Abstract
Immunogold staining and electron microscopy show that IL-2 receptor α-subunits exhibit nonrandom surface distribution on human T lymphoma cells. Analysis of interparticle distances reveals that this clustering on the scale of a few hundred nanometers is independent of the presence of IL-2 and of the expression of the IL-2R β-subunit. Clustering of IL-2Rα is confirmed by confocal microscopy, yielding the same average cluster size, ≈600–800 nm, as electron microscopy. HLA class I and II and CD48 molecules also form clusters of the same size. Disruption of cholesterol-rich lipid rafts with filipin or depletion of membrane cholesterol with methyl-β-cyclodextrin results in the blurring of cluster boundaries and an apparent dispersion of clusters for all four proteins. Interestingly, the transferrin receptor, which is thought to be located outside lipid rafts, exhibits clusters that are only 300 nm in size and are less affected by modifying the membrane cholesterol content. Furthermore, transferrin receptor clusters hardly colocalize with IL-2Rα, HLA, and CD48 molecules (crosscorrelation coefficient is 0.05), whereas IL-2Rα colocalizes with both HLA and CD48 (crosscorrelation coefficient is between 0.37 and 0.46). This coclustering is confirmed by electron microscopy. The submicron clusters of IL-2Rα chains and their coclustering with HLA and CD48, presumably associated with lipid rafts, could underlie the efficiency of signaling in lymphoid cells.
Keywords: IL-2 receptor, HLA glycoproteins, transferrin receptor, receptor clustering, electron microscopy
Cytokines regulating immune responses have their specific private receptor but may also share public receptors with other cytokines. IL-2 secreted by T lymphocytes when stimulated with antigen or mitogens is essential for T cell growth (1, 2). The private receptor for IL-2 is the IL-2Rα-subunit, exhibiting relatively low affinity for IL-2 compared with the IL-2Rαβγ heterotrimer, which is considered a fully functional receptor (3). We have recently shown in a fluorescence resonance energy transfer study that the IL-2Rα, β, and γ-subunits are preassembled even on the surface of unstimulated Kit 225 K6 T lymphoma cells and cannot, therefore, be considered a transient signaling assembly (4). It is still unclear how IL-2Rα is recruited to the less abundant β and γ chains to form the functionally active receptor.
Similar assemblies (nonrandom colocalization) of cell-surface antigens and receptors have been reported previously for lymphoid cells, as reviewed in ref. 5. Such supramolecular formations on the nanometer level have been explored primarily by using flow cytometric energy transfer (6), joined by other mostly fluorescence-based techniques that assess lateral or rotational mobility of membrane proteins or assemblies thereof (7–9). Possibly functional protein-association patterns were discovered, including the di/oligomerization of HLA I and II molecules on activated T cells and lymphoid cell lines (10–13), the heteroassociation between HLA I and HLA II glycoproteins (14), or HLA I and the IL-2Rα-subunit (15). These observations argue against independent freely moving membrane proteins postulated by the fluid mosaic membrane model; instead, a segregated “corralled” structure may be the valid hypothesis, with specific molecules confined to specific regions (5).
Various studies directed at the plasma membrane have provided evidence for the existence of such distinct domains in the submicron range (12, 16–18); for a most recent overview, see ref. 19. From the biochemical point of view, these domains appear as detergent insoluble/resistant glycolipid-enriched membrane domains [DRMs, DIGs, or GEMs (20)] and are often termed lipid rafts (21). Physically, they are expected to be represented by cell-surface patches found for both lipid and protein molecules (22). The physical and chemical forces giving rise to such domains are under intensive investigation (8, 23). One presumes that several intracellular, extracellular, and intramembrane constraints and forces influence the size and distribution of these clusters, one being the cholesterol content of the membrane area in question (21, 24). There is indeed evidence that changing the cholesterol composition of the cell membrane alters the association pattern and signaling properties of various molecules (24, 25). Such a change can be brought about by treating the cell membrane with filipin, a polyene antibiotic specifically complexing cholesterol (26), or by extracting cholesterol from the membrane by methyl-β-cyclodextrin (27).
The physiological significance of the lateral microdomain organization of biological membranes is not clear yet. One can assume that a larger-scale local accumulation of receptors and their signal transduction machinery (28, 29) may enhance the efficiency of transmembrane signaling by providing a focusing effect. We have previously shown patchy aggregation of platelet-derived growth factor receptors on glioblastoma cells (18) as well as assemblies of up to 1,000 erbB2 molecules on various mammary tumor cell lines (30). Also, in addition to the molecular association of MHC class I and class II glycoproteins on lymphoma cells, we have observed their submicron-scale clusters and coclusters (12, 16). Given this knowledge, as well as the evidence that MHC class I glycoproteins are in the molecular vicinity of IL-2Rα on some cell lines (15), in the present study we have undertaken to investigate the higher-order clustering of IL-2Rα on Kit 225 K6 and MT-1 lymphoma cells and the possible heteroassociation of IL-2Rα with MHC glycoproteins on the submicron–micron scale. To shed light on the possible role of lipid rafts (19, 21) in organizing these receptor assemblies, we have examined the colocalization of IL-2Rα and MHC molecules with CD48, a glycosylphosphatidylinositol-linked T cell membrane protein reported to associate with rafts (20) and with the transferrin receptor (TrfR) that is excluded from glycosphingolipid-enriched membrane microdomains (31, 32). Furthermore, we have tested the influence of membrane cholesterol composition on the submicron-scale clustering of these molecules by extracting cholesterol with methyl-β-cyclodextrin or complexing cholesterol in situ with filipin.
Materials and Methods
Cell Culture and Treatment.
Kit 225 K6 and MT-1 T cell lines were cultured in RPMI-1640 medium (5% CO2) supplemented with 10% FCS and antibiotics. To maintain the growth of Kit 225 K6 cells, 20 units/ml recombinant IL-2 was added every 48 h. Resting cells were obtained by culturing for 72 h in IL-2-free medium. Filipin III (Sigma; 0.1 mg/ml) was used to complex cholesterol in the cell membrane. Incubation (106 cells/ml) for 1 h at 37°C was followed by washing twice in PBS. Cholesterol extraction was achieved by treating 106 cells/ml with 7 mM methyl-β-cyclodextrin (Sigma) for 1 h at 37°C, and its efficiency was assessed from the decrease of fluorescence polarization of the trimethylamine-diphenylhexatriene membrane probe (25).
mAbs.
The IL-2Rα-subunit, MHC class I and II were labeled with αTac, W6/32, and L-243, respectively (4, 12). Anti-CD48 (MEM102) and anti-TrfR (MEM75) were a generous gift of V. Horejsi (Institute of Molecular Genetics, Academy of Science, Czech Republic). Fab fragments were prepared from mAbs as previously described (33). Whole mAbs or Fab fragments were conjugated with fluorescein or rhodamine succinimidyl esters having long linkers (XF or XR, Molecular Probes) or sulfoindocyanine–succinimidyl ester (Cy3, monofunctional, Amersham Pharmacia) as described earlier (14, 15).
Labeling of Cells with mAbs.
Cells (106/ml) suspended in ice-cold PBS were labeled in the dark for 40 min at 4°C. Optimal antibody concentrations were determined from saturation curves. Antibodies were air fuged (20,000 × g, 30 min) before labeling. Labeled cells were washed in cold PBS and either fixed with 1% formaldehyde, used unfixed immediately, or further processed for electron microscopy.
Labeling of Cells with Colloidal Gold for Electron Microscopy.
Labeling with the first Fab (see above) was followed by incubation with polyclonal secondary antibodies conjugated to gold beads of 10- or 30-nm diameter (Aurogamig G-10, against the heavy and light chains, or Aurogamig G-30 against the Fc fragment; from Amersham Pharmacia) for 40 min. After washing with PBS (10 min, 250 × g) the cells were fixed with 2% paraformaldehyde for 1 h and then with 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) over night on ice. For sequential double labeling, the first antibody was a Fab followed by Aurogamig G-10, and then the unused binding sites of the polyclonal antibodies were blocked for 5 min with the Fab fragments used for primary labeling. mAbs against the second target epitope were whole antibodies that were tagged with Aurogamig G-30 against the Fc fragment. These beads will label only that fraction of the second target epitope that is not hidden by the first cohort of beads tagging the first target epitope (16).
Electron Microscopy.
After immunogold labeling, cells were spread on poly-l-lysine-coated Formvar grids, dehydrated in ascending ethanol series, and air dried from ether. Gold beads were counted in a JEOL electron microscope (JEM 100 B microscope operated at 100 kV) on the periphery and thinner parts of cells, where transparency allowed a good contrast.
Calculation of Actual and Expected Cell-Surface Distribution of Gold-Labeled Antigens.
For a random distribution of the gold particles, the statistics of the number of beads per unit area should be Poissonian (12, 16). The average density of gold particles was calculated from their number and the area over which they were distributed. The area assigned to one bead on average was taken as the unit, and thus the parameter λ of the assumed Poisson distribution was taken as 1. The number of beads counted in each unit area on the cell surface was used to calculate the actual distribution.
Comparison of Characteristic Interparticle Distances of Colloidal Gold Labels.
Scanned images of electron micrographs were processed by using a custom-written program developed in the labview (National Instruments, Austin, TX) environment. The coordinates of recognized labeling particles were used to produce the distribution of all interparticle distances in the sample.
Confocal Microscopy.
Cells were labeled with fluorescent Fab fragments, fixed in 1% formaldehyde or left unfixed in control experiments, and attached to poly-l-lysine-coated slides. A Zeiss LSM 420 laser scanning confocal microscope was used for measurements. Cy3 and XR were excited at 543 nm, XF at 488 nm. For double-labeled samples in crosscorrelation studies, a 510- to 525-nm narrow bandpass emission filter was used to detect XF fluorescence instead of the usual 510 nm long pass. Confocal sections (512 × 512 pixel, 0.6 μm thin) were obtained with a pinhole setting of 25, ×5–8 zoom, through a ×100 (numerical aperture = 1.3) objective. The intensity distribution of surface labeling was generated from the three-dimensional reconstruction of sections by using a projection algorithm in NIH image (National Institutes of Health, Bethesda, MD). In addition to the reconstructed free surfaces of cells, single confocal sections of each cell, flattened against the glass slide, were also analyzed.
Determination of Cluster Size from Confocal Images.
Average cluster size was determined from both projected surface distributions and flattened single confocal images by using the two-dimensional autocorrelation function
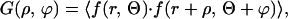 |
1 |
where the angle brackets indicate summation over the whole domain of the ρ radius and ϕ angle. The autocorrelation image was calculated by taking the inverse Fourier transform of the two-dimensional power spectrum matrix of the original images. Because in our case we do not expect the distribution to be anisotropic, G(ρ,ϕ) is independent of ϕ. Consequently, an angle-invariant autocorrelation function G(ρ) can be generated by averaging G(ρ,ϕ) over the range 0 ≤ ϕ < 2π. G(ρ) is fitted to the equation
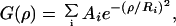 |
2 |
where the Ri characteristic radii serve as an adequate measure of the mean size (half-width at the 1/e height of a Gaussian distribution) of each class of clusters distinguishable on the basis of its size (22). Calculation of G(ρ) and fitting Eq. 3 by using the Levenberg–Marquardt algorithm were performed with a custom-written program developed in the labview environment. Using two exponentials gave a good fit with small residuals. The smaller value of Ri, generally in the 100- to 500-nm range, was taken as the radius of submicron-sized clusters, in coherence with individual cluster sizes measured directly on the images. The larger value of Ri, several microns in magnitude, was assumed to be characteristic for background fluctuation (30).
Determining Colocalization from Image Crosscorrelation.
Colocalization of pairs of cell-surface antigens was determined from confocal images of double-labeled cells. For a pair of images x and y, the crosscorrelation coefficient was calculated as
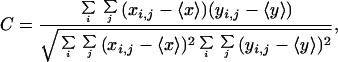 |
3 |
where xi,j and yi,j are fluorescence pixel values at coordinates i, j in images x and y. Only those pixels were used for the summation that were above detection threshold in both images. The theoretical maximum is C = 1 for identical images, and a value close to 0 implies disparate localization of the label. A program in labview was written to register and threshold image pairs and compute the crosscorrelation coefficient.
Results
Immunogold Labeling and Electron Microscopy Reveal a Submicron-Level Clustering of IL-2Rα-Subunits on K6 Cells.
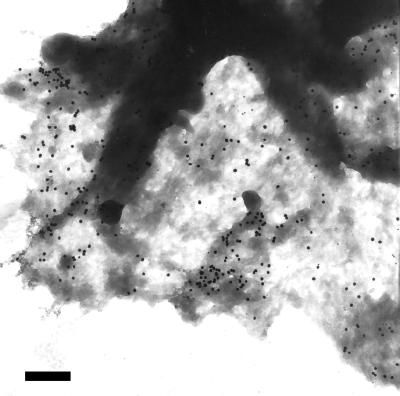
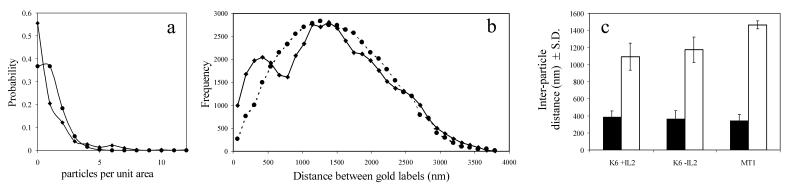
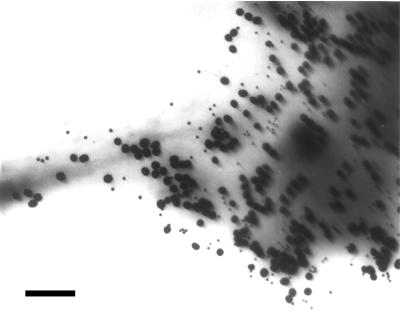
Fig. 1 shows immunogold-labeled IL-2Rα on Kit225 K6 cells. Clusters of several gold beads can be observed in addition to singly placed labels and larger areas with no label at all. Counting the labels, we constructed the actual probability distribution (Fig. 2a, diamonds) and compared it to the theoretical Poissonian (Fig. 2a, closed circles). It is clear that there are a larger number of unit areas with no label than that expected for a Poissonian, and that unit areas with four or more gold labels are more abundant as well. Because of this disproportion, unit areas close to the expected Poissonian parameter (i.e., those with one, two, and three labels) are lesser in number than predicted for a random distribution. The observed and expected distributions are different beyond a confidence level of 99.99 by using the χ2 test. Thus, the localization of labels follows a nonrandom distribution, manifesting as clustering on the submicron scale.
Figure 1.
Nonrandom distribution of IL-2Rα on Kit 225 K6 cells revealed by colloidal gold labels. Kit 225 K6 cells were labeled with anti-Tac as primary antibody and then with 30 nm colloidal gold conjugated to the secondary antibodies. A representative electron micrograph of the periphery of a cell is shown. The distribution of gold labels appears to be nonrandom. Bar = 470 nm.
Figure 2.
Quantitative analysis of the distribution of gold labels on IL-2Rα-subunits. (a) Gold labels shown in Fig. 1 were counted (n = 406), and the unit area was defined such that the expected value of gold labels per unit area was one. The image was divided into equal squares of one unit area each and the actual distribution of labels among the squares determined. The probability distribution of the particle density per unit area is plotted for the actual finding (♦) and compared to a Poisson distribution with parameter λ = 1(●). (b) The coordinates of all labels in Fig. 1 were determined, and the distribution of all interparticle distances was plotted (♦). A model distribution was also generated assuming Poissonian statistics (dashed line, ●). In contrast to the single peak of the expected random distribution, the measured distribution has two peaks. The first peak, around 400 nm, represents the characteristic distance of gold labels within clusters. (c) Characteristic distances for gold labels determined as in b are plotted for Kit 225 K6 (K6 + IL2), IL-2-starved Kit 225 K6 (K6-IL2), and MT-1 cells. Characteristic distances within clusters are represented by filled columns, and average distances within the whole sample area are shown with open columns. Data are mean ± SD from six independent experiments.
Quantitative Assessment of Interparticle Distances Reveals No Difference in the Higher-Level Clustering of IL-2Rα on IL-2 Fed and Starved K6 Cells and MT-1 Cells.
Because the receptor clusters are not expected to be anisotropic, the calculation of a relevant autocorrelation function that can be used to quantify cluster size simplifies to constructing the distribution histogram of all interparticle distances, without regard to the direction of localization. Such a distribution histogram is presented in Fig. 2b (diamonds) for the sample shown in Fig. 1. It is compared to a simulated distribution of the same number of particles randomly scattered over the same area (closed circles). The actual distribution is comprised of two peaks, whereas the randomly generated particle pattern shows a single Poissonian peak. The first peak of the actual distribution likely corresponds to the average interparticle distance within the small clusters of gold beads, whereas the second peak characterizes the average distance to labels outside the cluster. These average distances describe quantitatively the submicron-level receptor patterns on various T cells. In Fig. 2c, we see that in the case of Kit 225 K6 cells, the distance distribution is not influenced by IL-2 deprivation. Furthermore, the average distances within the clusters are the same in the case of MT-1 cells as for the K6 line.
Confocal Laser-Scanning Microscopy of Hydrated Samples Confirms the Presence of Submicron IL-2Rα Clusters on MT-1 and Kit225 K6 Cells.
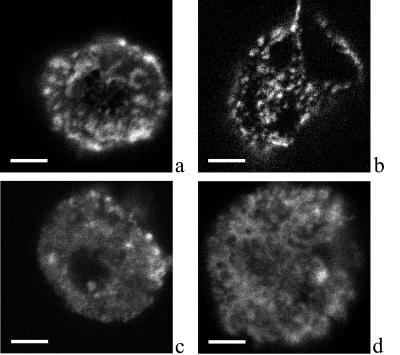
Confocal laser-scanning microscopy was used to confirm the presence of submicron IL-2Rα clusters on MT-1 and K6 cells. Cells were labeled on ice with Fab fragments to visualize receptors without inducing aggregation artifacts. Fig. 3 shows images of Kit 225 K6 cells labeled with Cy3-conjugated αTac (Fig. 3a) or XR-conjugated MEM-75 against the TrfR (Fig. 3b). A patchy receptor distribution is observed with clusters of 500–800 nm for the IL-2Rα and 200–300 nm for the TrfR. Controls on prefixed and live cells indicate that this clustering is not caused by the labeling procedure. Cluster diameter for the IL-2Rα determined from G(ρ) (Eqs. 1 and 2) was ≈600 nm (see also Fig. 4). This implies that the minimum and maximum distances between immunogold labels within a cluster would be 0 and 600 nm, averaging to ≈300, which corresponds well to the ≈380-nm average interparticle distance within clusters determined in electron microscopy (Fig. 2).
Figure 3.
Confocal laser-scanning microscopy of Cy3-labeled IL-2Rα and TrfR. Kit 225 K6 cells were labeled with Cy3-conjugated αTac Fab against the IL-2Rα-subunit (a, c, and d) or XR-conjugated MEM-75 against the TrfR (b). Cells in c and d were treated with filipin and methyl-β-cyclodextrin, respectively. Confocal slices of 0.6 μm thickness were obtained. Surface fluorescence distribution was reconstructed from z directional projection of image slices. Bar = 4 μm. A patchy receptor distribution can be observed with clusters of 200- to 1,200-nm diameter depending on the type of receptor and the treatment.
Figure 4.
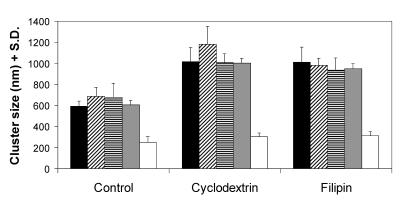
Cluster sizes of IL-2Rα, HLA class I and II, CD48, and TrfR and their modulation by membrane cholesterol content. Cluster sizes on Kit 225 K6 cells determined from the angle-averaged autocorrelation function are presented for IL-2Rα (filled columns), HLA class I (crosshatched columns), and class II (striped columns), CD48 (gray columns), and TrfR (open columns). The effect of modulating the cholesterol content of the membrane is also displayed: with the exception of TrfR, all receptor clusters exhibit a significant increase of cluster size on both cholesterol depletion by cyclodextrin or in situ complexation of cholesterol by filipin. (n > 9, from three independent experiments).
Cluster Size of IL-2Rα, HLA Glycoproteins, and CD48 Depends on the Integrity of Cholesterol-Rich Lipid Rafts.
Interestingly, clusters of the TrfR are significantly smaller in size (250 nm) than those of IL-2Rα. TrfR is thought to be localized outside rafts (31), whereas several src-family kinases that play a role in T cell activation are detected in association with rafts (21, 32). To check whether these cholesterol-rich regions could be held responsible for keeping together the islets of IL-2Rα-subunits, we have examined the change of cluster size after modifying the membrane cholesterol content with cyclodextrin or filipin. Fig. 3 c and d show that cluster boundaries become blurred, and their size increases on both treatments. Thus, modulation of cholesterol content seems to break up the tightness of IL-2Rα clusters. The increase in cluster size is significant in both cases (Fig. 4, filled columns) and is paralleled by a decrease of absolute fluorescence intensities, indicating dispersion of the labeled proteins. The same observations can be made for MT-1 cells (data not shown).
As Fig. 4 shows, both class I and II MHC glycoproteins and the raft marker CD48 behave similarly to IL-2Rα: their cluster size is comparable to that of IL-2Rα (control group, crosshatched, striped, and gray columns) and is dispersed significantly on cyclodextrin and filipin treatment. On the other hand, TrfR not only possesses smaller clusters in control cells, but its cluster size hardly changes on either of the treatments modifying membrane cholesterol (open columns). The same molecules behave similarly on the surface of MT-1 cells (data not shown).
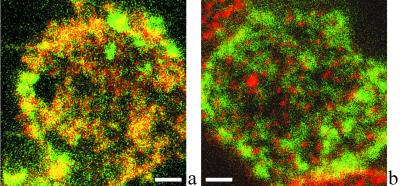
Crosscorrelation Analysis of Confocal Images Reveals Partial Coclustering of IL-2R with MHC and CD48 Molecules, but Not with TrfR.
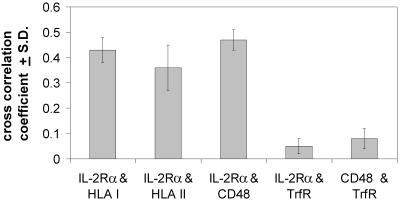
Because flow cytometric energy transfer measurements gave evidence of nanometer-level proximity between IL-2Rα and MHC antigens, we have used double labeling with fluorescent Fabs in confocal microscopy to investigate the colocalization of IL-2Rα with MHC glycoproteins. Fig. 5a shows the colocalization of IL-2Rα (green) and HLA class II (red). Because of the high degree of colocalization (crosscorrelation coefficient C = 0.37; see Fig. 6), many pixels of the image appear orange when the red and green channels are overlaid. Fig. 5b demonstrates that IL-2Rα (green) and TrfR (red) images from the same cell exhibit disparate localization of these two receptors. Accordingly, their crosscorrelation coefficient is very low (C = 0.05). In similar experiments, the crosscorrelation coefficient was measured for a set of receptor pairs (Fig. 6). While IL-2Rα colocalizes with HLA class I, II, and CD48, neither IL-2Rα nor CD48 colocalizes with TrfR. A similar colocalization pattern was observed on MT-1 cells. This is consistent with a selective association of IL-2Rα, MHC, and the raft marker CD48 with cholesterol-rich membrane rafts that are perturbed by filipin or cyclodextrin treatment and the observation that TrfR clusters are virtually unaffected by the modification of membrane cholesterol.
Figure 5.
IL-2Rα exhibits submicron-scale colocalization with MHC II but not with TrfR. A representative confocal fluorescence image of the colocalization of IL-2Rα and HLA class II is shown in a. IL-2Rα and HLA class II are labeled with XF (green) and XR (red), respectively. Because of the high degree of colocalization, many pixels appear orange when the two channels are fused. b demonstrates that IL-2Rα (green) and TrfR (red) codetected in a similar experiment are mostly localized at different areas of the plasma membrane. Bar = 2 μm.
Figure 6.
IL-2Rα staining crosscorrelates with MHC glycoproteins and CD48 but not with TrfR. Kit 225 K6 cells were double labeled with pairs of antibodies against IL-2Rα, HLA class I and II, CD48, and TrfR. The crosscorrelation coefficient is measured for the following receptor pairs: IL-2Rα and HLA class I, IL-2Rα and HLA class II, IL-2Rα and CD48, IL-2Rα and TrfR, and CD48 and TrfR. While IL-2Rα colocalizes with HLA class I and II and CD48, a raft marker, neither IL-2Rα nor CD48 colocalizes with TrfR (n = 7).
Sequential Immunogold Labeling and Electron Microscopy Confirm the Higher- Level Partial Colocalization of IL-2Rα and MHC.
Earlier we have developed a strategy to examine coclustering of class I and class II MHC antigens on lymphoid cells (16). Fig. 7 shows an example of IL-2Rα labeled with 10 nm gold followed by 30 nm gold tags on MHC-I molecules on the surface of Kit225 K6 cells. Although there are also some labels that are not proximal to the other labeling species, many of the small and large beads are seen coclustered. A similar partial colocalization of IL-2Rα and MHC-II was observed on both Kit 225 K6 and MT-1 cells (data not shown). Thus, the coclustering deduced from confocal images is supported by electron microscopic evidence.
Figure 7.
Electron microscopy confirms the partial coclustering of IL-2Rα and MHC molecules. Immunogold labeling followed the sequence: αTac Fab—10 nm AuroGamig—blocking by αTac Fab—W6/32 whole antibody—30 nm AuroGamig (anti-Fc). Electron microscopy shows that the selective labels against the IL-2Rα and the MHC are partially, although not completely, colocalized, thus confirming the confocal microscopic data (Bar = 200 nm).
When the surface density of 30-nm gold labels on IL-2Rα is averaged for several cells, 25/μm2 are seen. If IL-2Rα is labeled after covering MHC class II molecules with L243 Fab and 10-nm gold beads, the detectable 30-nm labels on IL-2Rα decrease to 13/μm2. This is consistent with the idea that the interaction of MHC-II and IL-2Rα at the molecular level allows detection of only about 50% of the IL-2Rα after having shielded those α-subunits that are in the vicinity of MHC-II antigens. A similar ≈50% coclustering ratio could be determined for IL-2Rα in relation to MHC class I molecules on these cells. This observation is in line with the crosscorrelation coefficient of 0.37–0.43 seen between fluorescent labels on IL-2Rα and MHC antigens.
Discussion
Recently, we have presented fluorescence resonance energy transfer efficiency data indicating spontaneous assembly of the α-, β-, and γc-subunits of the multisubunit IL-2 receptor on Kit 225 K6 T lymphoma cells even in the absence of IL-2 (4). Here we show by using immunogold labeling in electron microscopy that this molecular-level assembly of the IL-2 receptor is extended to a higher hierarchical level, i.e., on the several hundred nanometer scale in the plasma membrane of human T lymphoma cells. The distribution of colloidal gold labels attached to IL-2Rα was significantly different from a hypothetical random Poissionian pattern (P > 0.9999). The distribution of interparticle distances also showed two peaks, one close to that expected for the single peak of a Poissonian pattern and another at smaller distances, likely corresponding to the average cluster radius of 350–380 nm.
Clusters of similar size were observed both on Kit 225 K6 cells, which have an absolute requirement of IL-2 for their growth, and on MT-1 cells, which do not express the β-subunit of the IL-2 receptor and grow independently of IL-2. Also, IL-2 deprivation of Kit 225 K6 cells had no effect on cluster size. Thus the molecular interactions producing submicron-scale clusters seem to be largely independent of the presence of the IL-2R β-subunits or the specific ligand IL-2.
Confocal laser-scanning microscopy of both fixed and live cells confirmed the submicron clusters observed with electron microscopy. Also, the cluster size deduced from electron microscopy corresponded well to that seen in confocal slices and surface reconstructions. The cluster sizes measured are on the same order of magnitude as the areas of confined diffusion determined from single particle tracking for ganglioside GM1 and the Thy-1 antigen (34); furthermore, they are comparable to those assessed for ErbB2 (30) and platelet-derived growth factor receptor molecules (18) by scanning near-field optical microscopy.
Such high degree of receptor aggregation has been found both as a ligand-induced phenomenon (35) and as a stably maintained structure with yet undetermined lifetimes. The latter frequently occurs under polarizing conditions, especially in the nervous system (36) or in the neuromuscular junction (37). However, similar receptor clustering can also be found on cells that are nonpolarized, e.g., lymphoid cells. Earlier we have demonstrated higher hierarchical-level distribution patterns of the MHC class I (12) and class II molecules (16). These findings have been corroborated recently by data revealing anomalous diffusion of MHC I and II molecules (11, 38).
The present experiments significantly support the view that such above-nanometer-level codistribution patterns could be common among various receptor types even on nonpolarized cells. The existence of lipid rafts in the plasma membrane can easily have a central role in maintaining such receptor superstructures (21). Besides glycosphingolipids, cholesterol has been postulated as an important functional component of lipid rafts. Consistent with this, modifying membrane cholesterol content was shown to influence signaling by raft-associated molecular assemblies (24, 39). We found that specifically complexing cholesterol in situ with filipin or extracting it from the membrane with cyclodextrin changes the higher level colocalization patterns by dispersing and blurring the clusters of both IL-2Rα, HLA I, II, and the raft protein CD48. At the same time, flow cytometric energy transfer measurements have shown that IL-2Rα is in the nanometer-scale proximity of both MHC-I and MHC-II on Kit 225 K6 and MT-1 cells (fluorescence resonance energy transfer efficiency was in the range of 12–21%; unpublished data). These data together support the notion that IL-2Rα and MHC proteins may be partially confined to lipid rafts. In accordance with this hypothesis, a significant crosscorrelation of different color fluorescent labels on pairs of CD48 and these molecules was found, whereas the submicron clusters of the TrfR, which is not a constituent of rafts (31, 32), were significantly smaller and did not colocalize with either IL-2Rα or CD48.
The combined application of fluorescence resonance energy transfer, electron microscopy, confocal laser-scanning microscopy, and image processing suggests that both small receptor islands and larger rafts can accommodate the IL-2 receptor α-subunit together with HLA class I and class II glycoproteins. The importance of the organizing role of lipid rafts is underlined by the finding that caveola-like domains serve as concentrators of various signal transduction machineries (28, 40), and several small cytoplasmic kinases, a group vastly important in the signal transduction of T lymphocytes (20, 41), are bound to lipid rafts (21, 29, 42). Thus, the clusters on both the molecular and the submicron level could underlie the efficiency of signaling in lymphoid cells and might play a role in the directed secretion of lymphokines and in specific internalization pathways.
MHC class II, recently reported to coaggregate with lipid rafts on stimulation by crosslinking (43), and class I molecules seem to be partially recruited into the signaling platform of IL-2R subunits and CD48 formed by rafts. Although it is believed so far that posttranslational modification (fatty acylation, glycosylphosphatidylinositol linkage) is a specific predictor for targeting lipid rafts (20), for transmembrane proteins with (MHC molecules) or without (IL-2Rα) intracellular tails, it is hard to predict their partitioning behavior based on their sequence. In these domains, MHC molecules may provide a stabilizing effect through their direct cytoskeletal connections (44). In addition, MHC class I molecules may also contribute to this biochemical switchboard as potential regulators of IL-2 receptor signaling by an intracellular tyrosine phosphorylation crosstalk, as reported for the insulin receptor recently (45).
Coimmobilization of GPI-anchored raft proteins, including CD48, on T cells has been reported to inhibit recruitment of IL-2Rα chains with the signaling β and γ-subunits, but not the association of β and γ chains with the Janus kinases (46). In light of our data showing a “focusing” effect of rafts for IL-2Rα, this might be due to coimmobilization of IL-2Rα and/or a steric blocking of its interaction with the β and γ. Thus, T cell rafts related to cell-surface clusters of the proteins investigated here may promote IL-2R-mediated signaling by recruiting the α-chains into a signaling platform regardless of their ligand binding and, on the other hand, may also have a control on T cell growth (46) through the colocalized GPI-linked proteins (CD48 or Thy-1).
Acknowledgments
This research was supported by grants Hungarian National Research Fund (OTKA) F025210, T23873, T30411, T19372, T30399, T029947 and Fund for Higher Education Research (FKFP) 0518/99 and 327/2000.
Abbreviations
- TrfR
transferrin receptor
- Cy3
sulfoindocyanine–succinimidyl ester
References
- 1.Waldmann T A. Science. 1986;232:727–732. doi: 10.1126/science.3008337. [DOI] [PubMed] [Google Scholar]
- 2.Waldmann T A. J Biol Chem. 1991;266:2681–2684. [PubMed] [Google Scholar]
- 3.Nakamura Y, Russell S M, Mess S A, Friedmann M, Erdos M, Francois C, Jacques Y, Adelstein S, Leonard W J. Nature (London) 1994;369:330–333. doi: 10.1038/369330a0. [DOI] [PubMed] [Google Scholar]
- 4.Damjanovich S, Bene L, Matkó J, Alileche A, Goldman C K, Sharrow S, Waldmann T A. Proc Natl Acad Sci USA. 1997;94:13134–13139. doi: 10.1073/pnas.94.24.13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Damjanovich S, Gáspár R, Jr, Pieri C. Q Rev Biophys. 1997;30:67–106. doi: 10.1017/s0033583596003307. [DOI] [PubMed] [Google Scholar]
- 6.Damjanovich S, Trón L, Szöllősi J, Zidovetzki R, Vaz W L C, Regateiro F, Arndt-Jovin D J, Jovin T M. Proc Natl Acad Sci USA. 1983;80:5985–5989. doi: 10.1073/pnas.80.19.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edidin M, Zuniga M C, Scheetz M. Proc Natl Acad Sci USA. 1994;91:3378–3382. doi: 10.1073/pnas.91.8.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobson K, Sheets E D, Simson R. Science. 1995;268:1441–1442. doi: 10.1126/science.7770769. [DOI] [PubMed] [Google Scholar]
- 9.Cherry R J, Smith P R, Morrison I E, Fernandez N. FEBS Lett. 1998;430:88–91. doi: 10.1016/s0014-5793(98)00595-x. [DOI] [PubMed] [Google Scholar]
- 10.Bene L, Balázs M, Matkó J, Most J, Dierich M P, Szöllősi J, Damjanovich S. Eur J Immunol. 1994;24:2115–2123. doi: 10.1002/eji.1830240928. [DOI] [PubMed] [Google Scholar]
- 11.Cherry R J, Wilson K M, Triantafilou K, O'Toole P, Morrison I E, Smith P R, Fernandez M. J Cell Biol. 1998;140:71–79. doi: 10.1083/jcb.140.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damjanovich S, Vereb G, Schaper A, Jenei A, Matkó J, Starink J P, Fox G Q, Arndt-Jovin D J, Jovin T M. Proc Natl Acad Sci USA. 1995;92:1122–1126. doi: 10.1073/pnas.92.4.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matkó J, Bushkin Y, Wei T, Edidin M. J Immunol. 1994;152:3355–3360. [PubMed] [Google Scholar]
- 14.Szöllősi J, Damjanovich S, Balázs M, Nagy P, Trón L, Fulwyler M J, Brodsky F M. J Immunol. 1989;143:208–213. [PubMed] [Google Scholar]
- 15.Szöllősi J, Damjanovich S, Goldman C K, Fulwyler M J, Aszalós A, Goldstein G, Rao P, Waldmann T A. Proc Natl Acad Sci USA. 1987;84:7246–7250. doi: 10.1073/pnas.84.20.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenei A, Varga S, Bene L, Mátyus L, Bodnár A, Bacsó Z, Pieri C, Gáspár R, Jr, Farkas T, Damjanovich S. Proc Natl Acad Sci USA. 1997;94:7269–7274. doi: 10.1073/pnas.94.14.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenworthy A K, Edidin M. J Cell Biol. 1998;142:69–84. doi: 10.1083/jcb.142.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vereb G, Meyer C K, Jovin T M. In: Interacting Protein Domains, Their Role in Signal and Energy Transduction. NATO ASI Series. Heilmeyer L M G Jr, editor. H102. New York: Springer; 1997. pp. 49–52. [Google Scholar]
- 19.Jacobson K, Dietrich C. Trends Cell Biol. 1999;9:87–91. doi: 10.1016/s0962-8924(98)01495-0. [DOI] [PubMed] [Google Scholar]
- 20.Horejsi V, Cebecauer M, Cerny J, Brdicka T, Angelisova P, Drbal K. Immunol Lett. 1998;63:63–73. doi: 10.1016/s0165-2478(98)00054-6. [DOI] [PubMed] [Google Scholar]
- 21.Simons K, Ikonen E. Nature (London) 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 22.Hwang J, Gheber L A, Margolis L, Edidin M. Biophys J. 1998;74:2184–2190. doi: 10.1016/S0006-3495(98)77927-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edidin M. Curr Opin Struct Biol. 1997;7:528–532. doi: 10.1016/s0959-440x(97)80117-0. [DOI] [PubMed] [Google Scholar]
- 24.Rothberg K G, Ying Y S, Kamen B A, Anderson R G. J Cell Biol. 1990;111:2931–2938. doi: 10.1083/jcb.111.6.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bodnár A, Jenei A, Bene L, Damjanovich S, Matkó J. Immunol Lett. 1996;54:221–226. doi: 10.1016/s0165-2478(96)02677-6. [DOI] [PubMed] [Google Scholar]
- 26.de Kruijff B, Demel R A. Biochim Biophys Acta. 1974;339:57–63. doi: 10.1016/0005-2736(74)90332-0. [DOI] [PubMed] [Google Scholar]
- 27.Christian A E, Haynes M C, Phillips M C, Rothblat G H. J Lipid Res. 1997;38:2264–2272. [PubMed] [Google Scholar]
- 28.Liu P, Ying Y, Anderson R G. Proc Natl Acad Sci USA. 1997;94:13666–13670. doi: 10.1073/pnas.94.25.13666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harder T, Simons K. Eur J Immunol. 1999;29:556–562. doi: 10.1002/(SICI)1521-4141(199902)29:02<556::AID-IMMU556>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 30.Nagy P, Jenei A, Kirsch A K, Szöllősi J, Damjanovich S, Jovin T M. J Cell Sci. 1999;112:1733–1741. doi: 10.1242/jcs.112.11.1733. [DOI] [PubMed] [Google Scholar]
- 31.Smart E J, Ying Y S, Mineo C, Anderson R G. Proc Natl Acad Sci USA. 1995;92:10104–10108. doi: 10.1073/pnas.92.22.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xavier R, Brennan T, Li Q, McCormack C, Seed B. Immunity. 1998;8:723–732. doi: 10.1016/s1074-7613(00)80577-4. [DOI] [PubMed] [Google Scholar]
- 33.Matko J, Edidin M. Methods Enzymol. 1997;278:444–462. doi: 10.1016/s0076-6879(97)78023-6. [DOI] [PubMed] [Google Scholar]
- 34.Sheets E D, Lee G M, Simson R, Jacobson K. Biochemistry. 1997;36:12449–12458. doi: 10.1021/bi9710939. [DOI] [PubMed] [Google Scholar]
- 35.Ciruela F, Saura C, Canela E I, Mallol J, Lluis C, Franco R. Mol Pharmacol. 1997;52:788–797. doi: 10.1124/mol.52.5.788. [DOI] [PubMed] [Google Scholar]
- 36.Craig A M, Blackstone C D, Huganir R L, Banker G. Proc Natl Acad Sci USA. 1994;91:12373–12377. doi: 10.1073/pnas.91.26.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campanelli J T, Hoch W, Rupp F, Kreiner T, Scheller R H. Cell. 1991;67:909–916. doi: 10.1016/0092-8674(91)90364-5. [DOI] [PubMed] [Google Scholar]
- 38.Smith P R, Morrison I E, Wilson K M, Fernandez N, Cherry R J. Biophys J. 1999;76:3331–3344. doi: 10.1016/S0006-3495(99)77486-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keller P, Simons K. J Cell Biol. 1998;140:1357–1367. doi: 10.1083/jcb.140.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu C, Butz S, Ying Y, Anderson R G. J Biol Chem. 1997;272:3554–3559. doi: 10.1074/jbc.272.6.3554. [DOI] [PubMed] [Google Scholar]
- 41.Ihle J N, Kerr I M. Trends Genet. 1995;11:69–74. doi: 10.1016/s0168-9525(00)89000-9. [DOI] [PubMed] [Google Scholar]
- 42.Ko Y G, Liu P, Pathak R K, Craig L C, Anderson R G. J Cell Biochem. 1998;71:524–535. [PubMed] [Google Scholar]
- 43.Huby R D, Dearman R J, Kimber I. J Biol Chem. 1999;274:22591–22596. doi: 10.1074/jbc.274.32.22591. [DOI] [PubMed] [Google Scholar]
- 44.Geppert T D, Lipsky P E. J Immunol. 1991;146:3298–3305. [PubMed] [Google Scholar]
- 45.Ramalingam T S, Chakrabarti A, Edidin M. Mol Biol Cell. 1997;8:2463–2474. doi: 10.1091/mbc.8.12.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marmor M D, Bachmann M F, Ohashi P S, Malek T R, Julius M. Int Immunol. 1999;11:1381–1393. doi: 10.1093/intimm/11.9.1381. [DOI] [PubMed] [Google Scholar]