Abstract
Pseudoxanthoma elasticum (PXE), the prototypic heritable connective tissue disorder affecting the elastic structures in the body, manifests with cutaneous, ophthalmologic, and cardiovascular findings, with considerable morbidity and mortality. The molecular basis of PXE has remained unknown, but the disease locus has recently been mapped to an ≈500-kb interval on chromosome 16p13.1, without evidence for locus heterogeneity. In this study, we report pathogenetic mutations in MRP6, a member of the ABC transporter gene family, in eight kindreds with PXE. The mutation detection strategy consisted of heteroduplex scanning of coding sequences in the MRP6 gene, which were amplified by PCR by using genomic DNA as template, followed by direct nucleotide sequencing. A total of 13 mutant MRP6 alleles were disclosed in the eight probands with PXE. These genetic lesions consisted of either single base pair substitutions resulting in missense, nonsense, or splice site mutations, or large deletions resulting in allelic loss of the MRP6 locus. Examination of clinically unaffected family members in four multiplex families identified heterozygous carriers, consistent with an autosomal recessive inheritance pattern. Collectively, identification of mutations in the MRP6 gene provides the basis to examine the pathomechanisms of PXE and allows development of DNA-based carrier detection, prenatal testing, and preimplantation genetic diagnosis in families with a history of this disease.
Pseudoxanthoma elasticum (PXE) (Online Mendelian Inheritance of Man no. 177850 for the autosomal dominant, and no. 264800 for the autosomal recessive variant) is a systemic heritable connective tissue disorder affecting the elastic tissue network in the body, with primary manifestations in the skin, eyes, and the cardiovascular system (1, 2). The skin lesions manifest with yellowish papules which tend to coalesce into plaques of inelastic, redundant, and sagging skin. The histopathologic hallmark of PXE in the skin is accumulation of pleiomorphic, “elastotic” material, often appearing fragmented with perturbed ultrastructural morphology, suggesting functional alterations in the elastic fibers. Furthermore, a diagnostic histopathologic feature of PXE is calcification of the elastic structures, which can be demonstrated by special stains. The eye findings consist of angioid streaks, which result from pathological fractures in Bruch's membrane, an elastin-rich sheath on the back of the eye, that can cause varying degrees of visual impairment (3). Finally, the involvement of the cardiovascular system is manifested by intermittent claudication, rupture of blood vessels particularly within the gastrointestinal tract, and early myocardial infarction, with considerable morbidity and mortality (4).
PXE demonstrates marked clinical heterogeneity and apparently different modes of inheritance (2). The clinical variability is evident by observations that the involvement of all three major organ systems, i.e., skin, eyes, and the cardiovascular system, is encountered in some patients while others, even within the same family, have a limited involvement of one of these organs. Genetic heterogeneity has been suggested by reports on both autosomal dominant and autosomal recessive inheritance (5), but the majority of the cases appear to be sporadic (2). The assessment of inheritance is complicated by the clinical heterogeneity and highly variable age of onset of the disease. Specifically, in rare cases the manifestations are noted during infancy (6), whereas in most cases the clinical signs are not evident until the second or third decade of life (2).
The genetic basis of PXE has remained unknown. Considering the involvement of elastic structures in the affected tissues, initial efforts to elucidate the molecular basis of PXE revolved around elastin and elastin-associated gene/protein systems, as well as enzymes involved in elastin cross-linking; these included the elastin gene on chromosome 7q, fibrillins 1 and 2 on chromosomes 15q and 5q, respectively, and lysyl oxidase also on chromosome 5q. However, these candidate genes were excluded by genetic linkage analysis [see ref. 7, and unpublished data (A.M.C., Ph.D. thesis, Rutgers University, 1991). More recently, two studies provided strong evidence for linkage of both dominantly and recessively inherited forms of PXE to chromosome 16p13.1 by using positional cloning (8, 9), and subsequent multicenter collaborative efforts were able to narrow this interval to consist of ≈500 kb (10, 11). Examination of this interval revealed the presence of four candidate genes (MRP1, MRP6, pM5, and an unknown gene) potentially harboring mutations causing PXE. In this study we demonstrate pathogenetic mutations in the MRP6 gene, encoding a multidrug resistance-associated protein, in families with PXE.
Materials and Methods
Clinical.
A North American cohort consisting of eight families with PXE was subjected to study (Table 1). In four families there were two or more affected individuals, whereas in the other four families, with the exception of the proband, there was no history of PXE. Thus, a total of 13 affected and 20 unaffected family members were examined. Informed consent was obtained from all individuals. The diagnosis was based on dermatological examination in combination with ophthalmological and cardiovascular evaluation. In each proband, the diagnosis was confirmed by histopathology of a skin biopsy (hematoxylin-eosin) combined with special stains for elastin (Verhoeff-van Gieson) and/or for calcium (von Kossa). Total DNA from the affected individuals, their unaffected family members, and from 50 unrelated, unaffected controls was isolated from peripheral blood leukocytes by standard procedures.
Table 1.
MRP6 mutations in families with PXE
| Family | Age and sex of proband | Mutation | Exon | Consequence | Verification* |
|---|---|---|---|---|---|
| 1 | 53 F | 3421C → T | 24 | R1141X | BsiYI |
| 3803G → A | 27 | R1268Q | BstXI | ||
| 2 | 29 F | 3412C → T | 24 | R1138W | MspI |
| 3 | 40 F | 3421C → T | 24 | R1141X | BsiYI |
| Partial deletion | 24† | Allelic loss | D16S2720 | ||
| MRP6 | D16B9622 | ||||
| 4 | 53 F | 3736-1G → A | 27 | Altered splicing of exon 27 | AciI |
| Partial deletion | 27† | Allelic loss | D16S2720 | ||
| MRP6 | D16B9622 | ||||
| 5 | 60 M | 3413G → A | 24 | R1138Q | MspI |
| 3803G → A | 27 | R1268Q | BstXI | ||
| 6 | 28 F | 3421C → T | 24 | R1141X | BsiYI |
| 7 | 41 M | 3803G → A | 27 | R1268Q | BstXI |
| 8 | 25 F | 3421C → T | 24 | R1141X | BsiYI |
Mutations were verified in the proband and his/her family members by digestion with restriction enzyme, or in case of deletion, by microsatellite markers indicated.
Deletions extend beyond exons denoted.
Mutation Detection.
Total genomic DNA was used as a template for PCR amplification of coding sequences in the MRP6 gene using primer pairs placed on flanking intronic sequences. The linear sequence information (introns and exons) was obtained from the BAC clone A-962B4 (GenBank accession no. U91318), and the intron-exon borders for the 31 exons in MRP6 were deduced by comparison with the published cDNA sequence (ref. 12; GenBank accession no. AF076622). The PCR products, varying in size from 151 to 759 bp, were analyzed by heteroduplex scanning by using conformation-sensitive gel electrophoresis (CSGE) (13). In some cases, the PCR products from the family members were mixed in equal amounts with a PCR product from an unrelated, clinically healthy control. The PCR products displaying heteroduplexes were subjected to direct automated sequencing (Applied Biosystems model 377 sequencing system) after gel purification. The segregation of pathogenetic mutations in family members was tested by CSGE analysis, by restriction enzyme digestion, and/or by direct nucleotide sequencing. The presence of these nucleotide changes was also analyzed in 50 unrelated, unaffected control individuals by restriction enzyme digestions performed according to the manufacturers' recommendations. The digestion products were examined on 2% agarose or 6% polyacrylamide gels (6% TBE Gel; NOVEX, San Diego).
Results
Eight families with individuals affected with clinical features of PXE were subjected to a search for mutations within the 500-kb interval defined by linkage analysis on chromosome 16p13.1, which contained four candidate genes, including MRP1, MRP6, pM5, and an unknown gene. Direct sequencing of pM5 (GenBank accession no. X57398), which encodes an unknown protein with some similarities to the collagenase family (14), and of the unknown gene (found by sequencing of the BAC clone A-962B4; http://www.tigr.org) did not reveal evidence for pathogenetic mutations (C. Boyd and B. Struk, unpublished data). The two genes encoding the multiple drug resistance-associated proteins (MRP1 and MRP6) were then subjected to analysis. Examination of MRP1 (GenBank accession no. NM004996) revealed a neutral polymorphism (2031G→C; V677V) in one PXE family (family 4 in Fig. 1), and a heterozygous glycine-to-valine polymorphism (2012G→T; G671V) in another (family 2 in Fig. 1). However, these polymorphisms, both in exon 16, did not segregate with the PXE phenotype, and were also found in unrelated, unaffected controls. Subsequently, our attention was directed at MRP6, which consists of 31 exons, the coding sequence of 4.5 kb being dispersed within ≈73 kb of genomic DNA on chromosome 16p13.1 (12, 15). PCR primers, placed on flanking intronic sequences, were synthesized and used for amplification of individual exons by using genomic DNA as template. The PCR products were subjected to heteroduplex analysis by CSGE, followed by automated sequencing of those PCR products displaying heteroduplexes (see Fig. 2). Probands in eight families, four of them with more than one affected individual (families 1–4) and the remaining four families with only one affected individual (families 5–8), were screened by this strategy. The results revealed the presence of 13 mutant MRP6 alleles in the eight probands with PXE, representing seven distinct mutations. Among these mutations, there were three missense mutations, one nonsense mutation, one affecting a canonical splice site consensus sequence, and two consisted of large deletions affecting MRP6. None of these genetic lesions were found in DNA from 50 unrelated, unaffected controls.
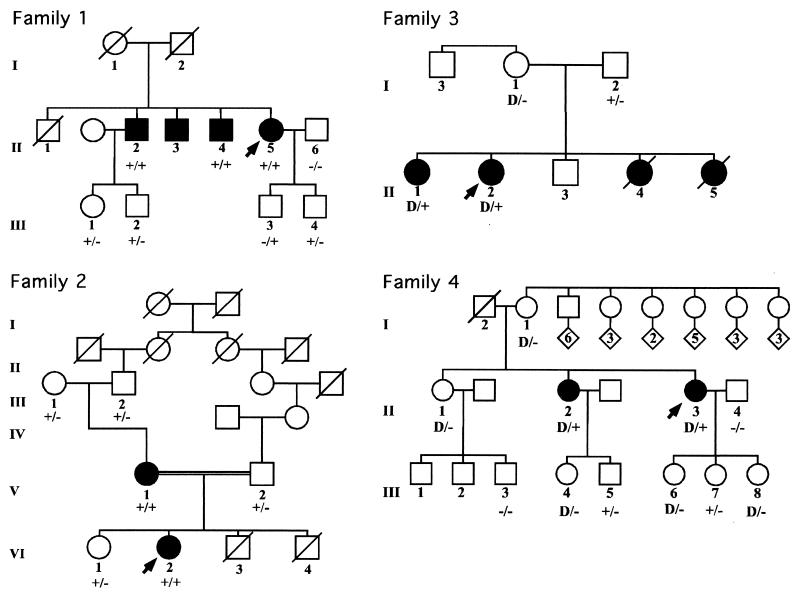
Figure 1.
Four multiplex pedigrees with PXE and mutations in MRP6. Clinically affected individuals are depicted with solid symbols, their unaffected family members are shown by open symbols, and the proband in each family is indicated by an arrow. The + sign refers to a single base pair substitution, whereas the − sign denotes the wild-type allele. D refers to large deletions resulting in allelic loss.
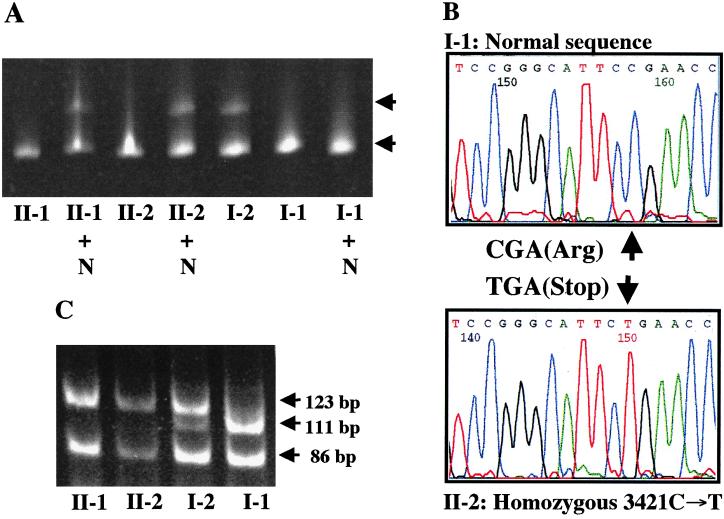
Figure 2.
Mutation detection in family 3. (A) CSGE analysis of PCR products spanning exon 24 in the family members reveals a heteroduplex band (upper arrow) in I-2 who is heterozygous carrier of a single base pair substitution (3421C→T). When initially tested by CSGE, II-1 and II-2 showed a homoduplex band only (lower arrow), but when mixed with a PCR product from a normal control (II-1 + N and II-2 + N), a heteroduplex band is evident, reflecting homozygosity of the mutation. (B) Nucleotide sequencing of the PCR products corresponding to exon 24 reveals that II-2 is homozygous for the 3421C→T transition mutation, whereas I-1 shows the wild type sequence only. (C) The 3421C→T substitution abolished a BsiYI site. The 327-bp PCR product is digested in normal allele to 111-, 86-, 64-, 54-, and 12-bp bands, whereas in case of the mutant allele a 123-bp band (111 + 12 bp) is evident. (The lower molecular weight bands are not shown.)
Family 1.
The proband, a 53-year-old female (II-5 in Fig. 1), had characteristic cutaneous, ophthalmologic, and cardiovascular findings of PXE; the diagnosis was confirmed by skin biopsy. The proband had three older brothers affected with PXE. The oldest brother (II-1) had died of a myocardial infarct at the age of 48 years and is also assumed to have been affected. Screening of DNA from the proband by CSGE revealed a heteroduplex band when sequences corresponding to exons 24 and 27 were examined. Sequencing of the corresponding PCR products revealed nucleotide substitutions 3421C→T and 3803G→A, respectively. The mutation in exon 24 resulted in substitution of a codon for arginine by a stop codon, a mutation designated R1141X. The mutation in exon 27 resulted in substitution of a codon for arginine by a codon for glutamine (R1268Q). Thus, the proband is a compound heterozygote for a nonsense and a missense mutation. No DNA was available from the parents to ascertain the parent of origin of each mutation. Subsequent evaluation of two affected brothers (II-2 and II-4) by restriction enzyme analysis (BsiYI for exon 24; BstXI for exon 27) revealed that they were also compound heterozygotes for the same mutations. Evaluation of the children of the affected individuals who did not show any signs of PXE revealed that all of them were heterozygous carriers of the mutations, as shown in Fig. 1.
Family 2.
The proband, a 29-year-old female (VI-2 in Fig. 1), had characteristic skin findings since age 7, as well as angioid streaks and weakened peripheral pulses. The diagnosis of PXE was confirmed by skin biopsy. The proband's 53-year-old mother (V-1) was similarly affected, but there were no other affected family members. The proband's parents were distant cousins (see Fig. 1). Evaluation of the proband's DNA revealed a homozygous nucleotide substitution 3412C→T in exon 24 of MRP6. This substitution resulted in the replacement of an arginine codon by a codon for tryptophan (R1138W). This nucleotide substitution abolished a restriction site for MspI, and evaluation of the rest of the family by MspI digestion revealed that the mother of the proband was also homozygous for the mutation R1138W. The proband's maternal grandparents (III-1 and III-2) were heterozygous carriers of this mutation, as was the proband's older sister (VI-I). The proband's two younger brothers (VI-3 and VI-4) had died at birth or shortly thereafter of unknown causes. Examination of the proband's father (V-2) also revealed the mutation R1138W in the heterozygous state. Thus, the proband had inherited one mutant allele from each parent, and the disease in this family is clearly recessively inherited with evidence of pseudodominance.
Family 3.
The proband (II-2 in Fig. 1) in the family was a 40-year-old female with severe skin, eye, and cardiovascular involvement. She had an older sister who, in addition to cutaneous and ophthalmologic findings, had a history of gastrointestinal bleeding and a coronary bypass operation in her forties. Examination of the proband's DNA by CSGE and nucleotide sequencing (Fig. 2 A and B) revealed a homozygous mutation in exon 24, 3421C→T, which resulted in a premature termination codon R1141X. This is the same mutation that was also noted in the compound heterozygous state in family 1. The older sister (II-1) was also homozygous for this mutation. Examination of the parents revealed that the father (I-2) was heterozygous for the mutation R1141X. However, the mother's DNA consisted of normal MRP6 alleles only, when analyzed by CSGE, by enzyme digestion with BsiYI, and by direct sequencing (Fig. 2). Haplotype analysis with 13 polymorphic markers on chromosome 16, together with 8 markers on chromosome 5, clearly established the maternal origin of DNA. Subsequent analysis of a microsatellite marker D16B9622, which resides ≈0.7 kb downstream from the translation stop codon at the 3′ end of the MRP6 gene, showed homozygosity for two different alleles in each parent, whereas both affected daughters were heterozygous (data not shown). However, the microsatellite D16S2720, which is located ≈13.3 kb upstream from exon 1 of MRP6, was homozygous in the daughters (II-1 and II-2) and the mother (I-1), while heterozygous in the father (I-2). These findings are consistent with the interpretation that the proband and her sister had inherited a nonsense mutation from the father and a deletion mutation affecting exon 24 of MRP6 from the mother, thus reducing the paternal mutation R1141X to hemizygosity. It is noteworthy that the proband's father had mild angioid streaks without visual impairment, as well as minimal skin findings on clinical examination, suggestive of PXE. A search for other mutations in the MRP6 gene was unyielding, and he may therefore manifest with mild features of PXE, reflecting his carrier status for the mutation R1141X.
Family 4.
The proband, a 53-year-old female (II-3 in Fig. 1), had characteristic skin lesions limited to the neck and arms, and the diagnosis of PXE was confirmed by skin biopsy. Ophthalmologic examination had not revealed the presence of angioid streaks, but she had macular degeneration. Examination of her DNA revealed a homozygous mutation 3736-1G→A, which affects the consensus splice site sequence ag-exon 27. This mutation is predicted to cause aberrant splicing involving exon 27. The proband had one older sister (II-2), who was clinically affected with PXE. In particular, she had more extensive skin involvement and vision impairment because of macular degeneration. Examination of her DNA by direct nucleotide sequencing revealed that she was also homozygous for the splice site mutation, 3736-1G→A. This mutation abolishes an EcoNI site, and this enzyme was used to study the segregation of the mutation in the other members of the family. On clinical examination, none of the individuals in generation III were thought to be affected by PXE. Genotype analysis revealed that individuals III-5 and III-7 were heterozygous carriers of the mutation. Interestingly, however, the remaining children of the affected individuals apparently had inherited only the normal alleles, despite the finding that the two mothers (II-2 and II-3) were homozygous for the mutation 3736-1G→A (family 4 in Fig. 1). Furthermore, the mother (I-1) of both affected individuals was not a carrier of this mutation, whereas DNA from the father (I-2), who was deceased, was not available. Haplotype analysis with 13 microsatellite markers spanning ≈5 cM of 16p, including the MRP6 locus, revealed that in generations II and III the inheritance of the mutation 3736-1G→A segregated with inheritance of a specific allele presumably inherited from individual I-2 (data not shown). This allele also contained the silent MRP1 polymorphism (V677V; see above). Collectively, these findings suggest that the mother (I-1) had an allelic loss, which, when inherited in trans with the paternal mutant allele by the two affected individuals (II-2 and II-3), resulted in a homozygous genotype for the mutation 3736-1G→A. This finding would also explain the fact that three children of the two affected mothers (II-2 and II-3) were hemizygous for the wild-type allele, because the other allele would harbor the maternally inherited deletion mutation (Fig. 1). This interpretation was supported by haplotype analysis by using microsatellites on the 16p13.1 region, including D16B9622, which revealed that the putative deletion does not extend beyond the 3′ end of MRP6 (data not shown).
Families 5–8.
Four additional patients with diagnostic features and biopsy-proven PXE were evaluated for possible mutations in MRP6. There was no family history of PXE, and DNA from other members of the family was not available. Therefore, it appears that these families represent sporadic cases of PXE. Evaluation of their DNA revealed that the patient in family 5 was compound heterozygous for nucleotide substitutions 3413G→A and 3803G→A in exons 24 and 27, which resulted in the amino acid substitutions R1138Q and R1268Q, respectively. In families 6–8, heterozygous nucleotide substitutions were discovered, resulting in the mutations R1141X (families 6 and 8), and R1268Q (family 7).
Discussion
In this study, we report pathogenetic mutations in PXE in the MRP6 gene on chromosome 16p13.1. Specifically, we examined eight probands with unequivocal diagnostic features of PXE. In four families, more than one affected individual was present, and in these families mutations in both alleles in each affected individual were discovered, supporting the notion that PXE was clearly inherited in an autosomal recessive fashion. It is of interest to note that in one of these families (family 2) the mode of inheritance was initially judged to be autosomal dominant because of inheritance of PXE from a mother to her daughter. However, mutation analysis in this family indicated autosomal recessive inheritance with a pseudodominant pedigree structure as a result of consanguinity in the family. Some mutations discovered in these autosomal recessive families resulted in a premature translation termination codon predicting either synthesis of a truncated polypeptide or reduced mRNA levels because of nonsense-mediated mRNA decay (families 1 and 3) (16). In either case, the level or functionality of MRP6 protein is expected to be reduced or absent. Affected individuals in one of the families showed a homozygous missense mutation (family 2: R1138W), whereas those in another family were compound heterozygous for a nonsense mutation and a missense mutation (family 1: R1141X/R1268Q). The nonconservative amino acid substitution mutations were not detected in 50 clinically unaffected, unrelated control individuals, and it is conceivable, therefore, that these missense mutations affect a critical amino acid residue within the MRP6 protein (see below). Nevertheless, these missense mutations were clearly recessive, as heterozygous carriers did not have clinical evidence of PXE. Finally, another family (no. 4) demonstrated a homozygous nucleotide substitution (3736-1G→A) that alters the conserved consensus sequence of the 3′-acceptor splice site at the intron 26/exon 27 border. This mutation predicts aberrant splicing, and if exon 27 would be deleted in its entirety, it would result in synthesis of a polypeptide internally shortened by 49 amino acids. This mutation is also autosomal recessive, as carriers of this mutation did not show signs of PXE. In addition to the single base substitutions, we discovered two large deletions in families 3 and 4, respectively. In these families, inheritance of the allele containing the deletion, when combined with a stop codon mutation (family 3) or with a splice site mutation (family 4) in trans, resulted in hemizygosity of the mutation in the MRP6 locus and caused PXE.
In addition to four extended families, four affected individuals without family history of PXE revealed mutations in MRP6. In one case (family 5) the proband was a compound heterozygote for two missense mutations, whereas only heterozygous mutations in one allele in the remaining three families (families 6–8) were detected. Although only one mutation in each of the latter three individuals was observed, it is likely that PXE in these cases was also autosomal recessive, because each of the two distinct mutations (R1141X and R1268Q) discovered in these three probands were also present in the multiplex families, and the heterozygous carriers did not show definitive signs of the disease. Collectively, the overwhelming genetic evidence in our study suggests that the majority of cases with PXE display autosomal recessive inheritance. This mode of inheritance is also supported by extensive genetic linkage analysis of these families by using informative microsatellite markers within the chromosomal region 16p13.1 (data not shown). The inability to detect second mutations in three individual patients may reflect the limitations of CSGE, as has been previously documented in search for mutations in other heritable diseases, as exemplified by the dystrophic forms of epidermolysis bullosa where this same technology detected only about 70–75% of all anticipated mutations in the COL7A1 gene (17, 18). On the other hand, it is plausible that the other mutations consist of type of genetic lesions that are not readily detectable by CSGE, including large insertions or deletions, intronic mutations beyond the region subjected to PCR amplification, or mutations affecting the transcriptional/translational regulatory regions at the 5′ end of the gene (19).
MRP6, which has also been described as MOAT-E (15), belongs to the MRP (multiple drug resistance associated proteins) family of the ABC (ATP-binding cassette) transmembrane transporters, which have been shown to be involved in drug resistance, particularly in association with cancer chemotherapy (20–22). MRP6 has been reported to be exclusively expressed in the kidney and the liver, based on Northern blot analysis of poly(A)+ RNA from a number of tissues (12, 15). Although a low level of expression has also been demonstrated in some tissues by RNase protection assay (12), Northern blot analyses have been negative in all such tissues, except the kidney and the liver (15). Furthermore, our attempts to amplify dermal fibroblast MRP6 mRNA by RT-PCR have been unsuccessful under standard PCR conditions in fibroblast cultures both from control individuals as well as from patients II-3 and III-4 in family 4 in Fig. 1. Immunohistochemistry of normal human skin with a polyclonal rabbit anti-human IgG Ab (kindly provided to us by Drs. M. Belinsky and G. Kruh, Fox Chase Cancer Center, Philadelphia, PA), recognizing the carboxyl-terminal end of MRP6, was negative for dermis in a wide range of dilutions, whereas human kidney and liver sections were brightly positive (data not shown). Similarly, immunofluorescence staining of human dermal fibroblasts or iliac artery smooth muscle cells was entirely negative. Thus, although PXE is a generalized connective tissue disorder with abnormalities in the elastic fiber network, the primary defect may reside in the liver and kidney, the sites of MRP6 expression.
The MRP6 mRNA, ≈6 kb, has a 4.5-kb ORF encoding a polypeptide of 1503 amino acids with a calculated molecular mass of 165 kDa (12, 15). The MRP6 protein has a high degree of identity (≈62%) with MRP1, the prototypic protein within this family of ABC transporter proteins (15). The MRP6 protein has been predicted to consist of three membrane spanning domains comprised of 5, 6, and 6 transmembrane segments, respectively (15). The intracellular portion of the protein depicts two nucleotide binding folds (NBF), both having conserved Walker A and B ATP-binding motifs, as well as a conserved C motif, critical for the function of the protein as a transmembrane transporter (15). The majority of the mutations disclosed in this study reside in exons 24 and 27 (Table 1). Exon 24 encodes the intracellular loop between the 4th and 5th helical transmembrane segment within the third membrane spanning domain, and exon 27 codes for beginning of the second NBF (15). It is conceivable that these intracellular segments of MRP6 are critical for the function of the protein, and mutations affecting these domains, including amino acid substitutions, result in phenotypic manifestations of PXE. In fact, single amino acid substitutions in either one of the NBF motifs entirely abolish the function of MRP6 (G. Kruh, unpublished data). In this context, it should be noted that mutations in another MRP transporter, MRP2, are associated with a clinical disease, the Dubin–Johnson syndrome (23, 24), an autosomal recessive disorder characterized by conjugated hyperbilirubinemia. In analogy with PXE mutations in MRP6, the mutations in MRP2 consist of large deletions, splice site mutations, and/or missense substitutions affecting the NBF. In addition to Dubin–Johnson syndrome, a number of recessive diseases are caused by mutations in other ABC transporter genes, including cystic fibrosis and the Stargardt disease (25–28).
The specific biological function of the MRP6 protein is currently unknown, and consequently, its relation to the phenotypic findings in PXE is unclear. MRP1, the best-characterized protein within the MRP family, functions in normal tissues as an efflux pump for amphipathic anionic conjugates, including glutathione-5 conjugates, as well as glucuronidated and sulfated compounds (22, 29). It has been suggested that the function of MRP6 also relates to cellular detoxification (15), and may thus result in accumulation of yet unidentified compounds that may lead to progressive calcification of elastic structures in the target tissues. Alternatively, the perturbed metabolism of cells lacking functional MRP6 may cause alterations in the cellular microenvironment, resulting in the synthesis of aberrant elastic fibers which become calcified in target tissues. In this context, it should be noted that not all elastic fibers undergo calcification in patients with PXE and, for example, the lungs, which are particularly rich in elastin, are only rarely affected (1, 2). These observations suggest that calcification of the elastic structures in PXE is a secondary phenomenon. Nevertheless, identification of mutations in the MRP6 gene provides the basis to examine the cellular and molecular events that lead to the phenotypic manifestations of PXE.
The definitive diagnosis of PXE is often hampered by extensive intrafamilial and interfamilial clinical heterogeneity (2, 5). Furthermore, many of the diagnostic criteria of PXE, such as the cardiovascular accidents, are prevalent in the general population, albeit at a later age (2, 30). Also, cutaneous findings mimicking PXE are frequently encountered in older individuals in sun-exposed areas (31). For these reasons, PXE may be overdiagnosed in individuals with a family history of PXE. At the same time, it has been suggested that heterozygous carriers of PXE mutations may manifest with minimal signs of the disease (2, 32, 33), a notion underscored by clinical findings in the individual I-2 in family 3. In this context, it is of interest to note that in families with Stargardt disease, an autosomal recessive macular dystrophy, the heterozygous carriers can also show evidence of mild disease (34). The diagnostic dilemma in PXE can now be overcome by identification of specific mutations that allows DNA-based carrier detection in families with a history of PXE, and the prediction of affected individuals before the age of onset. Finally, this progress will also permit development of prenatal testing and preimplantation genetic diagnosis in families at risk for recurrence of PXE.
Acknowledgments
We thank the PXE families for their willingness to participate in this study. Family 2 was referred to us by The National Association of PXE (NAPE). We appreciate helpful discussions with Drs. Gabriele Richard and Gary Kruh. Drs. James Nordlund and Leena Pulkkinen assisted in clinical aspects of the study. Drs. Charles Boyd and Berthold Struk kindly provided unpublished information. Carol Kelly provided excellent assistance in preparation of the manuscript. This study was supported by the U.S. Public Health Service, National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant RO1 AR 28450.
Abbreviations
- PXE
Pseudoxanthoma elasticum
- CSGE
conformation-sensitive gel electrophoresis
- MRP
multiple drug resistance-associated proteins
- ABC
ATP-binding cassette
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100041297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100041297
References
- 1.McKusick V A. Heritable Disorders of Connective Tissue. 4th Ed. St. Louis: Mosby; 1972. pp. 475–520. [Google Scholar]
- 2.Neldner K. Clin Dermatol. 1988;6:1–92. doi: 10.1016/0738-081x(88)90003-x. [DOI] [PubMed] [Google Scholar]
- 3.Clarkson J G, Altman R D. Surv Ophthalmol. 1982;26:235–246. doi: 10.1016/0039-6257(82)90158-8. [DOI] [PubMed] [Google Scholar]
- 4.Lebwohl M, Halperin I, Phelp R G. N Engl J Med. 1993;17:1237–1239. doi: 10.1056/NEJM199310213291705. [DOI] [PubMed] [Google Scholar]
- 5.Pope F M. Br J Dermatol. 1975;92:493–509. doi: 10.1111/j.1365-2133.1975.tb03117.x. [DOI] [PubMed] [Google Scholar]
- 6.Goodman R M, Smith E W, Paton D, Bergman R A, Siegel C L, Ottesen O E, Shelley W M, Pusch A L, McKusick V A. Medicine. 1963;42:297–334. [PubMed] [Google Scholar]
- 7.Christiano A M, Lebwohl M G, Boyd C D, Uitto J. J Invest Dermatol. 1992;99:660–663. doi: 10.1111/1523-1747.ep12668156. [DOI] [PubMed] [Google Scholar]
- 8.Struk B, Neldner K H, Rao V S, St. Jean P, Lindpaintner K. Hum Mol Genet. 1997;6:1823–1828. doi: 10.1093/hmg/6.11.1823. [DOI] [PubMed] [Google Scholar]
- 9.van Soest S, Swart J, Tijmes N, Sandkuijl L A, Rommers J, Bergen A A. Genome Res. 1997;7:830–834. doi: 10.1101/gr.7.8.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Saux O, Urban Z, Goring H H, Csiszar K, Pope F M, Richards A, Pasquali-Ronchetti I, Terry S, Bercovitch L, Lebwohl M G, et al. Genomics. 1999;62:1–10. doi: 10.1006/geno.1999.5925. [DOI] [PubMed] [Google Scholar]
- 11.Cai L, Struk B, Adams M D, Ji W, Haaf T, Kang H-L, Dho S H, Xu X, Ringpfeil F, Nancarrow J, et al. J Mol Med. 2000;78:36–46. doi: 10.1007/s001090000079. [DOI] [PubMed] [Google Scholar]
- 12.Kool M, van der Linden M, de Haas M, Baas F, Borst P. Cancer Res. 1999;59:175–182. [PubMed] [Google Scholar]
- 13.Ganguly A, Prockop D J. Proc Natl Acad Sci USA. 1994;90:10325–9. doi: 10.1073/pnas.90.21.10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Templeton N S, Rodgers L A, Levy A T, Ting K L, Krutzsch H C, Liotta L A, Stetler-Stevenson W G. Genomics. 1992;12:175–176. doi: 10.1016/0888-7543(92)90425-r. [DOI] [PubMed] [Google Scholar]
- 15.Belinsky M G, Kruh G D. Br J Cancer. 1999;80:1342–1349. doi: 10.1038/sj.bjc.6690527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maquat L E. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 17.Christiano A M, Hoffman G G, Zhang X, Xu Y, Tamai Y, Greenspan D S, Uitto J. Hum Mutat. 1997;10:408–414. doi: 10.1002/(SICI)1098-1004(1997)10:5<408::AID-HUMU12>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 18.Whittock N V, Ashton G H, Mohammedi R, Mellerio J E, Mathew C G, Abbs S J, Eady R A, McGrath J D. J Invest Dermatol. 1999;113:673–686. doi: 10.1046/j.1523-1747.1999.00732.x. [DOI] [PubMed] [Google Scholar]
- 19.Fujita Y, Nakata K, Yasui N, Matsui Y, Kataoka E, Hiroshima K, Shiba R I, Ochi T. J Clin Endocrinol Metab. 2000;85:425–431. doi: 10.1210/jcem.85.1.6247. [DOI] [PubMed] [Google Scholar]
- 20.Zeng H, Bain L J, Belinsky M G, Kruh G D. Cancer Res. 1999;59:5964–5967. [PubMed] [Google Scholar]
- 21.van der Kolk D M, Vellenga E, Muller M, Vries E G. Adv Exp Med Biol. 1999;457:187–198. doi: 10.1007/978-1-4615-4811-9_20. [DOI] [PubMed] [Google Scholar]
- 22.Deeley R G, Cole S P. Semin Cancer Biol. 1997;8:193–204. doi: 10.1006/scbi.1997.0070. [DOI] [PubMed] [Google Scholar]
- 23.Wada M, Toh S, Taniguchi K, Nakamura T, Uchiumi T, Kohno K, Yoshida I, Kimura A, Sakisaka S, Adachi Y. Hum Mol Genet. 1998;7:203–207. doi: 10.1093/hmg/7.2.203. [DOI] [PubMed] [Google Scholar]
- 24.Toh S, Wada M, Uchiumi T, Inokuchi A, Makino Y, Horie Y, Adachi Y, Sakisaka S, Kuwano M. Am J Hum Genet. 1999;64:739–746. doi: 10.1086/302292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riordan J R, Rommens J M, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou J L, et al. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 26.Ko Y H, Pedersen P L. J Bioenerg Biomembr. 1997;29:417–427. doi: 10.1023/a:1022402105375. [DOI] [PubMed] [Google Scholar]
- 27.Allikmets R, Singh N, Sun H, Shroyer N F, Hutchinson A, Chidambaram A, Gerrard B, Baird L, Stauffer D, Peiffer A, et al. Nat Genet. 1997;15:236–246. doi: 10.1038/ng0397-236. [DOI] [PubMed] [Google Scholar]
- 28.De La Paz M A, Guy V K, Abou-Donia S, Heinis R, Bracken B, Vance J M, Gilbert J R, Gass J D M, Haines J L, Pericak-Vance M A. Ophthalmology. 1999;106:1531–1536. doi: 10.1016/S0161-6420(99)90449-9. [DOI] [PubMed] [Google Scholar]
- 29.Jedlitschky G, Leier I, Buchholz U, Barnouin K, Kurz G, Keppler D. Cancer Res. 1996;56:988–994. [PubMed] [Google Scholar]
- 30.Mendelsohn G, Bulkley B H, Hutchins G M. Arch Pathol Lab Med. 1978;102:298–302. [PubMed] [Google Scholar]
- 31.Uitto J, Bernstein E F. J Invest Dermatol. 1998;3:41–44. doi: 10.1046/j.1523-1747.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- 32.Altman L K, Fialkow P J, Parker F, Sagebiel R W. Arch Intern Med. 1974;134:1048–1054. doi: 10.1001/archinte.134.6.1048. [DOI] [PubMed] [Google Scholar]
- 33.Bacchelli B, Quaglino D, Gheduzzi D, Taparelli F, Boraldi F, Trolli B, Le Saux O, Boyd C D, Pasquali-Ronchetti I. Mod Pathol. 1999;12:1112–1123. [PubMed] [Google Scholar]
- 34.Lewis R A, Shroyer N F, Singh N, Allikmets R, Hutchinson A, Li Y, Lupinski J R, Leppert M, Dean M. Am J Hum Genet. 1999;64:422–434. doi: 10.1086/302251. [DOI] [PMC free article] [PubMed] [Google Scholar]