Abstract
Cortical activity is the product of interactions among neuronal populations. Macroscopic electrophysiological phenomena are generated by these interactions. In principle, the mechanisms of these interactions afford constraints on biologically plausible models of electrophysiological responses. In other words, the macroscopic features of cortical activity can be modelled in terms of the microscopic behaviour of neurons. An evoked response potential (ERP) is the mean electrical potential measured from an electrode on the scalp, in response to some event. The purpose of this paper is to outline a population density approach to modelling ERPs.
We propose a biologically plausible model of neuronal activity that enables the estimation of physiologically meaningful parameters from electrophysiological data. The model encompasses four basic characteristics of neuronal activity and organization: (i) neurons are dynamic units, (ii) driven by stochastic forces, (iii) organized into populations with similar biophysical properties and response characteristics and (iv) multiple populations interact to form functional networks. This leads to a formulation of population dynamics in terms of the Fokker–Planck equation. The solution of this equation is the temporal evolution of a probability density over state-space, representing the distribution of an ensemble of trajectories. Each trajectory corresponds to the changing state of a neuron. Measurements can be modelled by taking expectations over this density, e.g. mean membrane potential, firing rate or energy consumption per neuron. The key motivation behind our approach is that ERPs represent an average response over many neurons. This means it is sufficient to model the probability density over neurons, because this implicitly models their average state. Although the dynamics of each neuron can be highly stochastic, the dynamics of the density is not. This means we can use Bayesian inference and estimation tools that have already been established for deterministic systems. The potential importance of modelling density dynamics (as opposed to more conventional neural mass models) is that they include interactions among the moments of neuronal states (e.g. the mean depolarization may depend on the variance of synaptic currents through nonlinear mechanisms).
Here, we formulate a population model, based on biologically informed model-neurons with spike-rate adaptation and synaptic dynamics. Neuronal sub-populations are coupled to form an observation model, with the aim of estimating and making inferences about coupling among sub-populations using real data. We approximate the time-dependent solution of the system using a bi-orthogonal set and first-order perturbation expansion. For didactic purposes, the model is developed first in the context of deterministic input, and then extended to include stochastic effects. The approach is demonstrated using synthetic data, where model parameters are identified using a Bayesian estimation scheme we have described previously.
Keywords: evoked response potentials, population dynamics, Fokker–Planck equation, generative models, synthetic data, system identification
1. Introduction
Neuronal responses are the product of coupling among hierarchies of neuronal populations. Sensory information is encoded and propagated through the hierarchy depending on biophysical parameters that control this coupling. Because coupling can be modulated by experimental factors, estimates of coupling parameters provide a systematic way to parametrize experimentally induced responses, in terms of their causal structure. This paper is about estimating these parameters using a biologically informed model.
Electroencephalography (EEG) is a non-invasive technique for measuring electrical activity generated by the brain. The electrical properties of nervous tissue derive from the electrochemical activity of coupled neurons that generate a distribution of current sources within the cortex, which can be estimated from multiple scalp electrode recordings (Mattout et al. 2003; Phillips et al. 2005). An interesting aspect of these electrical traces is the expression of large-scale coordinated patterns of electrical potential. There are two commonly used methods to characterize event-related changes in these signals: averaging over many traces, to form event-related potentials (ERP) and calculating the spectral profile of ongoing oscillatory behaviour. The assumption implicit in the averaging procedure is that the evoked signal has a fixed temporal relationship to the stimulus, whereas the latter procedure relaxes this assumption (Pfurtscheller & Lopes da Silva 1999).
Particular characteristics of ERPs are associated with cognitive states, e.g. the mismatch negativity in auditory oddball paradigms (Winkler et al. 1998, 2001). The changes in ERP evoked by a cognitive ‘event’ are assumed to reflect event-dependent changes in cortical activity. In a similar way, spectral peaks of ongoing oscillations within EEG recordings are generally thought to reflect the degree of synchronization among oscillating neuronal populations, with specific changes in the spectral profile being associated with various cognitive states (Pfurtscheller & Lopes da Silva 1999). These changes have been called event-related desynchronization (ERD) and event-related synchronization (ERS). ERD is associated with an increase in processing information, e.g. voluntary hand movement (Pfurtscheller 2001), whereas ERS is associated with reduced processing, e.g. during little or no motor behaviour. These observations led to the thesis that ERD represents increased cortical excitability, and conversely, that ERS reflects de-activation (Pfurtscheller & Lopes da Silva 1999; Pfurtscheller 2001).
The conventional approach to interpreting the EEG in terms of computational processes (Churchland & Sejnowski 1994) is to correlate task-dependent changes in the ERP or time-frequency profiles of ongoing activity with cognitive or pathological states. A complementary strategy is to use a generative model of how data are caused, and estimate the model parameters that minimize the difference between real and generated data. This approach goes beyond associating particular activities with cognitive states to model the self-organization of neural systems during functional processing. Candidate models developed in theoretical neuroscience can be divided into mathematical and computational (Dayan 1994). Mathematical models entail the biophysical mechanisms behind neuronal activity, such as the Hodgkin–Huxley model neuron of action potential generation (Dayan & Abbott 2001). Computational models are concerned with how a computational device could implement a particular task, e.g. representing saliency in a hazardous environment. Both levels of analysis have produced compelling models, which speaks to the use of biologically and computationally informed forward or generative models in neuroimaging. We focus here on mathematical models.
The two broad classes of generative models for EEG are neural mass models (NMM) (Wilson & Cowan 1972; Nunez 1974; Lopes da Silva et al. 1976; Freeman 1978; Jansen & Rit 1995; Valdes et al. 1999; David & Friston 2003) and population density models (Knight 1972a,b, 2000; Nykamp & Tranchina 2000, 2001; Omurtag et al. 2000; Haskell et al. 2001; Gerstner & Kistler 2002). NMMs were developed as parsimonious models of the mean activity (firing rate or membrane potential) of neuronal populations and have been used to generate a wide range of oscillatory behaviours associated with the EEG (Jansen & Rit 1995; Valdes et al. 1999; David & Friston 2003). The model equations for a population are a set of nonlinear differential equations forming a closed loop between the influence neuronal firing has on mean membrane potential and how this potential changes the consequent firing rate of a population. Usually, two operators are required: linking membrane responses to input from afferent neurons (pulse-to-wave) and the dependence of action potential density on membrane potential (wave-to-pulse; see Jirsa 2004; for an excellent review). They are divided into lumped models (Lopes da Silva et al. 1976; David & Friston 2003) where populations of neurons are modelled as discrete nodes or regions that interact through cortico-cortical connections or as continuous neural fields (Jirsa & Haken 1996; Wright et al. 2001, 2003; Rennie et al. 2002; Robinson et al. 2003), where the cortical sheet is modelled as a continuum, on which the cortical dynamics unfold. Frank (2000) and Frank et al. (2001) have extended the continuum model to include stochastic effects with an application to MEG data.
David & Friston (2003) and David et al. (2004) have recently extended a NMM proposed by Jansen & Rit 1995, and implemented it as a forward model to analyse ERP data measured during a mismatch negativity task (David et al. 2005). In doing this, they were able to infer changes in effective connectivity, defined as the influence one region exerts on another (Aertsen et al. 1989; Friston et al. 1995, 1997; Buchel & Friston 1998; Buchel et al. 1999; Friston 2001; Friston & Buchel 2000) that mediated the NMM. Analyses of effective connectivity in the neuroimaging community were first used with positron emission tomography and later with functional magnetic resonance imaging (fMRI) data (Friston et al. 1993; Harrison et al. 2003; Penny et al. 2004a,b). The latter applications led to the development of dynamic causal modelling (DCM) (Friston et al. 2003). DCM for neuroimaging data embodies organizational principles of cortical hierarchies and neurophysiological knowledge (e.g. time constants of biophysical processes) to constrain a parametrized nonlinear dynamic model of observed responses. A principled way of incorporating these constraints is in the context of Bayesian estimation (Friston et al. 2002). Furthermore, established Bayesian model comparison and selection techniques can be used to disambiguate different models and their implicit assumptions. The development of this methodology by David et al. for electrophysiological data was an obvious extension and continues with this paper.
An alternative to NMM are population density models (Knight 1972a,b, 2000; Frank 2000; Nykamp & Tranchina 2000, 2001; Omurtag et al. 2000; Frank et al. 2001; Haskell et al. 2001; Gerstner & Kistler 2002; Sirovich 2003). These models explicitly show the effect of stochastic influences, e.g. variability of pre-synaptic spike-time arrivals. This randomness is described probabilistically in terms of a probability density over trajectories through state space. The ensuing densities can be used to generate measurements, such as the mean firing rate or membrane potential of an average neuron within a population. In contrast, NMM models only account for the average neuronal state, and not for stochastic effects. Stochastic effects are known to be important for many phenomena, e.g. stochastic resonance (Wiesenfeld & Moss 1995). A key component of modelling population densities is the Fokker–Planck equation (FPE) (Risken 1996). This equation has a long history in the physics of transport processes and has been applied to a wide range of physical phenomena, e.g. Brownian motion, chemical oscillations, laser physics and biological self-organization (Haken 1973, 1996; Kuramoto 1984; Risken 1996). The beauty of the FPE is that, given constraints on the smoothness of stochastic forces (Risken 1996; Kloeden & Platen 1999), stochastic effects are equivalent to a diffusive process. This can be modelled by a deterministic equation in the form of a parabolic partial differential equation (PDE). The Fokker–Planck formalism uses notions from mean-field theory, but is dynamic, and can model transitions from non-equilibrium to equilibrium states.
ERPs represent the average response over millions of neurons, which means it is sufficient to model their population density to generate responses. This means the FPE is a good candidate for a forward or generative model of ERPs. Furthermore, the population dynamics entailed by the FPE are deterministic. This means Bayesian techniques that are already established for deterministic dynamical systems can be used to model electrophysiological responses. However, the feasibility of using population density methods, in the context of ERP/EEG data analysis, has not been established. What NMMs lack in distributional details, the FPE lacks in parsimony, which brings with it a computational cost. This cost could preclude a computationally efficient role in data analysis. The purpose of this paper was to assess the feasibility of using the FPE in a forward model of average neuronal responses.
(a) Overview
In the first section, we review the theory of the integrate-and-fire model neuron with synaptic dynamics and its formulation into a FPE of interacting populations mediated through mean-field quantities (see the excellent texts of Risken 1996; Dayan & Abbott 2001; Gerstner & Kistler 2002; for further details). The model encompasses four basic characteristics of neuronal activity and organization; neurons are (i) dynamic units, (ii) driven by stochastic forces, (iii) organized into populations with similar biophysical properties and response characteristics and (iv) multiple populations interact to form functional networks. In the second section, we briefly review the Bayesian estimation scheme used in current DCMs (Friston et al. 2002, 2003; David et al. submitted). In the third, we discuss features of the model and demonstrate the face-validity of the approach using simulated data. This involves inverting a population density model to estimate model parameters given synthetic data. The discussion focuses on outstanding issues with this approach in the context of generative models for ERP/EEG data.
2. Theory
(a) A deterministic model neuron
The response of a model neuron to input, s(t), has a generic form, which can be represented by the differential equation.
| (2.1) |
where . The state vector, x (e.g. including variables representing membrane potential and proportion of open ionic channels), defines a space within which its dynamics unfold. The number of elements in x defines the dimension of this space and specific values identify a coordinate within it. The temporal derivative of x quantifies the motion of a point in state space, and the solution of the differential equation is its trajectory. The right-hand term is a function of the states, x(t), and input, s(t), where input can be exogenous or internal, i.e. mediated by coupling with other neurons. The model parameters, characteristic time-constants of the system, are represented by θ. As states are generally not observed directly, an observation equation is needed to link them to measurements, y
| (2.2) |
where ϵ is observation noise (usually modelled as a Gaussian random variable). An example of an observation equation is an operator that returns the mean firing rate or membrane potential of a neuron. These equations form the basis of a forward or generative model to estimate the conditional density given real data, as developed in §2b.
Neurons are electrical units. A simple expression for the rate of change of membrane potential, V, in terms of membrane currents, Ii (ith source), and capacitance is
| (2.3) |
Figure 1 shows a schematic of a model neuron and its resistance-capacitance (RC) circuit equivalent. Models of action potential generation, e.g. the Hodgkin–Huxley neuron, are based on quantifying the components of the right-hand of equation (2.3). Typically, currents are categorized as voltage, calcium or neurotransmitter-dependent. The dynamic repertoire of a specific model depends on the nature of the different source currents. This repertoire can include fixed-point attractors, limit cycles and chaotic dynamics.
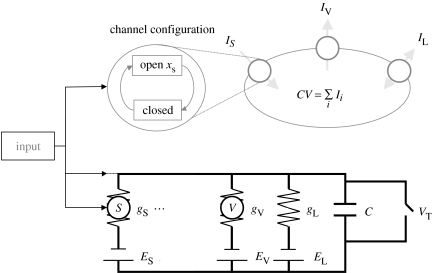
Figure 1.
Schematic of a single-compartment model neuron including synaptic dynamics and its RC circuit analogue. Synaptic channels and voltage-dependent ion channels are shown as a circle containing S or V respectively. There may be several species of channels, indicated by the dots. Equilibrium potentials, conductance and current owing to neurotransmitter (synaptic), voltage-dependent and passive (leaky) channels are ES, EV, EL, gS, gV, gL, IS, IV and IL, respectively. Depolarization occurs when membrane potential exceeds threshold, VT. Input increases the opening rate of synaptic channels. Note that if synaptic channels are dropped from a model (e.g. as in a SIF neuron), then input is directly into the circuit.
A caricature of a spiking neuron is the simple integrate-and-fire (SIF) model. It is one-dimensional as all voltage and synaptic channels are ignored. Instead, current is modelled as a constant passive leak of charge, thereby reducing the right-hand side of equation (2.3) to ‘leakage’ and input currents.
| (2.4) |
where gL and EL are the conductance and equilibrium potential of the leaky channel, respectively (see tables 1 and 2 for a list of all variables used in this paper). This model does not incorporate the biophysics needed to generate action potentials. Instead, spiking is modelled as a threshold process, i.e. once membrane potential exceeds a threshold value, VT, a spike is assumed and membrane potential is reset to VR, where VR≤EL<VT. No spike is actually emitted; only sub-threshold dynamics are modelled.
Table 1.
Variable description and symbols.
| variable description | symbol |
|---|---|
| membrane potential | V |
| inter-stimulus time | T |
| proportion of open channels (slow potassium) | xsK |
| proportion of open channels (AMPA) | xAMPA |
| proportion of open channels (GABA) | xGABA |
| proportion of open channels (NMDA) | xNMDA |
| state vector | |
| probability density | ρ |
| probability mode density | μ |
| dynamic operator | Q |
| right eigenvector matrix | R |
| left eigenvector matrix | L |
| eigenvalue matrix | D |
| observations | y |
| observation operator | M |
Table 2.
Parameter values used in simulations.
| parameter description | symbol | value/units |
|---|---|---|
| firing threshold | VT | −53 mV |
| reset voltage | VR | −90 mV |
| equilibrium potential (passive current) | EL | −73 mV |
| equilibrium potential (sK) | EsK | −90 mV |
| equilibrium potential (AMPA) | EAMPA | 0 mV |
| equilibrium potential (GABA) | EGABA | −70 mV |
| equilibrium potential (NMDA) | ENMDA | 0 mV |
| passive conductance | gL | 25 nS |
| active conductance (sK) | gsK | 128 nS |
| active conductance (AMPA) | gAMPA | 24 nS |
| active conductance (GABA) | gGABA | 64 nS |
| active conductance (NMDA) | gNMDA | 8 nS |
| membrane capacitance | C | 0.375 nF |
| time constant (sK) | τsK | 80 ms |
| time constant (AMPA) | τAMPA | 2.4 ms |
| time constant (GABA) | τGABA | 7 ms |
| time constant (NMDA) | τNMDA | 100 ms |
| background opening coefficient (AMPA) | pAMPA | 0.875 a.u. |
| background opening coefficient (GABA) | pGABA | 0.0625 a.u. |
| background opening coefficient (NMDA) | pNMDA | 0.0625 a.u. |
| diffusion coefficient (V) | w1 | 4 mV2 ms−1 |
| diffusion coefficient (t) | w2 | 0 ms−1 |
| diffusion coefficient (sK) | w3 | 0.125 ms−1 |
| diffusion coefficient (AMPA) | w4 | 0.125 ms−1 |
| diffusion coefficient (GABA) | w5 | 0.125 ms−1 |
| diffusion coefficient (NMDA) | w6 | 0.125 ms−1 |
(b) Modelling supra-threshold dynamics
First, we augment a SIF model with an additional variable T inter-spike time (IST). Typically, the threshold potential is modelled as an absorbing boundary condition, with re-entry at the reset voltage. However, we have chosen to model the IST as a state variable for a number of important reasons. First, it constrains the neuronal trajectory to a finite region of state space, which only requires natural boundary conditions, i.e. the probability mass decays to zero as state space extends to infinity. This makes our later treatment generic, because we do not have to consider model-specific boundary conditions when, for example, formulating the FPE or deriving its eigen-system. Another advantage is that the time between spikes can be calculated directly from the density of IST. Finally, having an explicit representation of the time since the last spike allows us to model time-dependent changes in the systems parameters (e.g. relative refractoriness) that would be much more difficult in conventional formulations. The resulting model is two-dimensional and automates renewal to reset voltage, once threshold has been exceeded. We will refer to this as a modified SIF model. With this additional state-variable, we have
| (2.5) |
The firing rate of this model, in response to different input, is shown in a later figure (figure 8) when we compare its response to a stochastic model neuron. A characteristic feature of this deterministic model is that the input has to reach a threshold before spikes are generated (see Appendix A for a brief derivation of this threshold), after which, firing rate increases monotonically. This is in contrast to a stochastic model that has non-zero probability of firing, even with low input. Given a supra-threshold input to the model of equation (2.5), membrane voltage is reset to VR using the Heaviside function (last term in equation (2.5)). This ensures that once V>VT, the rate of change of T with respect to time is large and negative (α=104), reversing the progression of IST and returning it to zero, after which it increases constantly for VR<V<VT. Membrane potential is coupled to T via an expression involving β, which is a Gaussian function, centred at T=0 with a small dispersion (γ=1 ms). During the first few milliseconds following a spike, this term provides a brief impulse to clamp membrane potential near to VR (cf. the refractory period).
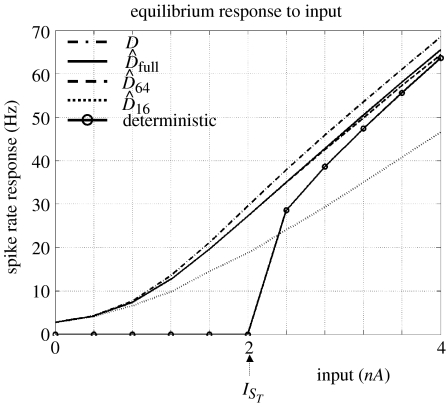
Figure 8.
Comparison of perturbation approximation of equilibrium response rates of a stochastic and deterministic model-neuron (equation (2.5)). All rates from diffusion equations taper off gradually as input falls below threshold, sT, in contrast to the deterministic model. The curve labelled D(s) is from an explicit re-calculation of the dynamic operator at each input, whereas , and are first-order approximations using 128, 64 or 16 modes (out of 128). is in good agreement with D, loosing some accuracy as input increases. is almost indistinguishable from . is less accurate, however, at low inputs it is still in reasonable agreement with and maintains the characteristic profile of the response curve.
(c) Modelling spike-rate adaptation and synaptic dynamics
Equation (2.5) can be extended to include ion-channel dynamics, i.e. to model spike-rate adaptation and synaptic transmission.
| (2.6) |
These equations model spike-rate adaptation and synaptic dynamics (fast excitatory AMPA, slow excitatory NMDA and inhibitory GABA channels) by a generic synaptic channel mechanism, which is illustrated in figure 1. The proportion of open channels is modelled by an activation variable, xi (i=sK (slow potassium), AMPA, GABA or NMDA), where 0≤xi≤1. Given no input, the ratio of open to closed channels will relax to an equilibrium state, e.g. p/(1+p) for GABA and NMDA channels. The rate at which channels close is proportional to xi. Conversely, the rate of opening is proportional to 1−xi. Synaptic input now enters by increasing the opening rate of AMPA channels instead of affecting V directly (see the RC circuit of figure 1).
(d) A stochastic model neuron
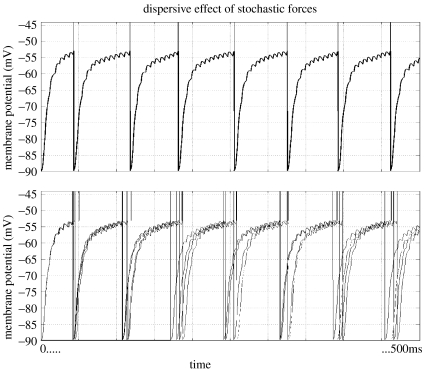
The response of a deterministic system to input is known from its dynamics and initial conditions. The system follows a well-defined trajectory in state-space. The addition of system noise, i.e. random input, to the deterministic equation turns it into a stochastic differential equation (SDE), also called Langevin's equation (Risken 1996). In contrast to deterministic systems, Langevin's equation has an ensemble of solutions. The effect of stochastic terms, e.g. variable spike-time arrival, is to disperse trajectories through state space. A simple example of this is shown in figure 2. The top figure shows just one trajectory and five are shown below, each with the same initial condition. The influence of variable input is manifest as a dispersion of trajectories.
Figure 2.
Dispersive effect of variable input on trajectories. The top figure shows one trajectory of the model of equation (2.6), whereas the lower figure shows five trajectories, each the consequence of a different sequence of inputs sampled from a Poisson distribution. Once threshold is exceeded, V is reset to VR. Vertical lines above threshold represent action potentials. Stochastic input is manifest as a dispersion of trajectories.
Under smoothness constraints on the random input, the ensemble of solutions to the SDE are described exactly by the FPE (Risken 1996; Kloeden & Platen 1999). The FPE enables the ensemble of solutions of a SDE to be framed as a deterministic dynamic equation, which models the dispersive effect of stochastic input as a diffusive process for which many solution techniques have been developed (Risken 1996; Kloeden & Platen 1999). This leads to a parsimonious description in terms of a probability density over state space, represented by ρ(x,t). This is important, as stochastic effects can have a substantial influence on dynamic behaviour (for example, the response profile of a stochastic neuron verses a deterministic model neuron in figure 8).
Population density methods have received much attention over the past decades as a means of efficiently modelling the activity of thousands of similar neurons. Knight (2000) and Sirovich (2003) describe an eigen-function approach to solving particular examples of these equations and extend the method to a time-dependent perturbation solution. Comparative studies by Omurtag et al. (2000) and Haskell et al. (2001) have demonstrated the efficiency and accuracy of population density methods with respect to Monte Carlo simulations of populations of neurons. The effects of synaptic dynamics have been explored (Haskell et al. 2001; Nykamp & Tranchina 2001) and Nykamp & Tranchina (2000) have applied the method to model-orientation tuning. Furthermore, Casti et al. (2002) have modelled bursting activity of the lateral geniculate nucleus. Below, we review briefly a derivation of the FPE and its eigen-solution (Knight 2000; Sirovich 2003). We have adopted some terminology of Knight and others for consistency.
Consider a 1-divisional system described by equation (2.1), but now with s(t) as a random variable.
| (2.7) |
where h is a discrete quantity representing the change in post-synaptic membrane potential owing to a synaptic event, and tn represents the time of the nth event, which is modelled as a random variable. Typically, the time between spikes is sampled from a Poisson distribution (see Nykamp & Tranchina 2000; Omurtag et al. 2000). Given the neuronal response function σ(t) (see Dayan & Abbott 2001), the mean impulse rate, r(t), can be calculated by taking an average over a short time-interval, T,
| (2.8) |
How does ρ(x,t) change with time, owing to variability in s(t)? To simplify the description we will consider a SIF model with an excitatory input only (without synaptic dynamics). See Omurtag et al. (2000) for further details.
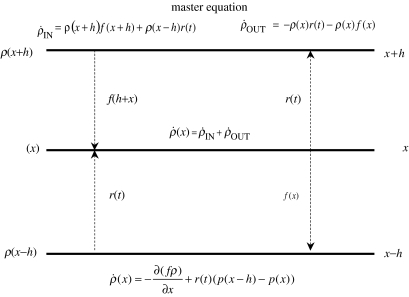
Consider the ladder diagram of figure 3. The Master equation (see Risken 1996), detailing the rate of change of ρ(x,t) with time, can be intuited from this figure,
| (2.9) |
Figure 3.
The master equation of an SIF model with excitatory input (no synaptic dynamics). Three values of the state x (x and x±h), the probability of occupying these states (ρ(x±h) and ρ(x)) and transition rates (f(x), f(x+h) and r(t)) are shown. The rate of change of ρ(x, t) with time (bottom centre formula) is attained by considering the rates in and out of x (formulae at the top left and right, respectively).
on rearranging
| (2.10) |
If we approximate the first bracketed expression using a first-order Taylor expansion, we obtain
| (2.11) |
which Omurtag et al. (2000) refers to as the population equation. The first right-hand term is due to leakage current and generates a steady flow towards EL. The second expression is due to input and is composed of two terms, which can be considered as replenishing and depleting terms, respectively. Given an impulse of input, the probability between x−h and x will contribute to flow at x, whereas ρ(x) flows away from x. An alternative derivation in terms of scalar and vector fields is given in Appendix B.
The replenishing term of equation (2.11) can be approximated using a second-order Taylor series expansion about x:
| (2.12) |
Substituting this into equation (2.11) and using w2=sh, where w is the diffusion coefficient, brings us to the diffusion approximation of the population equation, i.e. the FPE or the advection-diffusion equation:
| (2.13) |
or, more simply
| (2.14) |
where Q(x, s) contains all the dynamic information entailed by the differential equations of the model. Knight et al. (2000) refer to this as a dynamic operator. The first term of equation (2.13), known as the advection term, describes movement of the probability density owing to the system's deterministic dynamics. The second describes dispersion of density brought about by stochastic variations in the input. It is weighted by half the square of the diffusion coefficient, w, which quantifies the root-mean-squared distance travelled owing to this variability. Inherent in this approximation is the assumption that h is very small, i.e. the accuracy of the Taylor series increases as h→0. Comparisons between the diffusion approximation and direct simulations (e.g. Nykamp & Tranchina 2000; Omurtag et al. 2000) have demonstrated its accuracy in the context of neuronal models.
So far, the FPE has been interpreted as the statistical ensemble of solutions of a single neurons response to input. An alternative interpretation is that ρ(x, t) represents an ensemble of trajectories describing a population of neurons. The shift from a single neuron to an ensemble interpretation entails additional constraints on the population dynamics. An ensemble, or ‘mean-field’ type population, equation assumes that neurons within an ensemble are indistinguishable. This means that each neuron ‘feels’ the same influence from internal interactions and external input.
These assumptions constrain the connectivity among a population that mean-field approaches can model, as they treat the connectivity matrix, quantifying all interactions, as a random variable which changes constantly with time. The changing connectivity matrix can be interpreted as a consequence of miss firing (Abeles 1991; Omurtag et al. 2000). If, however, local effects dominate, such as local synchronous firing, then a sub-population will become distinguishable, thereby violating the basic premise of a mean-field assumption. Assuming this is not the case, a mean-field approximation of ensemble activity can then be developed, which enables the modelling of neuronal interactions by the mean influence exerted on one neuron from the rest. Given this, a population response is modelled by an ensemble of non-interacting neurons, but whose input is augmented by a mean interaction term.
The FPE equation of (2.13) generalizes to models with multiple states, such as those derived in the first section.
| (2.15) |
where P represents the probability distribution over a multi-dimensional state space and W is the co-variation matrix involving diffusion coefficients (the denominator, 2, is absorbed into this). An example of F(x, s) is equation (2.6).
(e) Solution of the Fokker–Planck equation
Generally, the FPE is difficult to solve using analytic techniques. Exact solutions exist for only a limited number of models (see Knight et al. 2000). However, approximate analytic and numerical techniques (Risken 1996; Kloeden & Platen 1999; Knight 2000; de Kamps 2003; Sirovich 2003) offer ways of solving a general equation. We have chosen a solution based on transforming the PDE to a set of coupled odes (ordinary differential equations) and projecting onto a bi-orthogonal set. This results in an equal number of uncoupled equations that approximate the original PDE. In turn, this enables a dimension reduction of the original set of equations and an approximation of input-dependent solutions in terms of a known solution.
The dynamic operator, Q(s), is generally input-dependent and non-symmetric. By diagonalizing Q(s), we implicitly reformulate the density dynamics in terms of probability modes. Two sets of eigenvectors (right and left) are associated with the dynamic operator, forming a bi-orthogonal set. This set encodes modes or patterns of probability over state-space. The right-eigenvectors are column vectors of the matrix R, where QR=RD and left-eigenvectors are row vectors of matrix L, where LQ=DL. Both sets of eigenvectors share the same eigenvalues in the diagonal matrix D, which are sorted so that λ0>λ1>λ2…. The left eigenvector matrix is simply the generalized inverse of the right-eigenvector matrix. The number of eigenvectors and values, n, is equal to the dimensionality of Q. After normalization Q can be diagonalized,
| (2.16) |
For the time being, we will keep input constant, i.e. s=0. Projecting the probability density ρ(x,t) onto the space L generates an equivalent density μ, but within a different coordinate system. Conversely, R projects back to the original coordinate system,
| (2.17) |
Substituting Q=RDL and the right expression of equation (2.17) into equation (2.14) results in a diagonalized system, , which has the solution
| (2.18) |
where μ(0) is a vector of initial conditions. This solution can be framed in terms of independent modes, i.e. columns of R, each of which contributes linearly to the evolution of the density. The expression of each mode is given by the coefficients in μ, whose dynamics are governed by equation (2.21). The rate of exponential decay of the ith mode is characterized by its eigenvalue according to τi=−1/λi, where τi is its characteristic time-constant. The key thing here is that, in many situations, most modes decay rapidly to zero, i.e. have large negative eigenvalues. Their contribution to the dynamics at longer time-scales is therefore negligible. This is the rationale for reducing the dimension of the solution by ignoring them. Another advantage is that the equilibrium solution, i.e. the probability density that the system relaxes to (given constant input), is given by the principal mode, whose eigenvalue is zero (λ0). An approximate solution can then be written as
| (2.19) |
where ρ(x, 0) is the initial density profile, Rm and Lm are the principal m modes and Dm contains the first m eigenvalues, where m≤n. The benefit of an approximate solution is that computational demand is reduced in modelling the population dynamics.
(f) A time-dependent solution
The dynamic operator, Q(s), is input-dependent, and ideally needs calculating for each new input. This is time consuming and can be circumvented by using a perturbation expansion around a solution we already know, i.e. for s=0. Approximating Q(s) with a Taylor expansion about s=0
| (2.20) |
where Q(0) is evaluated at zero input and is a measure of its dependency on s. Substituting and using D(0)=LQ(0)R, we have the dynamic equation for the coefficients μ, in terms of the bi-orthogonal set of Q(0) (see Knight 2000 for a full treatment)
| (2.21) |
For multiple inputs
| (2.22) |
We are assuming here that the input can be treated as constant during a small interval. This approximation enters equation (2.19) to provide density change over this interval. is re-evaluated and the process repeated to give the time-dependent evolution of the population density.
(g) The observation function
Expectations over ρ(x, t) are generated by an observation function y=Mρ, where y corresponds to predicted measurements. An example is the average firing rate yr(t).
| (2.23) |
where ρ(T, t) is the marginal distribution over IST.
(h) Multiple populations: interaction through coupling
Interactions within a network of ensembles are modelled by coupling activities among populations. Coupled populations have been considered by Nykamp & Tranchina (2000) in modelling orientation tuning in the visual cortex. Coupling among populations, each described by standard Fokker–Planck dynamics, via mean field quantities (e.g. mean firing rate) induces nonlinearities, and thereby extends the network's dynamic repertoire. This can result in a powerful set of equations that have been used to model physical, biological and social phenomena (see Kuramoto 1984; Frank 2000 and others therein). To model influences among source and target populations, an additional input, which is a function of population activity, sc(y, t), is needed. The combined input is now
| (2.24) |
A simple example is self-feedback in one population, where yi(t) is the rate of firing per neuron, and g is a gain function, which here is a constant,
| (2.25) |
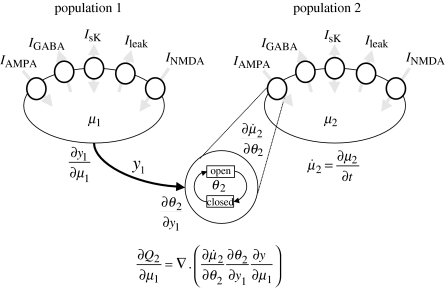
The total mean field now depends on the populations own activity, which has been described as a dynamic mean field (Omurtag et al. 2000). The notion can be extended to multiple populations, where g becomes a gain or coupling matrix. A schematic of two interacting populations of the model in equation (2.6) is shown in figure 4.
Figure 4.
Schematic of interacting sub-populations and terms used to estimate the coupling. The rate of change of density in μ2 over time, , depends on μ1 owing to coupling. The density in population 1 leads to a firing rate, y1, which modulates the synaptic channel-opening rate of population 2, parametrized by θ2, which in turn modulates . The coefficient of coupling is calculated using the chain rule.
Generally, coupling can be modelled as a modulation of the parameters of the target population (i) by inputs from the source (j). The effect of a small perturbation of these parameters is used to approximate the time-dependent network operator, which leads to a further term in equation (2.22). To a first-order approximation
| (2.26) |
The source-dependent change in the target operator follows from the chain rule,
| (2.27) |
where specifies the mechanism whereby synaptic input from the source population affects the biophysics of the target. The probability density over the source region μj causes a change in its output yj, which modulates synaptic channel-opening dynamics in the target region, parametrized by θi (e.g. θi=pAMPA). This leads to a change in the dynamics of the target region . These interactions among ensembles are scaled by the gain or coupling parameter gij, and are implemented by augmenting . Once equation (2.26) has been integrated, the output of the target region can be calculated,
| (2.28) |
It can be seen that self-coupling above is simply a special case of coupling in which g→gii.
3. Estimation and inference
The previous section has furnished a relatively simple dynamic model for measured electrophysiological responses reflecting population dynamics. This model can be summarized using equations (2.26) and (2.28)
| (3.1) |
This is a deterministic input-state-output system with hidden states μi controlling the expression of probability density modes of the ith population. Notice that the states no longer refer to biophysical or neuronal states (e.g. depolarization), but to the densities over states. The inputs are known deterministic perturbations s and the outputs are yi. The architecture and mechanisms of this system are encoded in its parameters. The objective is to estimate the conditional density of these parameters given some data. In the examples below we focus on the coupling parameters θ∈gij.
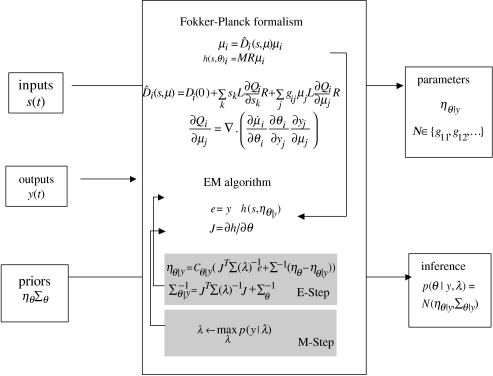
In this section, we review briefly the necessary inference and estimation procedure. This procedure has already been established for bilinear approximations to generative models of fMRI data and NMM of ERP. A full description can be found in our earlier papers. In brief, we use expectation maximization (EM) to identify the conditional moments of the parameters and the maximum likelihood estimates of the covariance components of the error term. This entails minimizing the variational free energy with respect to the conditional moments in an E-step and the covariance components in an M-step. This corresponds to a coordinate descent on the free energy (the negative log-likelihood of the model, plus the divergence between the estimated and true conditional density of the parameters). The conditional density depends on the prior density and the likelihood of the parameters, which are described next (see table 1).
(a) Prior assumptions
Priors have a dramatic impact on the landscape of the objective function (free energy) to be extremized: precise prior distributions ensure that the objective function has a global minimum that can be attained robustly. Under Gaussian assumptions, the prior distribution is defined by its mean and covariance. The mean ηθ corresponds to the prior expectation. The covariance Σθ encodes the amount of prior information about the parameters. A tight distribution (small variance) corresponds to precise prior knowledge. The log-prior density is
| (3.2) |
In our case, we can adopt very precise priors for biophysical constants, because these embody our knowledge of the intrinsic dynamics of neurons and synapses. The interesting parameters, which determine the large-scale functional architecture of the system, are the coupling parameters gij in equation (2.26). In the example below, we used relatively uninformative priors for these, and infinitely precise priors for the remaining parameters Σθ→0. In other words, we fix the remaining parameters to their prior expectations (see the values in table 2).
(b) Estimation and inference
Using the generative model of the §3a, the likelihood , where h(s,θ) is the predicted response, is easily computed under Gaussian assumptions about the error . λ are covariance parameters of the error. The vec operator arranges data from multiple channels and time bins into one large vector. This reflects the fact that we are treating the dynamic model as a static model that produces finite-length data sequences. We can do this because the model is, by design, deterministic.
The log-likelihood is based on the difference between the observed response and that predicted by integrating the generative model. This prediction is h(s,θ) (see Friston 2002 and Appendix B for details of this integration)
| (3.3) |
Having specified the prior and likelihood, the posterior or conditional expectation of the parameters can be computed using EM, where the conditional density .
The conditional moments (mean and covariance ) are updated iteratively using an expectation-maximization (EM) algorithm with a local linear approximation of equation (3.3) about the current conditional expectation. The E-step conforms to a Fisher-scoring scheme (Press et al. 1992) to update the conditional mean. In the M-step, the covariance parameters λ are updated to their maximum likelihood values, again using Fisher-scoring. The estimation scheme can be summarized as follows:
(i) Repeat until convergence
| (3.4) |
Under a Laplace or Gaussian approximation for the conditional density, the conditional covariance is an analytic function of the conditional expectation (Friston et al. 2002). See also figure 5.
Figure 5.
Bayesian estimation scheme summarizing the inputs, outputs and components of the generative model and its associated expectation maximization (EM) algorithm. The algorithm is given experimental inputs, e.g. experimental design, data and priors over parameters to be estimated (s(t), y(t), ηθ and Σθ). The forward model is used to predict the measured data, h(s,θ). The error and gradient, J, are used in an EM algorithm which performs a gradient descent on the free energy to optimize the conditional estimators of the parameters. Once this is achieved, inferences with regard to these estimates can be made.
Bayesian inference proceeds using the conditional or posterior density estimated by EM. Usually this involves specifying a parameter or compound of parameters as a contrast . Inferences about this contrast are made using its conditional covariance . For example, one can compute the probability that any contrast is greater than zero or some threshold, given the data. This inference is conditioned on the particular model specified. In other words, given the data and model, inference is based on the probability that a particular contrast is bigger than a specified threshold. In some situations, one may want to compare different models. This entails Bayesian model comparison.
4. Applications
In this section we illustrate the nature of the generative model of the previous sections and its use in making inferences about the functional architecture of neuronal networks. We focus first on a single population and the approximations entailed by dimension reduction. We then consider the coupling between two populations that comprise a simple network.
(a) Dynamics of a single population
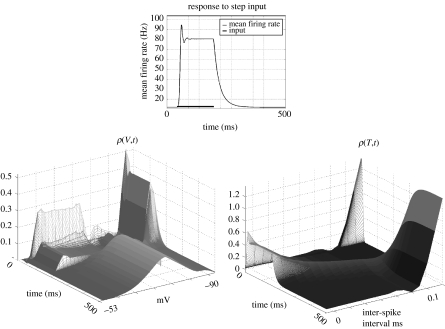
Figure 6 shows the response of a population of modified SIF neurons (cf. equation (2.5)) to an increase in exogenous input. The top figure shows the mean firing rate over time. The black bar indicates the duration of sustained input. The rate oscillates briefly before being damped, after which it remains constant at a new equilibrium-firing rate that is determined by the magnitude of the input. Once the input is removed, the rate decays to its background level. Below are two three-dimensional plots of the evolution of marginal distributions over V and T with time. The results were obtained by integrating equation (2.26) for a single population and single (boxcar) input, using a dynamic operator based on equation (2.5).
Figure 6.
Response of a single population of neurons (FPE based on the model dynamics of equation (2.5)) to a steep rise in input, i.e. a boxcar input. The top figure shows the mean firing rate per neuron within the ensemble. The horizontal bar indicates the duration of input. The population responds with an increase in firing that briefly oscillates before settling to a new equilibrium and returns to its original firing rate after the input is removed. Below are two three-dimensional images of the marginal distributions over V and T (left and right, respectively). Before input, the majority of probability over ρ(T, 0) is peaked close to 0.1 ms. However, input causes a shift in density towards shorter time-intervals, reflecting an increase in mean firing rate. This is also seen in the left figure, where input disperses the density towards the firing threshold and reset potential. After input is removed, both densities return to their prior distributions.
Just prior to input, there is very little probability mass at ISTs below 0.1 ms. This is seen as the large peak in ρ(T,t) at t=0 at the far right corner of the lower right figure. The inverse of the expected inter-spike interval corresponds to baseline-firing rate. After input, both distributions change dramatically. Density over the shorter ISTs increase as the population is driven to fire more frequently. This is also seen in ρ(V,t) (lower left), where density accumulates close to VT and VR, indicating a higher firing rate. These distributions return to their original disposition after the input returns to baseline.
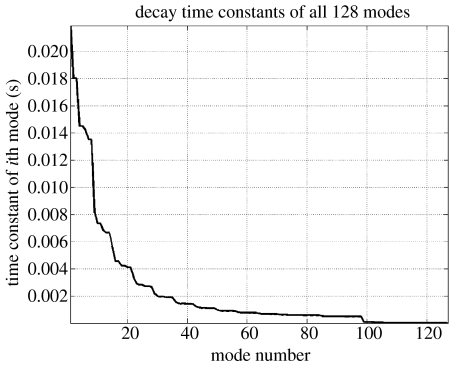
(b) Dimension reduction
The question now is how many probability modes are required to retain the salient aspects of these dynamics? An indication comes from the characteristic time-constants of each mode. These are shown for all modes, excluding the principle mode, which is stationary, in figure 7. The time-constants decrease rapidly with mode number. A comparison of approximations in terms of response to different levels of input is shown in figure 8. We considered approximations truncated at 16, 64 and 128 (, and , respectively) and the response curve for the deterministic model.
Figure 7.
Decay time-constants verses mode number. The model used in figure 6 was approximated using a system of 128 coupled ordinary differential equations (see text). This can be transformed into 128 uncoupled equations, where each equation describes the dynamics of a probability mode. The characteristic time to decay for each mode is shown, which illustrates a rapid decrease in decay time. Right most modes, i.e. short time constants, decay very rapidly and therefore will not contribute significantly to dynamics over a relatively longer time period. This is the rationale for approximating a solution by excluding modes with very short time constants.
First, the stochastic models exhibit a key difference in relation to the deterministic model, i.e. firing rate does not have an abrupt start at an input threshold. Instead, there is a finite probability of firing below threshold and the response curve tapers off with lower input. Second, the solution using all probability modes of the perturbation approximation compares well with D(s) computed explicitly at each time-point. The approximation is less accurate as input increases, however, it remains close to, and retains the character of, the true response curve. Third, the truncated approximation using 64 modes is almost indistinguishable from the full approximation. The solution using only 16 modes, despite loosing accuracy with larger inputs, still maintains some of the character of the response curve and, at low input levels, is a reasonable approximation. Given that this approximation represents an eightfold decrease in the number of modes, this degree of approximation is worth considering when optimizing the balance between computational efficiency and accuracy.
(c) Coupling among populations
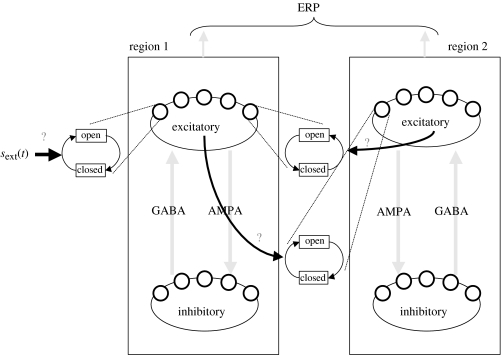
We next simulated a small network of populations. A schematic of the network is shown in figure 9. The model consisted of two identical regions, each containing sub-populations of excitatory and inhibitory neurons. The excitatory sub-population exerted its effect on the inhibitory through AMPA synaptic channels, while GABA channels mediated inhibition of the excitatory neurons. The regions were reciprocally connected, with region two being driven by one through fast excitatory AMPA channels, while region one was modulated by feedback from two, mediated by slow excitatory NMDA channels. Only region one was driven by external input. This conforms to a simple cortical hierarchy, with region two being supraordinate. These receptor-specific effects where specified by making non-zero for the rate of receptor-specific channel opening (see equation (2.6) and figure 9) and using the mean spike rate as the output yj from the source population. The dynamics of a population were approximated with 64 principal modes (see table 3).
Figure 9.
Schematic of the simulated network. Two regions were coupled via excitatory synaptic channels, and region one was driven by an external input. Each region was identical and composed of an excitatory and inhibitory population coupled via AMPA and GABA synaptic channels. Region two was driven by one via fast excitatory AMPA channels, while region one received excitatory feedback from two, mediated by slow NMDA channels. Local field potentials were modelled as measurements of mean membrane potential from the excitatory populations of both regions. Results from the simulation were used in an estimation scheme to identify the three coupling parameters indicated by question marks.
Table 3.
state variable ranges and number of bins used to grid state space.
| state variable | number of bins | range of values |
|---|---|---|
| V | 16 | [−92,−48] mV |
| T | 8 | [0,0.1] s |
| xsK | 4 | [0,1] a.u. |
| xAMPA | 2 | [0,1] a.u. |
| xGABA | 2 | [0,1] a.u. |
| xNMDA | 2 | [0,1] a.u. |
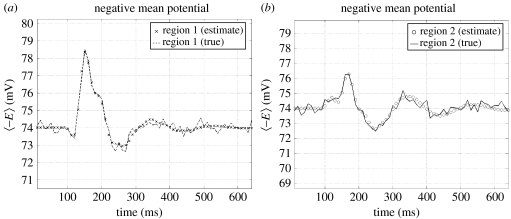
Event-related signals (mean depolarization of excitatory populations), generated by the network in response to an impulse of exogenous input, are shown in figure 10. These responses have early and late components, around 150 and 350 ms, respectively, which are characteristic of real evoked response potentials (ERPs).
Figure 10.
Comparison of simulated data (plus observation noise) and predicted responses after estimation (superimposed) of coupling parameters (see figure 11) from the network simulation. Mean membrane potential from the excitatory populations of each region (region one shown in a), in response to a brief input, are shown. The first 64 principal modes (out of 4096) were used to approximate the population dynamics of an ensemble.
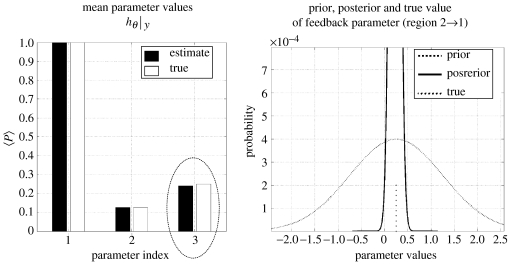
(d) Inverting the model to recover coupling parameters
Gaussian observation noise (approximately 10%) was added to the mean potentials from both regions to simulate data. The model was then inverted to estimate the known parameters using EM as described above. The predicted response of the generative model is compared with the synthetic data in figure 10. Three coupling parameters (mediating exogenous input to region one, from region one to two and from two to one—bold connections with question marks in figure 9) were given uninformative priors. These represent unknown parameters which the estimation scheme was trying to identify. Their conditional expectations (with prior and posterior densities of the feedback parameters) are shown with the true values in figure 11. They show good agreement and speak to the possibility of using this approach with real data.
Figure 11.
The conditional expectations of the three coupling parameters between input to region one, region one to two and backward coupling from two to one (parameter indices one to three, respectively) are shown next to the known parameters of the simulation. The prior, posterior and true value of the feedback parameter is shown on the right.
5. Discussion
The aim of this work was to establish the feasibility of using the FPE in a generative model for ERPs. The ensuing model embodies salient features of real neuronal systems; neurons are (i) dynamic units, (ii) driven by stochastic forces, (iii) organized into populations with similar biophysical properties and response characteristics and (iv) multiple populations interact to form functional networks. Despite the stochastic nature of neuronal dynamics, the FPE formulates the solution in terms of a deterministic process, where the dispersive effects of noise are modelled as a diffusive process. The motivation for using such a model is that its associated parameters have a clear biological meaning, enabling unambiguous and mechanistic interpretations.
We have reviewed well-known material on the integrate-and-fire model neuron with synaptic dynamics, which included fast excitatory AMPA, slow excitatory NMDA and inhibitory GABA mediated currents. The FPE was used to model the effect of stochastic input, or system noise, on population dynamics. Its time-dependent solution was approximated using a perturbation expansion about zero input. Decomposition into a bi-orthogonal set enabled a dimension reduction of the system of coupled equations, owing to the rapid decay of many of its probability modes. Interactions among populations were modelled as a change in the parameters of a target population that depended on the average state of source populations.
To show that the model produces realistic responses and, furthermore, it could be used as an estimation or forward model, separate ensembles were coupled to form a small network of two regions. The coupled model was used to simulate ERP data, i.e. mean potentials from excitatory sub-populations in each region. Signals were corrupted by Gaussian noise and subject to EM. Three parameters were estimated; input, forward and back connection strengths, and were shown to compare well to the known values. It is pleasing to note that the stochastic model produces signals that exhibit salient features of real ERP data and the estimation scheme was able to recover its parameters.
The key aspect of the approach presented here is the use of population density dynamics as a forward model of observed data. These models have been developed by Knight et al. and have been used to explore the cortical dynamics underlying orientation tuning in the visual cortex. Our hope is that these models may find a place in ERP and EEG data analysis. In these models, random effects are absorbed into the FPE and the population dynamics become deterministic. This is a critical point because it means system identification has only to deal with observation noise. Heuristically, the deterministic noise induced by stochastic effects is effectively ‘averaged away’ by measures like ERPs. However, the effect of stochastic influence is still expressed and modelled, deterministically, at the level of population dynamics.
There are many issues invoked by this modelling approach. The dimensionality of solutions for large systems can become extremely large in probability space. Given an N-dimensional dynamic system, dividing each dimension into M bins results in an approximation to the FPE with a total of MN ordinary differential equations. The model, used to simulate a network of populations, used 4096 equations to approximate the dynamics of one population. Dimension reduction, by using a truncated bi-orthogonal set is possible, however, as was demonstrated in figure 8, there is a trade-off between accuracy and dimension of the approximation. Generally, a more realistic model requires more variables, so there is a balance between biological realism and what we can expect from current computational capabilities.
The model neuron used in this paper is just one of many candidates. Much can be learnt from comparing models. For instance, is modelling the IST as a state variable an efficient use of dimensions? This may eschew the need for detailed boundary conditions, however, it may well be an extravagant use of dimensions given computational limitations. The current modelling approach is not limited to electrophysiological data. Any measurement which is coupled to electrical activity of a neuronal population could, in principle, also be included in the generative model, which could be used to combine measurements of electrical and metabolic origin, e.g. fMRI.
The use of Bayesian system identification enables the formal inclusion of additional information when estimating model parameters from data. We have not given the topic of priors much consideration in this paper, but it is an important issue. Analytic priors derived from stability or bifurcation analyses could be used to ensure parameter values which engender dynamics characteristic of the signals measured, i.e. stable fixed point, limit cycle or chaotic attractors. Empirical priors derived from data also have great potential in constraining system identification. A Bayesian framework also facilitates model comparison through quantifying the ‘evidence’ that a dataset has for a number of different models (Penny et al. 2004b).
Acknowledgments
We are very grateful to Pedro Valdes-Sosa and Rolf Kötter for their editorial guidance, two anonymous reviewers for their comments and the Wellcome Trust for funding this research.
Appendix A
Appendix A. Firing-rate of a deterministic, integrate-and-fire model-neuron
| (A1) |
where T(s) and f(s) are the period and frequency of firing. A plot of the last of these equations is shown in figure 8 to compare with the rate-response of a stochastic model. In the context of deterministic input, the neuron starts firing abruptly at threshold input and increases monotonically thereafter. Input threshold, ST, given by
| (A2) |
The current associated with the input s, in nano-ampere, is given by the relationship Is=s/gLRm, where total membrane resistance is Rm (see Dayan & Abbott 2001).
Appendix B
Appendix B. Derivation of population equation in terms of vector fields
For an alternative perspective on equation (2.11) we can think of the density dynamics in terms of scalar and vector fields. The probability density, ρ(x,t), is a scalar function which specifies the probability mass at x. This quantity will not be static if acted on by a force, whose influence is quantified by a vector field over x. This field represents the flow of probability mass within state space. The net flux within a volume dx is given by the divergence of the vector field ∇·J (where ∇ is the divergence operator). The net flux is a scalar operator, as it specifies how much the density at x changes per unit time. It contains all the information needed to determine the rate of change of probability mass with time:
| (B1) |
The negative sign ensures that probability mass flows from high to low densities. The two forces in our simplified model are the leakage current and excitatory input. The former moves a neuron towards its equilibrium potential, EL, while excitatory input drives V towards VT. Each force generates its own field of flux in state space, which are in opposite directions. The overall flux is the combination.
| (B2) |
The first term is the flux owing to leakage current, while the second is owing to input. Both are functions of probability density, which have the following form:
| (B3) |
Substitution of these expressions into (B 2) and (B 1) leads to equation (2.11) in the main text. This mathematical equivalence illustrates the convergence of different perspectives on this formulation of population dynamics.
Appendix C
Appendix C. Numerical solution of Fokker–Planck equation
The equation to solve is:
| (C1) |
First, grid state-space xi=ih, where i=1,…,N. Evaluate at all grid points. Calculate the operators −∇·(f+s) and w2/2∇2, where ∇2 is the Laplacian operator, required to construct an approximation to the dynamic operator Q
Equation (C 1) is a system of coupled differential equations that can be solved at discrete time points, where time is tn=nΔt (n=0,…,T).
| (C2) |
These are de-coupled using the eigenvectors and values of Q.
After discretizing state-space and approximating for one population, the eigenvectors and values were calculated using the Matlab function ‘eig’ and saved. Given these, a reduced or full model can be used to model a network of populations by specifying the connectivity among ensembles. The exponential matrix of the reduced or full model was calculated using the Matlab function ‘expm’ (see Moler & Van Loan 2003). This uses a (6, 6) Pade approximation of order 12. Explicit and implicit numerical integration schemes can be reformulated into a Pade approximation, e.g. (0, 1) approximation of order 1 is a forward Euler scheme, whereas (1, 1) approximation of order 2 is a Crank–Nicolson implicit scheme. As the ‘expm’ function uses an approximation of order 12 and is implicit, the scheme is accurate and unconditionally stable (see Smith 1985).
Footnotes
One contribution of 21 to a Theme Issue ‘Multimodal neuroimaging of brain connectivity’.
References
- Abeles M. Cambridge University Press; Cambridge: 1991. Corticonics: neural circuits of the cerebral cortex. [Google Scholar]
- Aertsen A, Gerstein G.L, Habib M.K, Palm G. Dynamics of neuronal firing correlation: modulation of “effective connectivity”. J. Neurophysiol. 1989;61:900–917. doi: 10.1152/jn.1989.61.5.900. [DOI] [PubMed] [Google Scholar]
- Buchel C, Friston K.J. Dynamic changes in effective connectivity characterized by variable parameter regression and Kalman filtering. Hum. Brain Mapp. 1998;6:403–408. doi: 10.1002/(SICI)1097-0193(1998)6:5/6<403::AID-HBM14>3.0.CO;2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchel C, Coull J.T, Friston K.J. The predictive value of changes in effective connectivity for human learning. Science. 1999;283:1538–1541. doi: 10.1126/science.283.5407.1538. [DOI] [PubMed] [Google Scholar]
- Casti A.R, Omurtag A, Sornborger A, Kaplan E, Knight B, Victor J, Sirovich L. A population study of integrate-and-fire-or-burst neurons. Neural Comput. 2002;14:957–986. doi: 10.1162/089976602753633349. [DOI] [PubMed] [Google Scholar]
- Churchland P.S, Sejnowski T.J.The computational brain1994MIT; Cambridge, MA [Google Scholar]
- David O, Friston K.J. A neural mass model for MEG/EEG: coupling and neuronal dynamics. NeuroImage. 2003;20:1743–1755. doi: 10.1016/j.neuroimage.2003.07.015. [DOI] [PubMed] [Google Scholar]
- David O, Cosmelli D, Friston K.J. Evaluation of different measures of functional connectivity using a neural mass model. NeuroImage. 2004;21:659–673. doi: 10.1016/j.neuroimage.2003.10.006. [DOI] [PubMed] [Google Scholar]
- David, O., Kiebel, S., Harrison, L., Mattout, Kilner, Friston, K. Submitted. Dynamic causal modelling of evoked responsed in EEG and MEG. NeuroImage. [DOI] [PubMed]
- Dayan P. Computational modelling. Curr. Opin. Neurobiol. 1994;4:212–217. doi: 10.1016/0959-4388(94)90075-2. [DOI] [PubMed] [Google Scholar]
- Dayan P, Abbott L. MIT Press; Cambridge, MA: 2001. Theoretical neuroscience. [Google Scholar]
- de Kamps M. A simple and stable numerical solution for the population density equation. Neural Comput. 2003;15:2129–2146. doi: 10.1162/089976603322297322. [DOI] [PubMed] [Google Scholar]
- Frank T.D. 2000. Stochastic properties of human motor control: nonlinear Fokker–Planck equations. Ph.D. thesis, Amstelveen, Netherlands. [Google Scholar]
- Frank T.D, Daffertshofer A, Beek P.J. Multivariate Ornstein–Uhlenbeck processes with mean-field dependent coefficients: application to postural sway. Phys. Rev. E. Stat. Nonlin. Soft Matter Phys. 2001;63:011 905. doi: 10.1103/PhysRevE.63.011905. [DOI] [PubMed] [Google Scholar]
- Freeman W.J. Models of the dynamics of neural populations. Electroencephalogr. Clin. Neurophysiol. 1978;(Suppl. 9–18) [PubMed] [Google Scholar]
- Friston K.J. Brain function, nonlinear coupling, and neuronal transients. Neuroscientist. 2001;7:406–418. doi: 10.1177/107385840100700510. [DOI] [PubMed] [Google Scholar]
- Friston K.J. Bayesian estimation of dynamical systems: an application to fMRI. NeuroImage. 2002;16:513–530. doi: 10.1006/nimg.2001.1044. [DOI] [PubMed] [Google Scholar]
- Friston K.J, Buchel C. Attentional modulation of effective connectivity from V2 to V5/MT in humans. Proc. Natl Acad. Sci. USA. 2000;97:7591–7596. doi: 10.1073/pnas.97.13.7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston K.J, Frith C.D, Frackowiak R.S.J. Time-dependent changes in effective connectivity measured with PET. Hum. Brain Mapp. 1993;1:69–79. [Google Scholar]
- Friston K.J, Ungerleider L.G, Jezzard P, Turner R. Characterizing modulatory interactions between areas V1 and V2 in human cortex: a new treatment of functional MRI data. Hum. Brain Mapp. 1995;2:211–224. [Google Scholar]
- Friston K.J, Buechel C, Fink G.R, Morris J, Rolls E, Dolan R.J. Psychophysiological and modulatory interactions in neuroimaging. NeuroImage. 1997;6:218–229. doi: 10.1006/nimg.1997.0291. [DOI] [PubMed] [Google Scholar]
- Friston K.J, Penny W, Phillips C, Kiebel S, Hinton G, Ashburner J. Classical and Bayesian inference in neuroimaging: theory. NeuroImage. 2002;16:465–483. doi: 10.1006/nimg.2002.1090. [DOI] [PubMed] [Google Scholar]
- Friston K.J, Harrison L, Penny W. Dynamic causal modelling. NeuroImage. 2003;19:1273–1302. doi: 10.1016/s1053-8119(03)00202-7. [DOI] [PubMed] [Google Scholar]
- Gerstner W, Kistler W. Cambridge University Press; Cambridge: 2002. Spiking neuron models. single neurons, populations, plasticity. [Google Scholar]
- Haken H. B. G. Teubner; Stuttgart: 1973. Synergetics: cooperative phenomena in multi-component systems. [Google Scholar]
- Haken H. Springer; Berlin: 1996. Principles of brain function. [Google Scholar]
- Harrison L, Penny W.D, Friston K. Multivariate autoregressive modeling of fMRI time series. NeuroImage. 2003;19:1477–1491. doi: 10.1016/s1053-8119(03)00160-5. [DOI] [PubMed] [Google Scholar]
- Haskell E, Nykamp D.Q, Tranchina D. Population density methods for large-scale modelling of neuronal networks with realistic synaptic kinetics: cutting the dimension down to size. Network. 2001;12:141–174. [PubMed] [Google Scholar]
- Jansen B.H, Rit V.G. Electroencephalogram and visual evoked potential generation in a mathematical model of coupled cortical columns. Biol. Cybern. 1995;73:357–366. doi: 10.1007/BF00199471. [DOI] [PubMed] [Google Scholar]
- Jirsa V.K. Connectivity and dynamics of neural information processing. Neuroinformatics. 2004;2:183–204. doi: 10.1385/NI:2:2:183. [DOI] [PubMed] [Google Scholar]
- Jirsa V.K, Haken H. Field theory of electromagnetic brain activity. Phys. Rev. Lett. 1996;77:960–963. doi: 10.1103/PhysRevLett.77.960. [DOI] [PubMed] [Google Scholar]
- Kloeden, Platen . Springer; Berlin: 1999. Numerical solution of stochastic differential equations. [Google Scholar]
- Knight B.W. The relationship between the firing rate of a single neuron and the level of activity in a population of neurons. Experimental evidence for resonant enhancement in the population response. J. Gen. Physiol. 1972;59:767–778. doi: 10.1085/jgp.59.6.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight B.W. Dynamics of encoding in a population of neurons. J. Gen. Physiol. 1972;59:734–766. doi: 10.1085/jgp.59.6.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight B.W. Dynamics of encoding in neuron populations: some general mathematical features. Neural Comput. 2000;12:473–518. doi: 10.1162/089976600300015673. [DOI] [PubMed] [Google Scholar]
- Knight B.W, Omurtag A, Sirovich L. The approach of a neuron population firing rate to a new equilibrium: an exact theoretical result. Neural Comput. 2000;12:1045–1055. doi: 10.1162/089976600300015493. [DOI] [PubMed] [Google Scholar]
- Kuramoto Y. Springer; Berlin: 1984. Chemical oscillations, waves and turbulence. [Google Scholar]
- Lopes da Silva F.H, van Rotterdam A, Barts P, van Heusden E, Burr W. Models of neuronal populations: the basic mechanisms of rhythmicity. Prog. Brain Res. 1976;45:281–308. doi: 10.1016/S0079-6123(08)60995-4. [DOI] [PubMed] [Google Scholar]
- Mattout J, Pelegrini-Issac M, Bellio A, Daunizeau J, Benali H. Localization estimation algorithm (LEA): a supervised prior-based approach for solving the EEG/MEG inverse problem. Inf. Process Med. Imaging. 2003;18:536–547. doi: 10.1007/978-3-540-45087-0_45. [DOI] [PubMed] [Google Scholar]
- Moler C, Van Loan C. Nineteen dubious ways to compute the exponential of a matrix, twenty five years later. SIAM Rev. 2003;45:3–49. [Google Scholar]
- Nunez P. The brain wave equation: a model for the EEG. Math. Biosci. 1974;21:279. [Google Scholar]
- Nykamp D.Q, Tranchina D. A population density approach that facilitates large-scale modeling of neural networks: analysis and an application to orientation tuning. J. Comput. Neurosci. 2000;8:19–50. doi: 10.1023/a:1008912914816. [DOI] [PubMed] [Google Scholar]
- Nykamp D.Q, Tranchina D. A population density approach that facilitates large-scale modeling of neural networks: extension to slow inhibitory synapses. Neural Comput. 2001;13:511–546. doi: 10.1162/089976601300014448. [DOI] [PubMed] [Google Scholar]
- Omurtag A, Knight B.W, Sirovich L. On the simulation of large populations of neurons. J. Comput. Neurosci. 2000;8:51–63. doi: 10.1023/a:1008964915724. [DOI] [PubMed] [Google Scholar]
- Penny W.D, Stephan K.E, Mechelli A, Friston K.J. Modelling functional integration: a comparison of structural equation and dynamic causal models. NeuroImage. 2004;23(Suppl. 1):264–274. doi: 10.1016/j.neuroimage.2004.07.041. [DOI] [PubMed] [Google Scholar]
- Penny W.D, Stephan K.E, Mechelli A, Friston K.J. Comparing dynamic causal models. NeuroImage. 2004;22:1157–1172. doi: 10.1016/j.neuroimage.2004.03.026. [DOI] [PubMed] [Google Scholar]
- Pfurtscheller G. Functional brain imaging based on ERD/ERS. Vision Res. 2001;41:1257–1260. doi: 10.1016/s0042-6989(00)00235-2. [DOI] [PubMed] [Google Scholar]
- Pfurtscheller G, Lopes da Silva F.H. Event-related EEG/MEG synchronization and desynchronization: basic principles. Clin. Neurophysiol. 1999;110:1842–1857. doi: 10.1016/s1388-2457(99)00141-8. [DOI] [PubMed] [Google Scholar]
- Phillips C, Mattout J, Rugg M.D, Maquet P, Friston K.J. An empirical Bayesian solution to the source reconstruction problem in EEG. NeuroImage. 2005;24:997–1011. doi: 10.1016/j.neuroimage.2004.10.030. [DOI] [PubMed] [Google Scholar]
- Press W.H, Teukolsky S.A, Vetterling W.T, Flannery B.P. Cambridge University Press; Cambridge, MA, USA: 1992. Numerical recipes in C. [Google Scholar]
- Rennie C.J, Robinson P.A, Wright J.J. Unified neurophysical model of EEG spectra and evoked potentials. Biol. Cybern. 2002;86:457–471. doi: 10.1007/s00422-002-0310-9. [DOI] [PubMed] [Google Scholar]
- Risken H. Springer; Berlin: 1996. The Fokker–Planck equation. [Google Scholar]
- Robinson P.A, Rennie C.J, Rowe D.L, O'Connor S.C, Wright J.J, Gordon E, Whitehouse R.W. Neurophysical modeling of brain dynamics. Neuropsychopharmacology. 2003;28(Suppl. 1):S74–S79. doi: 10.1038/sj.npp.1300143. [DOI] [PubMed] [Google Scholar]
- Sirovich L. Dynamics of neuronal populations: eigenfunction theory; some solvable cases. Network. 2003;14:249–272. [PubMed] [Google Scholar]
- Smith G. Clarendon Press; Oxford: 1985. Numerical solutions of partial differential equations: finite difference methods. [Google Scholar]
- Valdes P.A, Jimenez J.C, Riera J, Biscay R, Ozaki T. Nonlinear EEG analysis based on a neural mass model. Biol. Cybern. 1999;81:415–424. doi: 10.1007/s004220050572. [DOI] [PubMed] [Google Scholar]
- Wiesenfeld K, Moss F. Stochastic resonance and the benefits of noise: from ice ages to crayfish and SQUIDs. Nature. 1995;373:33–36. doi: 10.1038/373033a0. [DOI] [PubMed] [Google Scholar]
- Wilson H.R, Cowan J.D. Excitatory and inhibitory interactions in localized populations of model neurons. Biophys. J. 1972;12:1–24. doi: 10.1016/S0006-3495(72)86068-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler I, Czigler I, Jaramillo M, Paavilainen P, Naatanen R. Temporal constraints of auditory event synthesis: evidence from ERPs. NeuroReport. 1998;9:495–499. doi: 10.1097/00001756-199802160-00025. [DOI] [PubMed] [Google Scholar]
- Winkler I, Schroger E, Cowan N. The role of large-scale memory organization in the mismatch negativity event-related brain potential. J. Cogn. Neurosci. 2001;13:59–71. doi: 10.1162/089892901564171. [DOI] [PubMed] [Google Scholar]
- Wright J.J, Robinson P.A, Rennie C.J, Gordon E, Bourke P.D, Chapman C.L, Hawthorn N, Lees G.J, Alexander D. Toward an integrated continuum model of cerebral dynamics: the cerebral rhythms, synchronous oscillation and cortical stability. Biosystems. 2001;63:71–88. doi: 10.1016/s0303-2647(01)00148-4. [DOI] [PubMed] [Google Scholar]
- Wright J.J, Rennie C.J, Lees G.J, Robinson P.A, Bourke P.D, Chapman C.L, Gordon E, Rowe D.L. Simulated electrocortical activity at microscopic, mesoscopic, and global scales. Neuropsychopharmacology. 2003;28(Suppl. 1):S80–S93. doi: 10.1038/sj.npp.1300138. [DOI] [PubMed] [Google Scholar]