Abstract
The architecture of the brain is characterized by a modular organization repeated across a hierarchy of spatial scales—neurons, minicolumns, cortical columns, functional brain regions, and so on. It is important to consider that the processes governing neural dynamics at any given scale are not only determined by the behaviour of other neural structures at that scale, but also by the emergent behaviour of smaller scales, and the constraining influence of activity at larger scales. In this paper, we introduce a theoretical framework for neural systems in which the dynamics are nested within a multiscale architecture. In essence, the dynamics at each scale are determined by a coupled ensemble of nonlinear oscillators, which embody the principle scale-specific neurobiological processes. The dynamics at larger scales are ‘slaved’ to the emergent behaviour of smaller scales through a coupling function that depends on a multiscale wavelet decomposition. The approach is first explicated mathematically. Numerical examples are then given to illustrate phenomena such as between-scale bifurcations, and how synchronization in small-scale structures influences the dynamics in larger structures in an intuitive manner that cannot be captured by existing modelling approaches. A framework for relating the dynamical behaviour of the system to measured observables is presented and further extensions to capture wave phenomena and mode coupling are suggested.
Keywords: neural dynamics, functional connectivity, multiscale decomposition, nonlinear bifurcations
1. Introduction
Understanding the nature of neural dynamics is central to the valid analysis and interpretation of functional neuroscience data. Theoretical and numerical models of neural activity may play an important role in this regard, as they can elucidate the mechanisms leading to spatial and temporal patterns in the data. In addition, models of neural systems can be used to give construct validity to novel empirical methods (e.g. Breakspear 2002; David et al. 2004) or constrain parameters in empirical algorithms (e.g. Robinson et al. 2001; Friston et al. 2003). At a deeper level, neural models may also aid understanding of the neural processes underlying perception (Kiebel & Friston 2004), cognition (Breakspear et al. 2004), behaviour (Daffertshofer et al. 1999) and evolution (Sporns et al. 2000). There are three ingredients to such models: (i) a set of evolution equations which embody key physiological processes in neural structures; (ii) an ‘architecture’, which determines how these neural structures are coupled together to form a neural ensemble; and (iii) a formalism that allows the dynamics of the model to inform our understanding of cognitive processes and neural complexity. In this paper, we introduce a novel wavelet-based approach that permits neural models to be built with a uniquely brain-like architecture.
Many authors have stressed that the modular architecture of the neuropil, repeated over a hierarchy of scales, is an overarching principle of brain organization (e.g. Nunez 1997). At microscopic scales, neurons represent distinct entities and fulfil functionally specific roles, such as feature-specific activation in the extrastriate cortex. At a scale of approximately 40 μm, clusters of inhibitory neurons are loosely bound to pyramidal cells to form cortical ‘microcircuits’. At mesoscopic scales (ca. 0.3–3 mm), the histologically distinct and functionally specific roles of cortical columns (Szentagothai 1983; Mountcastle 1997, 1998; Shephard 2004) and cortical subareas or ‘macrocolumns’ (Hilgetag et al. 2000) have been well studied. At macroscopic levels, the brain has been partitioned into regions, delineated according to coincident functional and anatomical criteria, such as the cortical areas of the CoCoMac database of primate cortex (www.cocomac.org; Stephan et al. 2001; Passingham et al. 2002). Cortical lobes and hemispheres comprise the coarsest scales of the hierarchy. At each level of the hierarchy, there exists an arrangement of small networks into subsystems with more complex and diffuse functions: these form the elementary units at the next scale. Such a view is also in agreement with graph-theoretical studies of cortical connectivity, which observe ‘small-world’ (Watts & Strogatz 1998) complex connectivity patterns (Sporns et al. 2000). That is, networks with high cluster coefficients (dense local connectivity) and relatively short path lengths (owing to random, sparse long-range connections) support the emergence of local structures across scales. Such characteristics have also been observed in the functional connectivity of human electroencephalography (EEG) and magnetoencephalography (MEG) (Stam 2004) and functional magnetic resonance imaging (fMRI) data (Salvador et al. 2005). However, constraints are required on the probability distribution of short to long-range configurations if the network is to exhibit structures at specific scales as described here.
At all scales (some more than others), the functional and anatomical boundaries of the modules are coincident. This is not surprising, as the function of a cortical area is largely determined by its extrinsic connections and intrinsic architecture (Passingham et al. 2002). For example, neurons within a cortical column share internal connectivity, afferent input/efferent target, function (such as response to specific visual stimuli) and have highly correlated activity (Szentagothai 1983). At the largest scales (particularly in heteromodal cortex), function becomes increasingly diffuse and delocalized.1 However, even at coarse scales and in heteromodal cortex, general functional and anatomical principles have been described (Mesulam 1998) and investigated (Wright et al. 1999). At the very coarsest scale, functional competition between the hemispheres has been well described in a variety of cognitive and perceptual tasks (e.g. Blake & Logethetis 2002; Stephan et al. 2003). The scales of this hierarchy form a geometric series in the sense that the number of subunits within each larger structure are of the same order of magnitude (ca. 102) across scales (Nunez 1997, p.111).
Various neural models exist at each of these scales. Models involving individual neurons provide insight into a variety of phenomena, such as local high-frequency rhythms (e.g. Lumer et al. 1997), but face numerical barriers to large-scale extrapolation. Models at larger scales typically average over the dynamics at smaller scales, assuming a ‘mean-field’ approach (e.g. Nunez 1974; Freeman 1975; Van Rotterdam et al. 1982; Jirsa & Haken 1996; Robinson et al. 1997, 2001; Frank et al. 2000; Breakspear et al. 2003; Harrison et al. 2005). Such models emphasize that the organizational principles at each scale have a strong influence on the dynamics at that scale. Large-scale models are able to incorporate information about physiology at smaller scales such as post-synaptic potential decay times (e.g. Jirsa & Haken 1996), and can predict macroscopic observables (e.g. Robinson et al. 2001). However, these models cannot elucidate the manner in which the dynamics at any given scale are influenced by the dynamics at other scales. Consequently, they cannot fully explain the interplay between observables at different scales, or the functional significance of emergence in neural systems.
The aim of the present work is to illustrate a novel modelling framework that addresses these issues by placing the neuronal dynamics within a multiscale architecture. There are essentially three steps: (i) At each scale, local ‘within-module’ dynamics are derived from the key neurobiological processes occurring at that scale. (ii) These local dynamics are then coupled together—with a function representing scale-specific neuronal connectivity—to give a scale-specific neuronal ensemble. Temporal evolution of such an ensemble has been previously shown to generate neuronal activity with specific parameter-dependent spatio-temporal patterns (Larter et al. 1999; Breakspear et al. 2003, 2004a). (iii) A multiscale hierarchy of such neuronal ensembles—with interdependences between scales—is then constructed using the wavelet transform. As is elucidated below, this yields a hierarchy of neuronal systems whose spatial scales are geometrically nested, thus capturing the neuropil architecture sketched above. Spatio-temporal ‘structures’ at small scales are projected through the wavelet transform, into the dynamical motions at larger scales. Hence, the emergent properties of dynamics at any scale function to facilitate interscale interdependences with dynamics at other scales. A further modification is shown to allow concurrent geometric nesting in the temporal domain.
Previous studies have reported transient and complex emergent neuronal dynamics (e.g. Friston 2000a,b; Stam et al. 2003). The unique contribution of the present formulation is the third of the above steps, which permits temporal evolution of such dynamics at a variety of spatio-temporal scales with interdependences between scales. Two ideas are critical for this: (i) The orthogonality of the wavelet basis functions implies that the projection of dynamics from one scale to another introduces spatio-temporal information from a smaller scale on to another with, and only with, the appropriate spatial variance. Put differently, the dynamics at a given scale are only influenced by a single-scale congruent harmonic of the dynamics at a smaller scale. (ii) The spatial localization of the wavelet coefficients (in contrast to Fourier components) additionally permits local features of the dynamics to be present in the between-scale coupling functions. That is, at each scale, there exist local and global (scale-specific) mean-field effects, which are both also present in the modelled between-scale interdependences.
The paper is organized as follows. In §2, we provide a brief overview of wavelet theory. The main framework is explicated mathematically in §3. In §4, we present select numerical realizations of the model that capture some phenomena that we believe are unique to the proposed framework. In §5, we sketch a general framework for relating the dynamical realizations to measured variables in both functional neuroimaging and electrophysiological data in a manner that also uniquely permits scale-specific effects. We then consider the Kuromoto model as a means of capturing our departure from standard mean-field modelling approaches in the light of the numeric realizations of §4. Specifically, we explain the critical difference between ‘scale-free dynamics’ and ‘multiscale modelling’. The paper concludes with a discussion of more elaborate potential developments, a comparison with other complex dynamical systems, and some limitations of the proposed framework.
2. A brief tour of wavelets for dynamical systems
In this paper, we model the dynamics of a set of dynamical variables, V(j)(x, t), defined across a hierarchy of spatial scales . The dynamics of the V(j) are determined by the action of scale-specific evolution operators F(j) which are elaborated in §3. As stated above, interdependences between scales j1, j2,… are introduced through the action of wavelet decompositions. We now briefly review the wavelet transform, and the notation adopted throughout the remainder of the paper is described. For a more extensive elucidation, the reader is referred to Daubechies (1992) and Mallat (1999). For the present purposes, the wavelet transform is explicated mathematically in the setting of spatio-temporal dynamical systems (see also Guan et al. 2002). Readers less familiar with mathematical notation may gain an intuitive understanding of the wavelet multiscale subspace projectors Λ(j), defined below, from the final paragraph of this section and by examining figures 1 and 2.
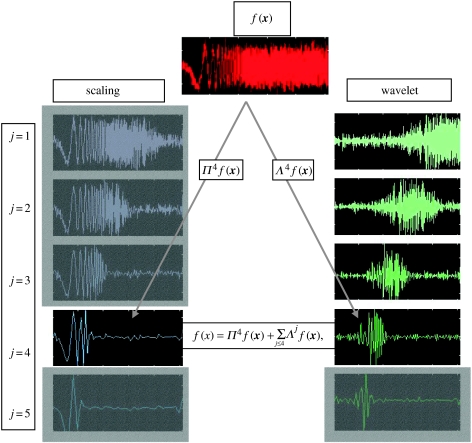
Figure 1.
Wavelet representation of a chirp, f(x). The chirp is shown as its centre frequency increases with time, showing a helpful time–frequency decomposition. Left column shows the projection of f on to a hierarchy of approximation subspaces through Π, and the right column shows the projection of f on to a hierarchy of wavelet subspaces through Λ. Note that the increasing frequency component (scales) is located progressively towards the end of the signal. The unmasked components represent an exemplar decomposition up to scale J=4.
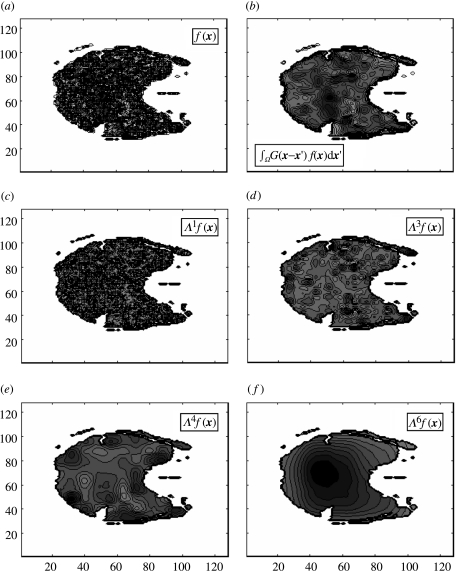
Figure 2.
Contour map of a single slice BOLD signal from a human subject (1.5 T). (a) Raw (unfiltered) data; (b) Gaussian smoothed data with confluence from all scales; and (c)–(f) four wavelet subspace projections, with scale-specific signal component.
In neural modelling, we are interested in the temporal evolution of a set of M dynamical variables across a spatial domain x∈N of dimension N≤3. The Vs embody the time varying physiological variables of interest, such as membrane potential, cell firing rates, ion channel conductance, and so on. In this study, we assume time t to be continuous but treat space x as discrete. For economy of expression, we begin with a one-dimensional spatial domain x={x1, x2,…,xn} with periodic boundary conditions xn+1=x1 (there are no ‘free edges’ in the brain). Because we are primarily interested in studying dynamics in a system with a multiscale spatial architecture, wavelet decomposition of the spatial dimension only is studied. Naturally, multiscale phenomena in the temporal domain may also be of crucial importance for neural systems; hence, relaxation of these constraints is also considered.
For any given dynamical variable at an instance in time, we thus have a spatially varying signal V=V(x). A ‘multi-resolution decomposition’ of this signal is an orthonormal representation of V across a hierarchy j=1,2,… of spatial scales (the smallest scale is j=1). At each scale, V(x) is projected on to two orthogonal subspaces: the detail (‘wavelet’) subspace D(J), which contains information about the fluctuations (variance) in signal intensity at that scale; and the approximation subspace A(J), which contains the residual of the signal after the details at that and all smaller scales j≤J have been removed. The decomposition is orthogonal across scales,
| (2.1) |
and complete so that
| (2.2) |
for a very broad class of (L2=‘square integrable’) real-valued functions. Denote the linear subspace projectors on to the detail and approximation subspaces at scale j as Λ(j) and Π(j), respectively. A multiscale decomposition—the lossless recovery of the original signal by linear superposition of the approximation of the signal at any scale J together with the details at that and all smaller scales—is thus
| (2.3) |
Wavelets are families of basis functions that permit such a decomposition. A family of wavelet functions {ψ(j,k)}j,k∈Z is generated through dilation (by scale factor j) and translation (by position factor k) of a single ‘mother’ wavelet function ψ(x),
| (2.4) |
The wavelet functions at scale J span the detail (‘wavelet’) subspace D(J), hence
| (2.5) |
| (2.6) |
where 〈〉 denotes the inner product and the c(j,k) are the ‘detail coefficients’. Note from equation (2.4) that the scale of adjacent subspaces scales with a factor of two; hence, the scale of the subspaces form a geometric series,
| (2.7) |
Uniquely associated with each mother wavelet function is a family of scaling functions ϕj,k generated from a ‘father’ scaling function ϕ(x); these span the approximation subspaces A(J). Hence, the wavelet and scaling functions provide complementary high-pass (wavelet) and low-pass (scaling) multiscale decompositions. There exist a growing library of suitable functions that differ according to their symmetry, width of support, and so on.
Temporal dynamics are accommodated by performing the wavelet projections at each point in time.2 The temporal evolution of the system in physical space V(x, t) defines a two-dimensional evolutionary sheet (the spatio-temporal equivalent of an orbit). Multi-scale decomposition yields a multiscale hierarchy of evolutionary sheets, Λ(j)V(x, t) where j=1,2,…,J. Examples are illustrated below.
In the interests of comprehensiveness, a rather technical description of the wavelet transform has been given here. For readers unacquainted with wavelet theory, the basic idea of the wavelet transform can be captured in graphical from. Figure 1 shows an example of a multiscale decomposition of a one-dimensional signal. Figure 2 presents a wavelet decomposition of a single slice fMRI BOLD signal expressed as a contour map, contrasting the raw signal S(x, t) (panel a), and a Gaussian-smoothed signal (panel b) with four subspace projections ΛjS(x, t) (panels c–f). Note that in the raw data, all scales are in confluence although the higher (spatial) frequencies are visually dominant. In the Gaussian-smoothed data, the larger scales are visually dominant. By contrast, a wavelet projection picks out the spatial variance at specific scales. The wavelet transform is analogous to a ‘mathematical microscope’ where the spatial frequency and resolution are naturally matched (Bullmore et al. 2004). Wavelet decompositions also bear a strong relationship to ‘detrended fluctuation analysis’ where lines of best fit fulfil the role of basis functions.
3. Multi-scale evolution equations
The central mathematical formalism of the multiscale model is described in this section. The basic dynamics within a subsystem (at any arbitrary scale) are explicated in §3a. The framework for coupling these together to form a single-scale ensemble is discussed in §3b. Examples from a particular model of coupled neural subsystems are given in order to illustrate the ‘evolutionary sheets’ introduced above. Decomposition into ‘multiscale evolutionary sheets’ is also visualized. Construction of the full spatio-temporal hierarchy is explicated in §3c.
(a) Dynamics within local modules
We commence with the evolution of a single local subsystem through the value of m independent real-valued variables V at spatial location xi. These represent the mean value of the corresponding physiological variable over the domain of the local ensemble. The dynamics are determined by an evolution equation,
| (3.1) |
where F:m→m is a continuous nonlinear function derived from the physiological behaviour and histological connectivity of the subsystem; a is a real-valued vector that smoothly parametrizes F and introduces physiological properties (such as the density of a class of membrane channels) that may slowly vary or be subject to external control; ϵ represents stochastic perturbations, which may be a result of noisy subcortical afferents.
The orbits of F are the solution curves V(xi, t) within the phase space spanned by m. Attractors are the set of all long-term solution curves (after initial transients) and may include fixed point (steady state), limit cycle (periodic oscillations), tori (quasi-periodic oscillations) or strange (chaotic) attractors. Mathematically, these are studied by setting ϵ=0 (see Breakspear & Terry 2002). While attractors represent the long-term system behaviour only, they also play a crucial role in shaping transient and itinerant dynamics (e.g. Ashwin 1995). However, the present model does not require that the dynamics have reached their asymptotic state.
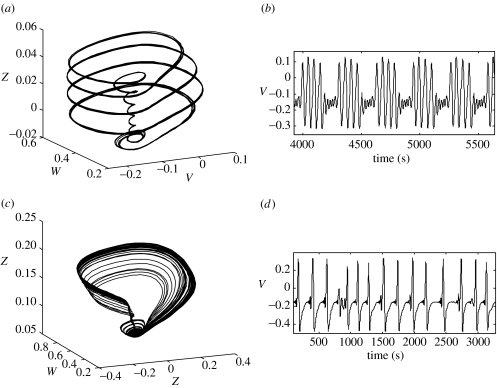
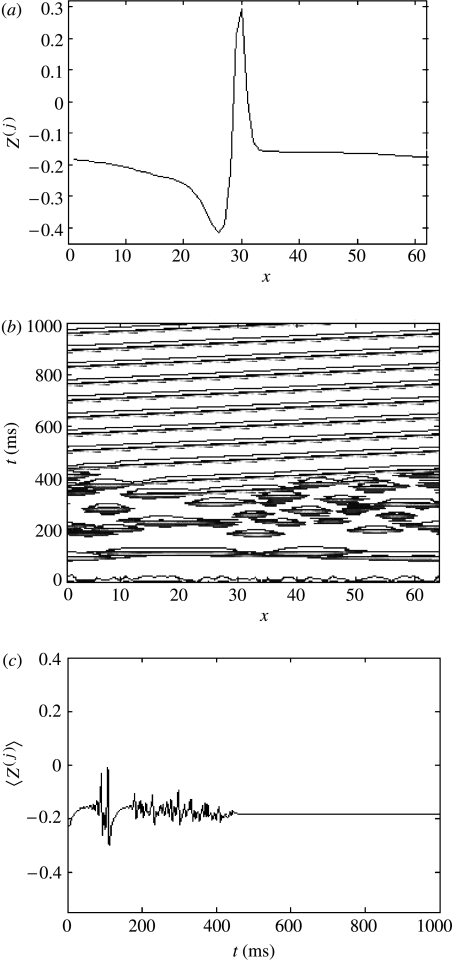
Equation (3.1) represents a general form for a dynamical neural system. The specific evolution equations employed in this study are adopted from Morris–Lecar dynamics for local population activity by Larter et al. 1999. They describe a local population of densely interconnected inhibitory and excitatory neurons whose behaviours are determined by voltage and ligand-gated membrane channels. Sodium and calcium channels display a nonlinear sigmoid-shaped graph of voltage-dependent conductance. Potassium channel conductance is modelled in a more complex manner, exponentially relaxing towards its voltage-dependent state. The equations are presented in the Appendix. Figure 3 presents exemplar attractors and time-series for this neural system. Complex periodic (panels a and b) and chaotic (panels c and d) are shown.
Figure 3.
Exemplar attractors and time-series for the neural system employed in this study for equation (3.1). Complex periodic (panels a and b) and chaotic (panels c and d) are shown. Panels (a) and (c) show the orbits and attractors in the system's phase space. Panels (b) and (d) show the corresponding time-series of the pyramidal cell potentials, V.
For the general multiscale framework, however, no specific form needs to be given to F.
(b) Within-scale dynamics
To construct a model for each scale of the hierarchy, the individual elements are coupled together to form an ensemble of nonlinear oscillators. This is a widely used approach for the modelling of neural systems (e.g. Frank et al. 2000; Strogatz 2001; Breakspear & Terry 2002; Breakspear & Terry 2004). The dynamics of an element at position xi are given by,
| (3.2) |
where the coupling function H introduces the influence of the entire ensemble V(x) on to each individual node, parametrized in strength by c. If the dynamics within the array are strongly nonlinear (chaotic), then a range of complex synchronous behaviours are possible, even if the function H is linear in V(xi) (Fujisaka & Yamada 1983; for review see Boccaletti et al. 2002). If the coupling is linear, then H can be written in matrix form and (3.2) reduces to
| (3.3) |
where Hij is the connection strength between nodes i and j. Note that the coupling still occurs within F (the local dynamics) so that the influences are injected into, rather than on to, the local evolution. This form can be used, for example, to model competitive agonistic action at post-synaptic glutamate receptors (Breakspear et al. 2003a, 2004a). If Hi,j=1 for all ij, then we have a globally coupled network, which is the most tractable case from an analytic perspective (Kaneko 1997). However, from a neurophysiological perspective, it is necessary to break the symmetry in a manner that embodies the nature of anatomical connectivity in real brains. For the present purposes, we wish to study coupled subsystems defined across a one- or two-dimensional array, x∈n n=1, 2. In this setting, the symmetry of the matrix must be broken in order to introduce a ‘metric’—a dependence of Hij on the distance between xi and xj. Hij can hence be viewed as a ‘synaptic footprint’—the dependence of coupling strength on distance. In the simulations of §4, we employ,
| (3.4) |
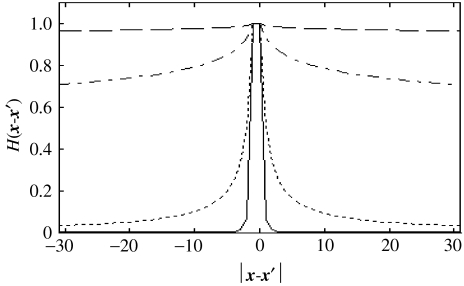
K>0 is a constant vector which parametrizes the shape of the synaptic footprint. When K is close to zero, Hij is almost constant, so that mean-field effects dominate. Increasing K decreases the effective support of Hij towards a neighbourhood of xi. Note that K is of the same dimension of the modelled array, in order to permit anisotropic coupling K1≠K2. Figure 4 illustrates the dependence of Hij on K. The constant h ensures that sum(H)=0 so that the choice of K has no influence on the overall interensemble feedback. A recent neuroimaging study reported that functional correlations in resting state data displayed the form of equation (3.4) with K=2 (Salvador et al. 2005).
Figure 4.
Dependence of the intrascale ‘synaptic footprint’ H on four choices of K, equation (3.4): K=4 (solid); K=1 (dotted); K=0.1 (dot-dashed); and K=0.01 (dashed).
Whereas the solution curves of individual ‘nodes’ are represented by orbits in the phase space m, the solutions of (3.2) can be thought of as evolutionary ‘sheets’ (surfaces) in the phase space m×n. For the present study, we consider five types of evolutionary sheets for a one-dimensional system n=1 and hence K=K. The number of coupled subsystems is set to n=64. The equations are given in the Appendix. Parameter values are given in table 1.
Table 1.
Parameter values unless otherwise stated.
| parameter | description | value |
|---|---|---|
| TCa | threshold value for Ca channels | −0.01 |
| δCa | variance of Ca channel threshold | 0.15 |
| gCa | conductance of population of Ca channels | 1.1 |
| VCa | Ca Nernst potential | 1 |
| TK | threshold value for K channels | 0.0 |
| δK | variance of K channel threshold | 0.30 |
| gK | conductance of population of K channels | 2.0 |
| VK | K Nernst potential | −0.7 |
| TNa | threshold value for Na channels | 0.3 |
| δNa | variance of Na channel threshold | 0.15 |
| gNa | conductance of population of Na channels | 6.7 |
| VNa | Na Nernst potential | 0.53 |
| VL | Nernst potential of leak channels | −0.5 |
| gL | conductance of population of leak channels | 0.5 |
| VT | threshold potential for excitatory neurons | 0.0 |
| δV | variance of excitatory threshold | 0.65 |
| ZT | threshold potential for inhibitory neurons | 0.0 |
| δZ | variance of inhibitory threshold | 0.70 |
| I | subcortical input strength | 0.30 |
| aee | excitatory to excitatory synaptic strength | 0.4 |
| aei | excitatory to inhibitory synaptic strength | 2 |
| aie | inhibitory to excitatory synaptic strength | 2 |
| ane | nonspecific to excitatory synaptic strength | 1 |
| ani | nonspecific to inhibitory synaptic strength | 0.4 |
| b | time constant scaling factor | 0.10 |
| φ | temperature scaling factor | 0.7 |
| τ | time constant for K relaxation time | 1 |
| rNMDA | ratio of NMDA to AMPA receptors | 0.25 |
| β | random modulation of subcortical input | 0 |
Adapted from Larter & Speelman (1999)
(i) Uncoupled array
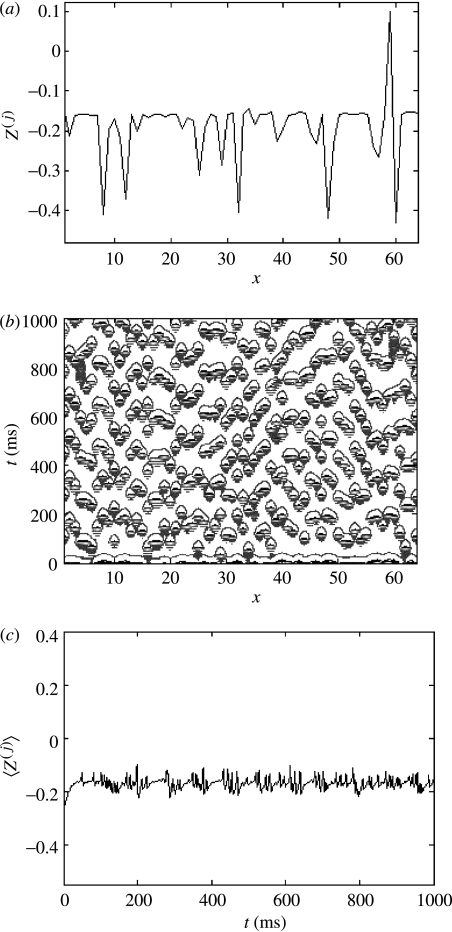
With c=0, the dynamics of each node are independent. A cross-section through the system's evolution (figure 5a) shows a lack of coordinated activity across the spatial domain. This is reflected in a contour plot of the system's spatio-temporal evolution (figure 5b), which illustrates the distinct lack of coordinated spatio-temporal activity. The mean field of the ensemble exhibits a low amplitude, disordered temporal evolution (figure 5c).
Figure 5.
Uncoupled array, c=0. (a) Cross-section through the system's evolution at an arbitrary time point; (b) contour plot of the system's spatio-temporal evolution; and (c) time-series of the mean field of the ensemble.
(ii) Mean-field effects and local spatio-temporal structures
With a broad synaptic footprint (K=0.01) and weak coupling (c=0.03), two distinct phenomena occur. Firstly, clusters of adjacent nodes exhibit synchronization (figure 6a), producing spatio-temporal structures in the system's contour plot (figure 6b). Secondly, synchronization occurs between such clusters across the domain, leading to large amplitude oscillations in the mean-field dynamics (figure 6c).
Figure 6.
Dynamics with a broad synaptic footprint (K=0.01) and weak coupling (c=0.03). (a) Cross-section through the system's evolution at an arbitrary time point; (b) contour plot of the system's spatio-temporal evolution; and (c) time-series of the mean field of the ensemble.
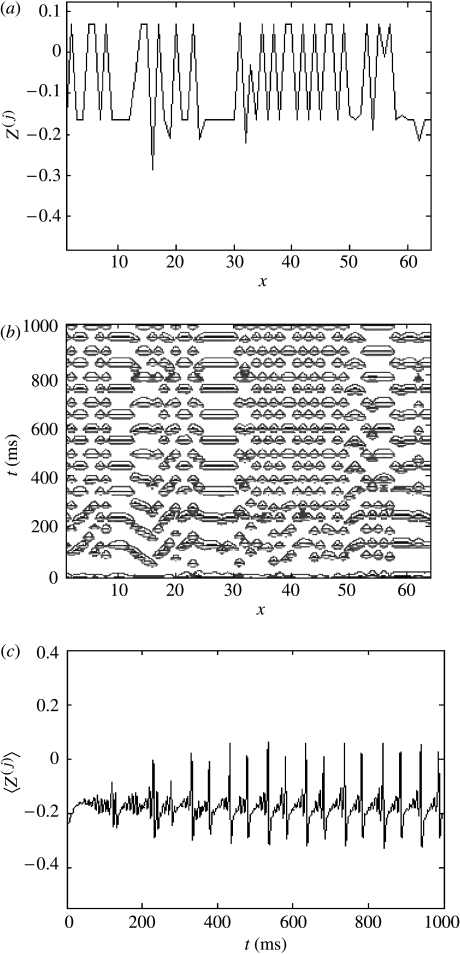
(iii) Local and scale-free structures
By narrowing the width of the synaptic footprint (K=1) while increasing the strength of the coupling (c=0.54), the system's evolution is biased towards the appearance of locally coherent spatio-temporal structures (figure 7a,b). Because coherence between such structures is weak, the mean-field term is somewhat diminished. However, the mean field remains considerably stronger than in the completely uncoupled case c=0 (figure 7c).
Figure 7.
Local and scale-free structures with a narrow synaptic footprint (K=1) and c=0.54. (a) Cross-section through the system's evolution at an arbitrary time point; (b) contour plot of the system's spatio-temporal evolution; and (c) time-series of the mean field of the ensemble.
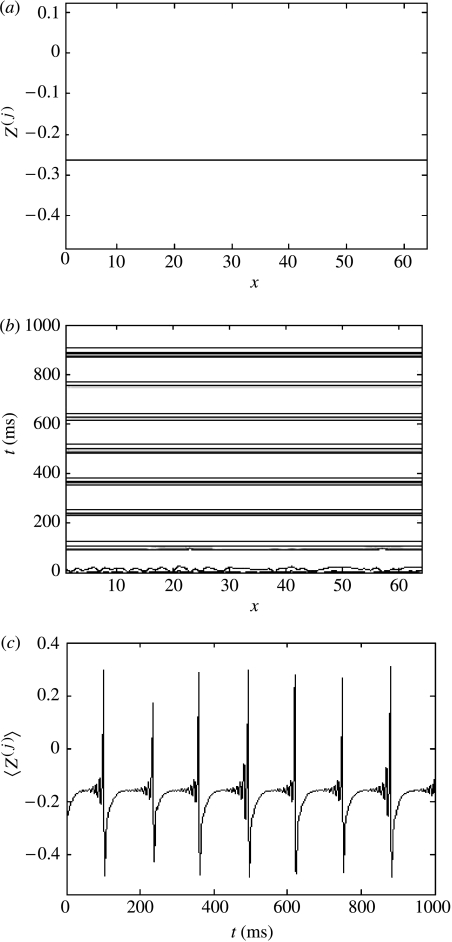
(iv) Complete synchronization
Increasing the coupling strength in either of the above scenarios (e.g. K=0.01, c=0.06, or K=1, c=0.6) leads to complete synchronization across the ensemble (figure 8a,b). This is associated with a significant increase in the amplitude of the mean-field oscillations, which becomes identical with each of the local subsystems (figure 8c).
Figure 8.
Complete synchronization, K=0.01, c=0.06. (a) Cross-section through the system's evolution at an arbitrary time point; (b) contour plot of the system's spatio-temporal evolution; and (c) time-series of the mean field of the ensemble.
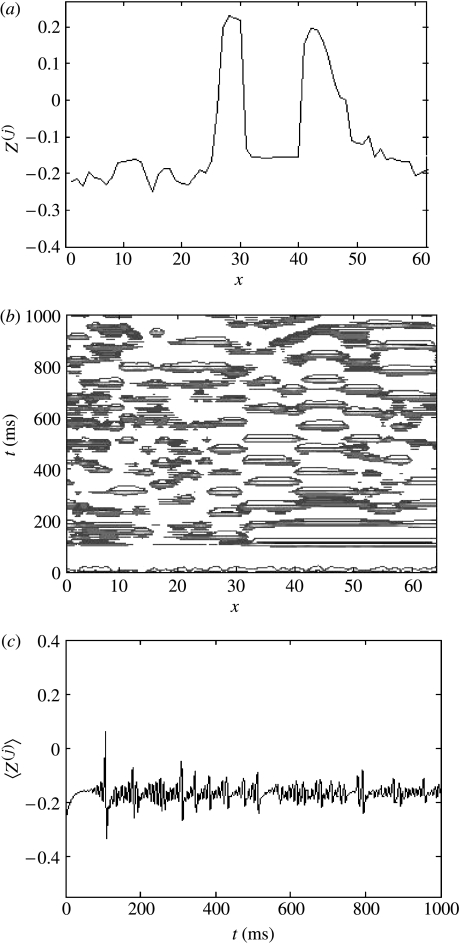
(v) Travelling waves
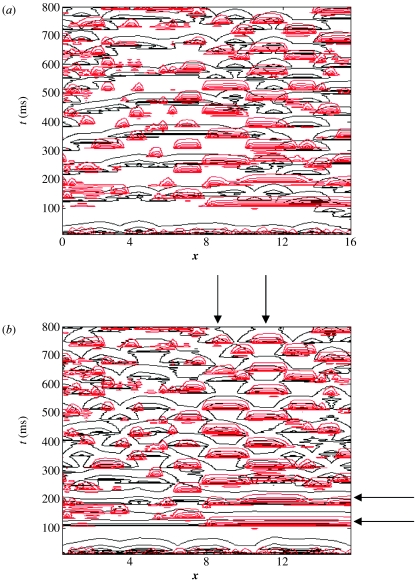
Systems whose parameter values are located in the region between those of §§3b(ii) and (iii), for example, K=0.1, c=0.12, exhibit dynamics that can be considered midway between those presented in figures 6 and 8 (i.e. increasing mean-field oscillations with decreasing K and increasingly isolated spatio-temporal structures plus decreasing mean-field effects with increasing K). However, for all systems where K<4, we observed a critical value of c at which the ensemble exhibited a transition to complete synchronization. At K∼4, a different phenomena is observed when coupling is increased: following an initial transient period, there appears the formation of spatially isolated travelling waves (figure 9a,b). Coincident with such travelling waves, oscillations in the mean-field term disappear (figure 9c).
Figure 9.
Travelling waves, K=16, c=3.54. The wave in (a) is moving from left to right. (a) Cross-section through the system's evolution at an arbitrary time point; (b) contour plot of the system's spatio-temporal evolution; and (c) time-series of the mean field of the ensemble.
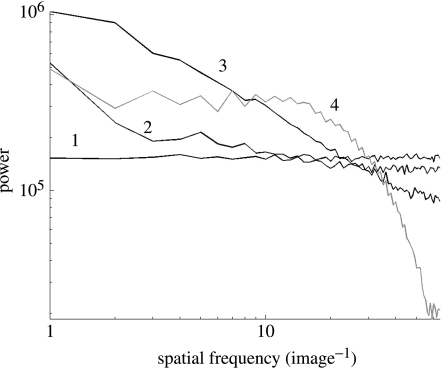
Hence, when the synaptic footprint is relatively broad (e.g. K<1), mean-field dynamics dominate over local structures. With a narrow footprint (e.g. K>4), local structures dominate and mean-field effects are suppressed.3 This can be seen by studying the spatial spectral density plots of the above dynamics, shown in figure 10. No coupling (line 1) is associated with uniform power across spatial frequencies. The introduction of coupling increases power at low frequencies (lines 2 and 3), causing the synchronized clusters and hence the appearance of log–log ‘scale-free’ structures.4 Travelling waves are associated with spectral peaks at the corresponding length-scales (line 4). These dynamics are also dependent upon the number of oscillators n in the array. If the ensemble is small (e.g. n=16) travelling waves do not occur. If the ensemble is large (e.g. n=128) multiple travelling waves may appear, although the transient time to reach this state increases. In Nakao et al. (2001), it is shown how such emergent phenomena are associated with a cascade of dynamical variance between spatio-temporal scales. For the present purposes, we use knowledge of these single-scale dynamics to inform the multiscale formalism.
Figure 10.
Spatial spectra of four coupled systems. (1) No coupling, (2) weak coupling c=0.03 and K=1, (3) moderate coupling c=0.54, K=1, (4) travelling wave c=3.54, n=16.
(c) Multi-scale modelling
We are now in a position to construct the multiscale hierarchy. Suppose we have two ensembles, V(j1) and V(j2), whose domains have spatial scales, j1 (fine) and j2 (coarse), so that j2=j1×2l, l∈Z; that is, j1 and j2 are dyadic (powers of 2). Define Λ(j2)V(j1) as the multi-scale projection of V(j1) on to scale j2. Then,
| (3.5) |
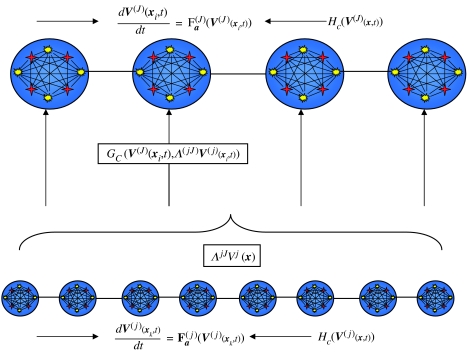
models the dynamics of V(j2) under the influence of the scale-congruent component of the dynamics at the finer scale V(j1) through G (and the rest of the same-scale ensemble through H). The between-scale coupling function G is parametrized in strength by C and injects Λ(j2)V(j1) ‘into’ the local dynamics. Owing to the spatial localization of the wavelet functions, this between-scale coupling is matched for spatial location; that is, as shown in figure 11, V(j2)(xi, t) is driven by Λ(j2)V(j1)(xi, t). Note also that G is dependent upon V(j2)(x, t), and is contained within the map F; that is, the coupling is state-dependent and ‘injected’. These characteristics will be shown in the numerical simulations below.
Figure 11.
Schema of the multi-scale model of equation (3.5). Two-tiered system with coarse-grained (J) and fine-grained (j) scales. Local ensemble at position xi in the coarse-grained system is governed by F(J). Within-scale coupling is introduced through H. The scale-congruent ‘harmonic’ of the dynamics at scale j are introduced through G, acting on the wavelet projection Λ(jJ)V(j) evaluated at xi.
The projection Λ(j2)V(j1) selects the spatial variance from V(j1) with scale j2. The spatial variance of the scale-congruent motions, however, can only have a disruptive effect on the dynamics of the coarser scale. If the variance is zero, then the coupling vanishes, whereas the coupling is strongest (and spatially varying) when the variance is maximal. This consideration motivates a modification of (3.5) so that an influence of the overall mean field of the fine-grained ensemble is also introduced,
| (3.6) |
where the overscore denotes the spatial average. There now exists three types of potential influence on V(j2)(x, t): (i) When there is complete synchronization within the fine-grained ensemble V(j1), then the dynamical variance within this ensemble at scale j2 is zero, and G introduces a uniform influence across the coarse-grained ensemble V(j2) through the mean-field term. That is, global synchronization within the fine-grained ensemble is projected as a spatially uniform oscillating influence on the coarse-grained ensemble, which will act to synchronize the motions within the coarse-grained ensemble. (ii) When there is no synchronization within the fine-grained ensemble, the mean-field term is negligible. The spatial variance within the fine-grained ensemble is uniform across scales (since there do not exist any large-scale structures), and the projection of this variance on to the coarse-grained scale will thus be small. Hence, G will introduce a weak and spatially incoherent influence into the dynamics of the coarse-grained ensemble. (iii) When spatio-temporal ‘structures’ within the fine-grained ensemble form—either through partial synchronization or the formation of travelling waves—then the influence of G on the dynamics of the coarse-grained ensemble will be dominated by these structures. The influence of these spatially varying inputs and the uniform mean-field term may be seen to be in competition with each other, and also with the dynamics within scale j2.
The dynamics of multiple spatially nested ensembles can be modelled recursively. In the case where within- and between-scale coupling functions are linear and diffusive, then at the scale J>1 the dynamics take the form,
| (3.7) |
where GiJ is the interscale coupling matrix. This is similar to equation (3.3), except the additional (third) term on the right-hand side injects the scale-congruent motions arising from smaller scales into the motions of the ensemble at scale J. Whereas the matrix H represents the ‘synaptic footprint’ for the within-scale dependence of coupling strength on separation, G represents a comparable coupling footprint between scales. The diffusive form of coupling is a first-order approximation of competitive agonism and additionally ensures that the state V(J)=V(j) is invariant under G and hence F (as long as a(J)=a(j)).
There may exist both physiological and histological mechanisms whereby the dynamics within the brain's ‘multiscale compartments’ become interdependent. The putative nature of these is considered in §7. For the numerical simulations below, we choose histological coupling. This was achieved by feeding the (scale-congruent) inhibitory activity of the fine-grained ensemble into the dynamics of the coarse-grained ensemble,
| (3.8) |
Before moving to the numerical examples, we offer a final generalization of equation (3.8). In many neural models, including the Morris–Lecar model, the time-scale for the system's evolution depends upon one of more ‘relaxation’ processes, such as the gradual closing of potassium channels towards their voltage-dependent resting state. At very small scales, the relaxation processes may reflect rapid atomic processes, whereas at larger scales, the relaxation speeds of entire neural membranes or macroscopic field potentials may be relatively slow. Hence, it is reasonable to expect that larger scale neural processes may exhibit slower characteristic temporal dynamics than small scale ensembles (see also Koenig et al. 2005, this issue). This can be modelled, following Fujimoto & Kaneko (2003), by introducing time-scale multipliers into the dynamics,
| (3.9) |
where the superscript Tj indicates the power exponent. J={j1, j2,…,J} is a vector denoting all scales ≤J and c={c, C} incorporates within- and between-scale coupling. T is a constant ‘time-scale’ multiplier that allows for explicit modelling of scale-specific temporal scales. Setting T>1 implies the spatially nested ensembles have geometrically nested internal time-scales. The time-scales of sequential layers of the multiscale system increase in concordance with their spatial scales. Adopting a power-series distribution ensures that the relationship between any adjacent scales, j and j+1 is identical (i.e. not dependent on j).
It is important to note that complex nonlinear processes typically exhibit broadband temporal spectra with nonlinear ‘mode-locking’ across different frequencies. The addition of the Tj has no influence on this. Rather, in a general setting, they merely shift the characteristic peak frequencies of each scale-specific dynamics to permit the temporal factors discussed above. In a specific model—that is, with specific evolution equations—such factors would typically be incorporated implicitly into the evolution equations through the various relaxation parameters at each scale.
We thus introduce equation (3.9), where F has the form of equation (3.7), as the ‘multiscale evolution operator’. Numerical examples are now given to elaborate on the structure of equation (3.9) and show some resulting phenomena. A systematic examination is to be the subject of future work.
4. Numerical examples
In this section, exemplar numerical simulations of equation (3.9) are provided. The simulations are realized in the specific form of the modified Morris–Lecar system as given in the Appendix. In §4a, we drive a coarse-grained ensemble J (N=16) with the fine-grained (j1) examples given in §3c. In §4b, a three-tier system is constructed by adding a very fine-grained (j2) level (N=256). We thus show how dynamics at the finest scale may influence those at the coarse scale. The simulations were performed in Matlab using purpose-built script files (available from MB on request). As a metric on the spatial order within the coarse-grained dynamics, we calculate the Shannon entropy of the system at each time point, and take the temporal average, denoted S(J), where J denotes the coarse-grained ensemble.
We use low-order wavelet basis functions (Daubechies wavelets of order 4) as they have short ‘footprints’ (small and compact support), which best preserve the localizing properties of the between-scale coupling functions. However, the general nature of the results does not critically depend upon the exact choice of basis functions.
(a) Two-tiered multiscale dynamics
We now drive a coarse-grained scale N(J)=16 with a fine-grained scale N(j)=64. We consider the cases where, in the absence of interscale coupling (C=0) the fine-scaled system has (i) no intrascale coupling (c(j)=0), (ii) strong intrascale coupling (c(j)=0.6), (iii) moderate intrascale coupling, and (iv) travelling waves. Hence, we introduce interscale influences from fine-scale dynamics which themselves differ in internal connectivity.
(i) Uncoupled fine-grained system, c(j)=0, k=1
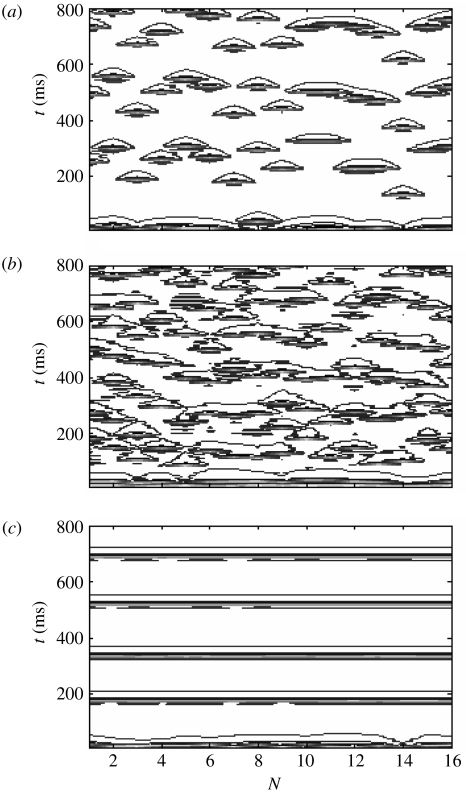
The Shannon entropy for an uncoupled array (figure 12a), random initial conditions and no interscale coupling C=0, is S(J)≅0.3. Introducing interscale coupling from a fine-grained array with no internal coupling c(j)=0 has the effect of increasing the entropy of the coarse scale. For example, setting C=0.15 yields S(J)=0.33. There is an approximately monotonic increase in this metric with C so that setting C=0.5 yields S=0.36, as shown in figure 12b.
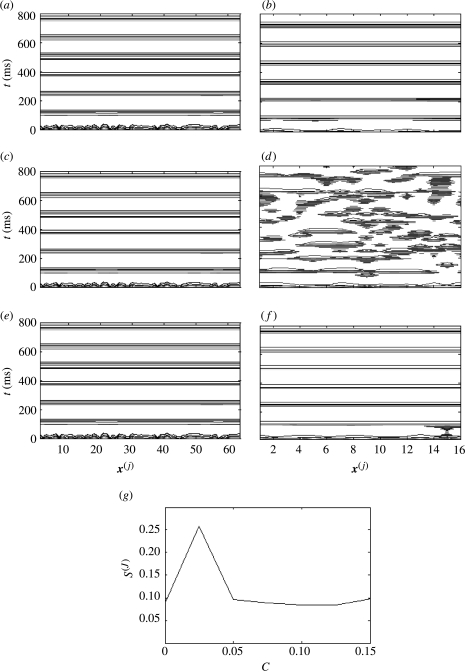
Figure 12.
Impact of uncoupled fine-grained dynamics on spatio-temporal structures within a coarse-grained scale c(J)=0. Contour plots of coarse-grained system with (a) no inter- or intrascale coupling; (b) strong input C=0.5 from an uncoupled fine-grained system; and (c) weak interscale coupling C=0.075 and strong intrascale coupling c(J)=0.6.
How is it possible that the introduction of interscale coupling decreases the spatial order of an already uncoupled array? The reason is that the influence of the disordered fine-grained array disrupts the time-series of each individual node in the coarse-grained array. This acts to increase the uniformity, and thus entropy, of the amplitude distribution of the entire ensemble.
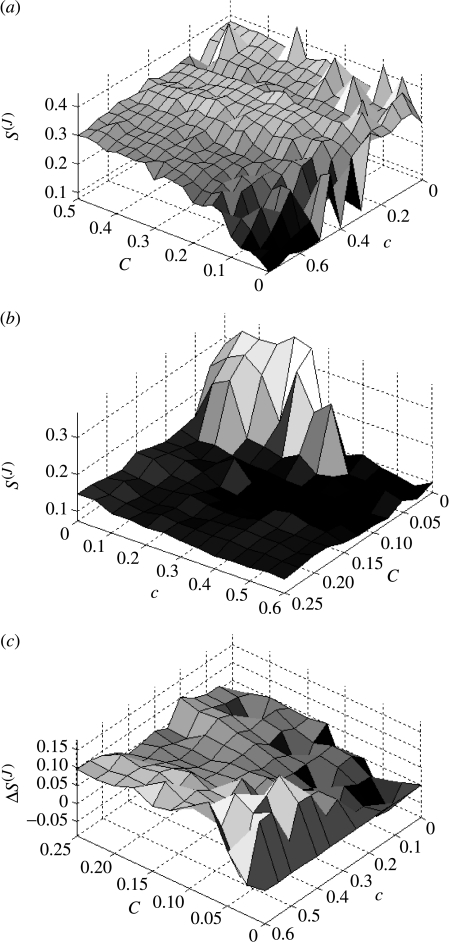
Increasing the intrascale coupling within the coarse-grained ensemble has the effect of counterbalancing the disruptive ‘upwards’ input from the fine-grained ensemble. For example, if the interscale coupling is weak (C=0.075) and the intrascale coupling moderately strong (c(J)=0.6), then the coarse-grained system exhibits complete synchronization (figure 12c). Figure 13a shows a contour plot of S(J) in the {C, c} parameter plane. It can be seen that S{J} dips to low values only in the region c≫C. Hence, within this scenario, there is competition between the intrascale coupling and the disruptive effect of interscale input from the discordant fine-scale system.
Figure 13.
Surface maps of entropy in coarse-grained system S(J) as a function of intra- (c) and inter- (C) scale coupling. (a) Input from an uncoupled fine-grained system; (b) input from a synchronized system; and (c) difference in entropy when input is from a fine-grained system with spatio-temporal structures (c(j)=0.03, k=0.01) compared with input from an uncoupled system. Note that for visualization, plot (b) is flipped in the {C, c}plane compared with (a) and (b).
(ii) Synchronized fine-grained system, c(j)=0.6, k=1
In contrast, interscale input from a synchronized fine-grained scale has a synchronizing influence on the coarse-grained system. In the case where C=0, such synchronization can be achieved if c(J)>0.3. If C is increased, then the influence of the synchronized fine-grained ensemble acts to synchronize the coarse-grained dynamics (figure 13b). Thus, for example, if C=0.075, then the coarse-grained system will synchronize with c(J)=0.15. Synchronization within the coarse-grained system occurs with no intrascale coupling (c(J)=0) with sufficient input (C>0.1) from the fine-grained system.
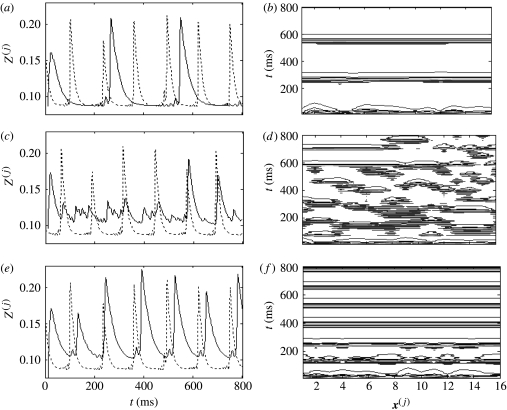
The introduction of interscale coupling modifies the relationship between the dynamics of the fine- and coarse-grained dynamics in a somewhat complex manner. With no interscale coupling (C=0) and sufficient intrascale coupling (e.g. c(J)=0.375), each system exhibits internal synchronization, although there is no synchrony across scales (figure 14a,b). The introduction of weak interscale coupling (C=0.025) disrupts the synchronization in the coarse-grained scale (figure 14c,d). Presumably, this reflects competition between the intrascale mean field and the ‘injected’ mean field from the fine-grained system. Hence, the entropy is increased in this region of {c(J),C} parameter space. A sufficient further increase in the interscale coupling (C=0.075) restores synchronization within the coarse-grained system; however, this time, the oscillations are also synchronous with those of the fine-grained system (figure 14e,f).
Figure 14.
Between-scales bifurcation. Panels (a), (c) and (e) show contour plots of synchronized fine-grained system. c(j)=0.6. Impact of this system on a coarse-grained system with c(J)=0.35 and (b) C=0, (d) C=0.025, (f) C=0.075. Panel (g) shows a cross-section through the surface plot of figure 13b at c(J)=0.35.
The transition between these two types of synchronous dynamics via a regime of disorder is reflected in the cross-sectional view of figure 13b at c(J)=0.35, which is given in figure 14g. The discontinuity between within-scale synchronous, but between-scale asynchronous, dynamics (C=0) and within- and between-scale synchronization (C=0.075) is evident as a local peak in S(J). We denote such a phenomenon as a ‘between-scales’ bifurcation. Stronger within-scales coupling (c(J)>0.45) results in a smooth transition between these dynamic states.
The situation becomes more complex again if the time-scales of the two scales are made dissimilar by setting T(j)≠T(J). Results of leaving T(j)=1 but setting T(J)=2 are shown in figure 15. Panels (a), (c) and (e) show the mean-field oscillations of the fine- (dashed) and coarse- (solid) grained systems. Panels (b), (d) and (f) show the dynamics of the coarse-grained ensemble. In panels (a) and (b), c(j)=0.6 and c(J)=0.35 but C=0, so that both systems exhibit within-scale synchronization. It can be seen that setting T(J)=2 has the result of slowing the coarse-grained ensembles oscillations. Introducing between-scale coupling C=0.025, (c) and (d), disrupts the synchronization within the fine-grained ensemble. Increasing the within-scale coupling of the coarse-grained system c(J)=0.75, (e) and (f), restores the synchronization, but in doing so, adjusts the mean-field dynamics to match those of the fine-grained system (through lag-synchronization). Hence, in this case, changing only the within-scale coupling causes both within-scale and between-scale bifurcations.
Figure 15.
Results of leaving t(j)=1 but setting t(J)=2. Panels (a), (c) and (e) show the mean-field oscillations of the fine- (dashed) and coarse- (solid) grained systems. Panels (b), (d) and (f) show the dynamics of the coarse-grained ensemble. In panels (a) and (b), c(j)=0.6 and c(J)=0.35 but C=0. Introducing between-scale coupling C=0.025, panels (c) and (d). Increasing the within-scale coupling of the coarse-grained system c(J)=0.75, panels (e) and (f).
(iii) Fine-grained system with travelling waves, c(j)=3.54, k=16
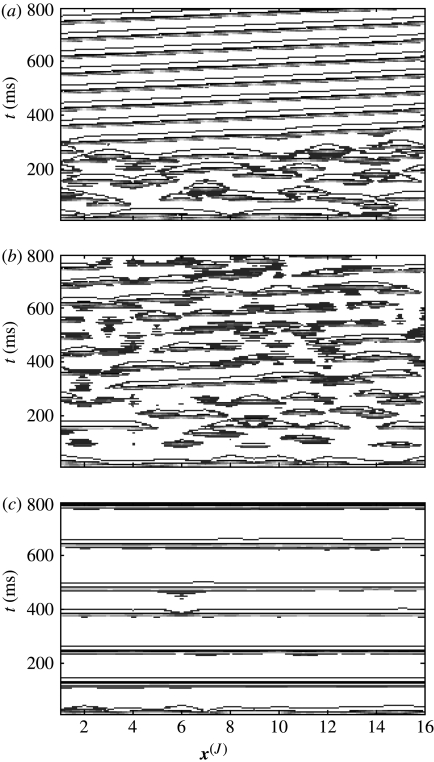
The effect of injecting input from a fine-grained system with travelling waves is shown in figure 16. As stated above, we do not observe travelling waves in an autonomous system with N=16. However, strong interscale input (C=0.4) with weak intrascale coupling in the coarse-grained system (c(J)=0.05) causes the appearance of travelling waves (panel (a)), and thus their interscale projection. Increasing the strength of the intrascale coupling (c(J)=0.3) has the effect of encouraging identical synchronization within the coarse-grained system, in competition with the ‘injected’ travelling waves. Hence, both local and travelling structures are seen (panel (b)). There exists a critical value of c(J), above which the projection of travelling structures is suppressed by global coherent dynamics (panel c).
Figure 16.
Injection of travelling waves from fine-grained system. Panel (a) strong interscale input (C=0.4) with weak intrascale coupling in the coarse-grained system (c(J)=0.05); panel (b) increasing the strength of the intrascale coupling (c(J)=0.3); and panel (c) increasing c(J)=0.6.
(iv) Fine-grained system with scale-free structures, c(j)=0.54, k=1; c(j)=0.03, k=0.01
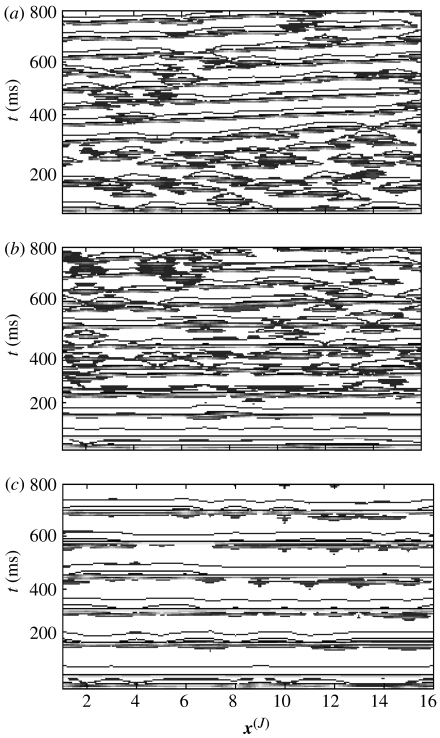
Introducing interscale coupling from fine-grained systems with spatio-temporal structures gives an entropy contour plot similar to that of the uncoupled fine-grained system illustrated in figure 13a. In figure 13c, the difference between S(J) under these two circumstances is illustrated. It shows that injecting dynamics from fine-grained systems with spatio-temporal structures has a greater influence on the disorder within the coarse-grained system than projections from uncoupled systems.
How can this be understood? As shown in figure 10, the appearance of coherent structures in weakly coupled systems leads to greater power at low spatial frequencies in comparison to uncoupled systems. Such structures arise irregularly according to the irregular nature of the systems’ internal dynamics, and thus have a discordant character when projected on to large-scale wavelet subspaces. Hence, they have a stronger disordering influence on the dynamics of the coarse-grained system than a system with no internal connectivity.
However, the projections do leave their ‘fingerprint’ on the coarse-grained system. In figure 17, the contour plots from the two scales have been overlaid. The fine-grained system V(j) is plotted in red over the coarse-grained system (V(J)=black), which has been matched in space. Both systems have internal connectivity (c(j)=0.54, c(J)=0.1). In the case of no interscale coupling C=0 (panel a), the two systems do not bear any obvious relationship. The introduction of interscale coupling C=0.4 is associated with coincident spatio-temporal structures in the two systems (arrows). In keeping with the general nature of the dynamics, such coincident structures (although evident in a wide variety of coupling combinations) are necessarily transient in any given realization. Moreover, they are typically confined in their spatial extent to match the congruence of the two scales (as implicit in the modelling approach). A quantitative measure of such subtle congruence is the subject of future work.
Figure 17.
Injection of spatio-temporal structures from fine-grained system. Both systems have internal connectivity (c(j)=0.54, c(J)=0.1). The fine-grained system is overlaid in red on the coarse-grained system. Panel (a) no interscale coupling C=0; and panel (b) interscale coupling, C=0.4, causes coincident spatio-temporal structures (arrows).
(b) Three-tiered multiscale dynamics
Clearly, there exists a large combination of possible intra- and interscale coupling coefficients when moving to a three-tiered system. For illustrative purposes, we focus on only two scenarios. Both involve the extra influence of a very fine-grained scale, j2 with N(j2)=256 on the previously studied two-tier examples. Hence, in this system we use an architecture that spans nearly three orders of magnitude.
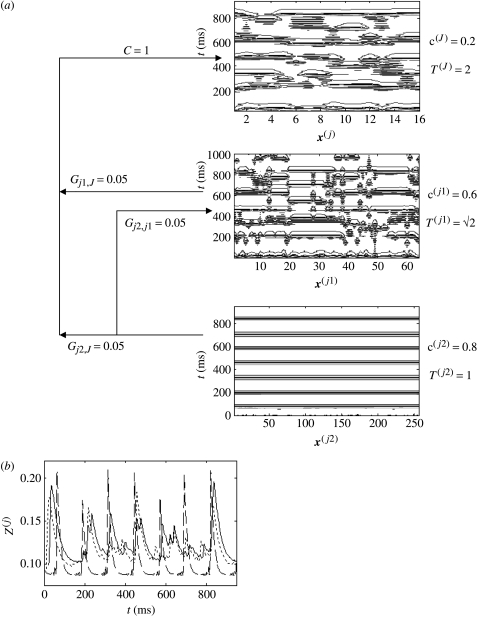
(i) Coarse-grained travelling waves
In the first simulation (figure 18), we return to the injection of travelling waves from a fine-grained system j1 with n=64 into a coarse-scaled system J with n=16. Intrascale (c(J)=0.2) and interscale (C=0.35) coupling were chosen to yield coarse-grained travelling waves in the absence of the third scale. We merely vary the relative interscale coupling matrix GjJ and the nature of the very fine-grained system.
Figure 18.
Three-tiered system with the injection of travelling waves from a fine-grained system j1 with n=64 into a coarse-scaled system J with n=16 and additional influence of a very fine-grained system. Intrascale (c(J)=0.2) and interscale (C=0.35) coupling were chosen to yield coarse-grained travelling waves in the absence of the third scale. Panel (a) no coupling in the very fine system c(j2)=0, with Gj1=0.33 and Gj2=0.67; (b) injection of a very fine-grained system with internal coupling (c(j2)=0.54) and Gj1=0.33, Gj2=0.67; and (c) effect of a completely synchronized very fine-grained system (c(j2)=0.8) and Gj1=0.33, Gj2=0.67.
With no internal coupling in the very fine scale (c(j2)=0) and with Gj1=Gj2=0.5 the finest scale system has no impact on the appearance of the coarse-grained travelling waves. Increasing the relative input from the finest scale Gj1=0.33, Gj2=0.67 results in a partial disruption of the travelling waves, figure 18a. A further bias towards the finest scale, Gj1=0.25, Gj2=0.75 results in the complete replacement of travelling waves with local spatio-temporal structures (not shown). Employing a very fine-grained system with internal coupling (c(j2)=0.54) achieves the same result (replacement of travelling waves with spatio-temporal structures) with relatively less input Gj1=0.33, Gj2=0.67 (figure 18b). In contrast, using a completely synchronized very fine-grained system (c(j2)=0.8) and Gj1=0.33, Gj2=0.67 leads to the replacement of travelling waves with global synchrony in the coarse-grained system (panel c), although some subtle non-uniform effects persist. Hence, in this system, there are multiple competing constraints between intra- and interscale functional connectivity.
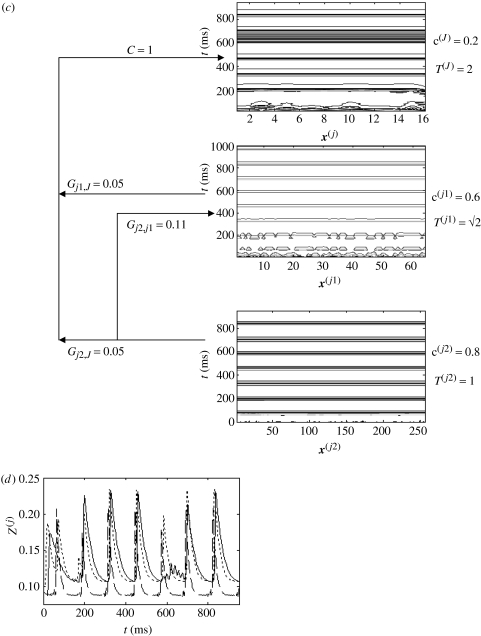
(ii) Spatial and temporal nesting
In these simulations, both fine scales were projected into the coarse-grained system although there was no interscale coupling between the fine scales, Gj1,j2=0. In a final simulation, coupling with Gj1,j2>0 is investigated in the setting where three systems are each internally synchronized but have different time-scales. This is achieved by setting the time-scale multiplier of (3.9), t=√2. The results are presented schematically in figure 19. Panels (a) and (b) show a very fine-grained system weakly driving a fine-grained system, both of which have internal coupling (c(j2)=0.8, c(j1)=0.6). As with §4a(ii) and figure 15, the mismatch in the two systems' time-scales (t(j2)=1, t(j1)=√2), in combination with weak interscale coupling (Gj2,j1=0.05), results in discordant dynamics in the medium-grained scale (j1). These two scales are also simultaneously injected into the dynamics of a coarse-grained system J which has both internal coupling (c(J)=0.2) and slow dynamics t(J)=T2=2. The strength of these interscale couplings is Gj1,J=Gj2,J=0.05. The outcome of such an injection is a discordant coarse-grained/macroscopic system. Panel (b) shows the mean-field oscillations of the each of the systems (very fine, dashed; fine, dotted; coarse, solid). The strength of these oscillations is seen to decrease as the scale increases, reflecting the increasingly discordant internal dynamics.
Figure 19.
Three-tiered system with different internal time-scales. Panel (a) contour maps with parameters as shown; (b) mean-field oscillations of very fine system (dashed), fine-grained system (dotted) and coarse-grained system (solid); and panels (c) and (d) the same system but Gj2j1 has been increased from 0.05 to 0.11.
The same system with only one change—stronger coupling between the two finer scales (Gj2,j1=0.11)—is presented in panels (c and d). The effect of such an increase is to adjust the dynamics within scale j1 to match those of the very fine scale j2. The flow-on effect is that the coarse-grained dynamics are now also synchronized, both internally (panel c) and between scales (panel d; except for the ‘missed’ oscillation at t=600 ms).
In §4a(ii), it was observed that a ‘between-scale bifurcation’ occurred with adjustment of a within-scales coupling parameter. In this example, we see that a coarse-grained system exhibits both within- and between-scale bifurcations following adjustment of coupling between two ‘deeper’ scales, and without any change to proximal intra- or interscale coupling. This illustrates the effect of emergent dynamics in small-scale systems on the nature of macroscopic neural activity.
5. Multi-scale dynamics, observables and functional connectivity
The relationship between neural activity and recorded data—such as functional neuroimaging or multichannel electrophysiological recordings—is by no means trivial (e.g. Friston et al. 2000, 2003; Zheng et al. 2002; Valdés-Sosa et al. 2005). In this section, we sketch a framework which relates the multidimensional neural activity V(x, t) to multivariate scalar observables Yj(t) in a manner that exploits the multiscale formalism V=V(1)⊕V(2)⊕…⊕V(N), and potentially assists interpretation of measures of functional connectivity. This framework is to be further elaborated in future work.
A generally accepted model is that neural activity V(x, t)∈N⊕ is mapped in a topologically distinct manner into the space of observables Yi(t)∈⊕ through spatio-temporal physiological transformations.5 These can be represented as
| (5.1) |
where Mx and Mt are the spatial and temporal observation functions. Note that activity centred at position x is mapped into the multivariate variable i (voxel/channel). Measurement noise is modelled as ϵi(t) which we assume to have static temporal and spatial structure at least up to first order (i.e. AR(1)). The measurement functions M may be quite complicated and involve temporal derivatives of ‘hidden’ variables such as blood flow and oxygen extraction (Buxton et al. 1998; Friston et al. 2000). Fortunately, these functions typically only depend upon a subset of all the dynamical variables Vm∈V such as the strong dependence of high-resolution BOLD data on local field fluctuations arising from dendritic membrane currents (Logothetis 2002). The M's are modality-specific, so that dependence on any subset of dynamical variables, including firing rates, could be modelled by equation (5.1). In many instances, it has proven sufficient to linearly approximate the measurement functions by convolutions with appropriate kernels such as the haemodynamic response functions. In this case, equation (5.1) is of the form,
| (5.2) |
For functional neuroimaging data, it is usually taken that Mx=[I] and for neurophysiological data Mt=[I], where [I] is the identity matrix (Valdés-Sosa et al. 2004). That is, the effects of spatial (fMRI) and temporal (EEG/MEG) smoothing are considered negligible (although Gaussian or Butterworth bandpass filtering is often applied as a preprocessing step). The differing effects of volume conduction in MEG compared with EEG data is incorporated by choosing different spatial kernels Mx in equation (5.2) for the two modalities.
Coupling between neural activity and observables, however, may have scale-specific dependences. For example, coupling between neuronal energy consumption and local vascular blood flow occurs at the scale of capillaries, probably mediated via a neural–glial–endothelial pathway (Riera et al. 2004). Changes in blood flow in the microvasculature, however, have upstream time-delayed effects owing to alterations in overall resistance of the local vascular tree. There are other scale-specific effects, such as a spatial decoupling of deoxy- and oxyhaemoglobin which may only be apparent at the micro-columnar scale (Riera et al. 2004). These considerations motivate the introduction of a scale-specific spatio-temporal measurement function:
| (5.3) |
This quite general form allows scale-dependent measurement functions/kernels and their parameters. Hence, neural dynamics are mapped into observation space in a scale-dependent manner. Modelling the dynamics and measurement processes in a multiscale space may hence be seen as a parsimonious solution to two important problems, namely, (i) modelling of neural activity in a brain-like architecture, and (ii) physiologically constrained state-space modelling of neural-vascular coupling. The recorded data are then considered a (weighted) mean of the scale-specific multivariate observables,
| (5.4) |
where the coefficients Aj permit scale-specific contributions to the emergent data (see Logethetis 2002 for a discussion of the contributions of vasculature at different scales to the BOLD signal).
If there exists a sufficient number of multivariate observables, then it is also possible to perform a multiscale decomposition of the experimental observables (as was depicted in figure 2) yielding scale-specific observations Ŷ(j)(t). This process, although unlikely to directly recover the same scale-specific observables generated according to (5.4), nonetheless motivates consideration of ‘multiscale functional connectivity’ as correlations within and between brain networks across spatio-temporal scales, arising through complex spatio-temporal neural dynamics. While this is offered as a measurement window into the dynamical architecture presented in this paper, caution is required when interpreting any such analysis.
As a final observation, given the spatio-temporal specificity of different brain imaging technologies (e.g. Nunez 1997), equation (5.2) could also be employed as a constraint for model fitting in the area of multimodal data fusion. That is, two datasets recorded concurrently using distinct imaging technologies can be seen as distinct weighted projections of the same multiscale neural dynamics, where the scale-specific measurement functions have a specific form for each imaging modality. Studying correlations between the observables can hence be constrained by their common (neural) source. As with other approaches, however, a unique backwards solution is not guaranteed.
6. Comparison with the Kuromoto model
We now use the Kuromoto model, which is a simple and well-known dynamical model, to re-frame the motivation for the present approach in the light of §4. Consider Kuromoto's equation for the phase evolution θi(t) of the ith node of a globally coupled phase oscillator,
| (6.1) |
where ωi is its natural (uncoupled) frequency and Γi,j couples the local dynamics to those of all the other oscillators in the array (Kuromoto 1984). When this takes the form of equally weighted, all-to-all, purely sinusoidal coupling,
| (6.2) |
then a simple change of variables allows (6.1) to be rewritten as
| (6.3) |
where r and φ are the ensemble average radius and phase respectively, given by
| (6.4) |
In the formulation of (2.1), each oscillator is coupled directly to the motions of all others. However, as Strogatz (2000) notes, by introducing the mean-field ‘order parameters’ r and φ ‘each oscillator appears to be uncoupled from all the others, although of course they are interacting, but only through the mean-field quantities r and φ.’ Although the evolution equations (and hence the dynamics) are identical, the two formulations (6.1) and (6.3) allow conceptual distinction between the constraining influence of direct node-to-node coupling and that of interactions only between the dynamical elements and the ensemble mean-field dynamics. The present study is an attempt to exploit this distinction by placing the dynamics in a multiscale architecture (and generalizing from phase to nonlinear oscillators). In essence, the symmetry expressed in equation (6.2) is broken in a manner so that equation (6.3) no longer holds. Instead, there exist three competing hierarchical constraints on local dynamics: (i) local nonlinear processes within each ‘dynamic module’ through inhibitory–excitatory connections; (ii) direct connections between modules within each scale, which decay over distance and, depending on the rate of decay can be thought of having ‘local’ and ‘mean-field’ (within scale) effects; and (iii) local mean-field effects projected up from smaller scales via the wavelet subspace projectors. Hence, in comparison to equation (6.1), the dynamical evolution of local units in the proposed model can be considered partially constrained by hierarchical global field effects.
If the coupling strength K exceeds a critical value, then the Kuromoto model exhibits a phase transition (which can be derived analytically in some cases): some of the oscillators spontaneously synchronize, while others remain incoherent. In such a state, the number of clusters and their respective sizes displays a (log–log) ‘scale-free’ relationship across spatial scales. If K is further increased, then the system eventually coalesces into a single cluster. The Kuromoto model thus illustrates the notion of circular causality and provides a remarkably simple model for the study of emergent, scale-free phenomena. However, the architecture of the model is a globally coupled, perfectly symmetrical group. The dynamical structure emerges through symmetry-breaking bifurcations (see Ashwin & Breakspear 2001 for such bifurcations in globally coupled logistic maps). The brain has a detailed and functionally significant architecture. We have attempted to model the interplay between scale-free dynamics and multiscale architectures.
7. Discussion
We have argued that the present formulation is uniquely ‘brain-like’, and may capture phenomena in an intuitive manner that cannot be achieved with existing dynamic models. As illustrated, its dynamics can display either interscale independence or interdependence, depending upon a variety of model settings. Prevailing neural models ignore such effects by focusing on one particular scale and averaging out the dynamics within other scales. On the other hand, ‘scale-free’ models, which are prevalent in the physical sciences (such as fluid dynamics), argue for spatio-temporal dynamics over a hierarchy of scales owing to complex internal dynamics (Bak et al. 1987). The scale-free approach allows the interaction of the system's components to give rise to spatial or temporal structures across many scales, but it does not permit unique dynamics within any scale or scale-independence under certain situations. In fact, the basic single-scale model elucidated above equation (3.4) can generate scale-free dynamics. Hence, it is the nesting of such a system within a multiscale structure that is the unique contribution here.
A related work is that of Wei et al. (2002) who formalize a standard coupled array of nonlinear (chaotic oscillators) with a linear coupling matrix (see also Ashwin 2003). The elements of this coupling matrix, although real-valued linear coefficients, are nonetheless modified in the wavelet domain. The dynamics are evaluated with the inverse-transformed linear coefficients. They show that modification of the low-pass filtered components of the decomposition (i.e. of the projection on to the approximation subspaces) is a parsimonious method of achieving global synchronization. Our approach differs in a number of ways: (i) the array is explicitly modelled over distinct spatial and temporal scales; (ii) the coupling function is not necessarily linear; and (iii) the coupling coefficients at each time point are evaluated as wavelet subspace projections of those representing fine scales on to those representing coarse scales. Nonetheless, in the presence of exact symmetry and linearity of all between-scale coupling functions, the two approaches have some mathematical similarities. However, the structure of the present formalism permits explicit study of neurodynamically interesting phenomena (§4) and modelling of explicit scale-specific mapping of the dynamics on to the observables (§5).
What is the biophysical basis for between-scale coupling? We propose two potential mechanisms: the first is physiological. Consider the effect of oscillating dendritic currents (and associated extracellular ion concentrations) on activity in a small-scale neural ensemble. If the ensemble's subunits are not synchronized, then it is reasonable to average out the local variations of resulting ion concentration when modelling the dynamics of a larger-scale neural ensemble. However, if the subunits become synchronized, then resulting large-scale fluctuations in ion concentration may influence the activity of the larger-scale ensemble. For example, if the potassium ‘field’ begins oscillating uniformly across some domain, then the effect of such oscillations on neural activity could no longer be considered a static parameter, but rather a dynamical variable or driving input. Hence, the emergent behaviour of synchronized dendritic currents acts like a ‘field’ which influences neural dynamics at the larger scale. A histological mechanism is the alternative discussed here. Within the vertical lamination of the cortex, there are distinctions between locally and distally projecting pyramidal cells and, additionally, between the relative functional strength of inhibitory feedback on to the different layers (e.g. Shephard 2004; Douglas & Martin 2004). Such local/long-range differences could hence facilitate the distinction between small and large-scale ensembles within the present formalism. Accordingly, the present model extends existing one- and two-dimensional models of neocortex by representing the third dimension (neocortical thickness) in the domain of scale. If it was shown that the dynamics of equation (3.7) confer some computational or metabolic benefit over a single-layered system, then we would have, parsimoniously, a theoretical explanation for the lamination of the neocortex. We return to this below.
There are a number of ways in which the current formulation could be modified or extended. The symmetrical form of H could be broken in a manner that introduces anatomically complex connectivity. Sporns et al. (2000), in an extensive study of coupled network activity (but with linear dynamics), provide a compelling argument for such considerations from perceptual, computational, empirical and evolutionary perspectives. The effect of introducing nonlinear dynamics has not yet been studied, but will presumably provide additional flexible and adaptive properties on to such systems. Conferring the within-scale coupling matrix H with a complex anisotropic geometry (‘network motifs’; see Sporns & Kotter 2004) may confer additional computational benefits.
Alternatively, if H is of a suitable form (Abramowitz & Stegun 1970), and the temporal derivatives of f are second order, then taking thermodynamic limit as N→∞ permits the within scale neural ensembles to be considered as wave-propagated continua. The dynamics could then be rewritten in terms of the propagating (scale-specific) field density terms ϕ(j):
| (7.1) |
where
| (7.2) |
is the spatio-temporal gradient operator, and space now varies continuously. Additional equations are required in order to close the system within each scale (see Jirsa & Haken 1996; Robinson et al. 1997, 2001). The subspace projectors in such a setting would be derived from a continuous wavelet transform of the V(j). In this framework, it would also be possible—through a Fokker–Planck formalism—to model the entire field density distribution (see Harrison et al. 2005, this issue), which may confer important additional neurobiological properties.
Finally, coupling in the ‘physical domain’ (of the naïve physical parameters) is not necessarily the only way to introduce interscale effects. Coupling of the wavelet modes themselves, that is, interscale influences that occur among the wavelet coefficients,
| (7.3) |
may be preferable (where we have now returned to the spatially discrete mean-field formalism). This step is analogous to the ‘decimation’ procedure adopted in the fluid dynamics literature (Zhou et al. 1997), whereby the emergence of complex scale-free spatio-temporal structures in turbulent flow is studied by truncation of the Fourier modes, and by introducing nonlinear coupling between the remaining harmonics. Adopting such a framework may allow pre-existing theoretical results to be exploited in order to achieve further insights into the proposed model.
This point brings us to the main limitation of the present work. Despite the symbolic explication of our framework, we have only offered numerical realizations. Although a more systematic evaluation of such realizations may be revealing (and is to be the subject of future work), it is probable that formal analytic results, such as within and between scale bifurcations, may be tractable only in restricted settings (such as strong symmetry assumptions in the coupling and simple chaotic motions within the local dynamics). To put this statement in context, a complete analytic understanding of equation (6.3) is far from trivial (see Strogatz 2000). The effect of multiscale symmetry breaking and introducing wavelet subspace projectors into the coupling matrix renders comparable analytic study more complex still. Such a cost can only be justified if the additional neurobiological plausibility is well justified and if additional insights into cognitive processes are achieved. We have argued that the multiscale structure of neuroanatomy is a critical feature of brain organization that requires functional explanation. The communication of activity from microscopic feature-specific neurons to large-scale distributed networks is a pressing neurocognitive problem that may be elucidated through the present framework.
However, the present model was motivated based on the architecture of the neuropil and not with respect to any specific neurocognitive problem. We have alluded to the explanatory potential for the model to describe the exchange of dynamical states between small-scale (feature-specific) neural systems and distributed, integrative cortical regions. Such explanatory power may be of significance for understanding processes such as the enrichment of a sensory stimulus into a perceptual experience (Mesulam 1998), and the interplay between working and long-term memory.
In conclusion, an important difference between scale-free structures in fluid motion and the current proposed framework is offered in order to underline the novel aspect of the present contribution. In fluid dynamics (and other comparable complex motions), the base dynamics are formulated at a single scale. Large-scale structures then arise through mode coupling of the resulting motions. There is no formal multiscale architecture of the phenomena studied prior to such effects. In contrast, motivated by the organization of the neuropil, the present framework begins with an a priori multiscale structure. The emergence of dynamical structures at any given scale—owing to complex dynamics within that scale—are then explicitly coupled to the motions at coarser scales, which may themselves have different inherent properties. In the absence of spatio-temporal structures within the finer scales, effective coupling across scales will not occur. Hence, there exists a synergism between the a priori architecture and the emergent dynamical structures in the proposed modelling framework. In a scale-free system, events occurring at different scales have essentially the same causes and characteristics. A multiscale model also allows for functionally specific processes at different scales. It also ensures, through topographically constrained tractograpy, that certain large-scale dynamical forms are expressed more often than by chance. Hence, we treat the brain as a constrained multiscale system in which emergent dynamics at any scale have a critical influence on the activity in larger scale structures.
For its size, the brain exhibits remarkable cognitive potential. As in all living systems, there exists an ongoing trade-off between increasing complexity and metabolic demand. Such an argument has been used to provide a compelling explanation of universal scaling laws in nature, such as the relationship between vessel (or airway) diameter and length (West & Brown 2004). Similarly, metabolic demands (which are easily higher in the brain than any other organ) may have acted concurrently with the cognitive potential of the evolving brain to constrain the expression and selection of neural architecture. Just as in a vessel, the competing demands of tissue penetration and intra-vascular volume are optimized, the brain finds a form which balances cognitive load and metabolic demand. It appears that during the evolutionary expanse of brain volume, there occurred a series of space-scale bifurcations, which are expressed in the neuropil's multiscale architecture. Does this form, in concert with information-rich coupled neural dynamics (Breakspear 2002), represent the optimal solution to such competing forces?
Acknowledgments
The objective to formalize a dynamical system with a multiscale architecture arose from discussions at the Functional Connectivity Workshop in Cambridge 2003, and was elaborated further at the subsequent 2004 Workshop in Havana through conversations with R. Kotter, A. R. McIntosh, B. Horwitz, P. A. Robinson, O. Sporns, and J. R. Terry. Section 5 arose from a suggestion by A. R. McIntosh elaborated in discussions with K. Friston, L. Harrison, W. Penny, K. Stephan, L. M. Williams, P. A. Valdés-Sosa and E. T. Bullmore. K. Stephan offered helpful suggestions on the manuscript and J. R. Terry assisted in preparation of the numerical code for single-scale systems. The authors are very grateful for such assistance and especially to the organizers of both workshops and two referees for their helpful comments on an earlier draft.
Appendix A
Appendix A. Equations for the multiscale neural system model
The equations model the behaviour of local ensembles of neurons, with dynamical variables V=[V W Z] representing ensemble averages over the extent of a local neural region. The dynamical variables incorporated are the mean membrane potential of pyramidal cells, V, and inhibitory interneurons, Z, and the average number of ‘open’ potassium ion channels, W. The evolution equations are adapted from a study of epileptic seizures in hippocampal slices (Larter & Speelman 1999), which in turn are derived from the model of Morris & Lecar (1981). The mean cell membrane potential of the pyramidal cells is governed by the conductance of sodium, potassium and calcium ion through voltage- and ligand-gated membrane channels,
| (A1) |
| (A2) |
where gion is the maximum conductance of each population of ion species if all channels are open, mion is the fraction of channels open, Vion is the Nernst potential for that ion species and QV(Z) is the firing rate of the excitatory (inhibitory) neurons. The fraction of open channels is determined by the sigmoid-shaped ‘neural activation function’,
| (A3) |
where δion incorporates the variance of this distribution. The fraction of open potassium channels is slightly more complicated, and is governed by W, with
| (A4) |
where ϕ is a temperature scaling factor and τ is a ‘relaxation’ time constant. Cell firing rate is also determined by sigmoid activation functions,
| (A5) |
where the Qmax are the maximum firing rate. An analogous term is also introduced for the inhibitory cells. The firing of these cell populations feeds back on to the ensemble through synaptic coupling to open ligand-gated channels and raises or lowers the membrane potential accordingly. In the case of excitatory-to-inhibitory and inhibitory-to-excitatory connections, this is modelled as additional inputs to the flow of ions across the membrane channel, weighted by functional synaptic factors, aei and aie. In the case of excitatory to excitatory connections, the rate of firing Qv is assumed to lead to a proportional release of glutamate neurotransmitter across the synapse, on to two classes of ligand-gated ion channels: (i) AMPA channels, which open an additional population of sodium channels; and (ii) NMDA receptors, which open an additional population of voltage-gated sodium and calcium channels. rNMDA incorporates the ratio of NMDA to AMPA receptors.
Each of the set of equations (2.1)–(2.5) govern the dynamics within each local cell assembly. Coupling between n nodes is introduced as competitive agonist excitatory action at the same populations of NMDA and AMPA receptors. Locating the ith node at position xi this is incorporated by modifying equation (A 1) to
| (A6) |
for i,j=1,…,n. c parametrizes the strength of excitatory coupling between cortical columns. If c=0, then the systems evolve independently. C>0 introduces interdependences between consecutive columns. c=1 corresponds to maximum coupling, with excitatory input from outside each column surpassing excitatory input from within each column.
All physiologically measurable parameters (conductances, threshold potentials and Nernst potentials) are set to their accepted values (Larter & Speelman 1999) and are given in table 1. The behaviour of this model is described in more detail in Breakspear et al. (2003).
For the present study, the same form of the equations is present in each of the separate scales. Abbreviating (A 6) as
| (A7) |
the interscale coupling function is introduced as,
| (A8) |
where C is the interscale coupling parameter and G the interscale coupling matrix. This models competitive agonist competition of feedback from inhibitory neurons on to pyramidal cells. Hence, this is an example of histological interscale coupling as discussed in §3c.
Endnotes
There exists a variety of categorization schemes across all scales within the brain. Some (e.g. lobes) are used widely in clinical neuroscience, whereas others (e.g. columns) are more conclusively supported. For the present purpose, we merely require the existence of structures at different scales. For a more comprehensive list of candidate structures, see Stephan et al. (2001, p. 1165).
The intersection of the temporal dynamics and spatial structure arises through coupling functions in the evolution equations, and implicitly in the resulting dynamics.
Close to the bifurcation between these two types of behaviour, the ensemble shows a very long transition with apparent competition between local and global structures.
Such scale-free dynamics have been observed in neural cultures by Beggs & Plenz (2003).
At this point, ‘neural activity’ V refers to the complete vector of all neural variables, including membrane potentials and ion channel conductivity.
One contribution of 21 to a Theme Issue ‘Multimodal neuroimaging of brain connectivity’.
References
- Abramowitz M, Stegun I.A. Dover Publications; New York: 1970. Handbook of mathematical functions. [Google Scholar]
- Ashwin P. Attractors stuck on to invariant subspaces. Phys. Lett. A. 1995;209:338–344. [Google Scholar]
- Ashwin P. Synchronization from chaos. Nature. 2003;422:384–385. doi: 10.1038/422384a. [DOI] [PubMed] [Google Scholar]
- Ashwin P, Breakspear M. Anisotropic properties of riddled basins. Phys. Lett. A. 2001;280:139–145. [Google Scholar]
- Bak P, Tang C, Wiesenfeld K. Self organized criticality: an explanation of 1/f noise. Phys. Rev. Lett. 1987;59:381–384. doi: 10.1103/PhysRevLett.59.381. [DOI] [PubMed] [Google Scholar]
- Blake R, Logethetis N.K. Visual competition. Nat. Rev. Neurosci. 2002;3:1–11. doi: 10.1038/nrn701. [DOI] [PubMed] [Google Scholar]
- Beggs J.M, Plenz D. Neuronal avalanches in neocortical circuits. J. Neurosci. 2003;23:11 167–11 177. doi: 10.1523/JNEUROSCI.23-35-11167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletti S, Kurths J, Osipov G, Valladares D.L, Zhou C.S. The synchronization of chaotic systems. Phys. Rep. 2002;366:1–101. [Google Scholar]
- Breakspear M. Nonlinear phase desynchronization in human electroencephalographic data. Hum. Brain Mapp. 2002;15:175–198. doi: 10.1002/hbm.10011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breakspear M. ‘Dynamic connectivity’: theoretical and empirical considerations. Neuroinformatics. 2004;2:205–226. doi: 10.1385/NI:2:2:205. [DOI] [PubMed] [Google Scholar]
- Breakspear M, Terry J. Nonlinear interdependence in neural systems, motivation, theory and relevance. Int. J. Neurosci. 2002;112:1163–1184. doi: 10.1080/00207450290026193. [DOI] [PubMed] [Google Scholar]
- Breakspear M, Terry J, Friston K. Modulation of excitatory synaptic coupling facilitates synchronization and complex dynamics in a nonlinear model of neuronal dynamics. Netw. Comput. Neural Syst. 2003;14:703–732. [PubMed] [Google Scholar]
- Breakspear M, Williams L, Stam K. Topographic analysis of phase dynamics in neural systems reveals formation and dissolution of ‘dynamic cell assemblies’. J. Comput. Neurosci. 2004;14:703–732. doi: 10.1023/b:jcns.0000004841.66897.7d. [DOI] [PubMed] [Google Scholar]
- Bullmore, E. T., Fadili, J., Maxim, V., Şendur, L., Whitcher, B., Suckling, J., Brammer, M. & Breakspear, M. 2004. Wavelets and functional magnetic resonance imaging of the human brain. NeuroImage23(Suppl. 1), S234–S249. [DOI] [PubMed]
- Buxton R.B, Wong E.C, Frank L.R. Dynamics of blood flow and oxygen changes during brain activation: the balloon model. Magn. Reson. Med. 1998;39:855–864. doi: 10.1002/mrm.1910390602. [DOI] [PubMed] [Google Scholar]
- Daffertshofer A, van den Berg C, Beek P.J. A dynamical model for mirror movements. Physica D. 1999;132:243–266. [Google Scholar]
- Daubechies I. SIAM; Philadelphia: 1992. Ten lectures on wavelets. [Google Scholar]
- David O, Cosmelli D, Friston K.J. Evaluation of different measures of functional connectivity using a neural mass model. NeuroImage. 2004;21:659–673. doi: 10.1016/j.neuroimage.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Douglas R.J, Martin K.A.C. Neuronal circuits of the neocortex. Ann. Rev. Neurosci. 2004;27:419–451. doi: 10.1146/annurev.neuro.27.070203.144152. [DOI] [PubMed] [Google Scholar]
- Frank T, Daffertshofer A, Peper C, Beek P, Haken H. Toward a comprehensive theory of brain activity: coupled oscillators under external forces. Physica D. 2000;144:62–86. [Google Scholar]
- Freeman W. Academic Press; New York: 1975. Mass action in the nervous system: examination of the neurophysiological basis of adaptive behaviour through the EEG. [Google Scholar]
- Friston K.J. The labile brain. I. Neuronal transients and nonlinear coupling. Phil. Trans. R. Soc. B. 2000;355:215–236. doi: 10.1098/rstb.2000.0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston K.J. The labile brain. II. Transients, complexity and selection. Phil. Trans. R. Soc. B. 2000;355:237–252. doi: 10.1098/rstb.2000.0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston K, Mechelli A, Turner R, Price C. Nonlinear responses in fMRI: the balloon model, Volterra kernels, and other hemodynamics. NeuroImage. 2000;12:466–477. doi: 10.1006/nimg.2000.0630. [DOI] [PubMed] [Google Scholar]
- Friston K.J, Harrison L, Penny W.D. Dynamic causal modeling. NeuroImage. 2003;19:1273–1302. doi: 10.1016/s1053-8119(03)00202-7. [DOI] [PubMed] [Google Scholar]
- Fujimoto K, Kaneko K. How fast elements can affect slow dynamics. Physica D. 2003;180:1–16. [Google Scholar]
- Fujisaka H, Yamada T. Stability theory of synchronized motion in coupled-oscillator system. Prog. Theor. Phys. 1983;69:32–47. [Google Scholar]
- Guan S, Lai C.H, Wei G.W. A wavelet method for the characterization of spatiotemporal patterns. Physica. 2002;163D:49–79. [Google Scholar]
- Harrison L.M, David O, Friston K.J. Stochastic models of neuronal dynamics. Phil. Trans. R. Soc. B. 2005;360:1075–1091. doi: 10.1098/rstb.2005.1648. doi:10.1098/rstb.2005.1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgetag C.C, Burns G.A.P.C, O'Neill M.A, Scannell J.W, Young M.P. Anatomical connectivity defines the organisation of clusters of cortical areas in macaque monkey and cat. Phil. Trans. R. Soc. B. 2000;355:91–110. doi: 10.1098/rstb.2000.0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirsa V.K, Haken H. Field theory of electromagnetic brain activity. Phys. Rev. Lett. 1996;77:960–963. doi: 10.1103/PhysRevLett.77.960. [DOI] [PubMed] [Google Scholar]
- Kaneko K. Dominance of Milnor attractors and noise-induced selection in a multiattractor system. Phys. Rev. Lett. 1997;78:2736–2739. [Google Scholar]
- Keibel S, Friston K.J. Statistical parametric mapping for event-related potentials (II): a hierarchical temporal model. NeuroImage. 2004;22:503–520. doi: 10.1016/j.neuroimage.2004.02.013. [DOI] [PubMed] [Google Scholar]
- Koenig T, Studer D, Hubl D, Melie L, Strik W.K. Brain connectivity at different time-scales measured with EEG. Phil. Trans. R. Soc. B. 2005;360:1015–1023. doi: 10.1098/rstb.2005.1649. doi:10.1098/rstb.2005.1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromoto Y. Springer; Berlin: 1984. Chemical oscillations, waves and turbulence. [Google Scholar]
- Larter R, Speelman B. A coupled ordinary differential equation lattice model for the simulation of epileptic seizures. Chaos. 1999;9:795–804. doi: 10.1063/1.166453. [DOI] [PubMed] [Google Scholar]
- Larter R, Speelman B, Worth R.M. A coupled ordinary differential equation lattice model for the simulation of epileptic seizures. Chaos. 1999;9:795–804. doi: 10.1063/1.166453. [DOI] [PubMed] [Google Scholar]
- Logothetis N.K. The neural basis of the BOLD fMRI signal. Phil. Trans. R. Soc. B. 2002;357:1003–1037. doi: 10.1098/rstb.2002.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumer E, Edelman G, Tononi G. Neural dynamics in a model of the thalamocortical system. I. Layers, loops and the emergence of fast synchronous oscillations. Cereb. Cortex. 1997;7:207–227. doi: 10.1093/cercor/7.3.207. [DOI] [PubMed] [Google Scholar]
- Mallat S. Academic Press; London: 1999. A wavelet tour of signal processing. [Google Scholar]
- Mesulam M. From sensation to cognition. Brain. 1998;121:1013–1052. doi: 10.1093/brain/121.6.1013. [DOI] [PubMed] [Google Scholar]
- Morris C, Lecar H. Voltage oscillations in the barnacle giant muscle fiber. Biophys. J. 1981;35:193–213. doi: 10.1016/S0006-3495(81)84782-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountcastle V.B. The columnar organization of the neocortex. Brain. 1997;120:701–722. doi: 10.1093/brain/120.4.701. [DOI] [PubMed] [Google Scholar]
- Mountcastle V.B. Harvard University Press; Cambridge, MA: 1998. The cerebral cortex. [Google Scholar]
- Nakao H, Mishiro T, Yamada M. Visualization of correlation cascade in spatiotemporal chaos using wavelets. Int. J. Bifurcation Chaos. 2001;5:1483–1493. [Google Scholar]
- Nunez P.L. The brain wave equation: a model for the EEG. Math. Biosci. 1974;21:279–297. [Google Scholar]
- Nunez P.L. Oxford University Press; Oxford: 1997. Neocortical dynamics and human brain rhythms. [Google Scholar]
- Passingham R.E, Stephan K.E, Kotter R. The anatomical basis of functional localization in the cortex. Nat. Rev. Neurosci. 2002;3:606–616. doi: 10.1038/nrn893. [DOI] [PubMed] [Google Scholar]
- Riera J.J, Watanabe J, Kazuki I, Naoki M, Aubert E, Ozaki T, Kawashima R. A state-space model of the hemodynamic approach: nonlinear filtering of BOLD signals. NeuroImage. 2004;21:546–567. doi: 10.1016/j.neuroimage.2003.09.052. [DOI] [PubMed] [Google Scholar]
- Robinson P.A, Rennie C.J, Wright J.J. Propagation and stability of waves of electrical activity in the cerebral cortex. Phys. Rev. E. 1997;56:826–840. [Google Scholar]
- Robinson P.A, Rennie C.J, Wright J.J, Bahramali H, Gordon E, Rowe D.L. Prediction of electroencephalographic spectra from neurophysiology. Phys. Rev. E. 2001;63:021903. doi: 10.1103/PhysRevE.63.021903. [DOI] [PubMed] [Google Scholar]
- Salvador R, Suckling J, Coleman M.R, Pickard J.D, Menon D, Bullmore E. Neurophysiological architecture of functional magnetic resonance images of human brain. Cereb. Cortex. 2005 doi: 10.1093/cercor/bhi016. doi:10.1093/cercor/bhi016 Published online 5 January 2005. [DOI] [PubMed] [Google Scholar]
- Shephard G.M. Oxford University Press; London: 2004. The synaptic organization of the brain. [Google Scholar]
- Sporns O, Kotter R. Motifs in brain networks. PLoS Biol. 2004;2:e369. doi: 10.1371/journal.pbio.0020369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Edleman G. Theoretical neuroanatomy: relating anatomical and functional connectivity in graphs and cortical connection matrices. Cereb. Cortex. 2000;10:127–141. doi: 10.1093/cercor/10.2.127. [DOI] [PubMed] [Google Scholar]
- Stam C.J. Functional connectivity patterns of human magnetoencephalography recordings: a ‘small world’ network? Neurosci. Lett. 2004;355:25–28. doi: 10.1016/j.neulet.2003.10.063. [DOI] [PubMed] [Google Scholar]
- Stam C.J, Breakspear M, van Cappellen, van Walsum A.M, van Dijk B.W. Nonlinear synchronization in EEG and whole-head MEG recordings of healthy subjects. Hum. Brain Mapp. 2003;19:63–78. doi: 10.1002/hbm.10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan K.E, Kamper L, Bozkurt A, Burns G.A.P.C, Young M.P, Kötter R. Advanced database methodology for the collation of connectivity data on the macaque brain (CoCoMac) Phil. Trans. R. Soc. B. 2001;356:1159–1186. doi: 10.1098/rstb.2001.0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan K.E, Marshall J.C, Friston K.J, Rowe J.B, Ritzl A, Zilles K, Fink G.R. Lateralized cognitive processes and lateralized task control in the human brain. Science. 2003;301:384–386. doi: 10.1126/science.1086025. [DOI] [PubMed] [Google Scholar]
- Strogatz S.H. From Kuromoto to Crawford: exploring the onset of synchronization in populations of coupled oscillators. Physica. 2000;143D:1–20. [Google Scholar]
- Strogatz S.H. Exploring complex networks. Nature. 2001;410:268–276. doi: 10.1038/35065725. [DOI] [PubMed] [Google Scholar]
- Szentagothai J. The modular architectonic principle of neural centers. Rev. Phys. Biochem. Pharmacol. 1983;98:11–61. doi: 10.1007/BFb0033866. [DOI] [PubMed] [Google Scholar]
- Valdés-Sosa P.A, et al. Estimating brain functional connectivity with sparse multivariate autoregression. Phil. Trans. R. Soc. B. 2005;360:969–981. doi: 10.1098/rstb.2005.1654. doi:10.1098/rstb.2005.1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rotterdam A, Lopes da Silva F.H, van den Ende J, Viergever M.A, Hermans A.J. A model of the spatio-temporal characteristics of the alpha rhythm. Bull. Math. Biol. 1982;44:283–305. doi: 10.1007/BF02463252. [DOI] [PubMed] [Google Scholar]
- Watts D, Strogatz S. Collective dynamics of small world networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- Wei G.W, Zhan M, Lai C.H. Tailoring wavelets for chaos control. Phys. Rev. Lett. 2002;89:281403. doi: 10.1103/PhysRevLett.89.284103. [DOI] [PubMed] [Google Scholar]
- West G.B, Brown J.H. Life's universal scaling laws. Phys. Today. 2004;57:36–42. [Google Scholar]
- Wright I.C, Sharma T, Ellison P.K, McGuire P.K, Friston K.J, Brammer M.J, Murray R.M, Bullmore E.T. Supra-regional brain systems and the neuropathology of schizophrenia. Cereb. Cortex. 1999;9:366–378. doi: 10.1093/cercor/9.4.366. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Martindale J, Johnston D, Jones M, Berwick J, Mayhew J. A model of the hemodynamic response and oxygen delivery to brain. NeuroImage. 2002;16:617–637. doi: 10.1006/nimg.2002.1078. [DOI] [PubMed] [Google Scholar]
- Zhou, Y., McComb, W. D., Vahala, G. 1997 Renormalization group (RG) in turbulence: historical and comparative perspective. NASA CR-201718 ICASE Report No. 97-36: 60–118.