Abstract
Illegitimate recombination is a major cause of genetic instability in prokaryotes as well as in eukaryotes. This recombination usually occurs at a low frequency, but it is greatly enhanced by UV irradiation or other environmental stresses. DNA damages produced by these environmental stresses are thought to induce DNA double-strand breaks, leading to illegitimate recombination. In this paper we show that UV-induced illegitimate recombination is enhanced by mutations of nucleotide excision repair genes, uvrA or uvrB, and partially by uvrC mutation, but not by uvrD mutation. Unexpectedly, the recombination was enhanced by the uvrA uvrB double mutation even without UV irradiation, but the uvrB uvrC double mutation has not shown this effect, suggesting that illegitimate recombination is mostly suppressed by UvrA and UvrB. Moreover, illegitimate recombination was synergistically enhanced by the recQ uvrA double mutation. In addition, overproduction of the UvrA protein suppressed the hyperrecombination phenotype of the recQ or uvrB mutant, but it did not affect the UV-sensitive phenotype of the uvrB mutant. We concluded that the UvrAB complex suppresses illegitimate recombination in a pathway shared with RecQ helicase. In addition, UvrA protein alone can suppress illegitimate recombination in the pathway, in which RecQ helicase and UvrAB complex work. Possible functions of the proteins involved in these pathways are also discussed.
Keywords: chromosomal aberration, genomic instability, xeroderma pigmentosum, Werner's syndrome, Bloom's syndrome
Illegitimate recombination is a major cause of chromosomal aberration, along with duplication, deletion, insertion, and translocation. Illegitimate recombination normally occurs at a low frequency, but it is greatly enhanced by treatment with UV light or other environmental stresses (1). This observation indicates that DNA lesions introduced by UV light induce illegitimate recombination. However, the mechanism by which this occurs is not yet understood.
DNA lesions introduced by UV irradiation are mainly removed by nucleotide excision repair (NER) (reviewed in ref. 2). In Escherichia coli, UvrABCD proteins accomplish NER, and mutants defective in the corresponding genes exhibit enhanced sensitivity to UV light. NER involves recognition of DNA damage by UvrA and UvrB, incision of the damaged DNA strand by UvrB and UvrC (2), removal of the damaged region by UvrD helicase, followed by repair synthesis, which fills the gap by using the intact strand as a template, and is completed by ligation of the repaired section to the undamaged DNA. The relationship between NER and illegitimate recombination has remained unclear. We therefore studied the effect of uvr mutations on illegitimate recombination by using E. coli as a model organism.
Illegitimate recombination is a class of recombination that takes place between sequences of little or no homology and results in gene rearrangements. Illegitimate recombination can be classified into two classes, short homology-independent illegitimate recombination (SHIIR) and short homology-dependent illegitimate recombination (SHDIR) (3–7). SHIIR occurs between sequences with virtually no homology and is mediated by DNA topoisomerases (5–7). SHDIR is induced by UV irradiation or other DNA damaging agents and requires short regions of homology between recombination sites. These regions usually contain 4–10 bp of homologous DNA (3, 4, 6). It is also known that RecJ exonuclease promotes SHDIR, but RecQ helicase suppresses it (4, 8).
In this study, we examined the effect of uvr mutations on illegitimate recombination (SHDIR) in E. coli and found that UvrA and UvrB proteins suppress illegitimate recombination. Moreover, illegitimate recombination was synergistically enhanced by the uvrA uvrB and recQ uvrA double mutations. Overproduction of the UvrA protein suppressed the hyperrecombination phenotype detected in the recQ or uvrB mutant. We concluded that the UvrAB complex suppresses illegitimate recombination in a pathway shared with the RecQ helicase. In addition, UvrA protein alone also suppresses illegitimate recombination in the pathway, in which RecQ helicase and UvrAB complex work.
Materials and Methods
Bacterial Strains and Plasmids.
All strains in this study are derivatives of E. coli K12 AB1157, which contains one unit of the λ cI857 prophage, except for Ymel and P2 lysogen. HI2051 is the wild type, HI2106 contained the uvrA6 mutation (9), and HI2105 contained the uvrB5 mutation (10). HI2217 contained the uvrC279∷Tn10 mutation derived from N3024. AB1886 uvrA6, AB1885 uvrB5, and N3024 were obtained from the Genetic Stock Research Center (National Institute of Genetics, Mishima, Japan). HI2219 contained the uvrA277∷Tn10 whose mutation was from N3055 that is obtained from the E. coli Stock Center (Yale University, New Haven, CT). HI2652 contained the uvrB∷Mu(d) (lacZ Apr) mutation (11). HI2215 contained the ΔuvrD∷tet mutation (12). We used two uvr double mutants, HI2653 uvrA277 uvrB∷Mu(d) and HI2654 uvrB∷Mu(d) uvrC279. All recQ derivatives had the recQ1802 mutation (13). These were HI2126 recQ, HI2108 recQ uvrA6, HI2220 recQ uvrA277, HI2107 recQ uvrB5, HI2218 recQ uvrC279, and HI2216 recQ ΔuvrD∷tet. pJA61 is a derivative of pBR322 carrying the uvrA+ gene (9), and pNP10 is a derivative of pBR322 carrying the uvrB+ gene (14).
Measurement of Frequency of λ Spi− Phage Induced Spontaneously or by UV Irradiation.
E. coli λ cI857 or its derivative was grown to 1 × 108 cells/ml at 30°C. If necessary, 2 ml of the culture was irradiated for 10–20 s at a distance of 20 cm (a dose of 8–16 J/m2) with a 4-W UV lamp whose wavelength is 253.6 nm (Manaslu-Light, Tokyo). The heat induction of λ prophage was carried out by incubation at 42°C for 15 min with aeration. The culture was then incubated at 37°C for 2 h. The titer of λ Spi− phage was measured by E. coli WL95. The titer of total phage was measured by E. coli Ymel. The frequency of λ Spi− phage was obtained by dividing the titer of λ Spi− phage by the titer of total phage. Burst size was obtained by dividing the titer of total phage by the titer of total cell.
UV Sensitivity Test of E. coli Strains on Agar Plate.
E. coli strains were streaked on Luria–Bertani agar plate, and the plate was irradiated for at a distance of 52 cm (a dose of 20–60 J/m2) with an 15-W germicidal lamp (Stanley Electric, Tokyo). The plate was then incubated at 37°C for overnight.
Determination of Nucleotide Sequences of Recombination Junction in λ Spi− Phages.
To analyze recombination junctions, independent isolation of Spi− phages was carried out as described by Yamaguchi et al. (3). Location of recombination junctions was determined by PCR by using several primers described by Ukita and Ikeda (4). A recombination junction carried by a recombinant was sequenced with an Applied Biosystems automatic DNA sequencer.
Results
Effect of the uvrA or uvrB Mutation on UV-Induced Illegitimate Recombination.
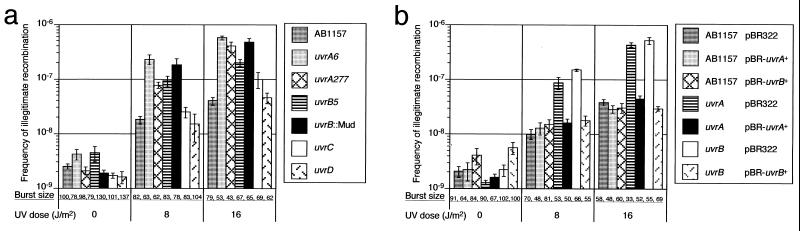
We have previously shown that UV irradiation enhances the frequency of illegitimate recombination (SHDIR) in E. coli (3). This finding indicates that this recombination is promoted by DNA damages induced by UV irradiation. To study the relationship between NER and UV irradiation-induced illegitimate recombination, we first examined the effect of uvr mutations on the recombination detected as the formation of λbio-transducing phages during prophage induction. λbio-transducing phages are formed by illegitimate recombination, lack both the red and gam genes, and can be distinguished from normal λ phage on the basis of a Spi− (sensitive to P2 interference) phenotype. These recombinant phages, which are called Spi− phages, can be positively detected as phages that grow in P2 lysogen of E. coli (1). On the other hand, normal λ phages cannot grow in P2 lysogen. Therefore, the frequency of illegitimate recombination can be calculated as a ratio of the number of Spi− phages to the number of total phages in a given lysate. By using this assay, we found that the frequency of illegitimate recombination was enhanced by the uvrA6 and uvrB5 mutations by about 5-to 15-fold at each dose of UV irradiation (Fig. 1a). The same results were also obtained for the uvrA277∷Tn10 and uvrB∷Mu(d) mutants (Fig. 1a). In contrast, the uvrC279∷Tn10 mutation only partially affected the frequency of illegitimate recombination and the uvrD∷tet mutation did not affect it (Fig. 1a). These results indicated that lesions introduced by UV light can induce illegitimate recombination, and that UvrA and UvrB proteins suppress UV irradiation-induced illegitimate recombination.
Figure 1.
(a) Effects of various uvr mutations on frequency of illegitimate recombination. We measured the frequency of formation of λ Spi− phages with or without UV irradiation. Burst size is the number of total λ phage per cell. (b) Complememtation test of the UvrA and UvrB defect by uvrA+ and uvrB+ plasmids. We measured the frequency of formation of λ Spi− phages with the strains carrying the uvrA plasmid, pJA61 (pBR322-uvrA+), the uvrB plasmid, pNP10 (pBR322-uvrB+), or control plasmid pBR322 with or without UV irradiation.
To confirm this result, plasmids expressing either UvrA (pJA61) or UvrB (pNP10) were introduced into the uvrA and uvrB mutants, and the frequency of λ Spi− phage formation was measured. Introduction of the Uvr plasmids reduced the frequency of enhanced illegitimate recombination in the uvrA and uvrB mutants under UV irradiation (Fig. 1b). These results confirmed that the UvrAB proteins play a role in suppression of UV irradiation-induced illegitimate recombination.
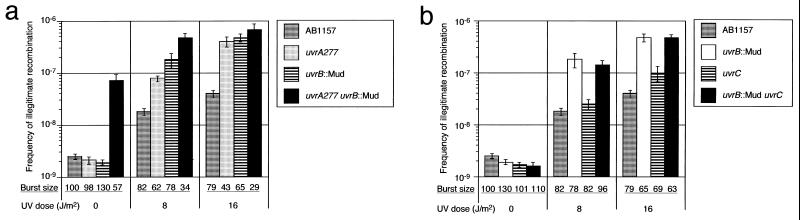
Effect of the uvrA uvrB Double Mutation on Spontaneous Illegitimate Recombination.
Next, we analyzed the effects of the uvrA uvrB and uvrB uvrC double mutations on illegitimate recombination. Unexpectedly, in the absence of UV irradiation, the uvrA uvrB double mutation increased the frequency of illegitimate recombination about 30-fold relative to the wild-type, the uvrA, or uvrB single mutant (Fig. 2a). In contrast, the effect of the uvrB uvrC double mutation without UV irradiation was comparable to that of the uvrB single mutation (Fig. 2b). Under UV irradiation, both double mutations increased the frequency of illegitimate recombination relative to the wild type, but only to the same extent as the uvrA or uvrB single mutations (Fig. 2a). These results indicate that the UvrA and UvrB proteins suppress illegitimate recombination even in the absence of UV irradiation. The double defects of UvrA and UvrB exhibited a synergistic effect, implying that UvrA and UvrB function in the same pathway but that they work at different steps.
Figure 2.
(a) Effects of uvrA uvrB double mutation on frequency of illegitimate recombination. We measured the frequency of formation of λ Spi− phages with or without UV irradiation as described in Fig. 1. (b) Effects of uvrB uvrC double mutation on frequency of illegitimate recombination.
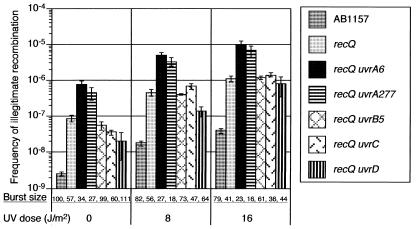
Effect of uvr recQ Double Mutations on Illegitimate Recombination.
In a previous study, we showed that RecQ helicase suppresses illegitimate recombination, with or without UV irradiation (8). To study whether UvrA and UvrB functions are involved in the same pathway as RecQ helicase functions, we examined the effects of uvr recQ double mutations on illegitimate recombination. The results indicated that only the uvrA recQ double mutations enhanced the frequency of illegitimate recombination synergistically with or without UV irradiation, but the uvrB recQ double mutation did not share this property (Fig. 3). The frequency of illegitimate recombination in the uvrA uvrB recQ triple mutation was comparable to that of the uvrA recQ double mutation (data not shown). These data indicate that at least the UvrA protein plays a role in the pathway shared by the RecQ helicase.
Figure 3.
Effect of various recQ uvr double mutations on illegitimate recombination. We measured the frequency of formation of λ Spi− phages with or without UV irradiation, as described in Fig. 1.
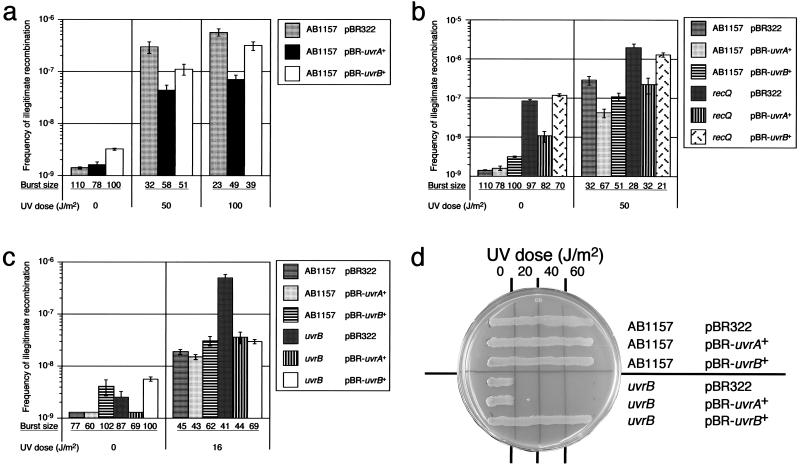
Overproduction of UvrA Suppresses the Hyperrecombination Phenotype of the recQ and uvrB Mutants.
In the previous sections, we showed that the combination between the uvrA and uvrB mutations or the combination between the uvrA and recQ mutations exhibits synergistic effects on suppression of illegitimate recombination. These results indicate a possibility that the UvrA protein alone can function as a suppressor of illegitimate recombination and works in the respective pathways in which UvrB and RecQ functions. Therefore, we examined the effect of overproduction of UvrA protein on illegitimate recombination. To examine this, plasmids expressing either UvrA (pJA61) or UvrB (pNP10) were introduced into the wild-type (AB1157), the recQ, and uvrB mutants, and the frequency of λ Spi− phage formation was measured. The frequency of illegitimate recombination in the wild type was reduced by overproduction of UvrA protein at each dose (50 Jm2 and 100 J/m2) of UV irradiation (Fig. 4a). Unexpectedly, overproduction of UvrA protein also suppressed the hyperrecombination phenotype of the recQ mutant with or without UV irradiation, but overproduction of UvrB could not share this function (Fig. 4b). In addition, overproduction of UvrA protein also suppressed the hyperrecombination phenotype of the uvrB mutant under UV irradiation (Fig. 4c). These results confirm that the UvrA protein alone can function as a suppressor of illegitimate recombination.
Figure 4.
(a) Effects of overproduction of the UvrA protein or the UvrB protein on frequency of illegitimate recombination in the wild type (AB1157). We measured the frequency of formation of λ Spi− phages with or without UV irradiation (50 J/m2 and 100 J/m2), as described in Fig. 1. (b) Effects of overproduction of the UvrA protein or the UvrB protein on the frequency of illegitimate recombination in the recQ mutant. (c) Effects of overproduction of the UvrA protein or the UvrB protein on the frequency of illegitimate recombination in the uvrB mutant. (d) UV sensitivity of the wild-type (AB1157) and the uvrB mutant carrying pBR322, pBR322-uvrA+ (pJA61), or pBR322-uvrB+ (pNP10).
To examine whether or not the effect of UvrA on illegitimate recombination is caused by activation of NER, we analyzed the UV sensitivity of the strain carrying the uvrA+ plasmid, pJA61, and found that overproduction of the UvrA protein did not suppress the UV-sensitive phenotype of the uvrB mutant (Fig. 4d). These results indicate that the UvrA protein by itself functions as a suppressor of illegitimate recombination independently of the NER pathway. Moreover, these results also indicate that the UvrA protein plays a role in the pathways shared by the UvrB protein and by the RecQ helicase, respectively.
Analysis of Recombination Junctions in UV-Induced λbio-Transducing Phages Derived from the uvrA or uvrB Mutant.
By using PCR analysis, we determined the distribution of recombination junctions of λ Spi− phages isolated independently from the wild-type or the uvr mutants following UV irradiation. Half of the transducing phages derived from HI2051 wild-type cells were formed at hotspot I, which was described by Yamaguchi et al. (3). In contrast, in the phages derived from the HI2106 uvrA or the HI2105 uvrB mutant, the frequencies at hotspot I were 74% and 63%, respectively (Table 1). Moreover, we found 11% of the transducing phages derived from the uvrB mutant resulted from recombination at hotspot III. In the previous study, we showed that hotspot II and hotspot III were formed in the recQ mutant (8). The frequency of recombination at hotspot III in the uvrB mutant was comparable to that in the recQ single mutant.
Table 1.
Distribution of recombination sites of λbio-transducing phages
| UV dose, J/m2 | Strain | Relevant mutation | Frequency of recombination (%) at:
|
|||
|---|---|---|---|---|---|---|
| Hot I | Hot II | Hot III | Non-hot | |||
| 16 | HI2051 | Wild type | 49 | 4.4 | 2.2 | 44 |
| 16 | HI2106 | uvrA6 | 71 | 4.1 | 2.0 | 23 |
| 16 | HI2105 | uvrB5 | 62 | 4.3 | 11 | 23 |
The distribution of recombination sites of recombinant phages derived from wild-type and uvrA and uvrB mutants were determined by PCR described in Materials and Methods. The junctions were classified into four classes: hotspot I (Hot I), II, III, and non-hotspot (Non-hot) sites. The nucleotide sequences of hotspot I, II, and III were described previously (refs. 3 and 8; see also Fig. 5). Numbers indicate percentages of total λbio-transducing phage tested.
Next, we determined nucleotide sequences at the recombination junctions of λ Spi− phages derived from the wild-type or uvr mutants. PCR products amplified with primers spanning the junctions were analyzed by direct sequencing. The nucleotide sequences of junctions derived from recombination hotspots I, II, and III, and non-hotspot sites showed that formation of λbio-transducing phages in the wild-type or uvr mutants requires short regions of homology between the E. coli and λ DNAs (Fig. 5) (average length of homology, 8.6 bp). These findings indicate that the UvrA and UvrB proteins suppress SHDIR.
Figure 5.
Nucleotide sequences of recombination junctions of λbio-transducing phages induced by UV. (a) Sequences of the hotspot I detected in the junctions of λbio-transducing phages WL3, AL1, and BL2, which were isolated from the wild-type, the uvrA mutant, and the uvrB mutant. The sequences shown in bold represent homology at the recombination sites. Map coordinates for phage and bacterial sequences are indicated. (b) Sequences of the hotspot II detected in the junctions of λbio-transducing phages WL28 and BL28. (c) Sequences of the hotspot III detected in the junctions of λbio-transducing phages AL40 and BL19. (d and e) Sequences of non-hotspot sites detected in the junctions of λbio-transducing phages derived from the wild type. (f and g) Sequences of non-hotspot sites derived from the uvrA mutant. (h and i) Sequences of non-hotspot sites derived from the uvrB mutant.
Discussion
SHDIR takes place spontaneously as a rare event, but it is greatly enhanced by UV irradiation in E. coli (1, 3). Lesions introduced by UV are mainly repaired by NER, but unrepaired lesions may induce illegitimate recombination. The roles of Uvr proteins in NER have been well established (2). UvrA2–UvrB1 complexes recognize DNA damage. Then, the UvrA proteins dissociate from the UvrB–DNA complex, and a UvrC protein binds to the UvrB–DNA complex (15), leading to incisions on either side of the damage. Finally, the UvrD helicase removes the damaged region. In this study, we showed that the frequency of illegitimate recombination induced by UV irradiation is enhanced by both uvrA and uvrB mutations, and partially by uvrC mutation, but not by uvrD mutation. Nucleotide sequence analysis of the recombination junctions derived from the uvrA or uvrB mutation showed that this recombination requires short regions of homology at the recombination sites. Therefore, the recombination enhanced by the uvrA or uvrB mutation is SHDIR. Unexpectedly, illegitimate recombination was enhanced in the uvrA uvrB double mutant even in the absence of UV irradiation. In addition, overproduction of UvrA protein reduced the frequency of illegitimate recombination in the wild-type and the recQ mutant, and also suppressed the hyperrecombination phenotype of the uvrB mutant under UV irradiation, but it did not suppress the UV-sensitive phenotype of the uvrB mutant. These results led us conclude that the suppression of illegitimate recombination by UvrA and possibly UvrB takes place independently of NER.
Illegitimate recombination is thought to occur in several steps (see refs. 3, 4, and 8). These steps include introduction of lesions in DNA, stalling of replication forks, formation of double-strand breaks, processing of broken ends, annealing of two DNA ends with short single-stranded overhangs, and gap-filling and ligation. Therefore, there are a number of possible explanations for the suppression of illegitimate recombination by UvrAB proteins. In this study, we found that overproduction of UvrA protein suppressed the hyperrecombination phenotype of the uvrB mutant under UV irradiation, suggesting that UvrA protein alone can suppress illegitimate recombination. Because UvrA protein has DNA binding activity (16, 17), suppression of illegitimate recombination may be caused by inhibition of the DNA double-strand break through binding with damaged DNA. Alternatively, the UvrA protein may inhibit processing of broken ends through DNA binding.
Next, we showed that a combination between the uvrA and uvrB mutations or between the uvrA and recQ mutations exhibits synergistic effect on illegitimate recombination. These findings indicate that the UvrA protein by itself plays roles in the pathways shared by UvrB protein and by RecQ helicase, respectively. On the other hand, we showed that the synergistic effect on illegitimate recombination is not detected between the recQ and uvrB mutations and that overproduction of UvrB protein does not suppress the hyperrecombination phenotype of the recQ mutant. These results indicate that UvrB protein functions in a pathway different from the one in which RecQ helicase works.
It is known that the UvrA protein has DNA binding activity (16, 17) and that the UvrAB complex has 5′-3′ helicase activity (18). This helicase activity may be involved in suppression of illegitimate recombination in a manner similar to that proposed for RecQ helicase (8). We and another group previously proposed a model in which RecQ helicase may disrupt a hydrogen-bonded intermediate formed by annealing of short single-stranded overhangs (8, 19). According to these models and the current results, we envisaged a new model, in which UvrA and UvrB proteins have dual functions for suppression of illegitimate recombination by virtue of DNA binding and DNA helicase activities (Fig. 6). In the first function, the UvrA protein by itself may bind damaged DNA and the UvrA–DNA complex may suppress introduction of double strand breaks. Alternatively, the UvrA protein may inhibit processing of broken DNA ends through DNA binding. In the second function, the UvrAB complex may work as a suppressor of illegitimate recombination, and their helicase activity may play a crucial role in the suppression in the pathway shared with RecQ helicase.
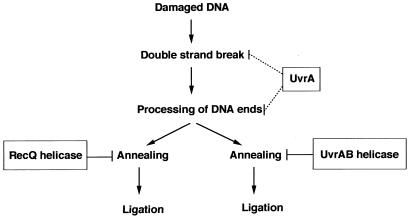
Figure 6.
Model for suppression of illegitimate recombination by UvrA and UvrB proteins. The UvrA protein by itself may bind damaged DNA, and the UvrA-DNA complex may suppress the introduction of DNA double-strand breaks. Alternatively, the UvrA protein may inhibit processing of broken DNA ends through DNA binding. In addition, the helicase activity of the UvrAB complex may be involved in suppression of illegitimate recombination in a manner similar to that proposed for RecQ helicase by Hanada et al. (8).
If the UvrA protein really works at both steps of illegitimate recombination, binding of UvrA with DNA and disruption of the recombination intermediate, then one can expect stronger effects of the uvrA single mutation or the uvrA and uvrB double mutation than that of the uvrB single mutation. But this was not the case. Possibly another helicase may be able to act in the absence of UvrAB helicase and may compensate its defect. This possibility is under investigation in our laboratory.
It should be noted that UvrAB helicase might act on substrates different from those, on which RecQ helicase does, because RecQ unwinds in the 3′→5′ direction (18, 20). This would explain how UvrAB helicase and RecQ helicase can share their roles in suppression of illegitimate recombination. It therefore indicates that UvrAB helicase functions as a by-path of the pathway in which RecQ helicase works.
In the previous study, we showed that illegitimate recombination is enhanced by overproduction of DnaB helicase (21). This recombination was also occurred between short regions of homology. We proposed a model for the initiation of illegitimate recombination, in which overproduction of DnaB helicase may excessively unwind DNA at replication forks and induce double-strand break, resulting in illegitimate recombination. In addition to this, a synergistic effect was shown between the RecQ defect and the overproduction of DnaB helicase on illegitimate recombination (21). This finding indicates that the DnaB helicase works in the same pathway as RecQ helicase does, but that they work at different steps. These studies suggest that DNA helicases have important roles for chromosome maintenance. DNA helicase II is coded by the uvrD gene, whose mutation did not affect illegitimate recombination (see Fig. 1a). We also tested the effect of a mutation in the helD gene, which encodes DNA helicase IV, and showed that it did not affect illegitimate recombination (K. Hanada and H. Ikeda, unpublished results).
Finally, patients with the rare skin disease, Xeroderma pigmentosum (XP), an inherited autosomal recessive genetic disorder characterized by defective repair of UV irradiation-induced DNA damage, are extremely sensitive to UV. Various types of skin cancer are very common among XP patients (22). Moreover, chromosomal aberrations such as translocation and deletion were often observed in fibroblast cultures taken from patients with XP (23, 24). UV irradiation-induced illegitimate recombination may be one of the causes of skin cancers in XP patients. In fact, enhancement of illegitimate recombination has been shown in ERCC1-deficient cell lines (25). Illegitimate recombination may play an important role in the formation of skin cancer in XP patients.
Acknowledgments
We thank Drs. A. Nishimura, A. Yasui, G. Nora, A. Sancar, and J. Kato for providing bacterial strains and plasmids. We are grateful to Drs. Y. Ishii and T. Nohmi for helpful comments. This work was supported by Grants-in-Aid for Scientific Research (B) and Scientific Research on Priority Areas (B) to H.I. from the Ministry of Education, Science, Sports, and Culture of Japan and the Uehara Memorial Foundation.
Abbreviations
- NER
nucleotide excision repair
- SHDIR
short homology-dependent illegitimate recombination
- SHIIR
short homology-independent illegitimate recombination
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100101297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100101297
References
- 1.Ikeda H, Shimizu H, Ukita T, Kumagai M. Adv Biophys. 1995;31:197–208. doi: 10.1016/0065-227x(95)99392-3. [DOI] [PubMed] [Google Scholar]
- 2.Sancar A. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 3.Yamaguchi H, Yamashita T, Shimizu H, Ikeda H. Mol Gen Genet. 1995;248:637–643. doi: 10.1007/BF02191702. [DOI] [PubMed] [Google Scholar]
- 4.Ukita T, Ikeda H. J Bacteriol. 1996;178:2362–2367. doi: 10.1128/jb.178.8.2362-2367.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimizu H, Yamaguchi H, Ikeda H. Genetics. 1995;140:889–896. doi: 10.1093/genetics/140.3.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimizu H, Yamaguchi H, Ashizawa Y, Kohno Y, Asami M, Kato J, Ikeda H. J Mol Biol. 1997;266:297–305. doi: 10.1006/jmbi.1996.0794. [DOI] [PubMed] [Google Scholar]
- 7.Bierne H, Ehrlich S D, Michel B. EMBO J. 1997;16:3332–3340. doi: 10.1093/emboj/16.11.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanada K, Ukita T, Kohno Y, Saito K, Kato J, Ikeda H. Proc Natl Acad Sci USA. 1997;94:3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandsma J A, de Ruijter M, Brouwer J, van de Putte P. J Bacteriol. 1988;170:1012–1014. doi: 10.1128/jb.170.2.1012-1014.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Backendorf C, Spaink H, Barbeiro A P, van de Putte P. Nucleic Acids Res. 1986;14:2877–2890. doi: 10.1093/nar/14.7.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brash D E, Haseltine W A. J Bacteriol. 1985;163:460–463. doi: 10.1128/jb.163.2.460-463.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendonca V M, Kaiser-Rogers K, Matson S W. J Bacteriol. 1993;175:4641–4651. doi: 10.1128/jb.175.15.4641-4651.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakayama K, Irino N, Nakayama H. Mol Gen Genet. 1985;200:266–271. doi: 10.1007/BF00425434. [DOI] [PubMed] [Google Scholar]
- 14.van den Berg E, Zwetsloot J, Noordermeer I, Pannekoek H, Dekker B, Dijkema R, van Ormondt H. Nucleic Acids Res. 1981;9:5623–5643. doi: 10.1093/nar/9.21.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orren D K, Sancar A. Proc Natl Acad Sci USA. 1989;86:5237–5241. doi: 10.1073/pnas.86.14.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazur S J, Grossman L. Biochemistry. 1991;30:4432–4443. doi: 10.1021/bi00232a009. [DOI] [PubMed] [Google Scholar]
- 17.Seeberg E, Steinum A L. Proc Natl Acad Sci USA. 1982;79:988–992. doi: 10.1073/pnas.79.4.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordienko I, Rupp W D. EMBO J. 1997;16:889–895. doi: 10.1093/emboj/16.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harmon F G, Kowalczykowski S C. Genes Dev. 1998;12:1134–1144. doi: 10.1101/gad.12.8.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Umezu K, Nakayama K, Nakayama H. Proc Natl Acad Sci USA. 1990;87:5363–5367. doi: 10.1073/pnas.87.14.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamashita T, Hanada K, Iwasaki M, Yamaguchi H, Ikeda H. J Bacteriol. 1999;181:4549–4553. doi: 10.1128/jb.181.15.4549-4553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robbins J H, Kraemer K H, Lutzner M A, Festoff B W, Coon H G. Ann Intern Med. 1974;80:221–248. doi: 10.7326/0003-4819-80-2-221. [DOI] [PubMed] [Google Scholar]
- 23.Huang C C, Banerjee A, Hou Y. Proc Soc Exp Biol Med. 1975;148:1244–1248. doi: 10.3181/00379727-148-38725. [DOI] [PubMed] [Google Scholar]
- 24.Seguin L R, Tarone R E, Liao K H, Robbins J H. Am J Hum Genet. 1988;42:468–475. [PMC free article] [PubMed] [Google Scholar]
- 25.Sargent R G, Rolig R L, Kilburn A E, Adair G M, Wilson H, Nairn R S. Proc Natl Acad Sci USA. 1997;94:13122–13127. doi: 10.1073/pnas.94.24.13122. [DOI] [PMC free article] [PubMed] [Google Scholar]