Abstract
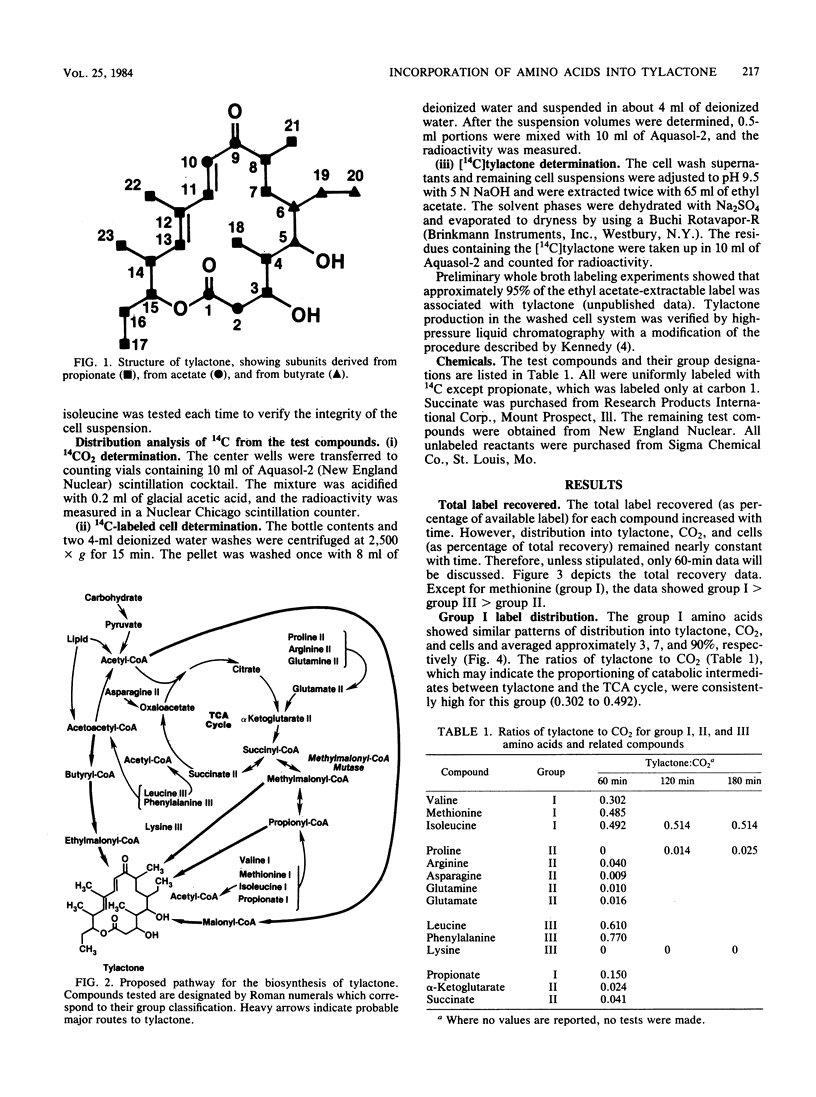
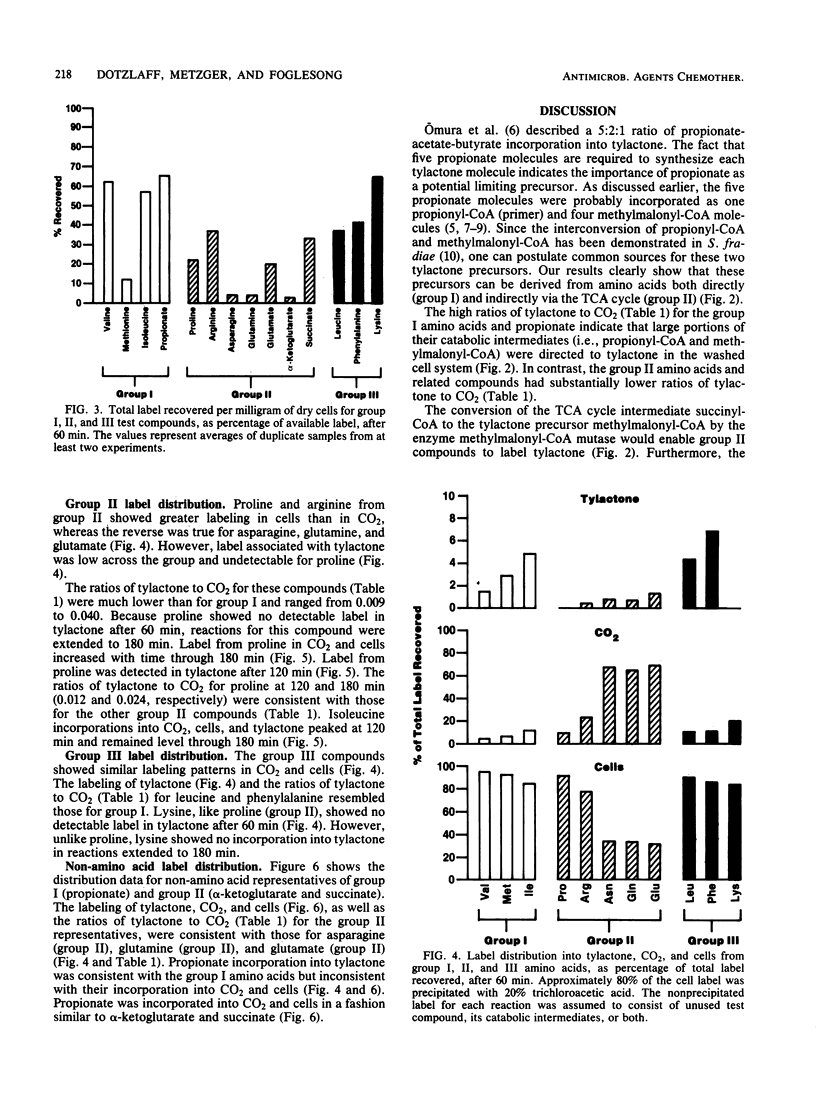
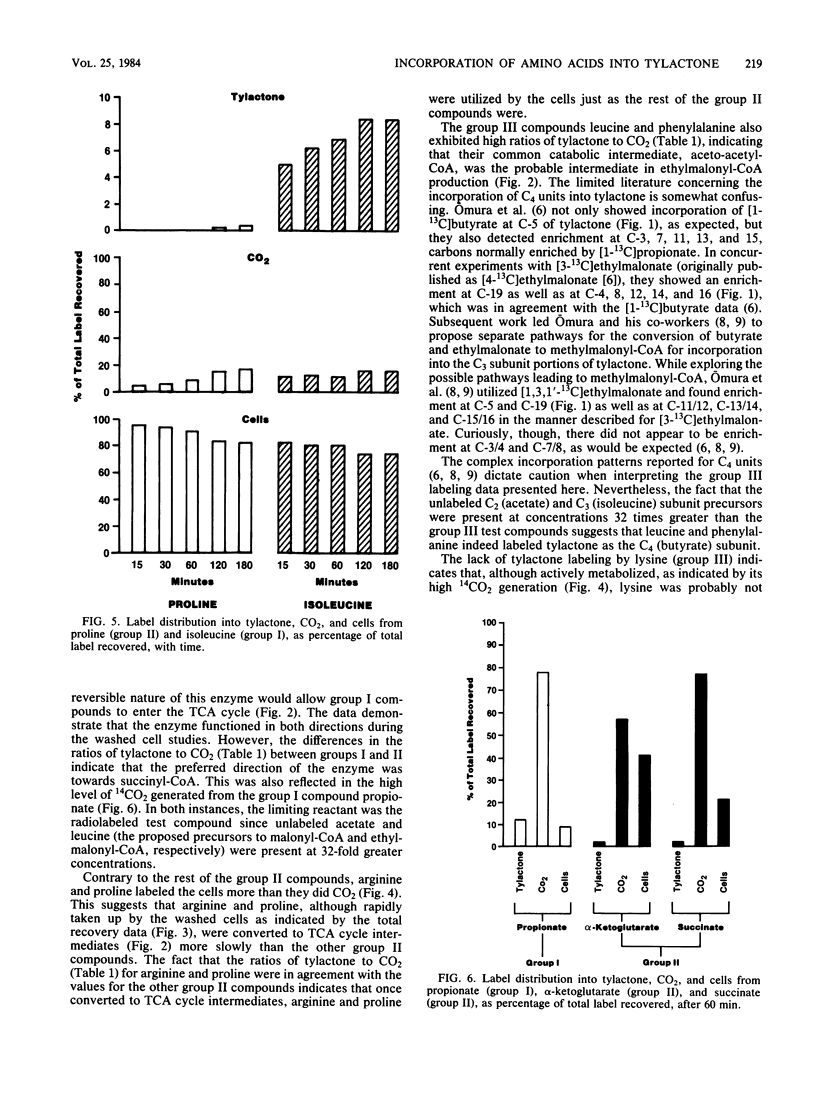
Washed cells from 72-h cultures of Streptomyces fradiae GS14 were used to examine the distribution of radiolabel from 14C-amino acids and related compounds into tylactone, CO2, and cells. Test compounds were categorized according to products of their oxidative degradation. Those compounds known to produce propionyl-coenzyme A by direct catabolic oxidation were designated as group I. Group II included those compounds oxidized to methylmalonyl-coenzyme A via succinyl-coenzyme A and the tricarboxylic acid cycle. Group III contained compounds known to be oxidized to acetoacetyl-coenzyme A. The total amount of label recovered after 60 min ranged from 3 to 65%. Although label from all test compounds except proline (group II) and lysine (group III) was incorporated into tylactone after 60 min, label from group I and group III compounds was incorporated at levels five times greater than label from group II compounds. From 55 to 75% of the recovered label from propionate (I), asparagine (II), glutamine (II), glutamate (II), alpha-ketoglutarate (II), and succinate (II) was recovered as 14CO2. From 75 to 95% of the recovered label from the remaining compounds tested was located in the cells. Based on the data, a pathway for the role of amino acids in the biosynthesis of tylactone is proposed.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baltz R. H., Seno E. T. Properties of Streptomyces fradiae mutants blocked in biosynthesis of the macrolide antibiotic tylosin. Antimicrob Agents Chemother. 1981 Aug;20(2):214–225. doi: 10.1128/aac.20.2.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz R. H., Seno E. T., Stonesifer J., Wild G. M. Biosynthesis of the macrolide antibiotic tylosin. A preferred pathway from tylactone to tylosin. J Antibiot (Tokyo) 1983 Feb;36(2):131–141. doi: 10.7164/antibiotics.36.131. [DOI] [PubMed] [Google Scholar]
- Kennedy J. H. High performance liquid chromatographic analysis of fermentation broths: cephalosporin C and tylosin. J Chromatogr Sci. 1978 Oct 10;16(10):492–495. doi: 10.1093/chromsci/16.10.492. [DOI] [PubMed] [Google Scholar]
- Omura S., Kitao C., Miyazawa J., Imai H., Takeshima H. Bioconversion and biosynthesis of 16-membered macrolide antibiotic, tylosin, using enzyme inhibitor: cerulenin. J Antibiot (Tokyo) 1978 Mar;31(3):254–256. doi: 10.7164/antibiotics.31.254. [DOI] [PubMed] [Google Scholar]
- Omura S., Takeshima H. Inhibition of the biosynthesis of leucomycin, a macrolide antibiotic, by cerulenin. J Biochem. 1974 Jan;75(1):193–195. doi: 10.1093/oxfordjournals.jbchem.a130375. [DOI] [PubMed] [Google Scholar]
- Omura S., Takeshima H., Nakagawa A., Miyazawa J., Piriou F., Lukacs G. Studies on the biosynthesis of 16-membered macrolide antibiotics using carbon-13 nuclear magnetic resonance spectroscopy. Biochemistry. 1977 Jun 28;16(13):2860–2866. doi: 10.1021/bi00632a009. [DOI] [PubMed] [Google Scholar]
- Vu-Trong K., Bhuwapathanapun S., Gray P. P. Metabolic regulation in tylosin-producing Streptomyces fradiae: regulatory role of adenylate nucleotide pool and enzymes involved in biosynthesis of tylonolide precursors. Antimicrob Agents Chemother. 1980 Apr;17(4):519–525. doi: 10.1128/aac.17.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]



