Abstract
Iron (Fe) deficiency is a worldwide agricultural problem on calcareous soils with low-Fe availability due to high soil pH. Rice plants use a well documented phytosiderophore-based system (Strategy II) to take up Fe from the soil and also possess a direct Fe2+ transport system. Rice plants are extremely susceptible to low-Fe supply, however, because of low phytosiderophore secretion and low Fe3+ reduction activity. A yeast Fe3+ chelate-reductase gene refre1/372, selected for better performance at high pH, was fused to the promoter of the Fe-regulated transporter, OsIRT1, and introduced into rice plants. The transgene was expressed in response to a low-Fe nutritional status in roots of transformants. Transgenic rice plants expressing the refre1/372 gene showed higher Fe3+ chelate-reductase activity and a higher Fe-uptake rate than vector controls under Fe-deficient conditions. Consequently, transgenic rice plants exhibited an enhanced tolerance to low-Fe availability and 7.9× the grain yield of nontransformed plants in calcareous soils. This report shows that enhancing the Fe3+ chelate-reductase activity of rice plants that normally have low endogenous levels confers resistance to Fe deficiency.
Keywords: ferrous iron, yeast Fe3+ chelate reductase, Fe-regulated transporter, transgenic rice
Iron (Fe) deficiency impairs chlorophyll biosynthesis and chloroplast development in both di- and monocotyledonous species. Therefore, Fe availability is directly correlated with plant productivity. Chlorosis due to the unavailability of Fe in calcareous soils with high pH is a major agricultural problem that results in reduced crop yields in ≈30% of cultivated soils worldwide (1).
To take up Fe, nongraminaceous plants use the Strategy I system, which consists of the induction of two processes under low-Fe conditions (2). First, the inducible activity of Fe3+ chelate reductase reduces Fe3+ to Fe2+ (3), which is the rate-limiting step for Fe acquisition from the soil (4). The generated Fe2+ is then transported into the plant by the Fe-regulated transporter [IRT1 (5)], which is the major Fe2+ transporter in the plant root (6–8).
Graminaceous plants take up Fe by the Strategy II system (9), which relies on Fe3+ chelation rather than reduction, similar to uptake in species of bacteria and fungi (10). In graminaceous plants, mugineic acid (MA) family phytosiderophores are synthesized and released into the soil, where they chelate Fe3+; they are then internalized in the Fe-bound state by specific transporters (11).
Rice plants also produce and secrete phytosiderophores under conditions of Fe deficiency, but in lower amounts than other graminaceous crops such as barley (12). We have shown that, in addition to a Strategy II Fe-uptake system, rice plants also possess a direct Fe2+-uptake system that uses the OsIRT1 Fe2+ transporter and low Fe3+ reduction (13, 14). In paddy fields, where rice plants are normally grown, there is abundant Fe2+ owing to the low redox potential. Rice plants do not need to reduce Fe3+ to Fe2+ under such conditions, which may have selected for the development of the functional Fe2+-regulated transporter and not the Fe3+ chelate reductase. The ancestors of cultivated rice, including Oryza rufipogon and Oryza nivara (15), are adapted to swamps and soils at the edges of rivers and lakes, and modern rice plants are also normally grown in paddy fields. Therefore, rice plants have developed in an edaphic environment, in which there is no need to reduce Fe3+ to Fe2+, and may have been selected for a functional Fe2+-regulated transporter rather than Fe3+ chelate reductase.
Under aerobic conditions, however, Fe in soil is present almost exclusively in its oxidized form (Fe3+), which is not readily available to rice plants thought to possess only the Strategy II system (2). Thus, rice plants tolerant of low-Fe availability have been produced only by strengthening the Strategy II system. In fact, the heterologous expression of barley nicotianamine aminotransferase genes has enabled transgenic rice plants to secrete large amounts of phytosiderophores, allowing survival in calcareous soils (16).
There are some reports obtaining Fe-deficiency tolerance by adding Fe3+ chelate reductase to Strategy I plants, such as tobacco and soybean (17, 18). The recent discovery of a direct Fe2+ transport system in rice plants has presented the possibility of obtaining Fe-deficiency tolerance by adding Fe3+ chelate reductase to rice plants, creating a complete Strategy I system. In the present study, we introduced the mutational reconstructed yeast Fe3+ chelate-reductase gene, refre1/372 (17), under the control of the OsIRT1 promoter, into rice plants. The yeast Fe3+ chelate-reductase gene (FRE1) was modified and completely reconstructed to produce full-size transcripts in plants (19). Furthermore, randomly mutagenized variants of refre1 were generated and screened for derivatives with high-Fe3+ chelate-reductase activity at alkaline pH. Transgenic rice plants showed higher Fe3+ chelate-reductase activity. An analysis using a positron-emitting tracer imaging system (PETIS) revealed that the initial Fe-uptake rate of transgenic rice plants was two times that of vector controls. Moreover, transgenic rice plants with the refre1/372 gene showed enhanced tolerance to low-Fe availability on calcareous soils. Although rice plants do possess the Fe2+ transport system, they have low levels of endogenous Fe3+ chelate-reductase activity. The work reported here demonstrates that enhancing the Fe3+ chelate-reductase activity of rice plants renders those plants resistant to Fe deficiency.
Results
Transgene Expression Analysis of Transgenic Rice Plants.
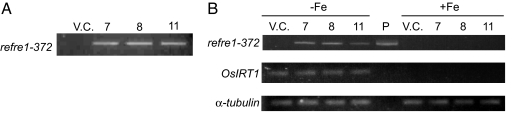
The mutational reconstructed yeast Fe3+ chelate-reductase gene, refre1/372 (17), in conjunction with the OsIRT1 promoter, was introduced into rice plants by using an Agrobacterium-mediated method. Three independent transformants were obtained and confirmed to carry the refre1/372 gene by using genomic PCR analysis (Fig. 1A). RT-PCR analysis confirmed the presence of a single DNA band corresponding to the expected size of the refre1/372 gene only in Fe-deficient roots of the transformants, and OsIRT1 expression was also observed in Fe-deficient roots (Fig. 1B). This result indicates that refre1/372 expression under the OsIRT1 promoter was tightly regulated in Fe-deficient roots and was consistent with native OsIRT1 expression (13, 14).
Fig. 1.
The refre1/372 expression analyzed by RT-PCR. (A) Genomic PCR of DNA prepared from each transformant (lines 7, 8, and 11) and the vector control (VC). (B) Transcript of refre1/372 levels in roots grown under Fe-deficient and Fe-sufficient conditions was detected by using 2 μg of total RNA for each transformant (lines 7, 8, and 11), the vector control (VC), and plasmid containing refre1–372 (P). α-tubulin and OsIRT1 were internal standards.
Root Fe3+ Chelate-Reductase Activity in the Transformants.
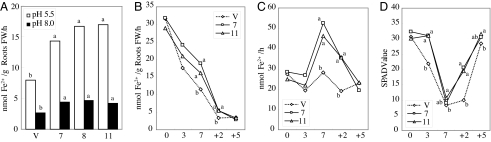
We examined Fe3+ chelate-reductase activity in the transformants compared with that in the vector control. Plants were grown for 2 weeks in Fe-sufficient culture solution and then transferred to Fe-deficient or Fe-sufficient culture solution for 5 days. The transformants had elevated Fe3+ chelate-reductase activity only when grown in Fe-deficient culture solution. The three transformant lines, 7, 8, and 11, displayed 1.8, 2.2, and 2.2 times the activity of the vector control, respectively, at pH 5.5; corresponding values at pH 8.0 were 1.6, 1.8, and 1.6 (Fig. 2A).
Fig. 2.
Assays of Fe3+ chelate-reductase activity in vector control and the transformants. (A) Transformants (lines 7, 8, and 11) and vector control (V) were grown on standard culture solution for 3 weeks and then transferred to Fe-deficient culture solution for 5 days before the assay. pHs of assay buffers were 5.5 or 8.0. (B) Fe3+ chelate-reductase activity per root fresh weight in roots surface of transformants (lines 7 and 11) and vector control (V). Rice plants were grown for 3 weeks in normal nutrient solution and then transferred to Fe-deficient culture; roots were harvested 0, 3, and 7 days after the transfer (+2 and +5 indicate the number of days after Fe resupply; n = 9). (C) Total Fe3+ chelate-reductase activity in whole roots surface of transformants (lines 7 and 11) and vector control (V) (n = 9). (D) Degree of chlorosis of the fully expanded youngest leaf by using a SPAD-502 chlorophyll meter. The values followed by different letters are statistically different according to a Student-Newman-Keuls test (P < 0.05).
Furthermore, we measured the time course of Fe3+ chelate-reductase activities at the surface of rice roots of the vector control and transgenic lines 7 and 11 under Fe-deficient conditions. Fe3+ chelate-reductase activities per roots fresh weight of transformants were two times higher as compared with that of the vector control 7 days after onset of Fe-deficiency treatment (Fig. 2B). Fe3+ chelate-reductase activities in whole transformants' roots was 1.7-fold as compared with that of the vector control 7 days after onset of Fe-deficiency treatment (Fig. 2C). Fe3+ chelate-reductase activities of the vector control and transformants were not different after Fe resupply.
The average SPAD-502 chlorophyll meter value in the leaves of the vector controls decreased, whereas SPAD values of transformants were not decreased 3 days after onset of Fe-deficiency treatment (Fig. 2D). These values of the vector controls and transformants were decreased 7 days after onset of Fe-deficiency treatment. After Fe resupply, the chlorophyll content of transformants, moreover, recovered faster as compared with vector control.
Fe-Uptake Rate of the Transformants.
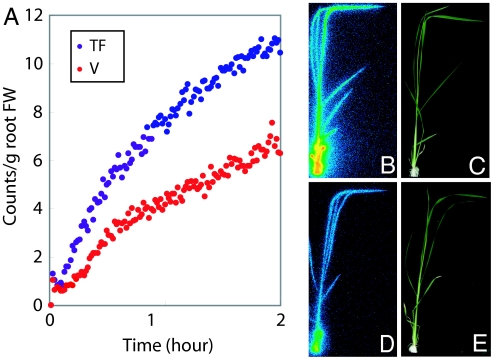
We conducted uptake experiments using PETIS, which is capable of visualizing real-time Fe translocation in plants, to determine the Fe-uptake rate in transformants and vector controls. Plants that were grown in Fe-sufficient culture solution for 2 weeks and then transferred to Fe-deficient conditions for 5 days were supplied with 52Fe3+-ethylenediamine-N,N,N′,N′-tetraacetic acid (EDTA; 0.43 MBq, 30.7 fmol) and Fe3+-EDTA (0.1 mM) for PETIS analysis. The relative Fe-uptake rate in the transformants was two times that in the vector controls (Fig. 3A). Images taken by using a Bio-imaging Analyzer system (BAS, FujiFilm, Minato-ku, Tokyo, Japan) confirmed that the transformants absorbed and translocated more Fe to the leaves than did the controls (Fig. 3 B–E). γ-ray spectrometry confirmed that the accumulation of Fe in the transformants was two times that in the vector controls for stems and leaves (data not shown).
Fig. 3.
Uptake and transport of Fe as monitored by using PETIS. (A) Relative Fe-transition rate of transformant (TF; line 7) and vector control (V) in stem. (B) Bio-Imaging Analyzer System (BAS Bioanalytical Systems) image of the transformant. (C) Photograph of the transformant. (D) BAS Bioanalytical Systems image of the vector control. (E) Photograph of the vector control. Colors from blue through green, yellow, orange, and red indicate increasing Fe uptake in B and D.
Tolerance to Fe Deficiency in the Transformants.
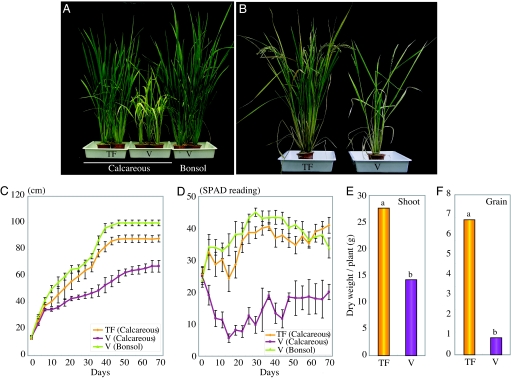
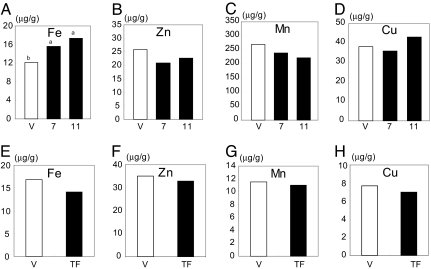
When plants were cultured in calcareous soil (pH 8.5) with low-Fe availability, the transformants showed remarkable tolerance to low Fe and grew better than the vector control plants (Fig. 4 A and B). The control plants showed reduced growth, and their leaves displayed chlorosis caused by the low-Fe supply. In contrast, the leaves of transformants remained green, similar to those of vector controls grown in bonsol (normal cultivation soil; pH 5.0). There were no other phenotypic differences between the transformants and controls grown in bonsol (data not shown). We determined the concentrations of Fe, Zn, Mn, and Cu in vector controls and transformants. There was a slight increase in Fe concentration in shoots of transgenic lines 7 and 11, as compared with vector control, whereas there was no difference in the concentrations of Zn, Mn, or Cu between transformants and vector control plants (Fig. 5 A–D).
Fig. 4.
Growth features and grain yield of transgenic rice plants containing the refre1/372 gene (line 7) and vector controls grown in calcareous soil (pH 8.5) and in bonsol (normal cultivated soil). (A) Transformant (TF, Left) and vector control (V, Center) after 4 weeks of growth in a calcareous soil; vector control (V, Right) in bonsol. (B) Transformant (TF, Left) and vector control (V, Right) after 17 weeks of growth in calcareous soil. (C) Plant height of transformant (TF; mean ± SD, n = 5) and vector control (V; mean ± SD, n = 3) for 70 days after transplanting into calcareous soil and bonsol. (D) SPAD-502 value (chlorophyll content) in leaves of the transformant (TF; mean ± SD, n = 5) and vector control (V; mean ± SD, n = 3) for 70 days after transplanting into calcareous soil and bonsol. (E) Dry weight of shoots (i.e., without grain) of transformant (TF; n = 5) and vector control (V; n = 3) after cultivation for 17 weeks in calcareous soil. (F) Grain yield of transformant (TF; n = 5) and vector control (V; n = 3) after cultivation for 17 weeks in calcareous soil. The values followed by different letters are statistically different according to a Student-Newman-Keuls test (P < 0.05).
Fig. 5.
Metal concentrations in transformants and vector control grown in calcareous soil. (A–D) Metal concentrations in shoots of transformants (lines 7 and 11, filled bars) and vector control (V, open bars). (E–H) Metal concentrations in seeds of transformants (TF; line 7, filled bars) and vector control (V, open bars).The concentrations of Fe, Zn, Mn, and Cu are expressed as micrograms per gram of dry weight (n = 3). The values followed by different letters are statistically different according to a Student-Newman-Keuls test (P < 0.05).
It was evident from the measurements of plant height that the transformant plants grew much better than the vector control plants (Fig. 4C). The average SPAD-502 value (leaf chlorophyll content), measured by a chlorophyll meter in the leaves of the transformants, was higher than that in control plants at the vegetative stage (Fig. 4D). The average shoot dry weight of transformants after 17 weeks of growth in calcareous soil was 1.9 times that of the controls, and the average grain yield per plant in transformants was 7.9 times that of the controls (Fig. 4 E and F). However, there was no difference in the concentrations in seeds of Fe, Zn, Mn, or Cu between transformants and vector control plants (Fig. 5 E–H).
Discussion
In this report, we have shown that transgenic rice plants with the refre1/372 gene display tolerance to low-Fe availability in calcareous soils (Fig. 4) by enhancing Fe3+ chelate-reductase activity (Fig. 2). These transformants took up Fe at a higher rate than did the vector controls under Fe-deficient conditions (Fig. 3), indicating that the transformants successfully reduced chelated Fe3+ to Fe2+ and took up Fe2+ by using an Fe2+ transporter, which is distinct from the Strategy II Fe-uptake system.
In rice plants, Fe3+ chelate-reductase activity is very low compared with that in other plants and does not increase even under Fe-deficient conditions (14). Although two Fe3+ chelate reductase homologs are present in the rice genome, these genes were not expressed in Fe-sufficient or Fe-deficient roots (14). This situation ties in with the evolutionary development of rice from plants adapted to soils of low redox potential, in which there is abundant Fe2+ (15). Rice plants, therefore, do not need to reduce Fe3+ to Fe2+ under such conditions, which may have been selected through the development of a functional Fe2+-regulated transporter, but not Fe3+ chelate reductase.
In previous work, we reported the generation of tobacco plants that are tolerant of Fe deficiency (19). Initially, we introduced FRE1 into tobacco plants. However, the transgenic tobacco only expressed truncated FRE1 transcripts and gained no additional Fe3+ chelate-reductase activity (19). The FRE1 gene was modified and completely reconstructed to produce full-size transcripts in plants (19). Moreover, we screened refre1 derivatives with higher reductase activity at high pH by using a mutagenesis method. The transgenic tobacco carrying one of the derivatives, refre1/372, showed additional Fe3+ chelate-reductase activity, especially in calcareous soils (17). Therefore, we decided that introducing refre1/372 into rice plants was appropriate to produce transgenic rice plants tolerant to Fe deficiency in calcareous soils.
Before this study, some trials had introduced Fe3+ chelate reductase into rice plants. The ArabidopsisFe3+ chelate-reductase gene with its own promoter was introduced into rice plants, but the transgene was not detected at the mRNA level (20). We therefore considered it crucial to use a preferred promoter for functional refre1.
In Arabidopsis, Fe3+ chelate reductase and IRT1 transcripts were undetectable by RNA gel blot analysis when the plants were grown under Fe-sufficient conditions, but were greatly induced in the epidermal cells of roots 24 h after transfer to Fe-deficient medium; protein levels were also under tight control because both Fe3+ chelate reductase and IRT1 are subject to posttranscriptional control (4, 8, 21). In rice plants, the functional Fe2+ transporter gene OsIRT1 is also highly expressed in Fe-deficient roots and is localized in root epidermal cells (14). Therefore, using the OsIRT1 promoter to drive the Fe3+ chelate-reductase gene seemed to be the best way to enhance Fe uptake by the Strategy I system in rice plants, as occurs in the Arabidopsis Fe uptake system. We finally succeeded in enhancing Fe uptake by the Strategy I system in rice plants, as in the Arabidopsis Fe uptake system by using the OsIRT1 promoter to drive refre1/372.
When refre1/372 was expressed in yeast, we detected strong Fe3+ chelate-reductase activity (17). However, Fe3+ chelate-reductase activity in transgenic rice plants was only approximately two times that in vector controls, which was similar to transgenic tobacco plants with refre1/372 under the control of the cauliflower mosaic virus 35S promoter (17). Despite their relatively low increase in Fe3+ chelate-reductase activity, transgenic rice plants gained resistance under low-Fe availability, suggesting that Fe3+ chelate-reductase activity may be regulated so as not to increase Fe uptake beyond that necessary.
A recent study has shown that transgenic rice containing nicotianamine aminotransferase genes from barley is more tolerant than nontransgenic rice to low-Fe availability in calcareous soil (16). This phenomenon occurs because transgenic rice plants secrete higher amounts of MAs than do nontransgenic rice plants. Introducing genes encoding the enzymes in the biosynthetic pathway of MAs, together with the OsIRT1-promoter refre1/372, has the potential for engineering rice plants that are even more tolerant to low-Fe conditions, thereby having increased productivity in calcareous soils.
Materials and Methods
Plant Material.
Transgenic rice T1 seeds were germinated for 2 weeks on Murashige and Skoog (MS) medium at 28°C under 16-h light/8-h dark conditions. After germination, the seedlings were transferred to a 20-liter plastic container containing a nutrient solution with the following composition: 0.7 mM K2SO4, 0.1 mM KCl, 0.1 mM KH2PO4, 2.0 mM Ca(NO3)2, 0.5 mM MgSO4, 10 μM H3BO3, 0.5 μM MnSO4, 0.2 μM CuSO4, 0.5 μM ZnSO4, 0.05 μM Na2MoO4, and 0.1 mM Fe-EDTA. The nutrient solution was adjusted daily to pH 5.5 with 1 M HCl and was renewed weekly. For the Fe-deficiency treatments, 3-week-old plants were transferred to a nutrient solution without Fe and grown for 5 more days.
RT-PCR.
Total RNA was extracted from rice plants grown under control or Fe-deficient conditions, and the RNA was treated with RNase-free DNase I (TaKaRa, Otsu, Shiga, Japan) to remove contaminating genomic DNA. First-strand cDNA was synthesized by using SuperScript II reverse transcriptase (Invitrogen, Minato-ku, Tokyo, Japan) by priming with oligo-d(T)30. The primers used for RT-PCR of refre1/372 were refre1/372 forward (5′-GCGCGCGGTACCTCTAGGATGGTT-AGAACCAGAGTC) and refre1/372 reverse (5′-CGC-GCGCTCGAGCCAAGTAAAACTCTCCTCCTCTA). The primers used for RT-PCR of OsIRT1 were OsIRT1 forward (5′-CGTCTTCTTCTTCTCCACCACGAC) and OsIRT1 reverse (5′-GCAGCTGATGATCGAGTCTGACC). The α-tubulin primers used for RT-PCR were α-tubulin forward (5′-TCTTCCACCCTGAGCAGCTC) and α-tubulin reverse (5′-AACCTTGGAGACCAGTGCAG). The sizes of the amplified fragments were confirmed by gel electrophoresis and by sequencing by using a Thermo Sequenase Cycle sequencing kit (Shimadzu, Kyoto, Japan) and a DNA sequencer (DSQ-2000L; Shimadzu).
Rice Transformation.
Plasmid pIG121Hm containing the OsIRT1 promoter-uidA gene was used (14). The construct had XbaI and SalI sites on the 3′ side of the 0.8-kb OsIRT1 promoter. Plasmid pOH5 (17) containing refre1/372 was digested at XbaI and SalI sites and subcloned to the 3′ side of the OsIRT1 promoter in pIG121Hm. Agrobacterium tumefaciens strain (C58) carrying the above construct was used to transform rice (Oryza sativa L. cv. Tsukinohikari) following the method of Ishimaru et al. (14). The T1 seeds obtained from the transformants were germinated on MS medium containing 50 mg/liter hygromycin B.
Root Fe3+ Chelate-Reductase Activity Assay.
The Fe3+ chelate-reductase activity was determined for whole intact root systems as described previously (17). In brief, roots were rinsed and submerged in 150 ml of assay solution (0.2 mM CaSO4, 5 mM 2-[4-(2-Hydroxyethyl)-1-piperazinyl]ethanesulfonic acid at pH 5.5 or 8.0, 0.1 mM Fe3+-EDTA, and 0.2 mM bathophenanthroline disulfonic acid disodium salt) (Wako Pure Chemical Osaka, Japan). After 1 h at 25°C, an aliquot of the assay solution was removed, and the absorbance of the solution at 535 nm was determined. The amount of Fe2+ produced was calculated from a calibration curve by using solutions of known Fe2+ concentrations.
Determination of Metal Concentrations.
We performed an elemental analysis of the vector control and transformants. These plants were dried for 1 week at 65°C. The plants (30–50 mg) were then wet-ashed with 2 ml of 11 M HNO3 for 5 h at 150°C. The metal concentrations were measured by using inductively coupled plasma atomic emission spectrometry (SPS1200VR; Seiko, Tokyo, Japan) at wavelengths of 238.204 (Fe), 213.856 (Zn), 293.930 (Mn), and 324.754 (Cu) nm (22).
Production of 52Fe.
The 52Fe (half life: 8.27 h) was produced by the natCr (α, 4n) 52Fe reaction by bombarding a 1.5-mm-thick Cr foil (natural isotopic composition, 99.9% purity; Goodfellow Metals Ltd., Huntingdon, U.K.) with a 100-MeV α-beam from the TIARA AVF cyclotron; ≈1 MBq of 52Fe was produced by using a beam current of 3 μA for 2 h. The radiochemical separation of 52Fe from the target was carried out as described by Watanabe et al. (23). The 52Fe3+ solution lacking nonradioactive Fe was adjusted to ≈pH 3.0 with 1 M KOH, and the 52Fe3+ was chelated with 1.12 μmol of EDTA in darkness for 2 h or reduced with 0.1 mM hydroxylamine for 2 h.
Measurements Using PETIS and Positron Multiprobe System (PMPS).
The PETIS used positron-emitting nuclides to monitor the movement of nutrients in plants in real time. Plant samples containing a positron-emitting nuclide were placed between two opposed 2D block detectors composed of Bi4Ge3O12 scintillator arrays. Two annihilation γ-rays from the decaying positrons were detected in coincidence by these detectors (24). The original position of the annihilation was localized at the intersection of the object plane with a line connecting the two detection points on the detectors. The field of view was 143 × 215.6 mm, and the spatial resolution was 2.4 mm. The resulting image was displayed on a monitor after automatic correction for a given half-life (8.27 h) and for relative detection efficiencies within the field of view. Two pairs of PMPS detectors were used for time-course tracer analysis of positron-emitting nuclides. The lack of the field of view in PETIS was supplemented with the PMPS detectors. The PETIS and PMPS detectors were calibrated as follows: 22Na (1 mm radius, 370 kBq) was placed at the center between the paired detectors, and radioactivity was counted by using these efficiency values. The analysis of the results was performed on a Macintosh (Cupertino, CA) computer by using the public-domain program National Institutes of Health Image (developed at the U.S. National Institutes of Health and available on the Internet by anonymous FTP at zippy.nimh.nih.gov or on floppy disk from the National Technical Information Service, Springfield, VA, part number PB95–500195GEI).
Quantitative Analysis by γ-Ray Spectrometry.
After PETIS analysis or image analysis by using a Bio-Imaging Analyzer System (BAS Bioanalytical Systems), as described below, the plants were cut into parts and analyzed for the absolute amount of radioactivity from the γ-rays of decaying positrons. γ-ray spectra were measured with an ORTEC HPGe detector of 38% relative efficiency and standard electronics. The detector and the sample, placed 11 cm from the detector's surface, were housed in a lead-shielded box. The radioactivity of the 52Fe was determined from the peak area at 168 keV.
Conditions for 52Fe Translocation in Plants.
Fe-deficient transgenic and vector control plants were supplied with 15 ml of culture solution lacking Fe in a polyethylene bag. The plants and bags were fixed on an acrylic board and placed between a pair of PETIS detectors in a chamber at 30°C under 65% humidity and a light density of 320 μmol m−2 s−1. Both EDTA-52Fe3+ (0.43 MBq, 30.7 fmol) and EDTA-Fe3+ (0.1 mM) were added to the culture solutions. After 6 h of PETIS analysis, the plants were removed from the polyethylene bags, and the roots were gently washed for 1 min in 100 ml of a solution of 50 μM EDTA lacking Fe. The plants were then placed under a Bio-Imaging plate inside a cassette. After 30 min, the plate was scanned by an image analyzing system (BAS, FujiFilm), and then a quantitative analysis by γ-ray spectrometry was performed with a maximum value set to 100. This experiment was repeated at least four times with different plants (V.C. and line 7) to confirm the reproducibility of the results.
Evaluation of Transgenic Rice for Tolerance to Fe Deficiency on Calcareous Soils.
Transgenic T1 rice seeds were germinated for 2 weeks on MS medium at 28°C under 16-h light/8-h dark conditions. After germination, the seedlings were transferred to 500 g of calcareous soil obtained from Takaoka City (Toyama, Japan) and containing 1 g of CFR-M2 fertilizer (25), and they were grown in a pot in a greenhouse under natural light conditions. For control soil conditions, bonsol (Sumitomo Chemical Company, Osaka, Japan) was used.
The degree of chlorosis of the fully expanded youngest leaf was determined by using a SPAD-502 chlorophyll meter (Minolta, Tokyo, Japan). The average SPAD-502 chlorophyll meter values are indicated as the mean ± SD of six assays.
Acknowledgments
We thank Dr. P. Blamey and Dr. K. Bashir for assistance with the English, and Dr. S. Nagasaka, R. N. Itai, Dr. H. Inoue, and Dr. M. Suzuki for variable discussion.
Abbreviations
- IRT1
Fe-regulated transporter
- FRE1
yeast Fe3+ chelate reductase
- MA
mugineic acid
- PETIS
positron-emitting tracer imaging system
- PMPS
positron multiprobe system
- refre1/372
reconstructed yeast Fe3+ chelate reductase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 7311.
References
- 1.Wallace A, Lunt O-R. J Am Soc Hortic Sci. 1960;75:819–840. [Google Scholar]
- 2.Marschner H, Romheld V, Kissel M. J Plant Nutr. 1986;9:695–713. [Google Scholar]
- 3.Robinson N-J, Procter C-M, Connolly E-L, Guerinot M-L. Nature. 1999;397:694–697. doi: 10.1038/17800. [DOI] [PubMed] [Google Scholar]
- 4.Connolly E-L, Campbell N-H, Grotz N, Prichard C-L, Guerinot M-L. Plant Physiol. 2003;133:1102–1110. doi: 10.1104/pp.103.025122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eide D, Broderius M, Fett J, Guerinot M-L. Proc Natl Acad Sci USA. 1996;93:5624–5628. doi: 10.1073/pnas.93.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henriques R, Jasik J, Klein M, Martinoia E, Feller U, Schell J, Pais M-S, Koncz C. Plant Mol Biol. 2002;50:587–597. doi: 10.1023/a:1019942200164. [DOI] [PubMed] [Google Scholar]
- 7.Varotto C, Maiwald D, Pesaresi P, Jahns P, Salamini F, Leister D. Plant J. 2002;31:589–599. doi: 10.1046/j.1365-313x.2002.01381.x. [DOI] [PubMed] [Google Scholar]
- 8.Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot M-L, Briat J-F, Curie C. Plant Cell. 2002;14:1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takagi S, Nomoto K, Takemoto T. J Plant Nutr. 1984;7:1–5. [Google Scholar]
- 10.Guerinot M-L, Yi Y. Plant Physiol. 1994;104:815–820. doi: 10.1104/pp.104.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curie C, Panaviene Z, Loulergue C, Dellaporta S-L, Briat J-F, Walker E-L. Nature. 2001;409:346–349. doi: 10.1038/35053080. [DOI] [PubMed] [Google Scholar]
- 12.Higuchi K, Kanazawa K, Nishizawa N-K, Mori S. Plant Soil. 1996;178:171–177. [Google Scholar]
- 13.Bughio N, Yamaguchi H, Nishizawa N-K, Nakanishi H, Mori S. J Exp Bot. 2002;53:1677–1682. doi: 10.1093/jxb/erf004. [DOI] [PubMed] [Google Scholar]
- 14.Ishimaru Y, Suzuki M, Tsukamoto T, Suzuki K, Nakazono M, Kobayashi T, Wada Y, Watanabe S, Matsuhashi S, Takahashi M, et al. Plant J. 2006;45:335–346. doi: 10.1111/j.1365-313X.2005.02624.x. [DOI] [PubMed] [Google Scholar]
- 15.Chang T-T. In: Rice: Origin, History, Technology, and Production. Smith CW, Dilday R-H, editors. Hoboken, NJ: Wiley; 2003. pp. 1–25. [Google Scholar]
- 16.Takahashi M, Nakanishi H, Kawasaki S, Nishizawa N-K, Mori S. Nat Biotechnol. 2001;19:466–469. doi: 10.1038/88143. [DOI] [PubMed] [Google Scholar]
- 17.Oki H, Kim S, Nakanishi H, Takahashi M, Yamaguchi H, Mori S, Nishizawa N-K. Soil Sci Plant Nutr. 2004;50:1159–1165. [Google Scholar]
- 18.Vasconcelos M, Eckert H, Arahana V, Graef G, Grusak M-A, Clemente T. Planta. 2006;224:1116–1128. doi: 10.1007/s00425-006-0293-1. [DOI] [PubMed] [Google Scholar]
- 19.Oki H, Yamaguchi H, Nakanishi H, Mori S. Plant Soil. 1999;215:211–220. [Google Scholar]
- 20.Vasconcelos M, Musetti V, Li C-M, Datta K-S, Grusak A-M. Soil Sci Plant Nutr. 2004;50:1152–1157. [Google Scholar]
- 21.Connolly E-L, Fett J-P, Guerinot M-L. Plant Cell. 2002;14:1347–1357. doi: 10.1105/tpc.001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishimaru Y, Suzuki M, Kobayashi T, Takahashi M, Nakanishi H, Mori S, Nishizawa N-K. J Exp Bot. 2005;56:3207–3214. doi: 10.1093/jxb/eri317. [DOI] [PubMed] [Google Scholar]
- 23.Watanabe S, Ishioka N-S, Osa A, Koizumi M, Sekine T, Kiyomiya S, Nakanishi H, Mori S. Radiochim Acta. 2001;89:853–858. [Google Scholar]
- 24.Kume T, Matsuhashi S, Shimazu M, Ito H, Fujimura T, Adachi K, Uchida H, Shigeta N, Matsuoka H, Osa A, Sekine T. Applied Radiot Isot. 1997;48:1035–1043. [Google Scholar]
- 25.Morikawa C-K, Saigusa M, Nakanishi H, Nishizawa N-K, Hasegawa K, Mori S. Soil Sci Plant Nutr. 2004;50:1013–1021. [Google Scholar]