Abstract
Background
Microorganisms and plants are able to produce tryptophan. Enzymes catalysing the last seven steps of tryptophan biosynthesis are encoded in the canonical trp operon. Among the trp genes are most frequently trpA and trpB, which code for the alpha and beta subunit of tryptophan synthase. In several prokaryotic genomes, two variants of trpB (named trpB1 or trpB2) occur in different combinations. The evolutionary history of these trpB genes is under debate.
Results
In order to study the evolution of trp genes, completely sequenced archeal and bacterial genomes containing trpB were analysed. Phylogenetic trees indicated that TrpB sequences constitute four distinct groups; their composition is in agreement with the location of respective genes. The first group consisted exclusively of trpB1 genes most of which belonged to trp operons. Groups two to four contained trpB2 genes. The largest group (trpB2_o) contained trpB2 genes all located outside of operons. Most of these genes originated from species possessing an operon-based trpB1 in addition. Groups three and four pertain to trpB2 genes of those genomes containing exclusively one or two trpB2 genes, but no trpB1. One group (trpB2_i) consisted of trpB2 genes located inside, the other (trpB2_a) of trpB2 genes located outside the trp operon. TrpA and TrpB form a heterodimer and cooperate biochemically. In order to characterise trpB variants and stages of TrpA/TrpB cooperation in silico, several approaches were combined. Phylogenetic trees were constructed for all trp genes; their structure was assessed via bootstrapping. Alternative models of trpB evolution were evaluated with parsimony arguments. The four groups of trpB variants were correlated with archeal speciation. Several stages of TrpA/TrpB cooperation were identified and trpB variants were characterised. Most plausibly, trpB2 represents the predecessor of the modern trpB gene, and trpB1 evolved in an ancestral bacterium.
Conclusion
In archeal genomes, several stages of trpB evolution, TrpA/TrpB cooperation, and operon formation can be observed. Thus, archeal trp genes may serve as a model system for studying the evolution of protein-protein interactions and operon formation.
Background
The synthesis of tryptophan is a common metabolic capability of microorganisms and higher plants, which is not provided by mammals. The prokaryotic trp operon encodes the enzymes catalysing the final and pathway-specific steps from chorismate to L-tryptophan. For more than 40 years, the enterobacterial operon has now been the classical model system for studying the evolutionary relation of genes and enzymes (see [1,2] and references therein) as well as gene regulation. Considering gene regulation, several, conceptually quite different mechanisms have been described for the trp operon. Most of them were elucidated in bacterial species (see e.g. [3-5], and references therein). However, regulation of trp operon expression has also been shown for the archaea Methanothermobacter thermoautotrophicus [6,7] and Thermococcus kodakaraensis [8]. The reason for an elaborated regulation may be the fact that tryptophan is one of the amino acids, whose biochemical synthesis is very expensive [9]. Besides regulation, other features of tryptophan biosynthesis have been studied extensively. The composition of the operon and several aspects of its evolution have been analysed [10], and for each enzyme, at least one 3D-structure has been determined. Taken together, the trp operon is besides the ribosomal protein operons one of the best-characterised gene clusters occurring in microorganisms. Its investigation has provided fundamental insights into many aspects of bacterial genetics and enzymology; see [2].
The canonical trp operon encodes seven enzymes responsible for the synthesis of L-tryptophan from chorismate. The first reaction is catalysed by the anthranilate synthase, a glutamine amidotransferase, which is a complex consisting of the larger synthase (TrpE) and a smaller glutaminase (TrpG) subunit. The anthranilate phosphoribosyl transferase (TrpD) provides the glutamine amidotransferase function that allows glutamine to serve as the amino donor in anthranilate formation. The two subsequent enzymes, TrpF and TrpC, catalyse the isomerisation of phosphoribosylanthranilate and the synthesis of indole-3-glycerol phosphate, respectively.
TrpA and TrpB constitute the αββα tryptophan synthase complex which catalyses the final reaction from indole-3-glycerole phosphate + L-serine to L-tryptophan + H2O. The α subunit (TrpA) cleaves indoleglycerol-3-phosphate to glyceraldehyde-3-phosphate and indole. The latter is transported through a hydrophobic tunnel to the associated β subunit (TrpB), where it is condensed with L-serine to yield L-tryptophan [11]. A sophisticated mechanism of allostery links the α and β monomers of the synthase; see e.g. [12].
Several Trp enzymes represent paradigmatically larger classes of proteins having similar function or protein architecture: TrpG is similar to HisH (an enzyme involved in histidine biosynthesis) and other glutaminases of type I glutamine amidotransferases [13]. TrpF, TrpC and TrpA are all (βα)8 barrels possessing similar phosphate binding sites [14]. The basic (βα)8 barrel is the most common enzyme fold in the PDB database of known protein structures [15].
For the bacterial trp genes, the following order was determined: large anthranilate synthase subunit (trpE), small anthranilate synthase subunit (trpG), anthranilate phosphoribosyl transferase (trpD), indole-3-glycerol phosphate synthase (trpC), phosphoribosyl anthranilate isomerase (trpF), tryptophan synthase β subunit (trpB) and tryptophan synthase α subunit (trpA), or abbreviated trpEGDCFBA [16]. The gene-fusions trpGD and trpEG have been observed in several species; moreover, in other genomes, the operon is broken up into several gene clusters. In archeal genomes, order of trp genes is highly variable. In Sulfolobus solfataricus, an intact operon trpBADFEGC is observed. In Haloferax volcanii, the trp operon is divided into two isolated clusters, trpCBA and trpDFEG, separated by more than 1200 kb. In the genome of Natronomonas pharaonis, there exist three homologs of trpD and two homologs of trpB, trpE and trpG each. Pyrococcus horikoshii completely lacks the genes for tryptophan synthesis (and for other aromatic amino acids).
The genes trpB, trpA and trpE, trpG are frequently in the same order and in close proximity, i.e. they comprise the linkage groups trpBA and trpEG. In both cases, the gene products constitute a bienzyme complex, whose active centres interact with each other. Because they occur in both bacterial and archeal genomes, these linkage groups have been identified as ancestral [16]. A reconstruction of the tentative ancestral trp operon is hampered by the observation that trp genes are poor phylogenetic reporters. Different rates of evolution, multiple gene duplications and convergent evolution, as a consequence of specific adaptation to environmental demands, may be the reason for inconsistencies seen in comparisons of phylogenies deduced from trp genes or rRNA [16]. Therefore, the evolution of each element of the trp operon has to be examined separately.
For evolutionary studies, tryptophan synthase is an especially interesting candidate. This enzyme has been analysed for decades in order to understand the structural basis and functional consequences of protein-protein interactions [17]. The isolated TrpA and TrpB proteins form stable, however poorly active α monomers and ββ homodimers, respectively [18,19]. Their assembly to the native αββα complex induces conformational changes in both subunit types, as shown by X-ray crystallography for the Pyrococcus furiosus synthase [18]. The result of this communication between the α and β subunits is a reciprocal activation by one to two orders of magnitude [20]. Conformational changes crucial for the allosteric communication between the active sites of the α– and β-subunits have been analysed in detail for the Salmonella typhimurium tryptophan synthase; see e.g. [21-24].
The role of the β-subunit is of particular importance for the evolution of Trp synthase. For archaea and bacteria, it is known that two variants of trpB genes occur, which can clearly be distinguished by their protein sequences [25]. The major group, harbouring proteins of type TrpB1 includes the enzymes of enterobacteria and Bacillus subtilis. The minor group (denoted TrpB2) contains many archeal proteins. Most prokaryotes like E. coli possess a single trpB1 gene. However, in several bacterial and archeal genomes, a combination of one trpB1 and one trpB2 gene occurs. In addition, some species exist, which have only one or two trpB2, but no trpB1 gene. This variety prompted us to characterise the evolution of TrpB and its interaction with TrpA in detail, both biochemically and in silico.
Based on biochemical findings, a model for the evolution of the tryptophan synthase complex has recently been introduced [26]. This model assumes the existence of an ancient and non operon-based trpB2. After duplication, only one trpB2 gene presumably has been integrated into the trp operon. Differences in evolutionary pressure may have been responsible for the divergence of non operon- and operon-based trpB genes. The coevolution with trpA may have led to a better adapted trpB1. The data on complex formation and subunit activation led us consider existing trpB variants as representatives of evolutionary steps in the postulated model.
In this study, I have assessed this model by phylogenetic methods. Two basic questions have been addressed: i) What is the evolutionary relationship of trpB1 and trpB2? ii) How did extant archeal trp operons evolve? Extending previous work [25], I will discuss novel hypotheses concerning the properties of TrpB2 and operon formation. Based on the content of 26 completely sequenced archeal genomes, comparative analyses of trp sequences, and their locations in genomes will be reported in order to reconstruct the evolution of TrpB-type subunits and of the coevolution of TrpA/TrpB. It will be shown that TrpB2 variants represent different stages of TrpA/TrpB cooperation and that TrpB2 is favoured over TrpB1 in certain environments. Moreover, TrpB2 has features of a more ancient TrpB variant.
Results and Discussion
Assessing the composition of trp gene clusters
In order to describe the composition of trp regulons in a quantitative manner and to compare their content in archeal and bacterial genomes, AMIGOS [27] was used. By comparing genomes, this program identifies gene clusters and rates each individual cluster element with a consCL-score. The consCL-score of an individual gene depends on i) the occurrence of this gene in a given gene cluster and ii) the global similarity of the genomes harbouring these clusters. Thus, individual scores assess both the relatedness of genomes and the frequency with which individual genes are members of a cluster. The higher a score, the more pronounced is the occurrence of an individual gene in a given gene cluster. Table 1 lists consCL-scores for elements of archeal and bacterial trp operons. The numbers indicate that in bacteria the clustering of trpA and trpB1 was stronger than that of all other trp genes. In archeal genomes, the clustering of trpE and trpG was most prominent. A reason for the lower score of trpB in archeal trp operons was the occurrence of two trpB variants (trpB1 and trpB2) in these species. The scores signalled that trpB1 was more frequently part of an trp operon than trpB2. Moreover, the score for trpA was lower than that of trpE or trpG. It follows for archaea that trpA and trpB are less strictly integrated into trp operons than in bacteria. This suggests that either evolutionary pressure responsible for operon formation is less pronounced or that additional selective forces disfavour the integration of trpA and trpB into certain archeal trp operons.
Table 1.
conscl scores for trp genes
| consCl – values | Protein | COG # | Function | |
| Archaea | Bacteria | |||
| 2.0 | 2.8 | TrpE | COG0147 | anthranilate/para-aminobenzoate synthases comp. I |
| 2.1 | 2.7 | TrpG | COG0512 | anthranilate/para-aminobenzoate synthases comp. II |
| 1.8 | 2.3 | TrpF | COG0135 | phosphoribosylanthranilate isomerase |
| 1.8 | 2.9 | TrpC | COG0134 | indole-3-glycerol phosphate synthase |
| 1.9 | 3.0 | TrpA | COG0159 | tryptophan synthase alpha chain |
| 1.4 | 3.0 | TrpB1 | COG0133 | tryptophan synthase beta chain |
| 0.6 | - | TrpB2 | COG1350 | paralogue of TrpB |
| 1.9 | 2.6 | TrpD | COG0547 | anthranilate phosphoribosyltransferase |
Note: The first two columns list score values deduced from representative sets of archeal and bacterial genomes. Columns three and four list protein names, COG numbers, and protein function. The conscl-scores were determined by using AMIGOS [27]. COG numbers indicate orthologous gene clusters as defined in the COG database [29].
It has been hypothesised that TrpB2 possesses a second function and acts as a serine deaminase [25]. This prediction has been deduced from the analysis of phyletic patterns, i.e. the absence of an encoded serine deaminase function in certain genomes. However, it has been shown that TrpB1 of Thermotoga maritima and TrpB2_o proteins of Sulfolobus solfataricus and T. maritima have poor serine deaminase activities [26]. An alternative method of non-homologous gene annotation is the exploitation of gene neighbourhoods [28], as e.g. implemented with AMIGOS. For trpB2, AMIGOS did not detect a second conserved gene neighbourhood besides the one constituting trp operons. Thus, no clues for an additional function besides tryptophan synthesis have been deduced for trpB2 by this approach.
A naming code for trpB genes
The two variants of trpB occur in various genomes in different combinations [25]. In order to facilitate the analysis of phylogenetic trees, a naming scheme was introduced. Names of genes and gene products were generated according to the scheme SPECIES_LOC|TYPE|TAX. Here, SPECIES is an abbreviation of the species name (see Materials). LOC indicates the position of the specific trpB gene relative to a putative trp operon (more precisely: relative to a trpA gene). If two trpB genes occur in a genome, they were labelled _i (if the gene was located inside the trp operon) or _o (if located outside the operon). If only a single trpB gene occurred in the genome, it was labelled _s, if the gene was linked to trpA, and it was labelled _S, if it was separate from trpA. TYPE indicates the gene type. It is 1 for trpB1 and 2 for trpB2. Finally, TAX gives the taxonomical classification. It is C for Crenarchaeota, E for Euryarchaeota and B for Bacteria. The following examples explain how to resolve sequence names: Aperni_o2C was used to name a trpB gene in the genome of Aeropyrum pernix (Aperni), which occurred outside the trp operon (_o) and was of type trpB2 (2). As A. pernix is a Crenarchaeota, the name ends with a C. The _o notation indicates that a second trpB gene exists in A. pernix. This gene was consequently named Aperni_i2C, as it is a trpB2 gene inside the trp operon. Note that also pairs like Tmarit_i1B and Tmarit_o2B exist indicating the occurrence of a trpB1 gene inside and a trpB2 gene outside the trp operon. Sacido_s2C is the designation of a trpB2 gene located inside the trp operon. As Sulfolobus acidocaldarius possesses only one trpB gene, it was labelled with a _s. Since Thermoplasma volcanium possesses only one trpB gene, which is non operon-based and of type trpB2, this gene was named Tvolc_S2E. Designations of the encoded proteins were assigned in a corresponding way.
Determining the occurrence of trpB genes
In order to determine the distribution of trpB variants, the COG [29] and the STRING database [30] were used. For all completely sequenced archeal and bacterial genomes, their occurrence was determined and their location was identified. Depending on the occurrence of trpB variants, archeal species were grouped into five categories, named species-types in the following. Note that these species-types characterise the content of genomes. Links to the above naming scheme for genes are gene location and type.
As Table 2 shows, there were six archeal genomes possessing a single trpB gene of class trpB1 (s1 or S1 species), four genomes with a single trpB gene of class trpB2 (s2 or S2 species), five genomes harbouring one operon-based and one additional, non operon-based trpB2 each (i2_o2 species), ten species of type i1_o2 (one operon-based trpB1 and one additional trpB2 gene) and one species possessing one operon-based and at least one non operon-based trpB1 gene (i1_o1 species). The most frequent combination (10 out of 26) was an operon-based trpB1 and a non operon-based trpB2 gene (i1_o2 species). N. pharaonis was the only archeal species of type i1_o1. All five completely sequenced Crenarchaeota possess exclusively genes of class trpB2.
Table 2.
Classifying known archeal genomes according to the occurrence of trpB genes
| S2 (3), s2 (1) | i2_o2 (5) | i1_o2 (10) | i1_o1 (1) | S1 (1), s1 (5) |
|
S. acidocaldarius, s2C 3, TA T. volcanium, S2E 3, TA T. acidophilum, S2E 3, TA P. horikoshii, S2E 2, HT |
A. pernix, i2C 4, o2C 2, HT P. aerophilum, i2C 3, o2C 4, HT P. torridus, i2E 2, o2E 2, TA S. solfataricus, i2C 3, o2C 3, TA S. tokodaii, i2C 3, o2C 4, TA |
A. fulgidus, i1E 2, o2E 6, HT M. acetivorans, i1E 1, o2E 5, MS M. barkeri, i1E 1, o2E 4, MS M. burtonii, i1E 1, o2E 4, MS M. hungatei, i1E 1, o2E 4, MS M. mazei, i1E 1, o2E 5, MS M. thermoautotrophicus, i1E 1, o2E 3, TP P. abyssi, i1E 3, o2E 2, HT P. furiosus, i1E 3, o2E 2, HT T. kodakaraensis, i1E 2, o2E 2, HT |
N. pharaonis, i1E 1, o1E 0, HP |
M. kandleri, S1E 4, YP Halobacterium, s1E 1, HP H. marismortui, s1E 1, HP M. maripaludis, s1E 1, MS M. stadtmanae, s1E 1, MS M. jannaschii, s1E 2, YP |
| 2.75 | i2: 3.0, o2: 3.0 | i1: 1.6, o2: 3.7 | i1: 1.0, o1: 0.0 | 1.6 |
Note: The occurrence and the location of trpB genes were coded according to the following scheme used in the top line: S2 species possess exactly one, non operon-based trpB2 gene, s2: ditto, the gene is located inside the trp operon. trpB1 was treated analogously. i2_o2 are species possessing an trpB2 gene inside and a second trpB2 outside the operon, i1_o2 are species with an operon-based trpB1 and a non operon-based trpB2, and i1_o1 are species possessing an operon-based and at least one non operon-based trpB1. The number of genomes having the same species-type is given in brackets. The acronyms following the species name classify trpB genes; see Results. Numbers give tryptophan codons occurring in the respective gene. The abbreviations indicate hyperthermophilic (HT), thermoacidophilic (TA), thermophilic (TP), mesophilic (MS), halophilic (HP), or hyperthermophilic + halophilic species (YP). The last line of the table gives mean values for the occurrence of tryptophan codons.
Bacterial species did not contribute species-types noticeably different from those observed among archaea (data not shown). Both Geobacter species represent special cases most plausibly explained by ongoing genomic rearrangements: Gsulfu_i2B is an operon-based trpB2 gene of type TrpB2_o. The trp operon of G. sulfurreducens harbours both a trpB1 and a trpB2 gene. According to the annotation, the trpB1 gene (Locus tag GSU2375) contains a frameshift and is annotated as a pseudogene [31]. A direct neighbour of trpB1 in G. metallireducens is a transposase, making a recent transfer of this gene plausible. In comparison to archaea, the occurrence of trpB2 was less frequent in bacterial genomes and none contained exclusively trpB2 genes.
Assessing phylogenetic relationship of trp genes
Sequences originating from all archaea and several representative bacteria were selected for a phylogenetic classification of trp genes. Multiple sequence alignments were created by using M-Coffee [32], and trees were constructed and evaluated using SplitsTrees [33]. Figures 1, 2, 3, 4 are plots of unrooted trees generated for protein sequences of TrpA, TrpB, TrpD, TrpE, and TrpG. In order to assess the statistical strength of individual edges, bootstrap resampling was used. For relevant edges, bootstrap values were plotted; see Figures 1, 2, 3, 4. The trees were analysed in detail, as follows.
Figure 1.
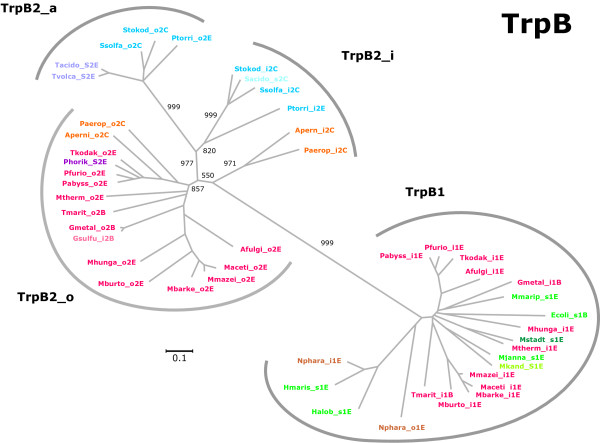
Phylogenetic tree of TrpB sequences. Using archeal and bacterial TrpB sequences, a multiple sequence alignment was generated and an unrooted phylogenetic tree was constructed. Proteins were labelled according to the naming scheme introduced in the Results section. Subtrees were marked according to the sequence type (TrpB1 or TrpB2). TrpB2 sequences span three subtrees; clustering is in agreement with the location of genes. TrpB2_o proteins are all encoded outside operons; 14 out of 16 originate from species that possess an operon-based trpB1 in addition. TrpB2_i proteins are encoded inside operons. Each of these genes is accompanied by a non operon-based trpB2. TrpB2_a sequences occur exclusively in genomes that have a single trpB2 gene or occur as a second trpB2 outside an operon in combination with a trpB2_i gene. The numbers are bootstrap values resulting from 1000 replications. Gene names are colour-coded. Blue colours indicate genes occurring in S2 (violet), s2 (light blue) and those i2_o2 species, which possess trpB2_a or trpB2_i genes (dark blue). Orange colours designate trpB2_i and trpB2_o genes. Red colours signify genes of i1_o2, S2, or s2 species, and green colours mark genes of s1 (light green) or S1 (dark green) species. The names of the two trpB1 copies occurring in N. pharaonis are printed in brown. For acronyms of species-types, see legend of Table 2. The length of the horizontal bar corresponds to 0.1 substitutions per site.
Figure 2.
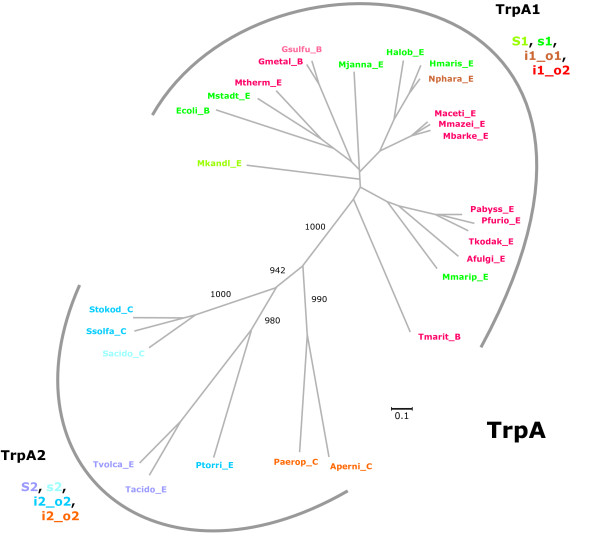
Phylogenetic tree of archeal and bacterial TrpA sequences. The two subtrees cluster genomes, which encode at least one trpB1 gene (S1, s1, i1_o1, or i1_o2 species) or which possess only genes of type trpB2 (S2, s2, i2_o2 species). The clusters were named TrpA1 or TrpA2, respectively. For abbreviations of sequence names, see Results. For colour code, see legend of Figure 1. For the acronyms of species-types, see legend of Table 2.
Figure 3.
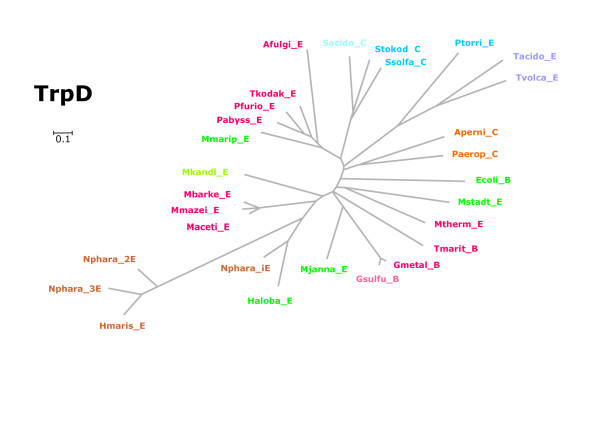
Phylogenetic tree of TrpD sequences. Archeal and bacterial protein sequences were used to construct the unrooted tree. The last letter of the acronyms indicates the taxonomical position of the species. "E" marks Euryarchaeota, "C" Crenarchaeota, and "B" bacterial species. The three TrpD sequences of N. pharaonis are designated as _1E, _2E, and _3E. For colour code and abbreviations, see legend of Figure 1.
Figure 4.
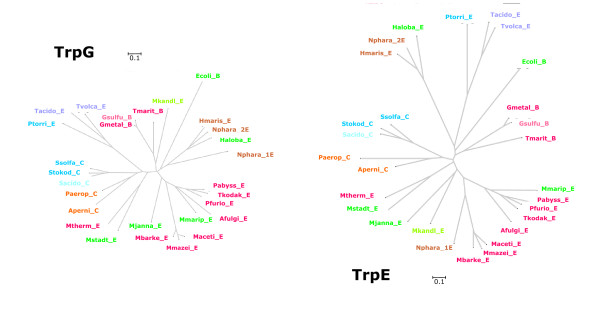
Phylogenetic trees for TrpG and TrpE sequences. Archeal and bacterial protein sequences were used to construct the unrooted tree. The last letter of the acronyms indicates the taxonomical position of the species. "E" marks Euryarchaeota, "C" Crenarchaeota, and "B" bacterial species. The two TrpE sequences of N. pharaonis are designated as _1E, _2E. For colour code and abbreviations, see legend of Figure 1.
TrpB
In agreement with previous findings [25], TrpB1 and TrpB2 clearly fall into two distinct groups. This distinction was supported by a high bootstrap value; see Figure 1. Moreover, among TrpB2 sequences a finer sub-clustering could be deduced, which was in agreement with the location of the genes. One group (labelled TrpB2_o) consisted of products of trpB2 genes not located in operons. 14 out of 16 elements were TrpB2 sequences originating from i1_o2 species, i.e. species possessing besides an isolated lying trpB2 an additional, operon-based trpB1. The genes Paerip_o2C and Aperni_o2C of the two i2_o2 species Pyrobaculum aerophilum and A. pernix belonged to this group too. These two species possess a trp operon containing a trpB2 gene. Bacterial TrpB2_o sequences, which originated from the i1_o2 species T. maritima and G. metallireducens did not form an isolated subtree. This finding argues for a common origin of bacterial and archeal trpB2_o genes.
The other two subgroups of TrpB2 variants were clearly distinct from the TrpB2_o cluster. The sequences of these clusters originated from archeal S2 (Thermoplasmataceae), s2 or i2_o2 species (Sulfolobaceae, Picrophilus torridus, A. pernix, P. aerophilum), i.e. species possessing exclusively one or two trpB2 genes. These sequences formed two clearly separated sets. The first set, named TrpB2_i, subsumes operon-based trpB2 genes, and harboured Stokod_i2C, Sacido_s2C, Ssolfa_i2C, Ptorri_i2E, Apern_i2C, and Paerop_i2C. The second set, named TrpB2_a, consisted of Ptorri_o2E, Stokod_o2C, Ssolfo_o2C, Tacido_S2E, and Tvolca_S2E, and subsumed trpB2 genes located outside trp operons. For Thermoplasma volcanium and Thermoplasma acidophilum, these trpB2 genes were the only trpB genes, for S. solfataricus, S. tokodaii and P. torridus, a second, however distinguishable trpB2 gene of type trpB2_i was part of the trp operon. Proteins of type TrpB2_i formed two finer subgroups: Those of P. torridus and the Sulfolobaceae resembled more sequences of TrpB2_a. Those of A. pernix and P. aerophilum, which possess a non operon-based trpB2_o gene, were different both from TrpB2_a and from TrpB2_o sequences; see Figure 1. All relevant edges separating these groups are due to their high bootstrap value statistically highly significant.
As a single exception, the genome of P. horikoshii did not follow the general classification scheme. It possesses a single trpB2 gene, which is of type trpB2_o and not – as expected – of type trpB2_a. However, this genome lacks all the other trp genes, which has been previously interpreted as reductive evolution [10]. The occurrence of a trpB2_o gene might be due to the loss of the complete trp operon after speciation of trpB2_i and trpB2_o. The fact that the P. horikoshii trpB2_o gene was not affected by the reduction has been considered as an argument for assigning to it an other selective function [25], which has not been identified yet. As noted above, the two bacterial Geobacter species represent special cases associated with the presumptive rearrangement of trp genes. Briefly, the trpB variants can be characterised as follows: trpB1 genes occur exclusively in trp operons. trpB2_o variants represent genes occurring outside operons in those species that have an operon-based trpB1. Several archeal species possess exclusively trpB2 genes: If only one trpB2 gene exists, it is of type trpB2_a, if two trpB2 genes occur, one is an operon-based trpB2_i, the second a trpB2_a, or a trpB2_o gene.
TrpA
Correlated with TrpB speciation, TrpA proteins showed a division into two, statistically highly significant subgroups; see Figure 2. The larger TrpA1 group consisted of TrpA sequences originating from genomes that possess a trpB1 gene. Most likely, TrpA1 proteins interact with the operon encoded TrpB1 and thus fall into the same class. The smaller TrpA2 group contained exclusively TrpA proteins of species-types S2, s2, or i2_o2, i.e. TrpA proteins whose putative interaction partner is exclusively a TrpB2 protein. The high bootstrap value of 1000 (≜100%) for the central edge emphasises the distinction made between TrpA1 and TrpA2. S2, s2, i2_o2 species formed three statistically significant subtrees; compare Figure 2. These harboured the TrpA sequences of (i) Sulfolobaceae, (ii) Thermoplasmatales (T. acidophilum, T. volcanium, P. torridus) and (iii) P. aerophilum, and A. pernix. The composition of these groups is in agreement with the TrpB2_a and TrpB2_i groups in Figure 1 and indicates the coevolution of trpB2 variants with trpA.
TrpD, TrpE, and TrpG
In all three trees (see Figures 3 and 4), both the proteins of Thermoplasmatales and of the three Sulfolobaceae constituted sub-clusters. The edges determined for TrpD or TrpE entries of these species have similar lengths as those calculated for TrpA or TrpB. Especially for the trpA and trpB genes of these species, an increased rate of evolution has been previously postulated [25]. However, the comparison of trees and edge lengths showed that in these species evolutionary divergence is similarly high for several proteins encoded by the trp operon. These findings argue against a specifically increased rate of trpA and trpB evolution. In general, smaller genomes evolve faster [34]. Therefore, a higher evolutionary rate in the trp genes of Thermoplasmatales is more plausible explained by a general trend, which is due to their smaller genome size.
Interestingly, no sub-clustering into smaller, distinctly separated groups was observed in TrpE and TrpG, which form like TrpA and TrpB a heteromeric complex. The above finding distinguishes the subunits of tryptophan synthase from those of anthranilate synthase. TrpG was characterised as the evolutionary most stable trp protein by the compactness of its phylogenetic tree; see Figure 4.
The three Euryarchaeota Halobacterium (s1), Haloarcula marismortui (s1) and Natronomonas pharaonis (i1_s1 species) constituted an isolated group in all five trees (Figures 1, 2, 3, 4); edge lengths were comparable to those of s2 or i2_o2 species. This congruence indicates an elevated evolutionary rate for all elements of these trp operons. Note that these operons harbour trpB1 genes.
Analysing typical differences in TrpA and TrpB sequences
The phylogenetic tree depicted in Figure 1 illustrates that all TrpB variants can be sorted into four, clearly separated groups. The tree did however not allow to deduce the degree of sequence similarity and to infer whether these differences were subtle sequence variations broadly distributed in the whole sequence or larger indels (inserts or deletions). Table 3 lists the results of pairwise sequence comparisons generated by using BLAST [35]. The selected sequences represent the species-types S2, i2_o2, i1_o2, s1, and S1. As expected, sequence similarity values are in agreement with tree composition. Importantly, for all pairwise comparisons, more than 25% identical residues were determined. Therefore, all TrpB variants should most probably have the same overall 3D-structure [36].
Table 3.
Pairwise sequence similarity values of TrpB proteins
| Ssolfa_i2C | Ssolfa_o2C | Paerop_i2C | Paerop_o2C | Afulgi_i1E | Afulgi_o2E | Tmarit_i1B | Tmarit_o2B | Mmarip_s1E | Mkand_S1E | Ecoli_s1B | |
| Tacido_S2E | 49, 72, 2 | 76, 88, 0 | 47, 65, 2 | 46, 67, 1 | 26, 44, 17 | 46, 64, 2 | 32, 43, 18 | 47, 66, 2 | 28, 43, 12 | 30, 46, 11 | 26, 42, 6 |
| Ssolfa_i2C | - | 54, 75, 2 | 56, 71, 3 | 53, 72, 1 | 34, 48, 14 | 57, 72, 1 | 34, 47, 12 | 54, 73, 1 | 30, 46, 15 | 30, 45, 12 | 28, 42, 14 |
| Ssolfa_o2C | - | 50, 66, 2 | 52, 71, 1 | 33, 50, 13 | 47, 68, 1 | 32, 46, 13 | 48, 70, 1 | 28, 43, 16 | 30, 45, 11 | 27, 40, 10 | |
| Paerop_i2C | - | 57, 68, 3 | 35, 49, 14 | 54, 63, 3 | 35, 50, 12 | 54, 65, 3 | 32, 48, 12 | 34, 46, 15 | 31, 43, 13 | ||
| Paerop_o2C | - | 30, 45, 13 | 63, 78, 1 | 32, 46, 13 | 60, 75, 1 | 28, 44, 16 | 33, 46, 11 | 28, 41, 16 | |||
| Afulgi_i1E | - | 31, 44, 11 | 65, 78, 0 | 36, 48, 10 | 65, 85, 0 | 65, 78, 0 | 59, 76, 1 | ||||
| Afulgi_o2E | - | 34, 47, 12 | 64, 76, 0 | 30, 45, 9 | 32, 45, 9 | 29, 40, 12 | |||||
| Tmarit_i1B | - | 35, 47, 11 | 61, 79, 1 | 64, 79, 1 | 58, 75, 1 | ||||||
| Tmarit_o2B | - | 34, 46, 9 | 32, 46, 8 | 29, 40, 12 | |||||||
| Mmarip_s1E | - | 59, 77, 0 | 58, 77, 0 | ||||||||
| Mkand_S1E | - | 57, 72, 0 |
Note: The sequences represent species-types S2, i2_o2, i1_o2, s1, and S1. All pairs were compared by using BLAST with default parameters. For each pair, the fraction of identical residues, similar residues, and inserted gaps is given in percent. For the generation of protein names, see Results.
In order to characterise sequence differences in detail, multiple sequence alignments (MSAs) were generated on the basis of a representative selection of TrpA and TrpB sequences. Figure 5 lists for TrpB the MSA, residue conservation, secondary structure and the location of the interface area. Residues interacting with ligands and residues, which are characteristic for TrpB1 and TrpB2 respectively, were labelled. 3D-data were deduced from the X-ray structure of Pfurio_i1E, i.e. the operon-based TrpB1 protein of P. furiosus [18], which has PDB code 1WDW. For Ssolfa_o2C, the 2D-structure was predicted by using Jpred [37]. SDPpred [38] was employed to identify those residues, which separated TrpB1 and TrpB2 due to their skewed or bimodal distribution. In the following, positions and residues are referenced according to the sequence Pfurio_i1E. Annotations referring active site residues and the interface originate from the PDBsum page and the Macromolecular Structure Database of the EMBL-EBI.
Figure 5.
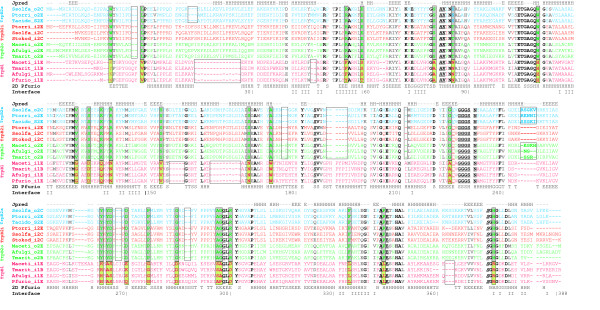
Multiple sequence alignment of TrpB sequences. Representatives of the four groups of TrpB sequences were aligned. Ssolfa_o2C, Ptorri_o2E, and Tacido_S2E represent TrpB2_a sequences. Ptorri_i2E, Ssolfa_i2C, and Stokod_i2C represent TrpB2_i, Maceti_o2E, Afulgi_o2E, and Tmarit_o2B represent TrpB2_o sequences. Maceti_i1E, Tmarit_i1B, Afulgi_i1E, and Pfurio_i1E represent TrpB1 sequences. The 2D-structural elements of Pfurio_i1E, as deduced from the PDB file 1WDW, are shown below the sequences, and residues involved in protein interaction with TrpA (I) are assigned under 'Interface'. The line Jpred (top) lists a 2D-prediction of Ssolfa_o2C generated by using the Jpred server [37]. Residues in bold face printing are conserved; black residues are strictly, grey residues are less strictly conserved. Active site residues are plotted in italics; residues in contact with ligands are underlined. These data were deduced from the PDBsum pages [56] and the PISA server [58] of the EMBL-EBI. Residues printed in boxes were predicted by SDPpred [38] as being specific for TrpB1 or TrpB2. See legend of Figure 1 for an explanation of sequence acronyms.
The MSA shows that nearly all differences between TrpB1 and TrpB2 are due to larger indels, in agreement with [25]. Interestingly, an insertion of 2 to 6 residues between positions 243 and 244 occurred coincidently in TrpB2_a and TrpB2_o sequences, i.e. exclusively in non operon-based proteins. All considered TrpB1 and TrpB2_i sequences lack this subsequence, which was not predicted as a well-defined 2D-element by Jpred. Several representatives belonging to these two sets of operon-based proteins were shown to interact with TrpA [26,39]. Therefore, it is probable that this putative loop influences the allosteric communication with TrpA. Most residues, which are in contact with ligands in the known TrpB1 structure, were strictly conserved among all TrpB1 and TrpB2 sequences. The only exception is residue C225, which is V225 in TrpB2_a sequences. The active site residues H81, K82, and S371 were strictly conserved, whereas active site residue K162 was conserved only in TrpB1 proteins and active site residue D300 (TrpB1) was an arginine in TrpB2. Several residues of the interface regions, adjacent to active sites and near sites interacting with ligands had a bimodal occurrence pattern distinguishing TrpB1 and TrpB2. Among these were residues 2 and 110, which were strictly conserved tryptophan residues in all TrpB2 proteins. Given its position near the gene start, W2 may assume a function in translation control. W110 succeeds a cluster of strictly conserved residues suggesting a role in stability or protein function.
Figure 6 lists the MSA generated for TrpA sequences. It shows that the active site residues E36, D47, and Y161 are strictly conserved in the TrpA sequences studied. Most evident was a three-residue insertion into TrpA2 sequences following position 125 (numbering deduced from TrpA of P. furiosus) as well as deletions at position 162 and between positions 172 and 174. Moreover, most positions showing a bimodal or skewed distribution specific for a trpA variant were located near interface regions. In summary, the deviations characterising the two TrpA variants were not as pronounced as those observed in TrpB sequences, however three indels distinguished TrpA1 from TrpA2.
Figure 6.
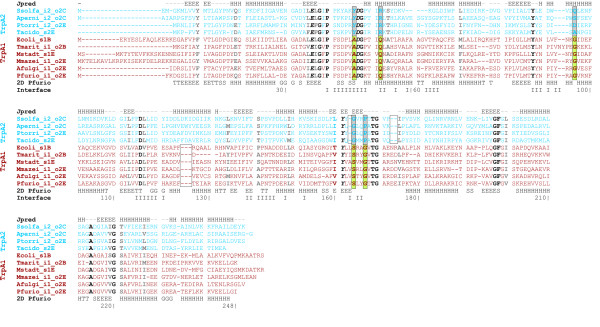
Multiple sequence alignment of TrpA sequences. Ssolfa_i2_o2C, Aperni_i2_o2C, and Ptorri_i2_o2C represent TrpA sequences from i2_o2 species possessing both an operon-based and a non operon-based trpB2. Tacido_s2E represents a species having exclusively an operon-based trpB2. Ecoli_s1B and Tmarit_i1_o2B represent bacterial TrpA proteins. Mstadt_s1E is from a species possessing exclusively an operon-based trpB1 gene, Mmazei_i1_o2E, Afulgi_i1_o2E and Pfurio_i1_o2E are TrpA sequences from i1_o2 species possessing an operon-based trpB1 and a non operon-based trpB2 gene. Presumably, these TrpA1 proteins interact with a protein of type TrpB1. Below the alignment, the 2D-structure of TrpA of P. furiosus (Pfurio_i1_o2E), and residues involved in protein interaction with its TrpB1 (I) are given. The line named Jpred lists a 2D-prediction of Ssolfa_i2_o2C generated by using the Jpred server [37]. Residues printed in bold are conserved; black residues are strictly, grey residues are less strictly conserved. Active site residues are plotted in italics. These data were deduced from the PDBsum pages [56] and the PISA server [58] of the EBI. Residues printed in boxes were predicted by SDPpred [38] as being specific for the two TrpA species.
Frequency of Trp codons in trpB genes
It has been postulated that the avoidance of tryptophan residues in enzymes for tryptophan synthesis provides a selective advantage [7] as has been shown for a number of amino acid biosynthetic enzymes [40]. This criterion was also applied to the trpB genes by assessing the frequency of tryptophan codons (Table 2). trpB1 genes contained one or two tryptophan codons with a mean value of 1.6 both for S1, s1, and i1_o2 species. trpB2 genes contained two tryptophan codons or more with a mean of 2.75 for S2 species, and 3.0 for i2_o2 species. Most pronounced was the difference for i1_o2 species. Here, trpB2 genes had a mean of 3.7, whereas trpB1 genes had a mean of 1.6 tryptophan codons. These trpB1 genes showed a habitat-specific imbalance of tryptophan codon occurrence with one in mesophilic species and at least two tryptophan codons in hyperthermophiles. In summary and according to the notion of tryptophan codon avoidance, trpB2 genes are less optimised than trpB1 genes.
The composition of archeal trp gene clusters
The evolution of individual genes and operon formation proceed in parallel. For the combined analysis of both processes, gene orders of relevant archeal and some bacterial trp operons were determined and plotted in Figure 7. In most operons, the gene orders trpBA and trpEG, respectively, were conserved; however, the arrangement of the linkage groups varied. Figure 7 is organised as six panels A – F. Panel A depicts the trp clusters of Thermoplasmataceae, which are of type trpA2DFEGC; trpB2 lies isolated. In Sulfolobaceae and P. torridus (Panel B), trpB2 is the first gene of the gene cluster trpB2ADFEGC, which matches the above trpA2DFEGC in all positions following trpB2. Panel C gives the gene clusters of A. pernix and P. aerophilum, which possess a trpB2_i and a trpB2_o gene. In the genome of A. pernix, two linkage groups trpA2B2FC and trpDEG occur; P. aerophilum possesses the cluster trpB2DEGA2. Panel D lists archeal genomes containing linkage groups trpCB1A1 and trpDFEG. In Methanosarcina mazei, these genes form a single cluster, resulting in trpCB1A1DFEG. In N. pharaonis, these groups are separated by more than 69kb. In panel E, operons are listed where trpB1 lies close to the 3'-terminal end. For Thermococcus kodakaraensis, Methanococcus maripaludis, Archaeoglobus fulgidus and Pyrococcus abyssi, gene order is trpCDEGFB1A1. The gene orders in Panel E resemble bacterial operons; two representative examples are plotted in panel F.
Figure 7.
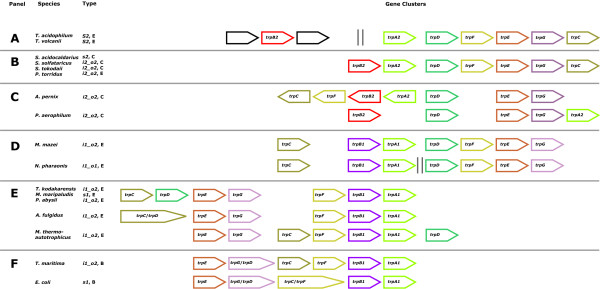
Gene organisation of archeal and bacterial trp gene clusters. Each panel A – F represents the occurrence and orientation of trp genes in the genomes of organisms listed in the second column. The third column gives the species-type and the taxonomical lineage. "E" marks Euryarchaeota, "C" Crenarchaeota, and "B" bacteria. A vertical double line (Panels A and D) indicates borders of gene clusters separately located in the genome. Open arrows represent hypothetical genes. The arrows are not to scale; gaps of arbitrary length were inserted between genes to allow the alignment of arrows. For acronyms of species-types, see legend of Table 2.
It has been argued that simple trp clusters may have been unstable until the complexity of regulation and the foundation of a metabolic theme had reached a certain level [10]. Gene clusters observed in s2 and i2_o2 species can be considered the less evolved stages of cluster organisation; compare Panels A – C. Moreover, the only archeal trp gene regulatory systems identified so far are part of the trp operons of M. thermoautotrophicus [7] and T. kodakaraensis [8], which both have a bacterial-like composition.
Besides Nanoarchaeum equitans, Thermoplasmata (T. volcanium, T. acidophilum, and P. torridus) possess the smallest archeal genomes sequenced so far. Most plausibly, strong selective pressure associated with the colonised habitat enforces the minimisation of genome size. However, both Thermoplasma species possess the gene cluster trpA2DFEGC. Therefore, the need for tryptophan synthesis can be taken for granted. The separation of trpB2 from the remaining trp genes is consistent with a demand for individual gene regulation and expression presumably due to an additional function of TrpB2. Most plausibly, under these constraints, trpB2 is the more optimal variant, which is in a specific environment favoured over trpB1.
What is the origin of trpB genes?
Recently, TrpA, Tmari_i1B and Tmari_o2B of T. maritima have been produced in E. coli, purified, and characterised [39]. It has been shown that recombinant TrpA forms an α-monomer, and that both recombinant TrpB proteins form β2-homodimers. However, only the operon-encoded Tmari_i1B – but not Tmari_o2B – associated with TrpA to constitute the conventional αββα tryptophan synthase complex in which both subunits reciprocally activate each other. An analogous experiment has been carried out for genes of S. solfataricus [26]. The results have shown that Ssolfa_i2C – but not Ssolfa_o2C – associates transiently with TrpA during catalysis to form a functional tryptophan synthase complex. However, in contrast to regular tryptophan synthases, the affinity between the two subunit-types was weak, and activation has been unidirectional from Ssulfo_i2C to TrpA. These results indicate the following ranking for the binding-affinity to TrpA: TrpB2_o < TrpB2_i < TrpB1.
In the course of modelling trpB evolution, the relationship between the trpB variants has to be made plausible. A possible explanation for the existence of two trpB variants would be convergent evolution, i.e. the independent development of trpB1 and trpB2 towards a trpB gene. In this case, few residues, which are critical for function, should correspond. However, one would expect these residues embedded into polypeptides, which are relatively dissimilar on the sequence level. In contrast, comparison of TrpB1 and TrpB2 sequences shows that on average 30% of the residues are identical and 40% are similar; compare Table 3. This finding and the conservation of indels makes convergent evolution highly improbable and argues for a common origin of trpB1 and trpB2 genes.
The most-widely accepted model for the evolution of novel protein functions postulates gene duplication and the generation of a redundant gene copy [41]. It is assumed that evolutionary stress for a copy is largely reduced thus facilitating the evolution of a paralogue with a novel function. This model is based on the notion that negative trade-offs dominate evolutionary processes [42]. According to this model of evolution, one of the trpB genes originates from a copy of the ancestral variant. Which of the two existing variants represents the more ancient gene? The arguments listed below suggest that trpB2 is the ancestral trpB gene.
i) trpB1 is not universally distributed among archaea. Crenarchaeota possess exclusively trpB2 genes. ii) A low frequency of amino acids in enzymes required for their synthesis provides selective advantage [40]. In general, trpB1 genes contain fewer tryptophan codons than trpB2 genes; in i1_o2 species, the ratio is 1.6/3.7 i.e. less than 0.5. Therefore, trpB1 is the more evolved gene. iii) The sophisticated inter-subunit communication suggests that the products of trpB1 and trpA1 of species-types s1 or i1_o2 are the most efficient enzymes; see [39] and references therein. Hence, TrpB1 is the more optimised and later evolved TrpB variant. iv) It has been postulated that ancient enzymes possess broad specificities [43]. The occurrence of trpB2 outside trp operons argues for either a new function or a broader specificity. In summary, it is plausible to regard trpB2 as representing the more ancient variant of trpB.
Modelling the evolution of TrpB
In order to reduce the number of possible alternative scenarios that have to be discussed for modelling the evolution of trpB, the following assumptions were made:
i) For bacteria, an ancestral trp operon of type trpEGDCFB1A1 is most likely [10]. Therefore, the existence of a trpB1 gene in the bacterial predecessor was taken for granted. In addition, it has been concluded for bacterial trp operons that horizontal gene transfer (HGT) did not affect the path of evolutionary history [44].
ii) trpB1, trpB2, trpA1 and trpA2 have been invented only once. The analysis of multiple sequence alignments (see Figures 5 and 6) shows that the main differences distinguishing the variants are conserved indels. It has been convincingly argued that conserved indels result less likely than e.g. point mutations from independent mutational events and provide useful milestones for the identification of evolutionary phases [45]. In addition, the strong coherence seen in the TrpB subtree argues against an independent evolution occurring in parallel for bacteria and archaea. Due to the existence of conserved indels, an evolutionary process trpB2_i → trpB1 → trpB2_o or vice versa is unlikely too.
iii) As has been deduced previously [46], the following order of importance was taken for the processes of genome evolution: gene loss > gene genesis > gene duplication > HGT.
iv) The integration of a trpB gene into the trp operon (or linkage group) was rated less probable than other translocations, gene duplications, gene loss, and mutations. It is presumably very rare that a particular gene gets integrated into a specific gene cluster [47], which is the trp operon in the considered case.
v) It is unlikely that several recent events of HGT explain the taxonomically widespread occurrence of trpB2 genes in bacteria. In bacteria, trpB2 genes were found in hyperthermophilic (Aquificae and Thermotogae) and mesophilic bacteria belonging to the taxonomical groups of Alpha- and Gammaproteobacteria and Bacteroides. The program SIGI [48] identifies genomic islands, i.e. gene clusters having a conspicuous codon usage indicating recent HGT events. In none of the considered genomes were trpB1 or trpB2 genes (both inside and outside operons) elements of such islands.
In order to model the evolution of tryptophan synthase, a phylogenetic tree based on archeal 16S rRNA sequence comparisons was plotted according to Fig. 2 from [49]. All considered species and their species-types were added. Using the above premises, the most plausible sequence-types of predecessors were determined. These types and evolutionary events needed to infer the modern species-types from the predecessors were added to the tree; see Figure 8.
Figure 8.
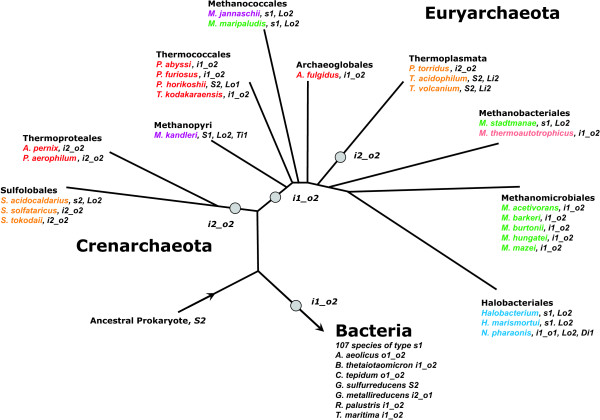
A parsimonious reconstruction of predecessors. The phylogenetic tree is based on 16S rRNA sequence comparisons (after Fig. 2 of [49]). For all modern species, their species-type and evolutionary events leading most plausibly from the ancestral predecessor to the current genome content are added. The most probable species-type of predecessors for Crenarchaeota, Euryarchaeota, and Bacteria is given next to the grey circles. Abbreviations for events changing genomic content: Li1 or Lo2, (L)oss of the operon-based trpB1 or the non operon-based trpB2 gene, respectively. Ti1 (T)ranslocation of the operon based trpB1, Di1 (D)uplication of the operon-based trpB1. The colour of species names indicates the habitat: Hyperthermophiles are given in red, thermoacidophiles in orange, thermophiles in pink, mesophiles in green, halophiles in blue, and species living in a both hyperthermophilic and halophilic environment are given in purple.
The most plausible predecessor of all Crenarchaeota is of type i2_o2; for Bacteria and for Euryarchaeota it is of type i1_o2. Assuming this and excluding Thermoplasmata (see below), of the 23 modern archeal species, 14 have the same species-type as their ancestor. Of the 9 species possessing a deviating type, 7 can be explained with a single gene loss, and for only 2 modern species a more complicated genomic rearrangement has to be postulated: Loss of trpB2 and dislocation of trpB1 has to be postulated for M. kandleri (representing Methanopyri), which is a S1 species. The replacement of trpB2_o with a copy of trpB1 is necessary to explain the i1_o1 genome of N. pharaonis. The only euryarcheal class requiring a more complex explanation than gene loss and translocation subsumes Thermoplasmata, which possess exclusively trpB2 and trpA2 genes. The composition of congruency groups (compare Figures 1 and 2) makes a common evolution with Sulfolobales or the acquisition of the same trp genes probable. The similarity of operon structures supports this assumption: operon structures of P. torridus and Sulfolobales are identical (compare Panel B of Figure 7). For T. acidophilum, a large amount of HGT with S. solfataricus, which is found in the same habitat, has been made plausible [50]. In summary, a common evolutionary history of trpB2 and trpA2 genes of Sulfolobales and Thermoplasmata is highly plausible, proposing for both taxonomical classes an ancestor of species-type i2_o2. Assuming an i2_o2 ancestor, gene loss is sufficient to explain the genome composition of all modern Thermoplasmata.
Based on these predecessors, three alternatives explaining the evolution and distribution of trpB species starting from the last universal common ancestor (LUCA) of bacteria and archaea were deduced (Figure 9, Panels A – C). In the following paragraph, the plausibility of these alternatives will be discussed. The rest of this paragraph is used to elucidate the three alternatives.
Figure 9.
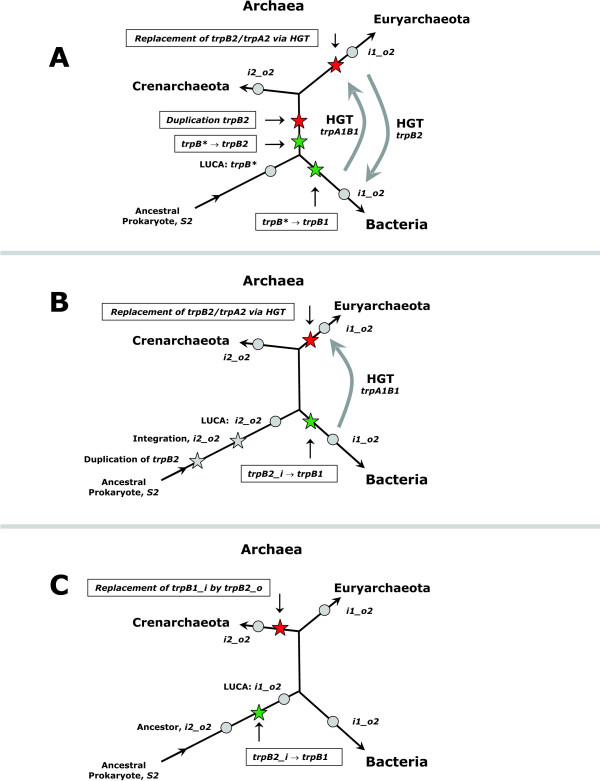
Alternative models of trpB evolution. Model A assumes that a single and intermediate trpB* gene existed in the last universal common ancestor (LUCA) of bacteria and archaea. The evolution of the trpB2 gene is considered an archeal and that of the trpB1 gene is considered a bacterial invention. The occurrence of trpA1 and trpB1 genes in archaea and of trpB2 genes in bacteria are explained by a twofold horizontal gene transfer (HGT). A duplication of trpB2 in an ancient archeal genome has been postulated to explain the existence of the non operon-based trpB2. Models B and C propose two alternatives for the evolution of the LUCA. Model B assumes that the evolution trpB2 → trpB1 occurred in an early bacterial species after the divergence of bacteria and archaea. The replacement of linkage group trpB2A2 by trpB1A1 via HGT was postulated to account for the euryarcheal predecessor of type i1_o2. Model C assumes that the evolution trpB2 → trpB1 occurred before the divergence of bacteria and archaea. Hence, the replacement of an operon-based trpB1 by a trpB2 gene and the evolution trpA1 → trpA2 was postulated for the crenarcheal ancestor. For acronyms of species-types, see legend of Table 2. Distances are arbitrary and do not represent evolutionary time intervals. Stars indicate events of genomic rearrangements, circles filled in grey represent ancient predecessors.
In Panel A of Figure 9, the existence of an ancestral trpB*, an intermediate of trpB1 and trpB2 was postulated for the LUCA. trpB* might then have diverged into a bacterial trpB1 and an archeal trpB2 variant. To explain the existence of a non operon-based trpB2 in archaea, a duplication of trpB2 is necessary. The advent of an euryarcheal i1_o2 predecessor requires the replacement of linkage group trpB2A2 with trpB1A1 via HGT from bacteria to archaea. The occurrence of trpB2 in bacterial genomes demands an early transfer of trpB2 from an archeal to a bacterial predecessor.
Panel B of Figure 9 depicts an alternative model for the evolution of the LUCA towards the bacterial and archeal ancestors. As introduced above, gene duplication is regarded the first step for evolving a novel gene function. In addition, trpB2 must be considered to represent the more ancient variant of trpB. Therefore, the evolution towards the LUCA of bacteria and archaea is most plausibly explained by the duplication of a non operon-based trpB2 gene, which was subsequently integrated into the trp operon and constituted an ancient linkage group trpB2A2. This makes a common ancestor of type i2_o2 plausible. These considerations are the basis for further reconstructing the evolution of predecessors. In Panels B and C, two alternatives are given.
In Panel B, it is assumed that the LUCA was of type i2_o2 and that the evolution trpB2 → trpB1 occurred in an early bacterial species. In this case, species-types of the LUCA and the crenarcheal predecessor are identical. To explain the advent of an euryarcheal predecessor of type i1_o2, an ancient event of HGT from Bacteria to Archaea has to be postulated for the acquisition of the linkage group trpB1A1, which replaced trpB2A2.
In Panel C, it is assumed that the LUCA was of type i1_o2, i.e. the evolution trpB2 → trpB1 occurred earlier than the speciation of Bacteria and Archaea. In this case, the species-types of the LUCA and the predecessors of Bacteria and Euryarchaeota are identical. However, a replacement of trpB2_i by trpB2_o is necessary to constitute the crenarcheal predecessor.
How plausible are these three models?
Model A requires at least two ancient events of HGT to explain the occurrence of trpB2 in Bacteria and of trpA1B1 in Euryarchaeota. The phenomenon of non-orthologous displacement in situ is well-characterised [51,52]. In addition to HGT, a duplication of the trpB2 gene is needed for the predecessor of Archaea. This model is not the most parsimonious one: Model B demands only one HGT event, the ancient acquisition of the linkage group trpB1A1 by an euryarcheal predecessor.
Model C postulates a LUCA of species-type i1_o2. The sophisticated inter-subunit communication clearly suggests that products of trpB1 and trpA1 genes are the most specialised and most recently evolved tryptophan synthases; see [39] and references therein. Thus, the replacement of trpB1 with trpB2, which is needed to explain the existence of a crenarcheal predecessor of type i2_o2, would – with respect to protein-protein interaction – lead to a less optimal tryptophan synthase. This seems unlikely, if one presumes the sustained need for tryptophan synthesis in Crenarchaeota.
In contrast, model B postulates the replacement of a (less evolved) trpB2_i by a trpB1 for the euryarcheal predecessor and does not require the replacement of a trpB1 by a trpB2 for the crenarcheal predecessor. The case of Thermoplasmata makes clear that in a thermophilic or hyperthermophilic environment trpB2 and trpA2 genes are favoured over trpB1 and trpA1. There is evidence that the LUCA was a thermophilic or hyperthermophilic species [34,53,54]. Therefore, it is more probable to expect a LUCA of species-type i2_o2. In summary, considering parsimony arguments and the assumption that negative trade-offs dominate evolutionary processes [42], model B is the likelier one. Figure 10 summarises the most parsimonious scenario explaining the composition of modern archeal trp operons: Assuming that the LUCA was of type i2_o2, and that trpB1 was a bacterial invention, besides gene loss, which is a frequent evolutionary event, two cases of ancient HGT are sufficient to explain the distribution of trpA and trpB species in current archeal genomes.
Figure 10.
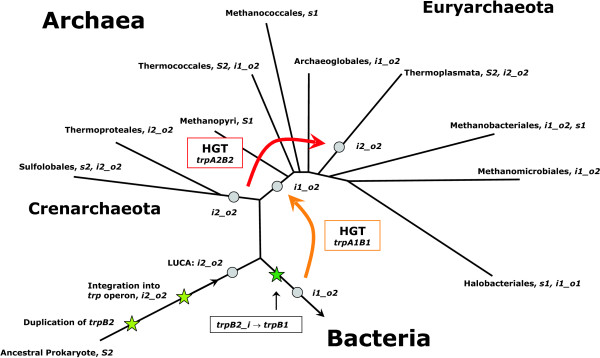
Composite model of trpB evolution. Upon duplication and integration of an ancient trpB2 gene into the trp operon, the last universal common ancestor (LUCA) of bacteria and archaea was of species-type i2_o2. In a bacterial ancestor, the evolution of a linkage group trpB1A1 occurred. Via horizontal gene transfer (HGT), an euryarcheal ancestor acquired this linkage group, which gave rise to a predecessor of type i1_o2. Thermoplasmata acquired trpA2 and trpB2 genes in an ancient event of HGT. For all taxonomical orders, species-types of current species are given. S2 species possess exactly one, non operon-based trpB2 gene, s2: ditto, the gene is located inside the trp operon. trpB1 was treated analogously. i2_o2 are species possessing a trpB2 gene inside and a second trpB2 outside the operon, i1_o2 are species with an operon-based trpB1 and a non operon-based trpB2, and i1_o1 are species possessing an operon-based and at least one non operon-based trpB1.
Conclusion
In archeal genomes, various stages of trpB function have been conserved. Most plausibly, trpB2 represents the ancestral variant of trpB genes. With respect to TrpA/TrpB communication and cooperativity, the situation observed in S2 species (T. acidophilum and T. volcanium) is probably the least complex one. Similarly archaic are the non operon-based trpB2 genes of Sulfolobaceae, whereas the operon-based trpB genes are more evolved. s1 and i1_o2 species possess highly cooperative synthases. Thus, the archeal tryptophan synthase (especially trpB variants) constitutes a model system for the study of protein complex formation. Due to different environmental conditions, several stages of cooperativity have been conserved, which allow to characterise the progress of trpA – trpB coevolution based on gene expression and on functional cooperativity.
Materials
Genomes and protein sequences
Genomic content was determined by analysing version 6.2 of the STRING database [30].
All protein sequences were downloaded via the "Genome Project" database of the NCBI [55], which allows to access completely sequenced genomes. Respective COG tables were consulted to determine the COG group of genes [29] and to download sequences. Genes originating from the following completely sequenced genomes were analysed (abbreviations used for Figures and accession numbers of genomes in brackets):
Crenarchaeota
Aeropyrum pernix K1 (Aperni, NC_00854), Pyrobaculum aerophilum str. IM2 (Paerop, NC_003364), Sulfolobus acidocaldarius DSM 639 (Sacido, NC_007181), Sulfolobus solfataricus P2 (Ssolfa, NC_002754), Sulfolobus tokodaii str. 7 (Stokod, NC_003106).
Euryarchaeota
Archaeoglobus fulgidus DSM 4304 (Afulgi, NC_000917), Haloarcula marismortui ATCC 43049 (Hmaris, NC_006396), Halobacterium sp. NRC-1 (Halob, NC_002607), Methanocaldococcus jannaschii DSM 2661 (Mjanna, NC_000909), Methanococcoides burtonii DSM 6242 (Mburto, NC_007955), Methanococcus maripaludis S2 (Mmarip, NC_005791), Methanopyrus kandleri AV19 (Mkandl, NC_003551), Methanosarcina acetivorans C2A (Maceti, NC_003552), Methanosarcina barkeri str. Fusaro (Mbarke, NC_007355), Methanosarcina mazei Go1 (Mmazei, NC_003901), Methanosphera stadtmanae DSM 3091 (Mstadt, NC_007681), Methanospirillum hungatei JF-1 (Mhunga, NC_007796) Methanothermobacter thermautotrophicus str. Delta H. (Mtherm, NC_000916), Natronomonas pharaonis DSM 2160 (Nphara, NC_007426), Picrophilus torridus DSM9790 (Ptorri, NC_005877), Pyrococcus abyssi GE5 (Pabyss, NC_000868), Pyrococcus furiosus DSM 3638 (Pfurio, NC_003413), Pyrococcus horikoshii OT3 (Phorik, NC_000961), Thermococcus kodakaraensis KOD1 (Tkodak, NC_006624), Thermoplasma acidophilum DSM1728 (Tacido, NC_002578), Thermoplasma volcanium GSS1 (Tvolca, NC_002689).
Bacteria
Escherichia coli K-12 (Ecoli, NC_000913), Geobacter metallireducens GS-15 (Gmetal, NC_007517), Geobacter sulfurreducens PCA (Gsulfu, NC_002939), Thermotoga maritima (Tmarit, NC_00853).
Methods
Generating multiple sequence alignments
For the generation of multiple sequence alignments (MSAs) the program M-Coffee [32] was used. It combines the output of nine individual MSA methods for the generation of a "meta"-MSA. M-Coffee has been shown to outperform all individual methods of MSA generation [32].
Annotating multiple sequence alignments
For each position in a MSA, residue conservation, secondary structure, the location of the interface area, active sites and residues, which are characteristic for sequence types, were determined and plotted. 3D-data were deduced from the PDB-file 1WDW, describing the TrpA/TrpB complex of P. furiosus [18]. For 2D-structure prediction, Jpred [37] was used. SDPpred [38] was utilised to identify those residues, which distinguished sequence groups due to their skewed or bimodal distribution. Annotations referring active site residues were deduced from the PDBsum page [56,57], interface residues were annotated according to the Protein interfaces, surfaces and assemblies service PISA [58,59]. Both services were located at the webserver of the European Bioinformatics Institute (EMBL-EBI).
Creating and evaluating phylogenetic trees
SplitsTrees4 [33], a frame-work for phylogenetic analyses, was used to generate and analyse phylogenetic trees. MSAs originating from M-Coffee were utilised to calculate maximum likelihood protein distance estimates based on a JTT [60] model. The bio-neighbour joining approach [61] was used to generate trees. Resulting trees were analysed by bootstrapping (1000 replications each).
Acknowledgments
Acknowledgements
I want to thank Reinhard Sterner and Rüdiger Schmitt for stimulating discussions and their help in preparing this manuscript.
References
- Yanofsky C. Using studies on tryptophan metabolism to answer basic biological questions. J Biol Chem. 2003;278:10859–10878. doi: 10.1074/jbc.X200012200. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. Advancing our knowledge in biochemistry, genetics, and microbiology through studies on tryptophan metabolism. Annu Rev Biochem. 2001;70:1–37. doi: 10.1146/annurev.biochem.70.1.1. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981;289:751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. The different roles of tryptophan transfer RNA in regulating trp operon expression in E. coli versus B. subtilis. Trends Genet. 2004;20:367–374. doi: 10.1016/j.tig.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Preciado A, Jensen RA, Yanofsky C, Merino E. New insights into regulation of the tryptophan biosynthetic operon in Gram-positive bacteria. Trends Genet. 2005;21:432–436. doi: 10.1016/j.tig.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Gast DA, Jenal U, Wasserfallen A, Leisinger T. Regulation of tryptophan biosynthesis in Methanobacterium thermoautotrophicum Marburg. J Bacteriol. 1994;176:4590–4596. doi: 10.1128/jb.176.15.4590-4596.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Reeve JN. Regulation of tryptophan operon expression in the archaeon Methanothermobacter thermautotrophicus. J Bacteriol. 2005;187:6419–6429. doi: 10.1128/JB.187.18.6419-6429.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Ezaki S, Fujiwara S, Takagi M, Atomi H, Imanaka T. The tryptophan biosynthesis gene cluster trpCDEGFBA from Pyrococcus kodakaraensis KOD1 is regulated at the transcriptional level and expressed as a single mRNA. Mol Gen Genet. 1999;262:815–821. doi: 10.1007/s004380051145. [DOI] [PubMed] [Google Scholar]
- Bentley R. The shikimate pathway--a metabolic tree with many branches. Crit Rev Biochem Mol Biol. 1990;25:307–384. doi: 10.3109/10409239009090615. [DOI] [PubMed] [Google Scholar]
- Xie G, Keyhani NO, Bonner CA, Jensen RA. Ancient origin of the tryptophan operon and the dynamics of evolutionary change. Microbiol Mol Biol Rev. 2003;67:303–342. doi: 10.1128/MMBR.67.3.303-342.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn MF, Aguilar V, Brzovic P, Drewe WF, Jr., Houben KF, Leja CA, Roy M. The tryptophan synthase bienzyme complex transfers indole between the a- and b-sites via a 25-30 Å long tunnel. Biochemistry. 1990;29:8598–8607. doi: 10.1021/bi00489a015. [DOI] [PubMed] [Google Scholar]
- Weber-Ban E, Hur O, Bagwell C, Banik U, Yang LH, Miles EW, Dunn MF. Investigation of allosteric linkages in the regulation of tryptophan synthase: the roles of salt bridges and monovalent cations probed by site-directed mutation, optical spectroscopy, and kinetics. Biochemistry. 2001;40:3497–3511. doi: 10.1021/bi002690p. [DOI] [PubMed] [Google Scholar]
- Zalkin H, Smith JL. Enzymes utilizing glutamine as an amide donor. Adv Enzymol Relat Areas Mol Biol. 1998;72:87–144. doi: 10.1002/9780470123188.ch4. [DOI] [PubMed] [Google Scholar]
- Wilmanns M, Hyde CC, Davies DR, Kirschner K, Jansonius JN. Structural conservation in parallel b/a-barrel enzymes that catalyze three sequential reactions in the pathway of tryptophan biosynthesis. Biochemistry. 1991;30:9161–9169. doi: 10.1021/bi00102a006. [DOI] [PubMed] [Google Scholar]
- Sterner R, Höcker B. Catalytic versatility, stability, and evolution of the (ba)8-barrel enzyme fold. Chem Rev. 2005;105:4038–4055. doi: 10.1021/cr030191z. [DOI] [PubMed] [Google Scholar]
- Brown JR, Doolittle WF. Archaea and the prokaryote-to-eukaryote transition. Microbiol Mol Biol Rev. 1997;61:456–502. doi: 10.1128/mmbr.61.4.456-502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles EW. Tryptophan synthase: a multienzyme complex with an intramolecular tunnel. Chem Rec. 2001;1:140–151. doi: 10.1002/tcr.4. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Ogasahara K, Ma J, Nishio K, Ishida M, Yamagata Y, Tsukihara T, Yutani K. Conformational changes in the tryptophan synthase from a hyperthermophile upon a2b2 complex formation: crystal structure of the complex. Biochemistry. 2005;44:11417–11427. doi: 10.1021/bi050317h. [DOI] [PubMed] [Google Scholar]
- Hioki Y, Ogasahara K, Lee SJ, Ma J, Ishida M, Yamagata Y, Matsuura Y, Ota M, Ikeguchi M, Kuramitsu S, Yutani K. The crystal structure of the tryptophan synthase b2 subunit from the hyperthermophile Pyrococcus furiosus. Investigation of stabilization factors. Eur J Biochem. 2004;271:2624–2635. doi: 10.1111/j.1432-1033.2004.04191.x. [DOI] [PubMed] [Google Scholar]
- Kirschner K, Lane AN, Strasser AW. Reciprocal communication between the lyase and synthase active sites of the tryptophan synthase bienzyme complex. Biochemistry. 1991;30:472–478. doi: 10.1021/bi00216a024. [DOI] [PubMed] [Google Scholar]
- Miles EW, Rhee S, Davies DR. The molecular basis of substrate channeling. J Biol Chem. 1999;274:12193–12196. doi: 10.1074/jbc.274.18.12193. [DOI] [PubMed] [Google Scholar]
- Schneider TR, Gerhardt E, Lee M, Liang PH, Anderson KS, Schlichting I. Loop closure and intersubunit communication in tryptophan synthase. Biochemistry. 1998;37:5394–5406. doi: 10.1021/bi9728957. [DOI] [PubMed] [Google Scholar]
- Weyand M, Schlichting I. Crystal structure of wild-type tryptophan synthase complexed with the natural substrate indole-3-glycerol phosphate. Biochemistry. 1999;38:16469–16480. doi: 10.1021/bi9920533. [DOI] [PubMed] [Google Scholar]
- Osborne A, Teng Q, Miles EW, Phillips RS. Detection of open and closed conformations of tryptophan synthase by 15N-heteronuclear single-quantum coherence nuclear magnetic resonance of bound 1-15N-L-tryptophan. J Biol Chem. 2003;278:44083–44090. doi: 10.1074/jbc.M308276200. [DOI] [PubMed] [Google Scholar]
- Xie G, Forst C, Bonner C, Jensen RA. Significance of two distinct types of tryptophan synthase beta chain in bacteria, archaea and higher plants. Genome Biol. 2002;3:RESEARCH0004. doi: 10.1186/gb-2001-3-1-research0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopoldseder S, Hettwer S, Sterner R. Evolution of multi-enzyme complexes: the case of tryptophan synthase. Biochemistry. 2006;45:14111–14119. doi: 10.1021/bi061684b. [DOI] [PubMed] [Google Scholar]
- Merkl R. AMIGOS: a method for the inspection of genomic organisation or structure and its application to characterise and dissect conserved gene arrangements. In Silico Biol. 2006;6:281–306. [PubMed] [Google Scholar]
- Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci U S A. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. The COG database: an updated version includes Eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, Jouffre N, Huynen MA, Bork P. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–7. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methe BA, Nelson KE, Eisen JA, Paulsen IT, Nelson W, Heidelberg JF, Wu D, Wu M, Ward N, Beanan MJ, Dodson RJ, Madupu R, Brinkac LM, Daugherty SC, DeBoy RT, Durkin AS, Gwinn M, Kolonay JF, Sullivan SA, Haft DH, Selengut J, Davidsen TM, Zafar N, White O, Tran B, Romero C, Forberger HA, Weidman J, Khouri H, Feldblyum TV, Utterback TR, Van Aken SE, Lovley DR, Fraser CM. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science. 2003;302:1967–1969. doi: 10.1126/science.1088727. [DOI] [PubMed] [Google Scholar]
- Wallace IM, O'Sullivan O, Higgins DG, Notredame C. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 2006;34:1692–1699. doi: 10.1093/nar/gkl091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander C, Schneider R. Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins. 1991;9:56–68. doi: 10.1002/prot.340090107. [DOI] [PubMed] [Google Scholar]
- Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- Kalinina OV, Novichkov PS, Mironov AA, Gelfand MS, Rakhmaninova AB. SDPpred: a tool for prediction of amino acid residues that determine differences in functional specificity of homologous proteins. Nucleic Acids Res. 2004;32:W424–8. doi: 10.1093/nar/gkh391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettwer S, Sterner R. A novel tryptophan synthase b-subunit from the hyperthermophile Thermotoga maritima. Quaternary structure, steady-state kinetics, and putative physiological role. J Biol Chem. 2002;277:8194–8201. doi: 10.1074/jbc.M111541200. [DOI] [PubMed] [Google Scholar]
- Alves R, Savageau MA. Evidence of selection for low cognate amino acid bias in amino acid biosynthetic enzymes. Mol Microbiol. 2005;56:1017–1034. doi: 10.1111/j.1365-2958.2005.04566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Heidelberg , Springer; 1970. [Google Scholar]
- Kondrashov FA. In search of the limits of evolution. Nat Genet. 2005;37:9–10. doi: 10.1038/ng0105-9. [DOI] [PubMed] [Google Scholar]
- Jensen RA. Enzyme recruitment in evolution of new function. Annu Rev Microbiol. 1976;30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- Xie G, Bonner CA, Song J, Keyhani NO, Jensen RA. Inter-genomic displacement via lateral gene transfer of bacterial trp operons in an overall context of vertical genealogy. BMC Biol. 2004;2:15. doi: 10.1186/1741-7007-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS. Protein phylogenies and signature sequences: A reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol Mol Biol Rev. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel B, Bork P, Huynen MA. Genomes in flux: The evolution of archaeal and proteobacterial gene content. Genome Res. 2002;12:17–25. doi: 10.1101/gr.176501. [DOI] [PubMed] [Google Scholar]
- Price MN, Arkin AP, Alm EJ. The life-cycle of operons. PLOS Genetics. 2006;2:e96. doi: 10.1371/journal.pgen.0020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkl R. SIGI: score-based identification of genomic islands. BMC Bioinformatics. 2004;5:22. doi: 10.1186/1471-2105-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber H, Hohn MJ, Stetter KO, Rachel R. The phylum Nanoarchaeota: present knowledge and future perspectives of a unique form of life. Res Microbiol. 2003;154:165–171. doi: 10.1016/S0923-2508(03)00035-4. [DOI] [PubMed] [Google Scholar]
- Ruepp A, Graml W, Santos-Martinez ML, Koretke KK, Volker C, Mewes HW, Frishman D, Stocker S, Lupas AN, Baumeister W. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature. 2000;407:508–513. doi: 10.1038/35035069. [DOI] [PubMed] [Google Scholar]
- Omelchenko MV, Makarova KS, Wolf YI, Rogozin IB, Koonin EV. Evolution of mosaic operons by horizontal gene transfer and gene displacement in situ. Genome Biol. 2003;4:R55. doi: 10.1186/gb-2003-4-9-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Ponomarev VA, Koonin EV. Two C or not two C: recurrent disruption of Zn-ribbons, gene duplication, lineage-specific gene loss, and horizontal gene transfer in evolution of bacterial ribosomal proteins. Genome Biol. 2001;2:RESEARCH 0033. doi: 10.1186/gb-2001-2-9-research0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giulio M. The universal ancestor and the ancestor of bacteria were hyperthermophiles. J Mol Evol. 2003;57:721–730. doi: 10.1007/s00239-003-2522-6. [DOI] [PubMed] [Google Scholar]
- Gribaldo S, Brochier-Armanet C. The origin and evolution of Archaea: a state of the art. Philos Trans R Soc Lond B Biol Sci. 2006;361:1007–1022. doi: 10.1098/rstb.2006.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI_Genomes NCBI complete microbial genomes http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi
- Laskowski RA, Chistyakov VV, Thornton JM. PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res. 2005;33:D266–8. doi: 10.1093/nar/gki001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PDBsum PDBsum home page http://www.ebi.ac.uk/thornton-srv/databases/pdbsum/
- Krissinel E, Henrick K. In: Detection of protein assemblies in crystals. Berthold MR, editor. Springer, Heidelberg; 2005. pp. 163–174. [Google Scholar]
- EBI_PISA EBI PISA home page http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]


