Abstract
In the most common form of stem cell leukemia (SCL) gene rearrangement, an interstitial deletion of 82 kb brings SCL under the control of regulatory elements that normally govern expression of the ubiquitously expressed SCL interrupting locus (SIL) gene, which is located directly upstream of SCL. To investigate the effect of this fusion in a mouse model, a bacterial artificial chromosome (BAC) clone containing both human SIL and SCL genes was isolated, and loxP sites were inserted into intron 1 of both the SIL and SCL genes, corresponding to the sites at which recombination occurs in human T-cell acute lymphocytic leukemia patients. This BAC clone was used to generate transgenic SILloxloxSCL mice. These transgenic mice were subsequently bred to Lck-Cre mice that express the Cre recombinase specifically in the thymus. The BAC transgene was recombined between the two loxP sites in over 50% of the thymocytes from SILloxloxSCL/Cre double-transgenic mice, bringing the SCL gene under the direct control of SIL regulatory elements. Aberrant SCL gene expression in the thymus was verified by reverse transcription-polymerase chain reaction. Using FACS analysis, we found that mice carrying both SILloxloxSCL and Cre transgenes have increased CD4-/CD8- thymocytes compared with transgene-negative mice. In the spleen, these transgenic mice show a marked reduction in the number of mature CD4+ or CD8+ cells. These results demonstrate that conditional activation of SCL under control of SIL regulatory elements can impair normal T-cell development.
Keywords: SCL, SIL, T-cell development, Cre-LoxP, T-ALL
Introduction
Activation of the stem cell leukemia (SCL) (also known as T-cell acute lymphoblastic leukemia 1 (TAL-1 or TCL5) transcription factor is a frequent gain-of-function mutation in pediatric T-cell acute lymphocytic leukemia (T-ALL) [1,2]. SCL is normally expressed in hematopoietic cells, endothelium, and the central nervous system, and has been shown to be important for blood vessel formation, endothelial development, and the control of normal hematopoiesis [3–6]. Mice with an SCL null genotype are nonviable, but can be rescued by the human SCL locus [7].
Normally, expression of SCL in the thymus is restricted to the DN1–DN2 subset of immature CD4-/CD8- thymocytes [8]. However, in the context of T-ALL, chromosomal translocations involving the SCL gene and chromosomes 3, 5, 7, and 14 have been associated with T-ALL and lead to unscheduled SCL expression [5,9–11]. In addition to the aforementioned chromosomal translocations, the most common event that deregulates SCL expression in T-ALL patients is an 82-kb interstitial deletion that occurs in approximately 25% of patients with T-ALL [2,5,12] and replaces SCL 5′ regulatory sequences with those of an upstream gene, known as SCL-interrupting locus (SIL) [1,2].
Attempts to model human T-ALL in mice have met with variable success. In two independent studies, expression of a full-length SCL gene under the control of the Lck promoter led to T-ALL at an advanced age in a fraction at Lck-SCL transgenic mice [13,14]. In contrast, other studies showed that deregulated SCL expression alone did not cause T-cell tumors [15–17]. The reasons for varying results among different studies are not clear, but may be due to differences in mouse strains, promoters used, or integration sites. The collective observations that T-ALL occurred at an advanced age, with incomplete penetrance, suggested that additional events were required to produce T-ALL in these mice. Indeed, several reports have indicated that aberrant SCL expression, in collaboration with activation of CKIIα, LMO1, or LMO2, leads to the development of aggressive T-cell malignancies in transgenic mice at an early age with a high degree of penetrance [14,16–19].
Transgenic mice that express either a full-length or an amino-terminal truncated SCL driven by a SIL promoter showed bony abnormalities and growth retardation, but did not develop T-cell malignancies [17].However, when crossed with mice that overexpress LMO1 in the thymus, these doubletransgenic mice formed aggressive T-cell malignancies with a high degree of penetrance at an early age [17,19]. Interestingly, abnormal T-cell development was noted in SCL/LMO1 double-transgenic mice before the onset of malignancy, whereas no abnormal immunophenotype for T-cell development was detected in mice transgenic only for SCL or LMO1 [8,19]. Consistent with these results, other groups also failed to detect a premalignant perturbation of T-cell differentiation in Lck-SCL or CD2-SCL mice before development of leukemia [13,14,16].
We previously demonstrated that SCL could inhibit E2A activity in a dose-dependent fashion in vitro [8,19]. We considered the possibility that misexpression of SCL under the control of SIL regulatory elements [17] did not lead to T-ALL in mice because the levels of SCL were insufficient to inhibit E2A (or the closely related HEB protein). Because high levels of SCL expression under the control of SIL regulatory elements seemed to be toxic to the developing embryo (reference 17 and unpublished data), we used Cre-loxP technology to generate mice that would conditionally express SCL under the control of SIL regulatory elements. Moreover, we reason that including additional SIL regulatory sequences might lead to higher levels of SCL expression compared to the 2.3 kb of SIL promoter sequence used for the pSIL/SCL mice [19].
Materials and Methods
Generation of Bacterial Artificial Chromosome Clone and SILloxloxSCL Transgenic Mice
We isolated a bacterial artificial chromosome (BAC) clone containing both human SIL and SCL genes as well as 47 kb of sequence immediately 5′ of SIL, and used “recombineering” [20] to introduce loxP sites into intron 1 of both SIL (nucleotide 47491426 of human chromosome 1; National Center for Biotechnology Information [NCBI] build 35 by the International Human Genome Sequencing Consortium, May 2004) and SCL (nucleotide 47409602), corresponding to the sites at which recombination occurs in human T-ALL patients. This BAC clone was linearized with PI-SceI and injected into fertilized FVB/N embryos to generate transgenic SILloxloxSCL mice.
Genomic DNA Polymerase Chain Reaction, Southern Blotting, and DNA Sequencing
All polymerase chain reaction (PCR) amplifications, unless otherwise indicated, were performed by using PCR SuperMix High Fidelity enzyme and buffers (Invitrogen, Carlsbad, CA). PCR primer sequences and annealing temperature are listed in Table 1. Mouse tail DNA was extracted for PCR analyses as follows. Two millimeters of tail was cut and added to 600 µl of 50 mmol/l NaOH, heated to 95°C for 1 hour, and neutralized with 50 µl of 1 mol/l Tris (pH 8). One microliter of this crude DNA preparation was used for mouse genotype analysis. For the human SIL gene, the primers were SIL5′F and 3′-SILR; for the human SCL gene, the primers were 3′SCLF and 3′SCLR. Cre-positive and -negative mice were identified by PCR using primers cre1083F and cre1085R. To identify SIL-SCL recombination at the LoxP site, sequences in the first intron of the SIL gene (SIL5′F) and the first intron of the SCL gene (SCLR-TARG) were used for PCR primers. To verify DNA quality, we amplified the mouse Scid locus by using primers SCIDA and SCIDB.
Table 1.
PCR Primers.
| Name | Sequence (5′ > 3′) | Annealing Temperature (°C) |
| SIL5′F (forward) | GCCCTGTAGTGGGTTCCGCCC | 60 |
| 3′-SILR (reverse) | TCCAGTCAAACTGAACTACTTGC | 60 |
| 3′of SCLF (forward) | GGCCTGGTGGGGAGGAGACAGC | 60 |
| 3′of SCLR (reverse) | GATACATACAACTGTCCCAGCC | 60 |
| cre1083F (forward) | GCGGCATGGTGCAAGTTGAATAA | 55 |
| cre1085R (reverse) | GTGAAACAGCATTGCTGTCACTT | 55 |
| SCLR-TARG (reverse) | GACACCACCCAAACACAGTCGC | 56 |
| SCIDA (forward) | GGAAGAGTTTTGAGCAGACAATG | 54 |
| SCIDB (reverse) | CATCACAAGTTATAACAGCTGGG | 54 |
| SILexon1 (forward) | GCTCCTACCCTGCAAACAGA | 60 |
| SCLexon3 (reverse) | GGCATATTTAGAGAGACCG | 60 |
| Tal1 (forward) | ATGGTGCAGCTGAGTCCTCC | 52 |
| Tal1 (reverse) | TCTCATTCTTGCTGAGCTTC | 52 |
| β-Actin F (forward) | GTGGGCCGCTCTAGGCACCAA | 58 |
| β-Actin R (reverse) | CTCTTTGATGTCACGCACGATTTC | 58 |
PCR products were cloned into the pGEM-T easy vector (Promega, Madison, WI) and transformed into DH-5α cells. Plasmid DNA was extracted (Qiagen, Valencia, CA) and sequenced (Napcore, The Children's Hospital of Philadelphia). All procedures were performed following the manufacturers' protocols.
For Southern blot studies, 10 mg of genomic DNA isolated from mouse tissue by use of a proteinase K/SDS salting out procedure [21] was digested with restriction enzymes and size fractionated using a 0.8% agarose gel. The DNA fragments were transferred to nitrocellulose membranes and hybridized to 32P-labeled probes. Probes used included a previously described human TCR Cβ2 probe [17], a human SIL exon 1 probe (nucleotides 47491736 to 47492065, human chromosome 1; NCBI build 35 by the International Human Genome Sequencing Consortium, May 2004) [22,23], and a human SCL probe from the 3′ untranslated region (929 bp, nucleotides 47394123 to 47393194, human chromosome 1; NCBI build 35 by the International Human Genome Sequencing Consortium, May 2004). Fragments were labeled with 32P by using Ready-To-Go DNA labeling beads (Amersham Bioscience, Piscataway, NJ). Hybridization was performed as previously described [21]. The washing conditions were 0.1% SDS/2 x SSC at 42°C for 20 minutes and 0.1% SDS/0.1 x SSC at 52°C for 40 minutes.
Reverse Transcription-PCR and Semiquantitative Reverse Transcription-PCR
Total RNA was isolated using Trizol reagent and the manufacturer's (Invitrogen) recommended protocol. To eliminate potential contamination of genomic DNA, all total RNA samples were treated with 2 U DNAse for 30 minutes (Ambion, Austin, TX). First-strand cDNA was synthesized from 1 mg of DNAse-treated RNA by using SuperScript First-Strand Synthesis System for reverse transcription (RT)-PCR (Invitrogen), following the manufacturer's protocol. Controls without reverse transcriptase were performed in all reactions to detect possible contamination of genomic DNA. PCR amplification of β-actin was used to verify that intact mRNA was present.
The primers for the SIL-SCL fusion mRNA were SILexon1 and SCLexon3. RT-PCR products were cloned into plasmid vectors and sequenced as described above. Semiquantitative RT-PCR was performed by serial 10-fold dilution of cDNA templates (1x, 0.1x, 0.01x, and 0.001x for each sample).
Immunophenotype Analysis
Cells from thymus and spleen were minced and homogenized using a loose-fitting ground-glass homogenizer as previously described [19] to generate a single-cell suspension in RPMI 1640 medium. The cell suspension was filtered through a cell strainer (BD, Bedford, MA) and treated with ACK lysing buffer (Biosource, Camarillo, CA) for 10 minutes. Viable cells (1 x 106) were used for subsequent antibody staining. The cells were incubated with 5 µl (0.5 mg/ml) of rat anti-mouse CD16/CD32 for 20 minutes as a blocking agent. The single-cell suspension was incubated with 5 µl (0.2 mg/ml) of fluorescein isothiocyanate- or phycoerythrinlabeled antibodies (murine CD4, CD8, CD25 or CD 44) for 30 minutes. All antibodies were purchased from BD Pharmingen (San Jose, CA). Ten thousand events per sample were scored using a FACSort flow cytometer (Becton-Dickinson, San Jose, CA).
Results
Generation of Mice Susceptible to SIL-SCL Recombination
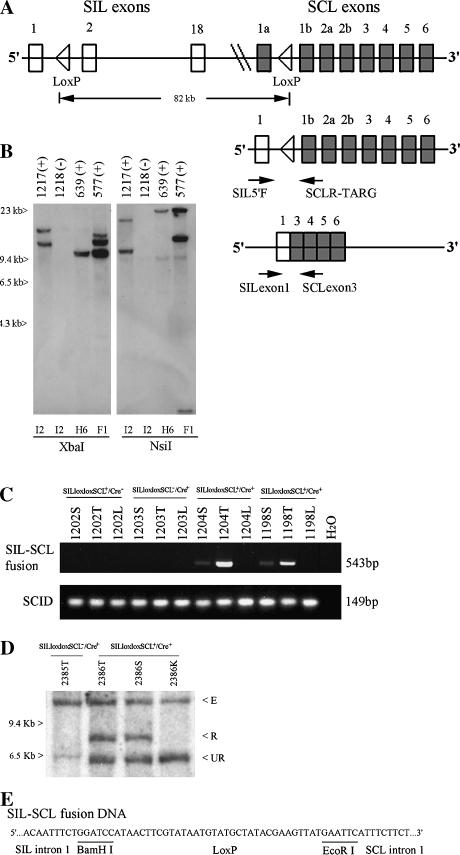
Three founder lines (F1, I2, and H6) that had incorporated at least one copy of the SILloxloxSCL BAC clone (Figure 1A) were identified by PCR analyses. We used Southern blot analysis of the BAC clone integration site to identify a founder that had integrated a single copy of the BAC clone (Figure 1B) and verified that this line had retained all of the SIL and SCL genomic sequences by PCR. To determine whether Cre-loxP-mediated SIL-SCL recombination could be achieved in vivo, we crossed offspring of the H6 founder, which had integrated a single copy of the SILloxloxSCL BAC clone, to mice that expressed the Cre recombinase under the control of the Lck promoter to produce SILloxloxSCL+/Cre+ double-transgenic mice.
Figure 1.
Generation and characterization of SILloxloxSCL mice. (A) Top panel shows the human SIL (18 exons), SCL genes (8 exons), and the introduced Lox P sites in the regions of intron 1. Not all SIL exons are shown. Middle panel shows the fusion of genomic DNA. Two arrows indicate the primers used for a detection of fusion SILloxSCL DNA. Bottom panel shows the fusion mRNA that is generated between exon 1 of SIL and exon 3 of SCL in double-transgenic mice. Two arrows indicate the primers used for a detection of SIL-SCL fusion mRNA. (B) Detection of SILloxloxSCL BAC clone integration site(s) by Southern blot. Genomic DNA from F1 offspring of founders I2, H6, or F1 was digested with XbaI or NsiI and hybridized to a probe from the SCL 3′ untranslated region located at one terminal of the BAC clone. Variable-sized fragments representing unique integration sites can be seen. A cross-hybridizing band is seen in the NsiI digest at 9.4 kb. Mouse numbers are indicated, as is transgene status (+ or -) (C) SIL-SCL genomic fusion can be detected in mice 1204 and 1198 by PCR. Mice 1202, 1203, and H2O were negative controls. PCR amplification of the scid locus was used as a DNA quality control. T, thymus; S, spleen; L, liver. (D) Detection of SIL-SCL fusion by Southern blot analysis of SstI-digested genomic DNA. 2385T, thymus from mouse without the SILloxloxSCL transgene. 2386T, 2386S, 2386K; thymus, spleen, and kidney from SILloxloxSCL+/Cre+ mouse. E, endogenous murine SIL; R, recombined SIL-SCL fusion; UR, unrearranged SILloxloxSCL transgene. (E) Nucleotide sequences of fusion SILloxSCL genomic DNA.
We used genomic DNA PCR to verify that recombination between the SIL and SCL loci had taken place in SILloxloxSCL+/Cre+ mice. A PCR product of 543 bp could be identified in thymus, and to a lesser extent in spleen, of the SILloxloxSCL+/Cre+ mice, but not the control genotypes, indicating a precise Cre-LoxP-mediated excision of 82 kb (Figure 1C).
To obtain a more quantitative estimate of the relative proportion of SILloxloxSCL alleles in the rearranged versus nonrearranged configuration, we used SIL exon 1 as a probe to detect recombination in the transgenic mice through Southern blot analysis. In SILloxloxSCL+/Cre+ mice, the ratio of recombined to unrecombined products is approximately 1 as seen in Figure 1D. As anticipated from the PCR results, these recombination events occurred only in the thymus and spleen of SILloxloxSCL+/Cre+ doubletransgenic mice. The sequence of the PCR product from thymus shows a SIL-SCL genomic fusion, with intervening LoxP sequence (Figure 1E).
SILloxloxSCL+/Cre+ Double-Transgenic Mice Do Not Develop T-Cell Tumors
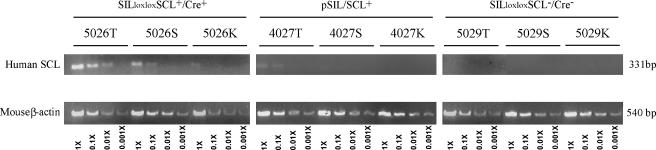
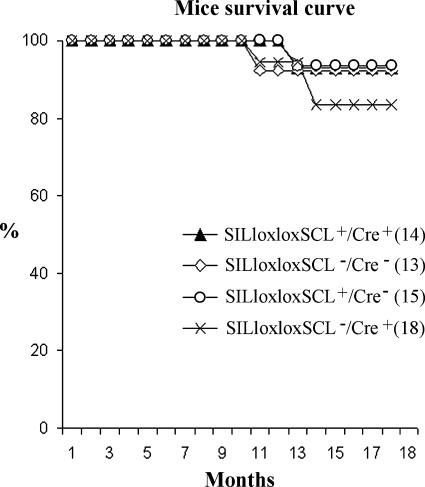
Using semiquantitative RT-PCR analysis, we demonstrated that SILloxloxSCL+/Cre+ mice that have undergone Cre-LoxP-mediated recombination in the thymus express ∼10- to 100-fold higher levels of SCL expression than those of the pSIL/SCL transgenic mice that express an SCL cDNA from 2.3 kb of SIL 5′ regulatory sequence [19] (Figure 2). We followed a cohort of SILloxloxSCL+/Cre+ mice for 19 months and compared them with three control groups: positive for either the SILloxloxSCL or the Cre only and negative for both transgenes. Their survival curves were similar; none of 14 double-transgenic mice or 46 control mice from this cohort developed T-cell malignancy (Figure 3).
Figure 2.
Higher level SCL expression in SILloxloxSCL+/Cre+ mice than pSIL/SCL mice. Mouse 5026, positive for both SILloxloxSCL and Cre transgenes, has a higher level of SCL expression than a pSIL/SCL transgenic mouse (4027); mouse 5029 is negative for both SILloxloxSCL and Cre transgenes. T, S, and K are thymus, spleen, and kidney, respectively. The cDNA templates were diluted to 1x, 0.1x, 0.01x, and 0.001x times in PCR reactions.
Figure 3.
SILloxloxSCL+/Cre+ mouse survival curve. Mice of the indicated genotypes were followed for 19 months and euthanized when morbid. There was no difference in survival for any group.
Mice Transgenic for SIL-SCL and Cre Demonstrate Impaired T-Cell Differentiation
Because SILloxloxSCL+/Cre+ double-transgenic mice expressed higher levels of SIL-SCL fusion mRNA in the thymus than did the pSIL/SCL mice (Figure 2), we searched for evidence that these higher levels of ectopically expressed SCL might affect T-cell growth and differentiation. To achieve this purpose, we performed T-cell subset analyses on the thymus and spleen of mice aged 6 to 15 months.
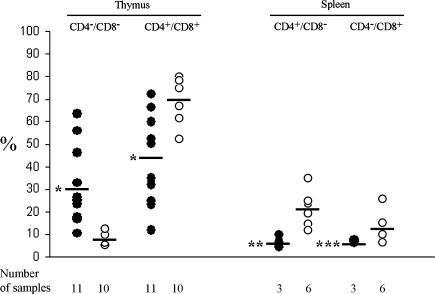
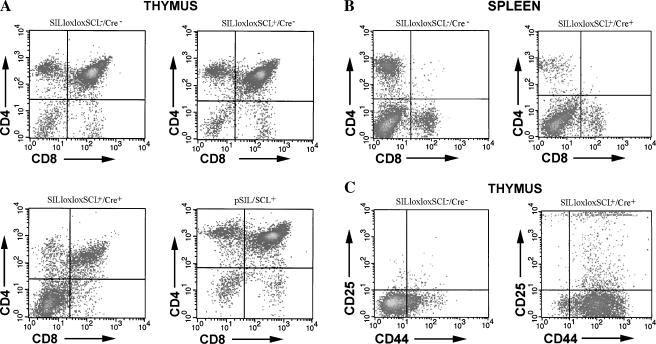
The SILloxloxSCL+/Cre+ mice showed a variable, and in some cases quite dramatic, increase in CD4-/CD8- (DN) cells and a corresponding decrease in CD4+/CD8+ (DP) cells in the thymus (Figures 4 and 5A). On average, the SILloxloxSCL+/Cre+ mice had 30.3 ± 17.4% DN cells and 44.5 ± 19.9% DP cells, compared with control mice, which had 8.0 ± 2.5% DN and 69.8 ± 8.5% DP cells (P < .001). Consistent with the thymocyte data, mice with decreased DP cells in the thymus had decreased percentages of mature single positive (SP) CD4+ (P < .01) and CD8+ (P < .05) cells in the spleen, compared with wild-type controls (Figures 4 and 5B). In addition, we searched for immature CD44+CD25- cells in the thymus of some SILloxloxSCL+/Cre+ mice. As shown in Figure 5C, SILloxloxSCL+/Cre+ mice had a marked increase in this population compare with wild-type controls.
Figure 4.
Aberrant T-cell differentiation in SILloxloxSCL+/Cre+ mice. Thymocytes and splenocytes from SILloxloxSCL+/Cre+ (closed circles) or SILloxloxSCLa or SILloxloxSCL+/Cre- mice (open circles) stained with CD4 + CD8. The number of samples analyzed is indicated. The means are indicated with a solid line. *P < .001; **P < .01; ***P < .05.
Figure 5.
FACS profiles demonstrating aberrant T-cell development. Thymocytes (A) and splenocytes (B) from clinically healthy mice with the indicated genotype were stained with CD4 and CD8. Note the increased DN population in the SILloxloxSCL+/Cre+ thymocytes, and decreased SP population in SILloxloxSCL+/Cre+ splenocytes. (C) Thymocytes from above mice were stained with CD44 and CD25; note increased CD44+CD25- population in SILloxloxSCL+/Cre+ sample.
To determine whether the SILloxloxSCL+/Cre+ mice had a clonal or oligoclonal expansion of thymocytes, genomic DNA from SILloxloxSCL+/Cre+ mice was digested with SstI, blotted, and hybridized with a TCR Cβ2 probe. No clonal TCRβ gene rearrangements were detected in SILloxloxSCL+/Cre+ transgenic mice nor in the control wild-type or single-transgenic mice (data not shown).
Discussion
The fusion of the SIL gene promoter region with the downstream SCL gene is a common rearrangement detected in patients with T-ALL [2,12,22]. Surprisingly, pSIL/SCL mice that express SCL under the control of SIL regulatory elements [19] did not develop T-ALL, nor did these mice show any abnormalities in T-cell differentiation. However, when crossed to transgenic mice that expressed LMO1 in the thymus, the SCL/LMO1 double-transgenic mice developed T-ALL preceded by abnormalities in thymocyte differentiation, including an oligoclonal expression of DN cells [17,19].
To determine if an SCL expression construct that more closely resembled the situation seen in human T-ALL patients could cause T-ALL, we isolated a human BAC clone that contained both the human SIL and SCL genes, and introduced LoxP sites into intron 1 of the SIL and SCL genes at the sites where recombination occurs in human T-ALL patients [12,24]. This clone was then used to generate transgenic SILloxloxSCL mice. After crossing the SILloxloxSCL mice to Lck-Cre mice, which expressed the Cre recombinase in the thymus, we were able to detect a SILloxSCL genomic fusion and a SIL-SCL fusion mRNA, thus mimicking the common form of SCL gene activation seen in T-ALL patients. Of note, although it is well established that the Cre recombinase efficiently catalyzes recombination between LoxP sites located less than 5 kb apart [25] and that the Cre recombinase can catalyze rare recombination between LoxP sites on different chromosomes [26], we show here that Cremediated recombination can efficiently act over a distance of 82 kb and cause recombination of approximately half of the SILloxloxSCL alleles.
In our previous studies, both T-ALL and abnormal T-cell development were detected in SCL/LMO1 double-transgenic mice, but not in pSIL/SCL transgenic mice that expressed a full-length SCL protein [19]. Using a semiquantitative RT-PCR assay, we show that SILloxloxSCL/Cre transgenic mice have higher levels of SCL expression than those of pSIL/SCL transgenic mice. This difference may be due to incorporation of additional SIL regulatory elements (′47 kb of sequences 5∼ of SIL exon 1), which may affect both timing and level of SCL expression in transgenic mice. Using Lck to direct SCL expression in the mouse thymus, two laboratories have induced T-ALL in mice but did not observe any evidence of a premalignant perturbation of T-cell development [13,14].
Four populations of developing T-cells can be identified through immunophenotype analysis: CD4-CD8- (DN), the CD4+CD8+ (DP) and CD4-CD8+ (CD8 SP), or CD4+CD8- (CD4 SP). The DN population can be further subdivided into CD44+CD25- (DN1), CD44+CD25+ (DN2), CD44-CD25+ (DN3), and CD44-CD25- (DN4) stages [27]. To determine whether expression of the SIL-SCL fusion generated in our current study affects T-cell development, we performed FACS analysis of thymocytes from SILloxloxSCL+/Cre+ mice. The fraction of DN thymocytes in SILloxloxSCL+/Cre+ mice was significantly increased compared with those of the control groups. Furthermore, we noted an increase in the percentage of immature DN1 cells in the thymus of SILloxloxSCL+/Cre+ transgenic mice, which were not detected in either SILloxloxSCL-/Cre- or SILloxloxSCL+/Cre- mice, indicating impaired T-cell development at the DN1 stage. We also found that CD4 SP and CD8 SP cells were decreased in the spleen of SILloxloxSCL+/Cre+ mice, consistent with the decrease in DP cells seen in SILloxloxSCL+/Cre+ mice thymus.
Tumorigenesis is a multistep process [28]. Although the development of T cells was impaired in SILloxloxSCL+/Cre+ mice, we did not detect any clonal TCRβ gene rearrangements in these mice, suggesting that there was no clonal or oligoclonal expansion. However, because TCRβ gene rearrangement typically occurs in the DN2 population, it is possible that clonal expansion, undetectable by TCRβ gene rearrangement, has occurred in the samples with a predominant DN1 population. None of the 14 mice with both the SIL-SCL and the Cre transgenes developed T-cell malignancies over a 19-month observation period. Despite the higher level of SCL expression and perturbations of thymocyte development observed in these mice, aberrant expression of SCL in the thymus was insufficient to induce leukemia.
There are several possible explanations for the lack of leukemic transformation in the SILloxloxSCL+/Cre+ mice that had undergone SIL-SCL recombination. First, the lack of leukemic transformation may be due to subtle, undetected mouse strain differences. Second, the SCL transgene may not be expressed at a “leukemogenic” point in thymocyte differentiation, as SCL expression is dependent on expression of the Cre recombinase under the control of Lck promoter. Lastly, it is possible that the SIL-SCL fusion is a primary oncogenic event and requires additional events for complete leukemic transformation [13,14].
In summary, we designed and generated a genetic event that faithfully recapitulates the interstitial deletion leading to SIL-SCL fusion gene seen in human T-ALL patients. We demonstrated that expression of SCL, under the control of SIL regulatory elements, can perturb normal T-cell development, which might provide an abnormal environment in which additional genetic events occur, and result in complete malignant transformation.
Acknowledgements
We thank IIan Kirsch and Michael Kuehl for helpful discussion.
Abbreviations
- SCL
stem cell leukemia
- SIL
SCL-interrupting locus
- BAC
bacterial artificial chromosome
- T-ALL
T-cell acute lymphocytic leukemia
Footnotes
This research was supported by the Intramural Research Program of the National Institutes of Health and National Cancer Institute.
References
- 1.Aplan PD, Lombardi DP, Ginsberg AM, Cossman J, Bertness VL, Kirsch IR. Disruption of the human SCL locus by “illegitimate” V-(D)-J recombinase activity. Science. 1990;250:1426–1429. doi: 10.1126/science.2255914. [DOI] [PubMed] [Google Scholar]
- 2.Brown L, Cheng JT, Chen Q, Siciliano MJ, Crist W, Buchanan G, Baer R. Site-specific recombination of the tal-1 gene is a common occurrence in human T cell leukemia. EMBO J. 1990;9:3343–3351. doi: 10.1002/j.1460-2075.1990.tb07535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shivdasanl RA, Mayer EL, Orkin SH. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature. 1995;373:432–434. doi: 10.1038/373432a0. [DOI] [PubMed] [Google Scholar]
- 4.Visvader JE, Fujiwara Y, Orkin SH. Unsuspected role for the T-cell leukemia protein SCL/tal-1 in vascular development. Genes Dev. 1988;12:473–479. doi: 10.1101/gad.12.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Begley CG, Green AR. The SCL gene: from case report to critical hematopoietic regulator. Blood. 1999;93:2760–2770. [PubMed] [Google Scholar]
- 6.Hall MA, Curtis DJ, Metcalf D, Elefanty AG, Sourris K, Robb L, Göthert JR, Jane SM, Begley CG. The critical regulator of embryonic hematopoiesis, SCL, is vital in the adult for megakaryopoiesis, erythropoiesis, and lineage choice in CFU-S12. Proc Natl Acad Sci USA. 2003;100:992–997. doi: 10.1073/pnas.0237324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sinclair AM, Bench AJ, Bloor AJC, Li J, Göttgens B, Stanley ML, Miller J, Piltz S, Hunter S, Nacheva EP, Sanchez MJ, Green AR. Rescue of the lethal scl-/- phenotype by the human SCL locus. Blood. 2000;99:3931–3938. doi: 10.1182/blood.v99.11.3931. [DOI] [PubMed] [Google Scholar]
- 8.Herblot S, Steff AM, Hugo O, Aplan PD, Hoang T. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-Tα chain expression. Nat Immunol. 2000;1:138–144. doi: 10.1038/77819. [DOI] [PubMed] [Google Scholar]
- 9.Finger LR, Kagan J, Christopher G, Kurtzberg J, Hershfield MS, Nowell PC, Croce CM. Involvement of the TCL5 gene on the chromosome 1 in the T-cell leukemia and melanoma. Proc Natl Acad Sci USA. 1989;86:5039–5043. doi: 10.1073/pnas.86.13.5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aplan PD, Raimondi SC, Kirsch IR. Disruption of the SCL gene by a t(1;3) translocation in a patient with Tcell acute lymphoblastic leukemia. J Exp Med. 1992;176:1303–1310. doi: 10.1084/jem.176.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Q, Yang CYC, Tsan JT, Xia Y, Ragab AH, Peiper SC, Carroll A, Baer R. Coding sequences of the tal-1 gene are disrupted by chromosome translocation in human T cell leukemia. J Exp Med. 1990;172:1403–1408. doi: 10.1084/jem.172.5.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aplan PD, Lombardi DP, Reaman GH, Sather HN, Hammond GD, Kirsch IR. Involvement of the putative hematopoietic transcription factor SCL in T-cell acute lymphoblastic leukemia. Blood. 1992;79:1327–1333. [PubMed] [Google Scholar]
- 13.Condorelli GL, Facchiano F, Valtieri M, Proietti E, Vitelli L, Lulli V, Huebner K, Peschle C, Croce CM. T-cell-directed TAL-1 expression induces T-Cell malignancies in transgenic mice. Cancer Res. 1996;56:5113–5119. [PubMed] [Google Scholar]
- 14.Kelliher MA, Seldin DC, Leder P. Tal-1 induces T cell acute lymphoblastic leukemia accelerated by casein kinase IIα. EMBO J. 1996;15:5160–5166. [PMC free article] [PubMed] [Google Scholar]
- 15.Robb L, Rasko JEJ, Bath ML, Strasser A, Begley CG. scl, a gene frequently activated in human T cell leukaemia, does not induce lymphomas in transgenic mice. Oncogene. 1995;10:205–209. [PubMed] [Google Scholar]
- 16.Larson RC, Lavenir I, Larson TA, Baer R, Warren AJ, Wadman I, Nottage K, Rabbitts TH. Protein dimerization between Lmo2 (Rbtn2) and Tal1 alters thymocyte development and potentiates T cell tumorigenesis in transgenic mice. EMBO J. 1996;15:1021–1027. [PMC free article] [PubMed] [Google Scholar]
- 17.Aplan PD, Jones CA, Chervinsky DS, Zhao XF, Ellsworth M, Wu CZ, McGuire EA, Gross KW. An scl gene product lacking the transactivation domain induces bony abnormalities and cooperates with LMO1 to generate T-cell malignancies in transgenic mice. EMBO J. 1997;16:2408–2419. doi: 10.1093/emboj/16.9.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chervinsky DS, Lam DH, Melman MP, Gross KW, Aplan PD. scid thymocytes with TCRβ gene rearrangements are targets for the oncogenic effect of SCL and LMO1 transgenes. Cancer Res. 2001;61:6382–6387. [PubMed] [Google Scholar]
- 19.Chervinsky DS, Zhao XF, Lam DH, Ellsworth M, Gross KW, Aplan PD. Disordered T-cell development and T-cell malignancies in SCL LMO1 double-transgenic mice: parallels with E2A-deficienct mice. Mol Cell Biol. 1999;19:5025–5035. doi: 10.1128/mcb.19.7.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aplan PD, Chervinsky DS, Stanulla M, Burhans WC. Sitespecific DNA cleavage within the MLL breakpoint cluster region induced by topoisomerase II inhibitors. Blood. 1996;87:2649–2658. [PubMed] [Google Scholar]
- 22.Aplan PD, Lombardi DP, Kirsch IR. Structural characterization of SIL, a gene frequently disrupted in T-cell acute lymphoblastic leukemia. Mol Cell Biol. 1991;11:5462–5469. doi: 10.1128/mcb.11.11.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colaizzo-Anas T, Aplan PD. Cloning and characterization of the SIL promoter. Biochim Biophys Acta. 2003;1625:207–213. doi: 10.1016/s0167-4781(02)00597-3. [DOI] [PubMed] [Google Scholar]
- 24.Aplan PD, Begley CG, Bertness V, Nussmeier M, Ezquerra A, Coligan J, Kirsch LR. The SCL gene is formed from a transcriptionally complex locus. Mol Cell Biol. 1990;10:6426–6435. doi: 10.1128/mcb.10.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orban PC, Chui D, Marth JD. Tissue- and site-specific DNA recombination in transgenic mice. Proc Natl Acad Sci USA. 1992;89:6861–6865. doi: 10.1073/pnas.89.15.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith AJH, De Sousa MA, Kwabi-Addo B, Heppell-Parton A, Impey H, Rabbitts PH. A site-directed chromosomal translocation induced in embryonic stem cells by Cre-LoxP recombination. Nat Genet. 1995;9:376–384. doi: 10.1038/ng0495-376. [DOI] [PubMed] [Google Scholar]
- 27.Godfrey DI, Kennedy J, Suda T, Zlotnik A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J Immunol. 1993;150:4244–4252. [PubMed] [Google Scholar]
- 28.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]