Abstract
Despite important advances in microarray-based molecular classification of tumors, its application in clinical settings remains formidable. This is in part due to the limitation of current analysis programs in discovering robust biomarkers and developing classifiers with a practical set of genes. Genetic programming (GP) is a type of machine learning technique that uses evolutionary algorithm to simulate natural selection as well as population dynamics, hence leading to simple and comprehensible classifiers. Here we applied GP to cancer expression profiling data to select feature genes and build molecular classifiers by mathematical integration of these genes. Analysis of thousands of GP classifiers generated for a prostate cancer data set revealed repetitive use of a set of highly discriminative feature genes, many of which are known to be disease associated. GP classifiers often comprise five or less genes and successfully predict cancer types and subtypes. More importantly, GP classifiers generated in one study are able to predict samples from an independent study, which may have used different microarray platforms. In addition, GP yielded classification accuracy better than or similar to conventional classification methods. Furthermore, the mathematical expression of GP classifiers provides insights into relationships between classifier genes. Taken together, our results demonstrate that GP may be valuable for generating effective classifiers containing a practical set of genes for diagnostic/prognostic cancer classification.
Keywords: Molecular diagnostics, biomarkers, prostate cancer, evolutionary algorithm, microarray profiling
Introduction
The development of high-throughput microarray-based technology will potentially revolutionize cancer research in a number of areas including cancer classification, diagnosis, and treatment. Expression profiling at the mRNA level can be used in the molecular characterization of cancer by simultaneous assessment of a large number of genes [1–5]. This approach can be used to determine gene expression alterations between different tissue types such as those obtained from healthy controls and patients with cancer. Analysis of such large-scale gene expression profiles of cancer will facilitate the identification of a subset of genes that could function as diagnostic or prognostic biomarkers. The development of molecular classifiers that allow segregation of tumors into clinically relevant molecular subtypes beyond those possible by pathologic classification may subsequently serve to classify tumors with unknown origin into different cancer types or subtypes. However, due to the large number of genes and the relatively small number of patient cases available from such studies, finding a robust gene signature for reliable prediction remains a challenge.
A number of computational and statistical models have been developed for molecular classification of tumors. Golub et al. [3] proposed a weighted voting scheme to identify a subset of 50 genes that can discriminate acute myeloid leukemia from acute lymphoblastic leukemia, and subsequently predict class membership of new leukemia cases. Using the same data set, Mukherjee et al. [6] developed a kernel-based support vector machine (SVM) classifier and achieved a higher performance in term of the accuracy of assigning leukemia samples into acute myeloid leukemia or acute lymphoblastic leukemia. van't Veer et al. [7] developed a 70-gene predictor that was highly correlated with breast cancer prognosis. Khan et al. [8] introduced an artificial neural network to classify different histopathologic types of small round blue cell tumors (SRBCTs) and identified 96 most discriminative genes for classification. A deterministic tree-based method was also introduced by Zhang et al. [9] in 2003 to construct random forests for classifications of leukemia and lymphoma. Although these methods have had some success in classifying tumors, they are often developed by using parametric statistical techniques and thus have difficulty in finding nonlinear relationships between genes. Alternatively, complex models are used that deliver “black box” solutions for classification and do not give insight into relationships between genes.
In this study, we present a machine learning approach called genetic programming (GP) for molecular classification of cancer. GP belongs to a class of evolutionary algorithms and was first introduced by Koza [10] in 1992. Recently, evolutionary algorithms have been used to analyze gene expression data and select molecular signatures for sample classification [11–17]. For example, GP has been shown to be a promising approach for discovering comprehensible rule-based classifiers from medical data [11,16] as well as gene expression profiling data [12,14,17–20]. However, the potential of GP in cancer classification has not been fully explored. For example, GP classifiers identified from one data set have not been validated in independent data sets. Here, we applied GP algorithm to cancer expression profiling data to identify potentially informative feature genes, build molecular classifiers, and classify tumor samples. We tested GP in one SRBCT, one lung adenocarcinoma, and five prostate cancer data sets and evaluated the generality of GP classifiers within and across data sets. In addition, we compared the performance of GP with that of other common classification techniques, such as linear discriminant analysis and SVMs, for prediction accuracy.
Materials and Methods
Data Sets
All data sets were obtained from ONCOMINE [21] or requested from the original authors. The SRBCT data [8] contained 88 samples from four types of cancer cells: neuroblastoma (NB), rhabdomyosarcoma (RMS), the Ewing family of tumors (EWS), and Burkitt lymphoma (BL). The entire data set, excluding five non-SRBCT samples, was divided into a training set (63 samples) and a validation set (20 samples) as described in the original study.
The lung adenocarcinoma data set [22] contains 86 lung cancer samples. The raw CEL files were preprocessed using RMAExpress (2003, UC Berkeley, http://rmaexpress.bmbolstad.com/) [23] and normalized so that each array had zero mean and unit variance. The samples were then subdivided into a high- or low-risk group as requested from the authors. Twenty-eight high-risk and 38 low-risk samples were included in the training set and the remaining 20 samples were considered as the validation set.
Three prostate cancer data sets [2,24,25] from the University of Michigan (UM), Stanford University (Stanford) and the University of Pittsburgh (Pittsburgh), respectively, were used to classify primary prostate cancer (PCA) from benign prostate samples. A total of 56 samples were randomly selected from Stanford data set as the training set. The rest of the Stanford samples and UM and Pittsburgh sets were treated as validation sets.
In addition, two prostate cancer data sets including the Pittsburgh and the LaTulippe et al. (Memorial Sloan-Kettering Cancer Center, MSKCC) data sets [25,26] were retrieved to distinguish metastatic prostate cancer (MET) and PCA. Detailed study information is shown in Table 1.
Table 1.
Gene Expression Data Sets Used in This Study.
| Class Description | Authors | Journal | Array Type | No. of Genes |
| Four classes: NB, RMS, BL, EWS | Khan et al. [8] | Nat Med 7:673 | cDNA | 2,308 |
| Two classes: high-risk group and low-risk group | Beer et al. [22] | Nat Med 30:41 | Affymetrix Hu6800 | 7,070 |
| Two classes: PCA and MET | LaTulippe et al. [26] | Cancer Res 62:4499 | Affymetrix HG_U95A | 3,547 |
| Yu et al. [25] | J Clin Oncol 22:2790 | Affymetrix HG_U95A | 3,547 | |
| Two classes: benign prostate samples (BENIGN) and PCA | Lapointe et al. [24] | Proc Natl Acad Sci USA 101:811 | cDNA | 4,168 |
| Dhanasekaran et al. [2] | Nature 412:822 | cDNA | 16,965 | |
| Yu et al. [25] | J Clin Oncol 22:2790 | Affymetrix HG_U95A | 12,558 |
Genetic Programming for Classification
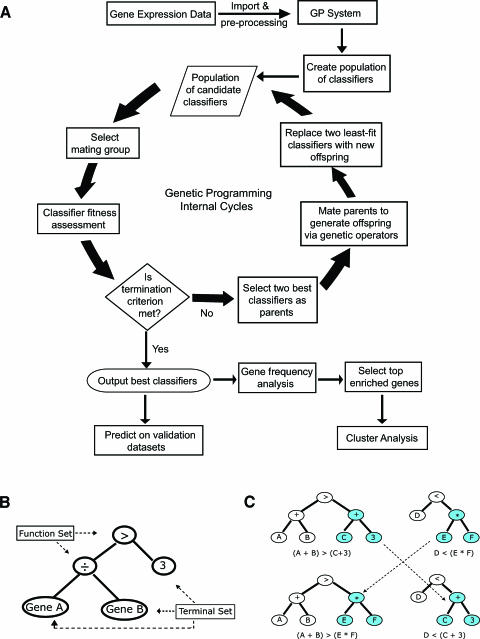
Genetic programming [10] is an evolutionary algorithm that simulates natural selection and population dynamics to search for intelligible relationships among the constituents in a system (classifiers in this study). A basic flowchart for the GP system is given in Figure 1A. Briefly, the system randomly selects inputs such as gene identifiers and constant values, which are used to represent the expression values of corresponding genes. Such selected inputs are then combined with the function operators such as arithmetic or Boolean operators to compose tree-based GP classifiers, an example of which is given in Figure 1B. Such classifiers are eventually accumulated to form an initial population, where a small subgroup of classifiers is then selected to create a “mating group.” Each classifier in this mating group is assessed by a fitness function defined as the area under the receiver operating characteristic curve (ROC-AUC), which is widely used to assess the accuracy of a diagnostic test that yields continuous test results in clinical research areas. The two fittest classifiers are then selected as “mating” parents by a tournament selection scheme and “mated” to produce “offspring” through selective genetic operators such as crossover, or mutation. The crossover operator exchanges a subtree of one parent with the other to generate offspring (Figure 1C), whereas the mutation operator probabilistically chooses a node in a subtree and replaces it with a new created subtree randomly. The generated offspring then replaces the least-fit parent classifiers in the population. Once new offspring fully replaces parent classifiers in the entire population, a new generation that in general contains better classifiers is created. This process of mating pool selection, fitness assessment, mating, and replacement is repeated over generations, progressively creating better classifiers until a termination criterion is met (e.g., a perfect classifier with a fitness score of 1 or the maximum number of “generations” is reached).
Figure 1.
(A) Flowchart for the GP process. Briefly, a population of tree-based classifiers is first created by randomly choosing gene expression data or constant values and combining with arithmetic or Boolean operators. An example of tree-based classifiers is represented in (B). A small subgroup of classifiers is then selected as a “mating group” and each classifier in this mating group is assessed by a fitness function, which is defined as the area under the ROC-AUC in this study. The two fittest classifiers are then selected as “mating” parents and “mated” to produce “offspring” by genetic operators (crossover or mutation). The generated offspring then replace the least-fit parent classifiers within the population. A new generation of populations is generated once the offspring fully replaced parent classifiers in the population. This process of mating pool selection, fitness assessment, mating, and replacement is repeated over generations, progressively creating better classifiers until a completion criterion is met. After the best classifiers are outputted, post-GP analyses are carried out to compute gene occurrence in the classifiers as well as to predict on new unknown samples. (B) The representation of a GP tree structure for an exemplified classifier, Gene[A]/Gene[B] > 3. In general, a GP classifier is represented as a tree-based structure composed of the terminal set and function set. The terminal set, in tree terminology, is composed of leaves (nodes without branches) and may represent as genes or constants. The function set is a set of operators such as arithmetic operators (+, -, -, H) or Boolean operators (AND, OR, NOT), acting as the branch points in the tree, linking other functions or terminals. (C) The representation of a crossover operator of GP tree.
Table 2 shows an example of primary GP parameters used to analyze the prostate cancer data set from the LaTulippe et al. (MSKCC) study [26]. Given the limited sample size of each data set, we used n-fold cross-validation procedure to estimate the generalization of classifiers in predicting samples with unknown class membership. For example, when a data set is selected as the training set, it is randomly subdivided into n parts (or folds), wherein classifiers are developed as described in the above GP process using samples in n-1 folds. These classifiers are then tested on samples in the left-out fold to assess their potential generalization capability because such samples are not involved in the development of the classifiers. A good classifier is expected to classify well in the training samples as well as the samples in the left-out fold. This process is repeated n times with each fold taking turns as the testing fold and the best classifiers are then selected based on overall performance on the training folds and the test fold.
Table 2.
Settings for Primary GP Parameters Used to Analyze the Latulippe et al. Prostate Cancer Data.
| Parameter | Setting | Description* |
| Terminal set | All inputs including gene expression values, and constant values | A set where all end (leaf) nodes in the parse trees representing the programs must be drawn. A terminal could be a variable, a constant, or a function with no arguments. |
| Function set | Boolean and floating point operators: <, >, <=, =>, *, /, +, - | A set of operators, e.g., +, -, *, +. These act as the branch points in the parse tree, linking other functions or terminals. |
| Selection | Generational, tournament size 5 | An evolution is called “generational” when the entire existing population of classifiers is replaced by a new created population at every generation. Tournament selection is a mechanism for choosing classifiers from a population. A group of classifiers are selected at random from the population and the best ones are chosen. |
| Elitism | 1 | Property of selection methods, which keeps the best classifier(s) into the next generation. |
| Initial population | Each tree was created by ramped half-and-half | Ramped half-and-half operates by creating an equal number of trees with each depth between a predetermined minimum and maximum. |
| Population size | 20,000 | The number of candidate classifiers in a population. |
| Number of demes | 12 | A deme is a separately evolving subset of the whole population. The subsets may be evolved on different computers. Emigration between subset may occur every generation. |
| Crossover probability | 0.2 | The probability of creating a new individual from parts of its parents. |
| Mutation probability | 0.2 | The probability of a subtree replaced by another, some or all of which is created at random. |
| Termination criteria | Fitness score reaches 1 or max generations (50) | A statement or condition to stop the GP cycle. |
| Initial tree depth | 3 | The initial distance of any leaf from the root of a tree. |
| Initial node count | 3 | The initial number of nodes in a tree. |
| Maximum tree depth | 7 | The maximum distance of any leaf from the root of a tree. |
| Maximum node count | 8 | The maximum number of nodes in a tree. |
| Number of folds | 4 | The number of parts a training set will be subdivided into. |
| Deme migration frequency | Every generation | The frequency of moving classifiers between isolated demes. |
| Deme migration percentage | 5% of individuals | The percentage of classifiers moving between two demes. |
| Fitness | ROC-AUC | A process that evaluates a member of a population and gives it a score or fitness. |
Source of some term descriptions: Langdon WB (1998). Genetic Programming and Data Structures: Genetic Programming + Data Structures = Automatic Programming! Amsterdam: Kluwer.
Overall Methodology for Classifier Validation
In this study, we used GP to discover classifiers that are capable of classifying samples into different cancer types based on gene expression patterns. Typically, a classifier is evolved from a training data set and then validated against independent validation sets to assess its prediction capacity on samples with unknown labels. A generic GP classifier-based prediction is shown as: IF '(GENE[A]/GENE[B]-GENE[C]) > D' THEN 'TARGET CLASS', where the IF clause is generated by GP, “TARGET CLASS” is predefined in the initial configuration file, D is a constant, and GENE[A], GENE[B], and GENE[C] represent the expression levels of genes A, B, and C, respectively. A continuous prediction score is preferred for certain analysis like the ROC curve analysis. To implement that, the classifier can be converted to the form of 'GENE[A]/GENE[B]-GENE[C]', where the calculated mathematical expression values can be then treated as a continuous variable for ROC test.
Implementation and Running Time
We implemented parallel GP algorithm in C (patented by Genetics Squared, Inc., Ann Arbor, MI; http://www.genetics2.com). The analyses were performed on a parallel computer cluster (7 Dell 1850 1U racks with 2x 3.2-GHz Xeon processor and 1 Dell 1750 1U rack with 1x 3.06-GHz Xeon processor; Dell Inc., Round Rock, TX; http://www.dell.com) running the Debian Linux operating system. The running times for different data sets varied from a few minutes to a few days, depending on a large number of parameters such as the complexity of the problem, size of population used in the evolution, number of generations, cost of fitness calculation, number of classifiers, and size of the data set. For the LaTulippe et al. [26] prostate cancer data set with the parameters listed in Table 2, it took approximately three and a half hours to complete a set of 1000 classifiers.
Results
Robust Selection of Feature Genes by GP Evolution
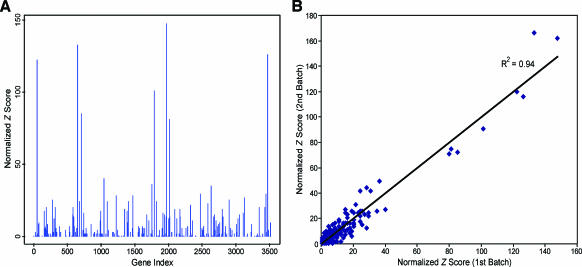
To investigate the ability of GP to robustly select feature genes, we examined gene occurrences across classifiers generated from our GP system. Our results revealed that a small set of genes was frequently selected. For example, an analysis of feature genes in a set of 1000 best classifiers from GP to distinguish PCA from metastatic samples on LaTulippe et al. (MSKCC) prostate data set [26] indicated a high tendency of GP in selecting certain genes across classifiers (Figure 2 and Table W1). Figure 2A presents the normalized z score [13] of the frequency of each gene in the 1000 classifiers that contains a total of 2000 gene occurrences, with the X-axis representing gene index. As shown in the figure, only 261 of the total 3547 genes used for this study occurred at least once. Interestingly, 46 of them occurred at least 12 times (z score ≥15, P < .0001, Table 3). That this small set of genes has dominated the generated classifiers implies that such genes may be truly important for prostate cancer metastasis and may serve as discriminative biomarkers for cancer progression. As GP is stochastic and may give different solutions in each run, it is interesting to examine the reproducibility of gene selection across independent runs. Thus, we created another independent set of 1000 classifiers by using identical GP parameters on the same training set. A total of 264 genes occurred at least once in this set of GP classifiers. Notably, 206 of them were common in both sets and a highly positive correlation of z scores of these gene between the two sets was observed (R2 = 0.94, P < 1 x 10-5; Figure 2B).
Figure 2.
(A) The statistical z score of each of the 3547 genes occurring in the 1000 classifiers generated from the LaTulippe et al. [26] prostate cancer study by GP based on the parameters listed in Table 2. Let Z = [Xi-E(Xi)]/σ, where Xi is the frequency times gene i is selected, E(Xi) is the expectation of frequency times gene i is selected, and σ is the standard deviation of this binomial model. Let, n = 1000; P, the probability of gene i being selected randomly, is approximately equal to the total counts of frequency in 1000 classifiers divided by the number of classifiers (1000), then divided by the total number of genes (3547), then E(Xi) = np, and σ = √[np(1 - p)]. (B) Correlation between commonly occurring genes on two independent sets of classifiers. Each set contains 1000 classifiers.
Table 3.
Frequency of Gene Occurrences in the 1000 Classifiers for the Latulippe et al. Prostate Study.
| Probe Set | Gene Symbol | Gene Title | Count* | Z Score |
| 37407_s_at | MYH11 | Myosin, heavy polypeptide 11, smooth muscle | 112 | 147.59 |
| 32582_at | MYH11 | Myosin, heavy polypeptide 11, smooth muscle | 101 | 133.02 |
| 767_at | MYH11 | Myosin, heavy polypeptide 11, smooth muscle | 96 | 126.40 |
| 1197_at | ACTG2 | Actin, gamma 2, smooth muscle, enteric | 93 | 122.42 |
| 36931_at | TAGLN | Transgelin | 77 | 101.23 |
| 32755_at | ACTA2 | Actin, alpha 2, smooth muscle, aorta | 65 | 85.34 |
| 37576_at | PCP4 | Purkinje cell protein 4 | 62 | 81.36 |
| 774_g_at | MYH11 | Myosin, heavy polypeptide 11, smooth muscle | 61 | 80.04 |
| 34203_at | CNN1 | Calponin 1, basic, smooth muscle | 31 | 40.30 |
| 36834_at | MOXD1 | Monooxygenase, DBH-like 1 | 28 | 36.33 |
| 39333_at | COL4A1 | Collagen, type IV, alpha 1 | 27 | 35.01 |
| 773_at | MYH11 | Myosin, heavy polypeptide 11, smooth muscle | 24 | 31.03 |
| 38834_at | TOPBP1 | Topoisomerase (DNA) II binding protein 1 | 23 | 29.71 |
| 685_f_at | LOC112714 | Similar to alpha tubulin | 23 | 29.71 |
| 34878_at | SMC4L1 | SMC4 structural maintenance of chromosomes 4-like 1 | 22 | 28.38 |
| 35970_g_at | MPHOSPH9 | M-phase phosphoprotein 9 | 22 | 28.38 |
| 41137_at | PPP1R12B | Protein phosphatase 1, regulatory (inhibitor) subunit 12B | 21 | 27.06 |
| 1884_s_at | PCNA | Proliferating cell nuclear antigen | 20 | 25.73 |
| 40407_at | KPNA2 | Karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | 20 | 25.73 |
| 32662_at | MDC1 | Mediator of DNA damage checkpoint 1 | 19 | 24.41 |
| 34376_at | PKIG | Protein kinase (cAMP-dependent, catalytic) inhibitor gamma | 19 | 24.41 |
| 35742_at | C16orf45 | Chromosome 16 open reading frame 45 | 19 | 24.41 |
| 36987_at | LMNB2 | Lamin B2 | 19 | 24.41 |
| 39145_at | MYL9 | Myosin, light polypeptide 9, regulatory | 18 | 23.09 |
| 38430_at | FABP4 | Fatty acid binding protein 4, adipocyte | 17 | 21.76 |
| 1599_at | CDKN3 | Cyclin-dependent kinase inhibitor 3 | 16 | 20.44 |
| 2012_s_at | PRKDC | Protein kinase, DNA-activated, catalytic polypeptide | 16 | 20.44 |
| 32305_at | COL1A2 | Collagen, type I, alpha 2 | 16 | 20.44 |
| 418_at | MKI67 | Antigen identified by monoclonal antibody Ki-67 | 16 | 20.44 |
| 651_at | RPA3 | Replication protein A3, 14kDa | 16 | 20.44 |
| 35474_s_at | COL1A1 | Collagen, type I, alpha 1 | 15 | 19.11 |
| 37749_at | MEST | Mesoderm specific transcript homolog (mouse) | 15 | 19.11 |
| 38031_at | DDX48 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 48 | 15 | 19.11 |
| 39990_at | ISL1 | ISL1 transcription factor, LIM/homeodomain (islet-1) | 15 | 19.11 |
| 1505_at | TYMS | Thymidylate synthetase | 14 | 17.79 |
| 33924_at | RAB6IP1 | RAB6 interacting protein 1 | 14 | 17.79 |
| 35694_at | MAP4K4 | Mitogen-activated protein kinase kinase kinase kinase 4 | 14 | 17.79 |
| 32306_g_at | COL1A2 | Collagen, type I, alpha 2 | 13 | 16.46 |
| 32847_at | MYLK | Myosin, light polypeptide kinase | 13 | 16.46 |
| 37305_at | EZH2 | Enhancer of zeste homolog 2 (Drosophila) | 13 | 16.46 |
| 41081_at | BUB1 | BUB1 budding uninhibited by benzimidazoles 1 homolog | 13 | 16.46 |
| 32272_at | K-ALPHA-1 | Tubulin, alpha, ubiquitous | 12 | 15.14 |
| 36627_at | SPARCL1 | SPARC-like 1 (mast9, hevin) | 12 | 15.14 |
| 37347_at | CKS1B | CDC28 protein kinase regulatory subunit 1B | 12 | 15.14 |
| 39519_at | KIAA0692 | KIAA0692 protein | 12 | 15.14 |
| 40845_at | ILF3 | Interleukin enhancer binding factor 3, 90 kDa | 12 | 15.14 |
Count is the number of occurrences of each gene in 1000 rules.
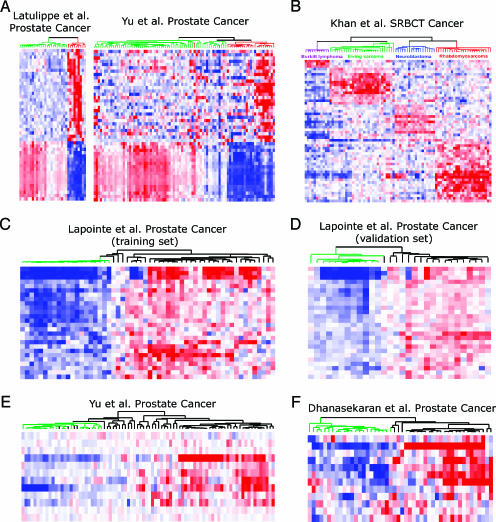
Next we examined the 46 most frequently occurring feature genes in the above analysis (Table 3). Strikingly, the top 3 probes represented the same gene, MYH11, which has been reported to be downregulated in multiple metastatic cancers [27]. Another top-listed gene was EZH2, encoding a polycomb group protein that we and others have previously characterized as overexpressed in aggressive epithelial tumors [28,29]. We therefore hypothesized that the top frequently occurring genes might serve as a multiplex signature to distinguish MET from PCA. To test this, hierarchical clustering was performed to group cancer samples based on the expression patterns of these genes. As shown in Figure 3A, these top 46 genes clustered tumor samples into their corresponding diagnostic classes (MET or PCA), each with a unique expression signature. Interestingly, the same set of genes also successfully classified the independent Yu et al. [25] (Pittsburgh) prostate cancer data set. Similar results were observed when samples of the SRBCT data set were clustered based on the top 54 frequent feature genes (z score ≥14) derived from the training samples of this data set (Figure 3B). In addition, we also selected the top 26 feature genes (z score ≥40) from the 2000 classifiers developed from the Lapointe et al. [24] (Stanford) prostate cancer training data set. Hierarchical clustering based on the expression pattern of these genes grouped tumors of four independent prostate cancer data sets with high classification accuracy (Figure 3, C–F).
Figure 3.
Top feature genes derived from GP separate tumors into their corresponding diagnostic classes. (A) Hierarchical clustering using the top 46 most frequent genes derived from the 1000 classifiers generated for the LaTulippe et al. (MSKCC) data set [26]. Genes were ranked by the frequency of their occurrences in the classifiers. The top 46 frequent genes (z score ≥15, see Figure 2) were selected for hierarchical clustering. The left panel is the clustering of metastatic samples and PCA samples for the MSKCC data, and the right panel is for the Yu et al. [25] (Pittsburgh) validation data set. Rows represent genes and columns represent samples. The green lines in the dendrogram indicate PCA and the red lines represent MET samples. (B) Hierachical clustering of the entire SRBCT data set using the top 54 feature genes obtained from the training set. (C–F) The top 26 most frequent genes from the 2000 classifiers generated from Lapointe et al. [24] (Stanford) training set was used to separate 34 benign/control prostate samples from PCA samples in the Stanford training set (C), Stanford validation set (D), Yu et al. [25] (Pittsburgh) validation set (E), and Dhanasekaran et al. [2] (UM) validation set (F), respectively. The green lines in the dendrogram indicate benign/control prostate samples and the black lines represent PCA samples.
To further investigate whether such feature genes can be used to predict class memberships of validation samples, we carried out class prediction of the SRBCT data set by diagonal linear discriminant analysis (DLDA) and k-nearest neighbor analysis (kNN, k = 3). The top 54 frequent genes selected from the 2000 classifiers generated from the training samples of SRBCT data were used as a gene signature to predict the validation samples. Both DLDA and kNN analysis predicted all of the 20 validation samples with 100% accuracy (data not shown), confirming that the frequent genes derived from GP are truly discriminative genes and capable of predicting unknown samples.
GP Classifiers Successfully Predict Validation Sample Sets
Next we sought to examine the performance of GP classifiers comprising only a handful of feature genes. We first evaluated the ability of GP classifiers to accurately classify four diagnostic classes of cancers (NB, RMS, EWS, and BL) within the SRBCT data set [8]. A set of 63 training samples was used byGPto generate distinguishing classifiers through cross-validation. Classification was performed in a binary mode (target vs. nontarget class). For each target class, the top 10 best classifiers (Table W2) were selected and used to predict a validation set of 20 samples. Most of the classifiers achieved 100% sensitivity and specificity on the training set. Similar prediction accuracy was observed when these classifiers were applied to the 20 blinded validation samples. The best classifiers (Table 4) perfectly predicted all of the validation samples. The average prediction accuracy of the top 10 classifiers for each target class was 98.5% for BL (95% confidence interval [CI], 0.97–1.00), 92.5% for EWS (95% CI, 0.89–0.96), 95.5% for NB (95% CI, 0.91–1.00), and 95.5% for RMS (95% CI, 0.92–0.99). Overall, GP classifiers achieved comparable classification and prediction performance as the method described in the original study, although using much less genes. This high prediction accuracy, however, might be partially due to the fact that the four cancer types here are much more heterogeneous than the subtypes of any single cancer.
Table 4.
Classifiers That Distinguish Different Cancer Classes of SRBCT, Subtypes of Prostate Cancer, or Lung Cancer.
| Analysis | Classifier | Training Errors |
Test Set Errors |
||
| FN | FP | FN | FP | ||
| Small, round blue-cell tumor | IF (HCLS1-GSTA4 > XPO6) THEN BL | 0 | 0 | 0 | 0 |
| IF (PTPN13/COX8A > CDK6) THEN EWS | 0 | 0 | 0 | 0 | |
| IF (SATB1 > CSDA^2) THEN NB | 0 | 0 | 0 | 0 | |
| IF (CDH17/FGFR4 <= MYL4) THEN RMS | 0 | 0 | 0 | 0 | |
| Primary prostate cancer vs. metastatic prostate cancer | IF (ARL6IP > MYH11) THEN MET | 0 | 0 | 0 | 4 |
| IF (MYH11 < MYH11) THEN MET | 0 | 0 | 0 | 4 | |
| Lung cancer (high risk vs. low risk) | IF (LTBP2-IARS) <= (ADM + (CCT2 * FCGR2A)) THEN High-Risk | 0 | 1 | 1 | 0 |
| IF (GYPB-MN1) < (ADM + (MCFD2 + CKS2)) THEN High-Risk | 1 | 0 | 3 | 0 | |
Only one or two classifiers per class per analysis are listed in the table. FN, number of false negatives; FP, number of false positives.
Thus, we next examined GP in classifying subtypes of lung adenocarcinoma, wherein samples were designated as “high risk” or “low risk” based on the original publication information [22]. One hundred classifiers were generated by GP from 66 training samples and the top 5 were found to have the highest training accuracy of 98.5%. When these 5 classifiers were applied to the 20 test samples, we found a maximal prediction accuracy of 98.5% and an average prediction rate of 84.0% (95% CI, 0.70–0.99), comparable with that of other classification methods as described in the later session.
A more challenging work is to validate classifiers across independent data sets. We thus investigated whether GP could distinguish molecular subtypes of a single cancer class from independent data sets. Two prostate cancer data sets (Pittsburgh and MSKCC sets, respectively) were used to evaluate GP in classifying PCA or MET. Genes within each data set were standardized to have zero mean and unit variance, given that similar proportion of metastatic samples was observed in both data sets. The MSKCC samples were used as a training set to generate GP classifiers. The 20 classifiers that perfectly classified PCA from MET in the training set were selected for prediction. When these classifiers were applied to predict the independent Pittsburgh prostate cancer samples, the best classifiers (Table 4) correctly predicted all METs, and 58 of 62 clinically localized PCAs. This led to 100% sensitivity and 93.5% specificity. The average prediction accuracy of all of the 20 classifierswas 95.2%sensitivity (95%CI, 0.87–1.00) and 82.1% specificity (95% CI, 0.65–0.99).
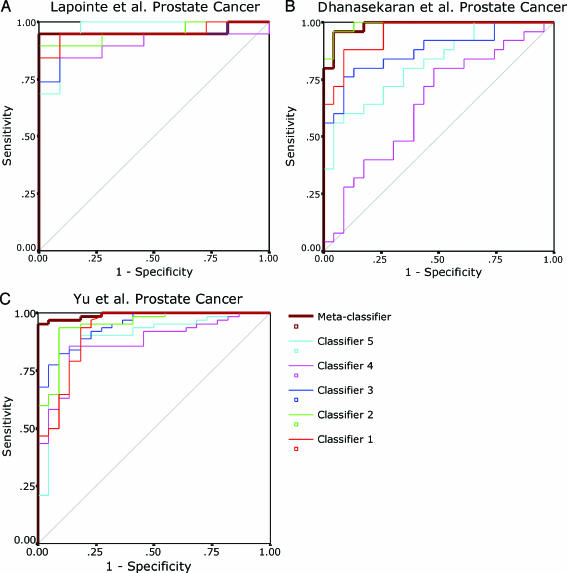
The above two prostate cancer data sets were hybridized by using the same AffymetrixHG-U95Av2 platform(Affymetrix Inc., Santa Clara, CA) and shared similar proportion ratios of target/nontarget samples. Next, we examined whether classifiers generated by GP could predict samples from independent studies that have used different microarray platforms. Three prostate cancer data sets [2,24,25] (UM, Stanford, and Pittsburgh) were used to test GP classifiers in predicting benign prostate and PCA samples. Among them, the Stanford and UM data sets used spot cDNA microarrays, whereas the Pittsburgh data used Affymetrix HG-U95Av2 oligonucleotide arrays. Two thirds of the Stanford samples were used as a training set to generateGPclassifiers, whereas the other one third, the UM and Pittsburgh samples, were all considered as validation samples. We used GP to generate 2000 classifiers and selected the top 26 frequently occurring genes (z score ≥40) as potential feature genes. To examine whether these genes are present in all three microarray platforms we cross-referenced them to the UM and Pittsburgh data sets using gene symbols. Of these 26 genes, 12 are present in all three data sets.We thus entered these 12 genes into the GP system to start a new round of five-fold crossvalidation on the Stanford training set. Five perfect classifiers were achieved and applied to the validation set. Prediction accuracy in the Stanford validation samples ranged from 84.4% to 90%. However, the classifiers performed poorly on UM and Pittsburgh data sets. We suspected that this might be due to the discrepancy in the proportion ratio ofPCA/benign samples and/or the probe intensity difference across array platforms, which led to divergence in the constant D of a classifier (e.g., GENE[A]/GENE[B]-GENE[C] > D). However, we believe that the relationships between the classifier genes, although with varying values of D,may still be predictive across studies, given that the classifier genes are putative discriminative genes. For instance, one of the classifiers, formulated as “IF (MYO6 + AMACR) >= -2.6776 THEN PCA” (see Table 5), did not predict well on the validation sets. However, the expression value of “MYO6 + AMACR” might still be predictive. To test this, we transformed the five classifiers individually as described in Materials and Methods and calculated a prediction score for each validation sample by computing the left side of each classifier inequality on a continuous scale. The predictive ability of each classifier on each validation set was then assessed by using the ROC-AUC. Notably, all classifiers were strongly significant (P < 5 x 10-4; Figure 4, Table 5) in both the Stanford and Pittsburgh validation sets. The lowest AUC values were 0.91 (95% CI, 0.80–1.00) and 0.87 (95% CI, 0.79–0.95), respectively. For UM data set, except for one classifier being marginally significant (AUC = 0.64, P = 0.09), all other classifiers were also strongly significant (P < 5 x 10-4).
Table 5.
Classifiers That Classify Benign Prostate and Primary Prostate Cancer.
| Classifier | ROC-AUC (95% CI) | ||
| Lapointe et al. Validation Set (Stanford) | Dhanasekaran et al. (UM) | Yu et al. (Pittsburgh) | |
| IF (ENC1 + GJB1) >= -0.8902 THEN PCA | 0.95 (0.87 – 1.00) | 0.95 (0.90 – 1.00) | 0.92 (0.85 – 1.00) |
| IF (MYO6 + AMACR) >= -2.6776 THEN PCA | 0.95 (0.88 – 1.00) | 0.99 (0.97 – 1.00) | 0.95 (0.90 – 1.00) |
| IF (TSPAN13 + PRKCBP1 >= -0.4172 THEN PCA | 0.94 (0.85 – 1.00) | 0.88 (0.78 – 0.98) | 0.94 (0.90 – 0.99) |
| IF (C20ORF74 + DAPK1) >= -0.7765 THEN PCA | 0.91 (0.80 – 1.00) | 0.64 (0.49 – 0.80) | 0.87 (0.79 – 0.95) |
| IF (IMAGE:396839 + ENC1) >= -0.5513 THEN PCA | 0.97 (0.91 – 1.00) | 0.82 (0.70 – 0.94) | 0.89 (0.81 –- 0.98) |
| Metaclassifier | 0.96 (0.87 – 1.00) | 0.99 (0.96 – 1.00) | 0.99 (0.98 – 1.00) |
Figure 4.
The ROCs of five classifiers and one metaclassifier for three prostate cancer validation sets. The classifiers were generated from the Lapointe et al. [24] (Stanford) training set to distinguish benign prostate from PCA. The ROCs are based on continuous prediction scores computed from the left side of the classifier inequality (see Materials and Methods). The scores of the metaclassifier are the mean values of prediction scores from each individual classifier. (A, B, C) ROC curves for Lapointe et al. [24] (Stanford), Dhanasekaran et al. [2] (UM), and Yu et al. [25] (Pittsburgh) validation set, respectively.
An ensemble “metaclassifier” combining multiple classifiers in general yields better prediction performance, as it involves more genes and multiple predictive signatures. Thus, we composed a metaclassifier based on the above five classifiers. For each sample, the calculated prediction scores of the five classifiers were totaled to an overall prediction score, which was then defined as the prediction score of the metaclassifier for that sample. As expected, this metaclassifier revealed higher AUCs in each data set (0.96, 0.99, and 0.99 for the Stanford, UM, and Pittsburgh set, respectively; P < 5 x 10-4; see Figure 4 and Table 5).
Feature Genes of a Classifier Are Correlated with Each Other
Examination of classifier genes have revealed that GP classifiers (Tables 4 and 5) are much simpler than predictors reported by other approaches [1,3,5,6,8,22,30–32], where more than 10 genes are often required to build an effective predictor. GP, by contrast, can use only 2 to 5 genes to produce effective classifiers and achieve high prediction power. This simplicity may owe to the relatively strict expression constraints (Table 2) and the use of a nonparametric method in selecting informative genes rather than usual parametric statistical techniques. Furthermore, unlike some other nonparametric approaches such as neural networks and SVMs, GP is transparent in that the entire procedure for classifier generation and evolution is readily available for inspection and adjustment.
A major difference between GP and other machine learning techniques is its mathematical connections between genes within a classifier. Studying the specific genes used by a classifier and the relationship between these genes may provide valuable information about gene interactions, transcriptional regulatory pathways, and clinical diagnosis. A quick examination of the classifiers revealed a consistent relationship between the expression levels of classifier genes. That is, a high (positive/negative) correlation between genes is preferred for a classifier. For example, within the classifier of “IF [MYO6] + [AMACR] >= -2.6776 THEN 'Primary Prostate Cancer',” the expression level of MYO6 was highly positively correlated with AMACR (r = 0.68, P < .0001). Furthermore, a two-sample t test revealed that both MYO6 and AMACR were significantly overexpressed in PCA relative to benign prostate (both Ps < .001 by t test), demonstrating that genes selected by the GP system were indeed potential discriminative biomarkers. Similar relationships were also observed in other classifiers.
GP Outperforms Other Molecular Classification Methods
One important criterion to assess a classification approach is how it performs in comparison to other commonly used algorithms in the same research area. To evaluate the performance of GP, the Burkitt lymphomas (“BL”) in the SRBCT data set and the high-risk class in the lung adenocarcinoma data were chosen as the target classes, and five classification methods including compound covariate predictor, 3-nearest neighbors, nearest centroid, SVMs, and DLDA were selected as comparing counterparts of the GP method. To produce a fair comparison, we took into account the small number of genes used by GP classifiers and conducted the comparison tests based on either 5- or 10-gene classifiers.
The same training and validation sets as described previously were used to evaluate the performance of each classification method. The basic procedure was defined by two steps: 1) two-sample Student t test was conducted for each gene in the training set, and the 5 or 10 genes with the smallest P values were selected as test classifiers; 2) expression data of the selected 5 or 10 genes across the training samples were used to build a training model, which was subsequently applied to the validation samples. Each individual validation sample was predicted as either “target class” or “nontarget class.” Misclassification rate was defined as the percentage of validation samples that were misclassified by a test classifier. Because GP generates multiple classifiers, the average of the misclassification rates of the top GP classifiers derived from the training set was used to represent the misclassification rate of a typical wellperforming GP classifier. For the SRBCT data, we used the averaged misclassification rate of the top 10 classifiers because there were 10 perfect classifiers generated from the training samples to classify “BL” and “non-BL.” Similarly, the 5 classifiers having the least classification error in the training set were used for the lung adenocarcinoma data set. As shown in Table 6, the error rates were comparable across different methods. The GP system ranked the second and the third in the SRBCT and the lung adenocarcinoma data sets, respectively, when 5-gene classifiers were evaluated. We believe that this may reflect the general prediction strength of GP system when only a small number of genes are chosen. Considering the additional advantages of GP in predicting samples across data sets and in revealing interrelationships between classifier genes, we concluded that GP outperforms other classification method.
Table 6.
Misclassification Error Rates of GP and Other Common Classification Models.
| Algorithm | Error Rate (%) |
|||
| SRBCT (BL vs. NON-BL) |
Lung Cancer (High Risk vs. Low Risk) |
|||
| 5 Genes | 10 Genes | 5 Genes | 10 Genes | |
| GP | 1.5 | NA | 16 | NA |
| Compound covariate predictor | 5 | 5 | 20 | 25 |
| 3-Nearest neighbors | 5 | 5 | 15 | 30 |
| Nearest centroid | 5 | 5 | 20 | 25 |
| SVMs | 0 | 5 | 20 | 25 |
| DLDA | 5 | 5 | 10 | 20 |
NA, not available.
Discussion
Although molecular classification has been successfully used to group tumors based on their gene expression patterns in retrospective research [1,3,6–9,15,22,32], its application in clinical settings has been greatly hindered. This is in part due to the large number of feature genes required to build a discriminative classifier. Therefore, there is a strong need to build molecular classifiers made of a small number of genes, especially in clinical diagnosis, where it would not be practical to have a diagnostic assay evaluate hundreds of genes in one test. In this study we developed a GP system to generate effective classifiers with a handful of genes.
An intrinsic advantage of GP is that it automatically selects a small number of feature genes during “evolution” [19]. The evolution of classifiers from the initial population seamlessly integrates the process of gene selection and classifier construction. By contrast, gene selection must be performed in a separate stage for many other classification algorithms such as kNN, weighted voting, and DLDA. Moreover, it is relatively easier for GP to keep the number of genes used in one classifier small. As GP searches a larger space than most traditional classification approaches, there is an increased chance of GP in finding a better performing classifier. By identifying and using a small number of genes and developing transparent and human-comprehensible rule-based classifiers, GP stands as a good algorithm of choice.
For each run of GP, it can generate tens to hundreds of classifiers. However, our data have shown that most of these classifiers used a limited set of feature genes (Figure 2). In addition, classifiers generated by different runs of GP also used a similar set of feature genes. Furthermore, we have observed that a number of these feature genes have been previously reported to be disease associated. Interestingly, the expression profile of the top selected feature genes easily classified cancer samples into their corresponding diagnostic groups as well as predicted validation sets of samples with high accuracy. These results strongly indicate that GP is very likely selecting the true discriminative feature genes, supporting the strong power of GP in feature selection.
The challenge of the field of molecular classification lies in the tradeoff of prediction power and the number of genes used. We have therefore stringently tested GP classifiers, composed of five genes or fewer, in achieving high prediction accuracy in data sets with varying levels of classification complexity. Unlike other studies [17,33] validating their classifiers using cross-validation within a data set, our result not only demonstrated that the top GP classifiers easily classified and predicted the SRBCT data set, which contained four classes of physiologically heterogeneous cancers, with 100% accuracy, but also showed optimal performance in classifying and predicting subtypes of prostate cancer for samples either of the same study or of a different study that used the same microarray platform. In addition, GP-selected feature genes stay discriminative even for cancer samples examined in different studies that used greatly different microarray platforms. Because of this robustness and stability of GP feature genes, we expect GP classifier to be highly applicable to clinical diagnosis.
We have found that GP identifiers have prediction accuracy comparable with small-gene-number classifiers generated by other classification methods. However, GP has added advantages over other algorithms. Its special features include the following: 1) the ability to automatically select a small number of genes as potential discriminative genes, 2) the ability to combine such genes and construct a simple and comprehensible classifier, and 3) the capability to generate multiple candidate classifiers.
A major issue in GP as well as other machine learning systems is data overfitting due to a large number of variables and a small number of cases in microarray profiling. This occurs when the classifier is strongly biased toward the training set and generates poor prediction generality in validation samples. To address this, our study restricted the complexity of classifiers and adopted an n-fold crossvalidation strategy. By limiting the size and complexity of classifiers using the minimum description length principle of risk minimization [34], the system was forced to generate the most salient features likely to be the most general solutions [35]. By resampling using n-fold cross-validation, classifiers derived from the training samples were reexamined in the test-fold samples to test how well the learning algorithm could be generalized. If the fitness on the training data in one fold is significantly better than the fitness on the test data, it may indicate that there is an issue of overfitting in the data. Therefore, a careful examination of the samples may be necessary.
Another issue for GP is that it is computationally intensive. The estimated running time increases along with the complexity of the problem and the number of variables. This can be partially resolved by using parallel processing, which segments the problem into parts running on different processors simultaneously and then synchronizes among them. In addition, variable prefiltering may also reduce the running time. As described in the Result section, GP typically selects those inherently discriminative genes and usually a small set of genes dominates the selection. Thus, a prefiltering, such as excluding genes with small variances, may significantly reduce the running time yet not affect the performance of classifiers.
Taken together, in this study we systematically evaluated the feasibility of GP in feature selection and cancer classification. By examining the feature genes used by GP classifiers we have demonstrated that GP is able to robustly select a set of highly discriminative genes. In addition, the mathematical expression of GP classifiers reveals interesting quantitative relationships between genes. By testing GP classifiers generated from training sets in validation sets, we have shown that GP classifiers can successfully predict tumor classes and outperform most of other classification methods when only a limited number of genes are allowed to build a classifier. Our work suggests that GP may be useful for feature selection and molecular classification of cancer using a practical set of genes.
Supplementary Material
Footnotes
This research was supported in part by the National Institutes of Health (R01 CA97063 to A.M.C. and D.G., U54 DA021519-01A1 to A.M.C., Prostate SPORE P50CA69568 to A.M.C.), the Early Detection Research Network (UO1 CA111275 to A.M.C. and D.G.), the National Institutes of General Medical Sciences (GM 72007 to D.G.), the Department of Defense (W81XWH-06-1-0224 to A.M.C., PC060266 to J.Y.), and the Cancer Center Bioinformatics Core (support grant 5P30 CA46592 to A.M.C.). A.M.C. is supported by a Clinical Translational Research Award from the Burroughs Welcome Foundation.
References
- 1.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 2.Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 3.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 4.Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi OP, et al. Geneexpression profiles in hereditary breast cancer. N Engl J Med. 2001;344:539–548. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
- 5.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 6.Mukherjee S, Tamayo P, Mesirov JP, Slonim D, Verri A, Poggio T. Support Vector Machine Classification of Microarray Data. CBCL: MIT; 1999. [Google Scholar]
- 7.van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 8.Khan J, Wei JS, Ringner M, Saal LH, Ladanyi M, Westermann F, Berthold F, Schwab M, Antonescu CR, Peterson C, et al. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med. 2001;7:673–679. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H, Yu CY, Singer B. Cell and tumor classification using gene expression data: construction of forests. Proc Natl Acad Sci USA. 2003;100:4168–4172. doi: 10.1073/pnas.0230559100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koza JR. Genetic Programming: On the Programming of Computers by Means of Natural Selection. Cambridge: MIT Press; 1992. [Google Scholar]
- 11.Bojarczuk CC, Lopes HS, Freitas AA. Proceedings Intelligent Data Analysis in Medicine and Pharmacology ({IDAMAP}-2001) 2001. Data mining with constrained-syntax genetic programming: applications to medical data sets. [Google Scholar]
- 12.Hong JH, Cho SB. The classification of cancer based on DNA microarray data that uses diverse ensemble genetic programming. Artif Intell Med. 2006;36:43–58. doi: 10.1016/j.artmed.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Li L, Weinberg CR, Darden TA, Pedersen LG. Gene selection for sample classification based on gene expression data: study of sensitivity to choice of parameters of the GA/KNN method. Bioinformatics. 2001;17:1131–1142. doi: 10.1093/bioinformatics/17.12.1131. [DOI] [PubMed] [Google Scholar]
- 14.Mitra AP, Almal AA, George B, Fry DW, Lenehan PF, Pagliarulo V, Cote RJ, Datar RH, Worzel WP. The use of genetic programming in the analysis of quantitative gene expression profiles for identification of nodal status in bladder cancer. BMC Cancer. 2006;6:159. doi: 10.1186/1471-2407-6-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ooi CH, Tan P. Genetic algorithms applied to multi-class prediction for the analysis of gene expression data. Bioinformatics. 2003;19:37–44. doi: 10.1093/bioinformatics/19.1.37. [DOI] [PubMed] [Google Scholar]
- 16.Tan KC, Yu Q, Heng CM, Lee TH. Evolutionary computing for knowledge discovery in medical diagnosis. Artif Intell Med. 2003;27:129–154. doi: 10.1016/s0933-3657(03)00002-2. [DOI] [PubMed] [Google Scholar]
- 17.Ho SY, Hsieh CH, Chen HM, Huang HL. Interpretable gene expression classifier with an accurate and compact fuzzy rule base for microarray data analysis. Biosystems. 2006;85:165–176. doi: 10.1016/j.biosystems.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Langdon WB, Buxton BF. Genetic programming for mining DNA chip data from cancer patients. Genetic Programming and Evolvable Machines. 2004;5:251–257. [Google Scholar]
- 19.Moore JH, Parker JS, Hahn LW. Symbolic discriminant analysis for mining gene expression patterns. Lecture Notes Artif Intell. 2001;2167:372–381. [Google Scholar]
- 20.Moore JH, Parker JS, Olsen NJ, Aune TM. Symbolic discriminant analysis of microarray data in autoimmune disease. Genet Epidemiol. 2002;23:57–69. doi: 10.1002/gepi.1117. [DOI] [PubMed] [Google Scholar]
- 21.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 23.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 24.Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 26.LaTulippe E, Satagopan J, Smith A, Scher H, Scardino P, Reuter V, Gerald WL. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- 27.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 28.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 30.Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 31.Gruvberger SK, Ringner M, Eden P, Borg A, Ferno M, Peterson C, Meltzer PS. Expression profiling to predict outcome in breast cancer: the influence of sample selection. Breast Cancer Res. 2002;5:23–26. doi: 10.1186/bcr548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RC, Gaasenbeek M, Angelo M, Reich M, Pinkus GS, et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- 33.Bhattacharyya C, Grate LR, Jordan MI, El Ghaoui L, Mian IS. Robust sparse hyperplane classifiers: application to uncertain molecular profiling data. J Comput Biol. 2004;11:1073–1089. doi: 10.1089/cmb.2004.11.1073. [DOI] [PubMed] [Google Scholar]
- 34.Rissanen J. Modeling by shortest data description. Automatica. 1978;14:465–471. [Google Scholar]
- 35.Vapnik VN. The Nature of Statistical Learning Theory. Berlin: Springer-Verlag; 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.