flanked by 50-bp DNA segments found immediately upstream and downstream of the rnc gene encoding RNaseIII. The rnc gene is thought to be non-essential (25); therefore, we tested whether an exact substitution of the cat coding region for the rnc coding region (from AUG to codon 224) could be made using our recombination technique. In this construct, cat is transcribed from the rnc promoter, and the 5′ primer used to generate cat started at the cat initiation codon. Following procedures used for galK, CmR colonies were found, but again only in the induced culture. The CmR colonies tested had a Rnc mutant phenotype (see Materials and Methods). Two other rnc<>cat recombinants were made. One replaced sequence from the AUG start to codon 126 of rnc, and the other from the AUG start to codon 192 of rnc. These two recombinants generate cat∷rnc gene fusions with an Rnc mutant phenotype. We chose different sets of primers to detect unambiguously the wild-type and/or recombinant alleles. This PCR procedure follows guidelines set forth by yeast researchers in characterizing chromosomal replacements in yeast (26). The PCR analysis (data not shown) of the recombinants verified the loss of the rnc+ gene and the predicted structures of the three rnc<>cat gene replacements.
The Effect of Induction Time on Targeting Efficiency.
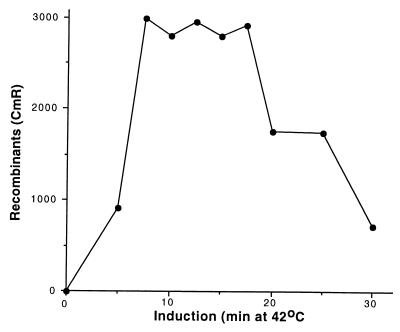
We found while crossing mutations into the pL operon (27) (H. R. Wilson, J.-G. Zhou, and D.L.C., unpublished work) that induction of pL operon expression for only 5 min enhanced recombination activity to the cell. The results shown in Fig. 3 reveal that, although a 5-min induction may be limiting for recombination, by 7.5 min, a maximum efficiency is reached. This maximum level is maintained for induction times from 7.5 to 17.5 min, with some reduction occurring for times longer than 17.5 min. Expression of the pL operon for periods longer than 60 min causes cell death (23).
Figure 3.
Effect of induction time on recombination. The strain DY330 was grown at 32°C to OD600 = 0.4–0.6, was induced at 42°C for the times indicated, and then was made electrocompetent. A linear cat cassette (10 ng) was used to target the cIII kil gam genes of the prophage. Total CmR recombinants were plotted vs. the time of induction.
The Effect of the Concentration of Linear DNA on Targeting Efficiency.
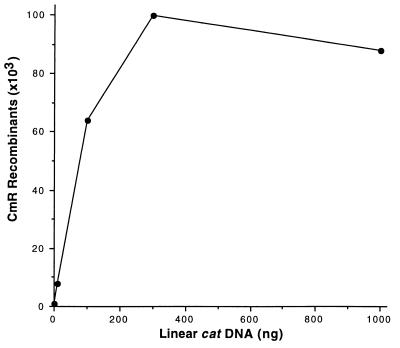
We determined the concentration at which linear DNA becomes saturating for generating recombinants. Targeting efficiency increased in a near linear relationship with increasing concentration of donor DNA in the range from 109 (1 ng) to 1011 (100 ng) molecules per electroporation (Fig. 4). A saturating level of linear DNA is reached at 3 × 1011 molecules, yielding 7.5 × 104 recombinants per ≈2 × 108 cells electroporated.
Figure 4.
Effect of concentration of the linear DNA cassette on recombination. The strain DY330 was grown at 32°C to OD600 = 0.4–0.6, was induced at 42°C for 15 min, and then was made electrocompetent. Different amounts (1, 10, 100, 300, and 1,000 ng) of a linear cat cassette (1 kbp in length) were used to target the cIII kil gam genes of the prophage. Total CmR recombinants were plotted vs. the DNA amount at 42°C.
The Effect of Homology Length on Targeting Efficiency.
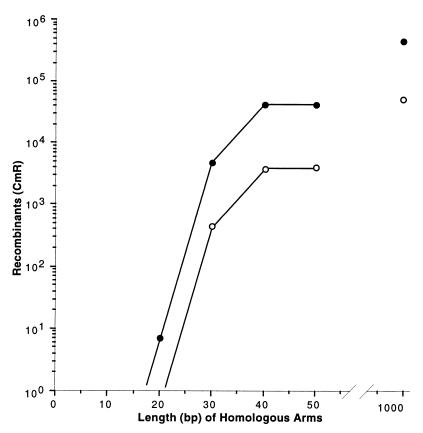
We made several pairs of primers to amplify the cat cassette for targeting the chromosome and designed each pair with a different length of flanking homology. The length of the homology segment on the primers varied by increments of 10 bases from 10 to 50 bases. A nested set of linear cat cassettes was made with the primers. We also constructed another linear cat cassette flanked by 1,000 bp of homology (see Fig. 5). This set of linear DNAs was then tested for recombinational targeting efficiency, as shown in Fig. 5. No recombinants were found with 10 bp of homology, and less than ten recombinants were found in each of three experiments with 20 bp of homology. From 20 bp to 40 bp of homology, homologous recombination increased by four orders of magnitude. From 40 bp to 1,000 bp of homology, recombination increased only 10-fold.
Figure 5.
Effect of homologous arm length on recombination. The strains DY330 (filled circles) and DY331 (open circles) were grown at 32°C, were induced at 42°C for 15 min, and then were made relectrocompetent. A linear cat cassette (100 ng) was used to target the cIII kil gam genes of the prophage. The homologous arm length of the cassette was varied from 0 to 1,000 bp. The primers containing the 0- to 50-bp homologies were chemically synthesized as described (Fig. 2). The cassette containing 1,000-bp homologous arms was made by PCR using primers 1,000 bp away on each side of an existing (cIII kil gam)<>cat disruption in the cell. Total CmR recombinants were plotted vs. the homologous arm length.
Gene Replacement on Plasmids: In Vivo Cloning.
We wanted to determine whether this method could be used to modify plasmid DNA. Following the same procedures described for the chromosome (see Fig. 2), we modified plasmid pGB2, a derivative of pSC101. A cat cassette was synthesized in vitro and recombined in vivo with pGB2 to replace the spectinomycin resistance gene with cat conferring CmR on the cell carrying the recombinant plasmid.
The same experiment, performed on pBR322 derivatives, generated recombinants, but they were joined in tandem to nonrecombinant plasmids as dimers and higher multimers. Induction of Gam expression from our prophage inactivates RecBCD nuclease. In the absence of RecBCD, pBR322 derivatives replicate by a rolling circle mode (28), and the plasmid converts from monomers to multimers. This is specific for pBR322 type replicons, as the pGB2 type did not form multimers (data not shown). To generate simple recombinants of pBR322 derivatives, we modified our protocol by coelectroporating the recA− strain DY331 with circular plasmid DNA (0.1 ng) and a linear drug cassette. Recombinant plasmid monomers were readily selected and isolated (data not shown).
The Requirement of RecA for Targeted Recombination.
The λ recombination system can function in cells lacking the bacterial RecA function (29). However, in replication-blocked studies (30) or in another linear DNA recombination study (9), the recombination in recA mutants is reduced more than 50-fold relative to levels in recA+ cells. We tested the requirement for RecA in targeted recombination by repeating the experiment described in Fig. 5 but using a recA− strain. Recombination efficiency was depressed about 10-fold in the recA mutant for the arm lengths tested (Fig. 5). Thus, only a moderate requirement for RecA is seen.
Determination of the λ Genes Required for Targeted Recombination in Wild-Type E. coli.
To determine which λ genes are required for targeted recombination, we generated a set of replacement deletions in the pL operon of the prophage using cat and amp cassettes. Each of these newly made deletions was verified structurally by PCR analysis and was tested for targeted recombination of a tet gene cassette into galK. Table 3 shows that only exo, bet, and gam deletions affected galK<>tet targeted recombination. Deletion of any one of these three genes eliminated the recombination, whereas deletion of all other genes in the operon had little if any effect. To show that the gam<>cat substitution was not polar on bet and exo, the gam gene was expressed in trans and shown to complement the defect (data not shown). Thus, although we used the entire pL operon in studies described here, three gene functions provide most of the activity.
Table 3.
Genetic requirements on λ
| Strain* | Prophage† | Recombinant‡ |
|---|---|---|
| DY330 | wild-type | 4,100 |
| DY392 | (hin-int)<>amp | 2,000 |
| DY351 | (sieB-kil)<>cat | 4,400 |
| DY386 | (hin-int)<>amp | 1,650 |
| (sieB-kil)<>cat | ||
| DY349 | (gam)<>cat | 0 |
| DY360 | (bet)<>cat | 0 |
| DY359 | (exo)<>cat | 0 |
Competent cells were induced for 15 min and electroporated with 10 ng of linear galK<>tet.
Parenthesis indicate deleted prophage genes (Fig. 1).
Total recombinants per electroporation are shown.
Discussion
We have characterized homologous recombination between linear DNA and the bacterial chromosome that depends on λ recombination functions, involves very short homologies, and is very efficient. We examined several parameters to establish a maximal efficiency for phage-mediated recombination with short homologies. Maximal recombination levels are achieved with induction times from 7.5–17.5 min at 42°C, and a homology segment of 40–50 bp. Recombination saturates at a linear DNA substrate concentration of about 300 molecules per cell.
The fact that 30- to 50-bp homologies are able to recombine in vivo opens a vast array of new possibilities for generating recombinant DNA. Several steps normally involved in generating recombinant DNA molecules are eliminated. Restriction enzyme digests are not required to generate DNA fragments, and DNA ligase reactions are not required to join different DNA fragments at novel junctions. PCR amplification followed by electroporation of the linear DNA into cells is required, but the cell generates the completed recombinant precisely joined through homologous recombination.
By using optimal conditions, the efficiency of recombination approaches 0.1% of surviving cells from a standard electroporation. At this efficiency, unselected colonies could be screened for recombinant DNA using colony hybridization techniques, eliminating the need for selection steps. Thus, this recombination protocol makes the bacterial chromosome and plasmid DNA amenable to almost any type of desired change. This includes directed mutagenesis of a gene, a gene segment, or even a base.
By using homologous recombination, several different genes on the bacterial chromosome and on plasmids have been substituted with drug resistance markers. However, it is also possible to create recombinants in which the final product does not include a selectable marker. In experiments not described here (E.-C. Lee and D.Y., unpublished work), genes have been fused to cassettes encoding specific tags such as the green fluorescent protein. Fusion tags can be placed precisely in the gene to be modified by either of two strategies. In one, the unselected cassette is joined to a selectable drug marker, and both are recombined into the chosen location selecting for drug resistance. In the other, the cassette is recombined into its location by substituting it for a counterselectable marker like sacB (13). The latter strategy requires recombining a counterselected marker like sacB along with a drug marker into position first. This strategy should permit cloning of any DNA into a plasmid or the chromosome.
In general our analyses of recombinant products show that the expected genetic and structural changes caused by the recombination occurred. However, some recombination products were unexpected. In two cases, we attempted to knock out essential genes, and surprisingly were able to select a few rare recombinants. These turned out to be diploid for the region of the targeted gene because they carried the wild-type and the mutant copy of the gene as determined by PCR analysis (data not shown). Rare diploid regions of the bacterial chromosome are known to occur spontaneously in growing cells at a frequency of about 0.1% (31). Because we targeted an essential gene, these rare diploids were selected. This was only possible because the recombination is so efficient.
Restriction endonuclease strategies of genetic engineering impose practical limitations, especially when one wants to manipulate segments on large DNAs like the bacterial chromosome, F plasmid derived bacterial artificial chromosomes (BACs), or P1 plasmid derived PACs. Homologous recombination presents an alternative and much more precise way to engineer DNA that does not rely on restriction site location or number. BAC plasmids are used to clone genomic DNA (>100 kbp) from human and other species (32). The ability to modify or to engineer these huge plasmids is highly desirable. BAC clones have been efficiently modified by using the induced λ prophage system described here to provide recombination potential in derivatives of the DH10B recA mutant (E.-C. Lee and D.Y., unpublished data). λ recombination functions expressed from a plasmid have been used previously to modify a BAC (33). However, gene modification using our prophage system is orders of magnitude more efficient, supporting a rationale for using prophage induction to generate recombination potential. In this respect, BAC clones containing eukaryotic DNA often include repetitive sequences making them vulnerable to rearrangement. The ability to induce efficient recombination activity in recA− hosts while maintaining tight control on expression of that activity is therefore highly desirable.
Our findings may provide fresh insights into how Exo and Beta work. We demonstrated that Exo and Beta can use very short homologies on the ends of linear DNA. We expect that Exo first resects each of the 5′ ends, allowing Beta to bind to the 3′ exposed single strands. Studies have demonstrated that Beta binds stably to DNA strands 36 bases long but not to shorter sequences (34). Coincidentally, recombination in our system drops exponentially when homology is reduced below 40 bp (Fig. 5). Exo presumably resects beyond the short homology segments to provide a full binding site for Beta even if homology is limiting. The length of the homologous strand may be important to find and pair effectively to its complement. If, during the recombination event, it is important for Beta to be removed from the homologous strand, then strong pairing of strands may also be important in Beta release from the 3′ end. If homology is too short (<40 nt) and pairing is not efficient, then Beta may remain bound causing inefficient recombination. A similar requirement for 35–50 bp of homology has been demonstrated for the λ recombination system during intron homing (35).
Expression of Exo and Beta functions from the chromosome as opposed to a plasmid has been shown to increase the efficiency of linear transformation (9). Similarly, our defective prophage construct offers significant advantages over the use of plasmid expressed recombination functions. When pL (or pR) is present on a plasmid, cI repression of pL depends on high concentrations of repressor. High repressor concentration makes it more difficult to turn on and off pL operon expression (36), as is true of other negatively and positively controlled promoter expression systems cloned on plasmids (37). In contrast, the prophage is present in one copy, and its expression is tightly controlled by the autoregulated, temperature sensitive cI-repressor. Despite being in one copy, expression from the pL promoter is strong enough to generate very efficient recombination without loss of cell viability.
Expressing the λ recombination functions from plasmids creates other dilemmas not presented by the prophage. Drug resistance is used to maintain plasmids in the cell. For experiments using gene replacement techniques, drug resistant markers often become a limiting factor. A plasmid system would also preclude using that particular plasmid type for recombinational cloning studies because of incompatibility. Additionally, ColE1-derived plasmids concatemerize in RecBCD-deficient conditions, causing plasmid instability and a loss of cell viability (28). The prophage avoids problems inherent in a plasmid-based system and serves as an excellent vehicle for providing recombination functions to the cell. The prophage can be easily moved to other strains by P1 co-transduction with a linked TetR marker, and can be removed from a strain by prophage-targeted homologous recombination (see Materials and Methods). We are attempting to adapt the complete system to other bacterial species.
Acknowledgments
We thank T. Baker for excellent technical assistance in generating gene “knockouts” and D. Friedman, N. P. Higgins, C. McGill, J. Sawitzke, and H. Wilson for providing suggestions and comments.
Abbreviation
- BAC
bacterial artificial chromosome
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100127597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100127597
References
- 1.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 2.Yamamoto T, Moerschell R P, Wakem L P, Ferguson D, Sherman F. Yeast. 1992;8:935–948. doi: 10.1002/yea.320081104. [DOI] [PubMed] [Google Scholar]
- 3.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lafontaine D, Tollervey D. Nucleic Acids Res. 1996;24:3469–3471. doi: 10.1093/nar/24.17.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benzinger R, Enquist L W, Skalka A. J Virol. 1975;15:861–871. doi: 10.1128/jvi.15.4.861-871.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jasin M, Schimmel P. J Bacteriol. 1984;159:783–786. doi: 10.1128/jb.159.2.783-786.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliner J D, Kinzler K W, Vogelstein B. Nucleic Acids Res. 1993;21:5192–5197. doi: 10.1093/nar/21.22.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi N K, Kusano K, Yokochi T, Kitamura Y, Yoshikura H, Kobayashi I. J Bacteriol. 1993;175:5176–5185. doi: 10.1128/jb.175.16.5176-5185.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy K C. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi N, Kobayashi I. Proc Natl Acad Sci USA. 1990;87:2790–2794. doi: 10.1073/pnas.87.7.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thaler D S, Stahl M M, Stahl F W. J Mol Biol. 1987;195:75–87. doi: 10.1016/0022-2836(87)90328-7. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Buchholz F, Muyrers J P, Stewart A F. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 13.Blomfield I C, Vaughn V, Rest R F, Eisenstein B I. Mol Microbiol. 1991;5:1447–1457. doi: 10.1111/j.1365-2958.1991.tb00791.x. [DOI] [PubMed] [Google Scholar]
- 14.Parkinson J S, Houts S E. J Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 16.Court D. In: Control of Messenger RNA Stability. Belasco J, Brawerman G, editors. New York: Academic; 1993. pp. 71–116. [Google Scholar]
- 17.Court D, Oppenheim A B. In: Lambda II. Hendrix R W, Roberts J W, Stahl F W, Weisberg R A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1983. pp. 251–277. [Google Scholar]
- 18.Greer H. Virology. 1975;66:589–604. doi: 10.1016/0042-6822(75)90231-7. [DOI] [PubMed] [Google Scholar]
- 19.Little J W. J Biol Chem. 1967;242:679–686. [PubMed] [Google Scholar]
- 20.Carter D M, Radding C M. J Biol Chem. 1971;246:2502–2512. [PubMed] [Google Scholar]
- 21.Patterson T A, Costantino N, Dasgupta S, Court D L. Gene. 1993;132:83–87. doi: 10.1016/0378-1119(93)90517-7. [DOI] [PubMed] [Google Scholar]
- 22.Weisberg R A, Gallant J A. J Mol Biol. 1967;25:537–544. doi: 10.1016/0022-2836(67)90204-5. [DOI] [PubMed] [Google Scholar]
- 23.Kourilsky P, Knapp A. Biochimie. 1974;56:1517–1523. [PubMed] [Google Scholar]
- 24.Adhya S. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Vol. 2. Washington, DC: Am. Soc. Microbiol.; 1987. pp. 1503–1512. [Google Scholar]
- 25.Takiff H E, Chen S M, Court D L. J Bacteriol. 1989;171:2581–2590. doi: 10.1128/jb.171.5.2581-2590.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winzeler E A, Shoemaker D D, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke J D, Bussey H, et al. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 27.Wilson H R, Kameyama L, Zhou J G, Guarneros G, Court D L. Genes Dev. 1997;11:2204–2213. doi: 10.1101/gad.11.17.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feiss M, Siegele D A, Rudolph C F, Frackman S. Gene. 1982;17:123–130. doi: 10.1016/0378-1119(82)90064-6. [DOI] [PubMed] [Google Scholar]
- 29.Brooks K, Clark A J. J Virol. 1967;1:283–293. doi: 10.1128/jvi.1.2.283-293.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stahl F W, McMilin K D, Stahl M M, Crasemann J M, Lam S. Genetics. 1974;77:395–408. doi: 10.1093/genetics/77.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haack K R, Roth J R. Genetics. 1995;14:1245–1252. doi: 10.1093/genetics/141.4.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shizuya H, Birren B, Kim U J, Mancino V, Slepak T, Tachiiri Y, Simon M. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muyrers J P, Zhang Y, Testa G, Stewart A F. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mythili E, Kumar K A, Muniyappa K. Gene. 1996;182:81–87. doi: 10.1016/s0378-1119(96)00518-5. [DOI] [PubMed] [Google Scholar]
- 35.Parker M M, Court D A, Preiter K, Belfort M. Genetics. 1996;143:1057–1068. doi: 10.1093/genetics/143.3.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lieb M. J Virol. 1979;32:162–166. doi: 10.1128/jvi.32.1.162-166.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hasan N, Szybalski W. Gene. 1987;56:145–151. doi: 10.1016/0378-1119(87)90167-3. [DOI] [PubMed] [Google Scholar]