Abstract
R-Ras is a Ras-family small GTPase that regulates various cellular functions such as apoptosis and cell adhesion. Here, we demonstrate a role of R-Ras in exocytosis. By the use of specific anti-R-Ras antibody, we found that R-Ras was enriched on both early and recycling endosomes in a wide range of cell lines. Using a fluorescence resonance energy transfer-based probe for R-Ras activity, R-Ras activity was found to be higher on endosomes than on the plasma membrane. This high R-Ras activity on the endosomes correlated with the accumulation of an R-Ras effector, the Rgl2/Rlf guanine nucleotide exchange factor for RalA, and also with high RalA activity. The essential role played by R-Ras in inducing high levels of RalA activity on the endosomes was evidenced by the short hairpin RNA (shRNA)-mediated suppression of R-Ras and by the expression of R-Ras GAP. In agreement with the reported role of RalA in exocytosis, the shRNA of either R-Ras or RalA was found to suppress calcium-triggered exocytosis in PC12 pheochromocytoma cells. These data revealed that R-Ras activates RalA on endosomes and that it thereby positively regulates exocytosis.
INTRODUCTION
R-Ras is a Ras-family GTPase and its amino acid sequence is 55% identical to those of the classical types of Ras (H-, K-, N-Ras, collectively referred to hereafter as “Ras”) (Lowe et al., 1987). As is the case with the other Ras-family GTPases, R-Ras is regulated primarily by two classes of protein, guanine nucleotide exchange factor (GEF) and GTPase-activating protein (GAP). Reflecting the high sequence similarity among Ras-family GTPases, many GEFs and GAPs for R-Ras catalyze other Ras-family GTPases as well (Ohba et al., 2000). Furthermore, R-Ras is known to interact with many effectors of Ras, such as Raf-1, Ral GEFs, and the p110α subunit of phosphoinositide 3-kinase (PI3K) (Rey et al., 1994; Spaargaren and Bischoff, 1994; Spaargaren et al., 1994; Marte et al., 1997). Despite this redundancy between R-Ras and Ras, R-Ras exhibits various properties that are distinct from those of Ras. For example, R-Ras preferentially activates Ral GEFs and PI3K, but it does not activate Raf (Huff et al., 1997; Rodriguez-Viciana et al., 2004). The transforming activity of constitutively active R-Ras is substantially less potent than that of the constitutively active Ras (Cox et al., 1994), although it should be noted that a recent report has suggested the involvement of R-Ras in human gastric cancer (Nishigaki et al., 2005). Meanwhile, R-Ras is known to regulate cell adhesion, cell spreading, and phagocytosis through the activation of integrin (Zhang et al., 1996; Keely et al., 1999; Berrier et al., 2000; Self et al., 2001). R-Ras-null mice have recently been shown to exhibit excessive vascular responses, in spite of the fact that they are otherwise normal (Komatsu and Ruoslahti, 2005). This phenotype seems to reflect higher levels of expression of R-Ras in smooth muscle cells, including blood vessel cells. The results obtained with R-Ras-null mice have also demonstrated that an R-Ras defect can be almost entirely compensated for by other gene products.
Ral GEFs, effectors of Ras-family GTPases, are activators of the two Ral proteins, RalA and RalB, which are also Ras-family GTPases (Wolthuis and Bos, 1999; Quilliam et al., 2002; Rodriguez-Viciana et al., 2004). It has been suggested that this Ral GEFs-Ral pathway is more important in the Ras-dependent oncogenesis of human cells than are other Ras-dependent pathways, such as those involving Raf and PI3K (Hamad et al., 2002; Rangarajan et al., 2004; Lim et al., 2005; Gonzalez-Garcia et al., 2005). The activated Ral then binds to various Ral-binding proteins and thereby regulates various cellular functions (Feig, 2003). The characterization of such Ral effector proteins has suggested that Ral may be involved in vesicular trafficking. For example, the Ral-binding protein RalBP1 is thought to regulate endocytosis, suggesting the involvement of Ral in endocytosis (Nakashima et al., 1999). Ral may also regulate exocytosis, because Ral binds to two components of the exocyst complex, Sec5 and Exo84 (Moskalenko et al., 2002, 2003).
The exocyst complex was originally identified by genetic and biochemical studies as a cluster of molecules required for exocytosis in budding yeast, and it was later characterized in a wide range of eukaryotes. The exocyst complex consists of eight subunits: Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70, and Exo84 (Lipschutz and Mostov, 2002). These proteins are primarily involved in the tethering and/or docking process of trafficking vesicles, which occurs before the fusion process (Finger et al., 1998; Tsuboi et al., 2005). Due to their fundamental role in exocytosis, any malfunction of the proteins in the exocyst complex can disrupt various cellular events, such as the basolateral transport of vesicles in polarized epithelial cells, neurite outgrowth in PC12 cells, paraxial mesoderm formation in mice, and secretory vesicle-mediated abscission in Drosophila (Friedrich et al., 1997; Grindstaff et al., 1998; Vega and Hsu, 2001; Murthy et al., 2003; Gromley et al., 2005).
To gain a better understanding of the function of R-Ras and its potential role in Ral-mediated exocytosis, it will be essential to elucidate not only the subcellular localization but also the activity change of these proteins. Thus, we developed specific anti-R-Ras sera and a probe for R-Ras activity based on the principle of fluorescence resonance energy transfer (FRET), a technique that has been shown to be extremely useful for the spatiotemporal analysis of small GTPases (Kurokawa et al., 2004b). Using these tools, we found that endogenous R-Ras is enriched and activated at endosomes and that these R-Ras proteins promote exocytosis by activating RalA.
MATERIALS AND METHODS
Probes Based on FRET
FRET probes for R-Ras, designated as Raichu-R-Ras, were prepared essentially as described previously (Mochizuki et al., 2001; Takaya et al., 2004). From the amino terminus, Raichu-R-Ras consisted of a modified yellow fluorescent protein (YFP) designated as “Venus” (Nagai et al., 2002) (amino acids [aa] 1-239), a spacer (Leu-Asp), human R-Ras (aa 1-199), a 17-amino acid-spacer (Gly-Gly-Gly-Thr-Gly-Gly-Gly-Gly-Gly-Ser-Gly-Gly-Thr-Gly-Gly-Gly-Thr), the Ras- binding domain of RalGDS (aa 785-871), a spacer (Gly-Gly-Arg), a modified cyan fluorescent protein (CFP) designated as “SECFP” (Lys27Arg, Asp130Ala, Asn165His, Ser176Gly) (aa 1-237), a spacer (Gly-Arg-Ser-Arg), and the carboxy-terminal region of R-Ras (aa 195-218) (Figure 1A). The characterization of Raichu R-Ras was performed as described previously (Takaya et al., 2004).
Figure 1.
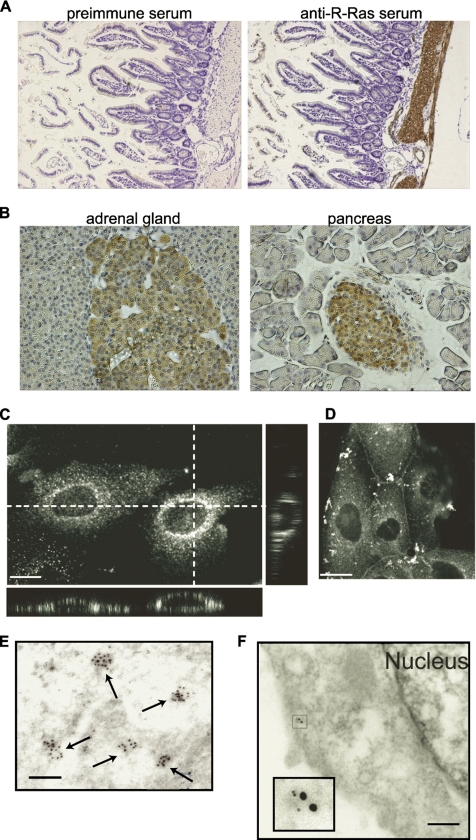
Tissue distribution and subcellular localization of R-Ras. (A) Immunohistochemistry analysis of mouse intestine with preimmune (left) or anti-R-Ras serum (right). Bound antibodies were detected with diaminobenzidine tetrahydrochloride, and the nuclei were counterstained with hematoxylin. Note the staining of the smooth muscle cell layer. (B) Immunohistochemistry of the mouse adrenal gland and pancreas. Note the staining of the adrenal medulla and Langerhans' islands. MDCK cells were fixed in 3.7% formaldehyde and methanol (C) or 4% paraformaldehyde (D) as described in the text, stained with the anti-R-Ras serum and the Alexa 488-coupled anti-rabbit IgG antibody, and observed with a confocal fluorescent microscope. Twenty-five XY images were obtained from the bottom to the top of the cells to prepare the stacked XY image. Cross sections are also shown at the dotted lines. Bar, 10 μm. (E) Immunoelectron micrograph of a MDCK cell stained with anti-R-Ras serum before detection with anti-rabbit IgG labeled with 10-nm gold particles. Bar, 100 nm. (F) Immunoelectron micrograph of a MDCK cell expressing an endosomal marker protein, AcGFP-Endo. Cells were double-stained with anti-R-Ras rabbit serum and anti-GFP mouse mAb JL-8, which were detected with anti-rabbit IgG labeled with 5-nm gold particles and anti-mouse IgG labeled with 10-nm gold particles, respectively. Inset depicts the magnified image of the gold particles. Bar, 100 nm.
Plasmids
pCXN2-mCFP, pCXN2-mRFP, and pCXN2-mCherry are expression vectors encoding a monomeric SECFP (Zacharias et al., 2002), a monomeric red fluorescent protein (RFP) (Kurokawa et al., 2004a), and mCherry, respectively. cDNA of mCherry was provided by R. Y. Tsien (University of California at San Diego). The pERedNLS and pERedMito expression vectors contain an internal ribosomal site followed by the cDNAs of DsRed-Express (Clontech, Mountain View, CA) with nuclear and mitochondrial localization signals, respectively (Aoki et al., 2005). pCXN2-5Myc and pCXN2-Flag are mammalian expression vectors containing Myc and Flag epitope tags, respectively. cDNAs of RalBP1 and Rgl were provided by A. Kikuchi (Hiroshima University, Hiroshima, Japan). cDNA of p110α and p85α were obtained from Y. Fukui (University of Tokyo, Tokyo, Japan), and cDNA of Rab5A was obtained from Y. Takai (Osaka University, Osaka, Japan). Rap1B cDNA was provided by N. Minato (Kyoto University, Kyoto, Japan). The cDNAs of K-Ras and N-Ras were obtained from L. A. Feig (Tufts University, Boston, MA). cDNAs of Rab7, Rab11A, and RalB were purchased from Guthrie cDNA Resource Center (Sayre, PA). pAcGFP1-Endo was purchased from Clontech. pVenus-N1-NPY was obtained from A. Miyawaki (The Brain Science Institute, RIKEN, Wako-shi, Japan) (Nagai et al., 2002). pHA-EYFP-GH1 has been described previously (Matsuno et al., 2005). pCXN2-Flag-CalDAG-GEFII, pCXN2-Flag-CalDAG-GEFIII, pCXN2-Flag-R-RasGAP, pCAGGS-Flag-p120RasGAP, pCXN2-Flag-rap1GAP1B, pCAGGS-RasGRF, pCAGGS-mSos1, and pEF-BOS-myc-Gap1m have been described previously (Yamamoto et al., 1995; Gotoh et al., 1997; Ohba et al., 2000). pCXN2-5Myc-R-Ras-rRNAi (RNA interference-resistant clone) encodes a R-Ras mutant resistant to the short hairpin RNA (shRNA) vector. For the preparation of the glutathione S-transferase (GST) fusion proteins, cDNAs were subcloned into pGEX vectors and recombinant proteins were prepared according to the manufacturer's protocol (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
Cells, Antibodies, and Reagents
293T cells were obtained from B. J. Mayer (University of Connecticut, Storrs, CT). The line of Cos7 cells used in this study was Cos7/E3, a subclone of Cos7 cells established by Y. Fukui. The PC12 cells were obtained from S. Kuroda (University of Tokyo). HeLa and Madin-Darby canine kidney (MDCK) cells were purchased from the Human Science Research Resources Bank (Sennan-shi, Osaka, Japan). The GH3 cells used here have been described previously (Matsuno et al., 2005). The PC12 cells were maintained in DMEM (Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal calf serum and 5% horse serum. GH3 cells were cultured in Ham's F-10 (Sigma-Aldrich) supplemented with 15% horse serum and 2.5% fetal calf serum. Other cells were maintained in DMEM supplemented with 10% fetal calf serum. GH3 cells stably expressing R-Ras were prepared essentially as described previously (Akagi et al., 2003). Anti-green fluorescent protein (GFP) rabbit serum was prepared in our laboratory. Anti-RalA and anti-RalB were purchased from BD Biosciences (San Jose, CA). Anti-FLAG M2 and tetradecanoyl phorbol-13-acetate (TPA) was purchased from Sigma-Aldrich. Anti-Myc 9E10 was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-GFP antibody was also purchased from Takara Bio (Otsu, Japan). Anti-Akt, anti-phospho Akt (Thr308), anti-phospho-mitogen–activated protein kinase kinase (MEK) 1/2 (Ser217/221), and anti-R-Ras were purchased from Cell Signaling Technology (Beverly, MA). Alexa 488 anti-rabbit immunoglobulin G (IgG), Alexa 488 anti-rat IgG, and Alexa 568 anti-mouse IgG were purchased from Invitrogen (San Diego, CA). To generate anti-R-Ras sera, three rabbits were injected with GST-R-Ras.
RNA Interference
Synthetic siRNAs against R-Ras and Ral proteins were prepared as described previously (Oinuma et al., 2004; Wozniak et al., 2005). siRNAs were transfected using Oligofectamine or Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. pSuper.retro.puro vector (OligoEngine, Seattle, WA) was used for short hairpin RNA. The shRNA sequences for rat R-Ras and RalA have been described previously (Oinuma et al., 2004; Vitale et al., 2005), and the sequence for rat RalB was 5′-GCCGACAGTTACAGAAAGA-3′. After transfection, the cells were incubated for at least 48 h before analysis.
Bos' Pull-Down Assay
Bos' pull-down assay for Ral proteins was performed essentially as described previously (Takaya et al., 2004). Briefly, the cells were lysed in Ral buffer (50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 2.5 mM MgCl2, 1% NP-40, 10% glycerol, 1 mM Na3VO4, 1 mM phenylmethylsulfunyl fluoride, 10 μg/ml aprotinin, and 10 μg/ml leupeptin) and were clarified by centrifugation. The supernatant was incubated with GST-Sec5-RBD or GST-RalBP1-RBD for 30 min at 4°C. The resulting complexes of Ral-GTP and GST fusion proteins were incubated with glutathione-Sepharose beads (GE Healthcare) for 1 h at 4°C, and after the bound proteins and cell lysates had been separated by SDS-polyacrylamide gel electrophoresis (PAGE), immunoblotting with anti-RalA or anti-RalB antibody was carried out. Bound antibodies were detected by an ECL chemiluminescence detection system (GE Healthcare), and binding was quantified with the aid of an LAS-1000 image analyzer (Fuji-Film, Tokyo, Japan). The pull-down assay for Ras, Rap1, and R-Ras was performed essentially as described above except for the use of GST-RalGDS-RBD.
Immunoprecipitation
Transfected Cos7 cells were harvested in ice-cold lysis buffer (50 mM Tris-HCl pH 7.5, 200 mM NaCl, 2.5 mM MgCl2, 1% NP-40, 0.5% sodium deoxycholate, 10% glycerol, 1 mM Na3VO4, 1 mM phenylmethylsulfunyl fluoride, 10 μg/ml aprotinin, and 10 μg/ml leupeptin). Anti-Myc antibody, GFP antiserum, or R-Ras antiserum was added to the cleared lysates. After the lysates were subjected to 1 h of rotation at 4°C with protein G-Sepharose or protein A-Sepharose (GE Healthcare), the beads were washed and boiled in sample buffer. The bound proteins were then subjected to immunoblot analysis.
Immunohistochemistry and Immunogold Electron Microscopy
Formalin-fixed, paraffin-embedded sections were deparaffinized with xylenes and rehydrated with ethanol. The sections were treated with normal goat serum and 1% H2O2 to quench endogenous peroxidase activity, and then they were incubated with primary antibody overnight at 4°C. After incubation of the sections with the biotinylated secondary antibody, immunopositive signals were visualized using 3,3′-diaminobenzidine tetrahydrochloride as a chromogen. For immunogold electron microscopy, the cells were fixed with 4% paraformaldehyde, 0.35% glutaraldehyde, and 0.2% picric acid at 4°C for 1.5 h, followed by fixation with 4% paraformaldehyde and 0.2% picric acid overnight at 4°C. After being washed with phosphate-buffered saline (PBS), the cells were dehydrated with ethanol and embedded in Lowicryl K4M (Polysciences; Tokyo, Japan). Ultrathin sections were placed on nickel grids and were immersed in a target retrieval solution (Dako Denmark, Glostrup, Denmark). Then, the samples were exposed to microwave radiation for 20 min, after which they were washed with distilled water. The grids were incubated with anti-R-Ras or preimmune rabbit serum at room temperature for 2 h, and then they were incubated for 1 h with anti-rabbit IgG labeled with 10-nm gold particles (GE Healthcare). After being washed and dried, the sections were stained with both uranyl acetate and lead citrate; the sections were then examined with a Hitachi H-800 transmission electron microscope (Hitachi High-Technologies, Tokyo, Japan). In some experiments, MDCK cells expressing an endosomal marker protein, AcGFP-Endo, were used for the analysis. Cells were double stained with anti-R-Ras rabbit serum and anti-GFP mouse monoclonal antibody (mAb) JL-8, which were detected with anti-rabbit IgG labeled with 5-nm gold particles and anti-mouse IgG labeled with 10-nm gold particles, respectively. As a control, anti-R-Ras rabbit serum preadsorbed to GST-R-Ras, preimmune rabbit serum, nonspecific rabbit IgG, or nonspecific mouse IgG was also used.
Immunocytochemistry
To stain the endogenous R-Ras protein, MDCK cells were fixed with 3.7% formaldehyde and then subjected to refixation with methanol at −20°C and permeabilization with 0.2% Triton X-100, followed by incubation in PBS containing 3% bovine serum albumin (BSA) and 0.02% Triton X-100 for 1 h. In some experiments, MDCK cells were fixed with 4% paraformaldehyde, immediately followed by permeabilization with 0.01% Triton X-100 for 1 min and incubation with 2% BSA in 50 mM NH4Cl-containing PBS. These fixed cells were incubated for 1 h at room temperature with anti R-Ras rabbit serum, washed with PBS, and then incubated for 30 min at room temperature with Alexa 488 anti-rabbit IgG. For the 3HA-tag and 5Myc-tag staining, Cos7 cells were fixed with 3% paraformaldehyde and subjected to permeabilization and staining as described above. Alexa 488 anti-rat IgG and Alexa 568 anti-mouse IgG were used to detect anti-hemagglutinin (HA) and anti-Myc, respectively. After being washed, the cells were imaged with an FV-500 confocal microscope equipped with an argon laser and with an HeNe laser microscope (Olympus, Tokyo, Japan). Twenty-five XY images scanned from the bottom to the top of the cells were obtained to prepare stacked images of XY and XZ sections.
Imaging of R-Ras and RalA Activity in Living Cells
R-Ras and RalA activity was visualized with Raichu-R-Ras or Raichu-RalA essentially as described previously (Mochizuki et al., 2001; Takaya et al., 2004). Expression plasmids were transfected into Cos7 cells by Polyfect (QIAGEN, Valencia, CA) or 293fectin (Invitrogen). More than 36 h after transfection, the cells were imaged with an Olympus IX70 inverted microscope equipped with an image splitter, Dual-View (Optical Insights, Santa Fe, NM) and an EMCCD camera, iXon DV887 (Andor Technology, Belfast, United Kingdom), and the imaging process was controlled by MetaMorph software (Molecular Devices, Sunnyvale, CA). In some experiments, cells were imaged with an Olympus IX81 inverted microscope equipped with a laser-based autofocusing system, IX2-ZDC, and an automatically programmable XY stage, MD-XY30100T-Meta, which allowed us to obtain the time-lapse images of several view fields in a single experiment. For dual-emission ratio imaging of the Raichu probes, we used previously described filter sets (Takaya et al., 2004), and we obtained images for CFP and FRET. After background subtraction was carried out, the FRET/CFP ratio was depicted using MetaMorph software, and this image was used to represent FRET efficiency. Confocal FRET images were obtained by an IX51 upright fluorescence microscope (Olympus) equipped with a CSU-10 spinning Nipkow disk confocal unit (Yokogawa, Tokyo, Japan), a W-view (Hamamatsu Photonics, Hamamatsu, Japan), and a diode-pumped solid state 430-nm laser (Melles Griot, Carlsbad, CA).
Exocytosis Assay
The exocytosis assay was carried out using Venus-tagged neuropeptide Y (NPY) as described previously (Nagai et al., 2002). pVenus-N1-NPY or pHA-EYFP-GH1 with or without additional expression vectors was transfected into PC12 cells or GH3 cells with Lipofectamine 2000 (Invitrogen). Sixty hours after transfection, the cells were washed in a low-potassium saline solution (145 mM NaCl, 5.6 mM KCl, 2.2 mM CaCl2, 0.5 mM MgCl2, 5.6 mM glucose, and 15 mM HEPES, pH 7.4). Then, the medium was exchanged for a high-potassium saline medium (95 mM NaCl, 56 mM KCl, 2.2 mM CaCl2, 0.5 mM MgCl2, 5.6 mM glucose, and 15 mM HEPES, pH 7.4) to depolarize the cells. After 20 min in the case of the PC12 cells or 10 min in the case of the GH3 cells, cell-free supernatants were collected and were stored as the secreted fraction. Cells remaining on the culture dishes were lysed in PBS containing 1% Triton X-100, and they were cleared by centrifugation to obtain the nonsecreted fraction. The fluorescence of NPY-Venus or EYFP-GH recovered in each fraction was measured by a FluoroSkan II fluorescence microplate reader (Global Medical Instrumentation, Ramsey, MN).
Online Supplemental Material
Time-lapse FRET images of Supplemental Figure S3J and Figure 3B are compiled into QuickTime videos available as supplemental material. Movie 1 shows Cos7 cells transfected with pRaichu-R-Ras and stimulated with TPA as described in the legend to Supplemental Figure S3K. Movie 2 shows Cos7 cells transfected with pRaichu-R-Ras as described in the legend to Figure 2B. Images were acquired every 30 s, and the video is displayed at 15 frames per second. Supplemental Figures 1–6 show characterization of anti-R-Ras serum, images of colocalization studies, basic property of Raichu-R-Ras probe, and results of coimmunoprecipitation studies.
Figure 3.
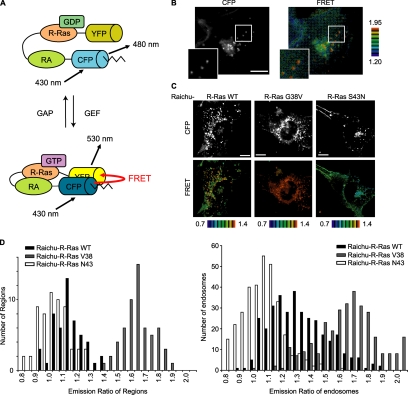
High R-Ras activity on the endosomes. (A) Schematic representation of the Raichu-R-Ras probe. Raichu-R-Ras consisted of a modified YFP designated as Venus, R-Ras, the RA domain (RA) of RalGDS, a modified CFP designated as SECFP, and the carboxy-terminal hypervariable region of R-Ras (indicated by the zigzag lines). On R-Ras activation, the intramolecular binding of R-Ras to the RA domain brings CFP into proximity with YFP, evoking YFP-derived fluorescence by the sensitized FRET. (B) A Cos7 cell expressing Raichu-R-Ras was imaged for CFP (excitation 430 nm/emission 480 nm) and YFP (excitation 430 nm/emission 530 nm). The images of the ratio (YFP versus CFP) were generated to represent the level of FRET. The upper and lower limits of the ratio range are shown on the right. Outlined regions are shown enlarged in the insets. Bar, 10 μm. (C) Cos7 cells expressing Raichu-R-Ras and its mutants were imaged with a confocal microscope. Bar, 10 μm. (D) FRET efficiency (YFP/CFP ratio) of Raichu-R-Ras, Raichu-R-Ras G38V, and Raichu-R-Ras S43N at the plasma membrane (left) and on the endosomes (right). For the endosomes, regions that exhibited higher CFP intensity than an appropriate threshold level were selected, and their YFP and CFP intensities were obtained. For the plasma membrane, three appropriate regions were arbitrarily selected, and the averages of their YFP and CFP intensities were obtained. Data from at least seven cells are shown in the histograms.
Figure 2.
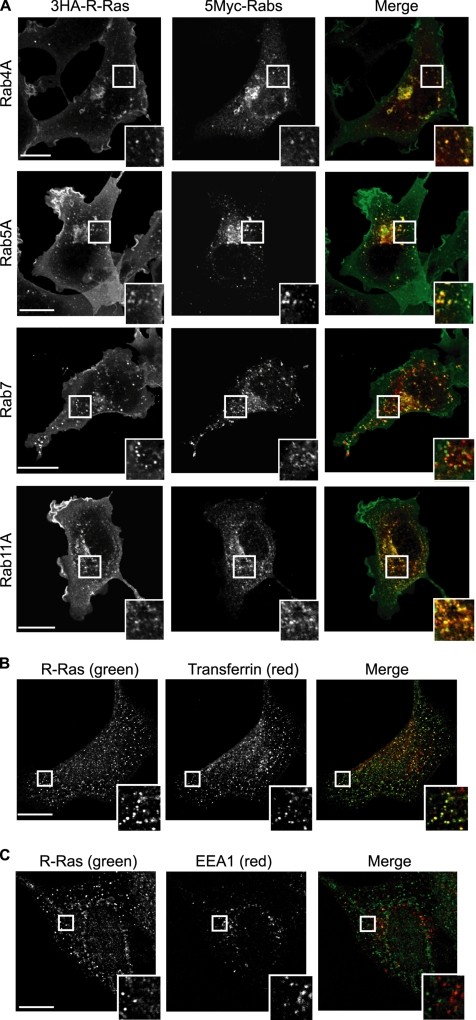
Colocalization of R-Ras with early and recycling endosome markers. (A) Cos7 cells expressing 3HA-tagged R-Ras and 5Myc-tagged Rab4A (early and recycling endosome marker), Rab5A (early endosome marker), Rab7 (late endosome marker), or Rab11A (recycling endosome marker) were double stained with anti-HA and anti-Myc antibodies and then observed with a laser-scanning confocal microscope. In the merged images, the red and green areas indicate anti-Myc and anti-HA antibodies, respectively. Bar, 10 μm. Outlined regions were enlarged and are shown in the insets. (B) Cos7 cells were incubated with Alexa-568-conjugated transferrin for 60 min and then stained with anti-R-Ras serum. (C) Cos7 cells were double stained with anti-R-Ras serum and anti-EEA1 antibody. Bar, 10 μm. Outlined regions were enlarged and are shown in the insets.
RESULTS
Tissue Distribution of Endogenous R-Ras
One of the reasons why the biological function of R-Ras remains elusive may be the lack of information concerning its tissue distribution and subcellular localization, which in turn is likely due to the lack of a specific antiserum. Therefore, we developed high-affinity anti-R-Ras sera that could be used for immunohistochemistry. One of the antisera was found to specifically recognize the endogenous R-Ras protein by immunoblotting analysis (Supplemental Figure S1). The obtained anti-R-Ras serum was used for immunohistochemical analyses. We found that R-Ras accumulated at high levels in the cytoplasm of smooth muscle cells, including those in the intestine and blood vessels, as has been reported recently (Komatsu and Ruoslahti, 2005) (Figure 1A). To a lesser extent, R-Ras expression was also observed in the neuroendocrine cells of the adrenal medulla and in islet cells in the pancreas (Figure 1B). Immunohistochemistry analysis in other tissues is described in Supplemental Figure S2.
Localization of Endogenous R-Ras on the Vesicular Structures Related to Early and Recycling Endosomes
To more closely analyze the subcellular distribution of R-Ras, we chose MDCK cells, which were found to express R-Ras most abundantly among the cell lines examined (Supplemental Figure S1C). Quantitative immunoblotting analysis revealed that the number of R-Ras molecules was 1.5 × 105/cell, which was about one fifth of the number of Ras molecules but similar to the numbers of RalA and RalB molecules (data not shown). In MDCK cells, endogenous R-Ras was enriched on the vesicles and/or on endosome-like structures, although weak staining of the plasma membrane was also clearly seen (Figure 1, C and D). Such vesicular structures were not observed with preimmune sera or anti-R-Ras serum preabsorbed with antigen (data not shown). To further investigate the nature of R-Ras–positive endosomes, R-Ras was coexpressed with endosomal markers (Figure 2). R-Ras localization was found to overlap significantly with that of transferrin (early and recycling endosomes), Rab4A (early and recycling endosomes), Rab5A (early endosome), and Rab11A (recycling endosome), but not with that of EEA1 (early endosome) and Rab7 (late endosome). Immunoelectron micrographs showed that R-Ras was localized on cytoplasmic vesicular structures that were ∼50 nm in diameter (Figure 1E). Furthermore, double staining showed the colocalization of R-Ras and AcGFP-Endo, an endosomal marker (Figure 1F). These results indicated that the R-Ras–loaded vesicles were related to early and recycling endosomes, but not to the late endosomes.
Development of a Probe for R-Ras, Raichu-R-Ras
The unexpected observation that R-Ras was enriched on the endosomes urged us to investigate the role played by R-Ras on endosomes. For this purpose, we developed a series of FRET probes for the live-cell imaging of R-Ras activity. For the sake of brevity, only the results obtained with the Raichu-205X probe (hereafter referred to as “Raichu-R-Ras”) are described here, because this probe performed best among those tested. From the amino terminus, Raichu-R-Ras includes a modified YFP designated as Venus, human R-Ras (aa 1-199), the Ras-association domain of RalGDS (aa 785-871), a modified CFP referred to as SECFP, and the carboxy-terminal hypervariable region of R-Ras (aa 195-218) (Figure 3A). Raichu-R-Ras fulfills most requirements for a FRET probe as described in the Supplemental Material and Supplemental Figure S3.
Activation of R-Ras on Endosomes
Using Raichu-R-Ras, we visualized R-Ras activity in living Cos7 cells. The distribution of the Raichu-R-Ras probe was indistinguishable from that of the authentic R-Ras: The probe was enriched on the endosomes, but also localized diffusely on the plasma membrane (Figure 3B, Supplemental Figure S4, and Supplemental Movie 2). R-Ras activity, as visualized by the FRET level, was higher on the endosomes than on the plasma membrane. The high R-Ras activity observed on endosomes was more clearly observed with a spinning disk confocal unit (Figure 3C). To exclude the possibility that the high FRET level on the endosomes was caused by accumulation of the probe, we used control probes, i.e., Raichu-R-Ras G38V and Raichu-R-Ras S43N, the properties of which are shown in Supplemental Figure S3. The FRET level was diffusely high on both the plasma membrane and on endosomes in cells expressing Raichu-R-Ras G38V (Figure 3, C and D). In contrast, cells expressing Raichu-R-Ras S43N showed low levels of FRET, both on the plasma membrane and on endosomes (Figure 3, C and D). The intensity of the probe on each endosome did not affect the emission ratio of sensitized FRET over CFP in either the Raichu-R-Ras wild type, G38V mutant, or S43N mutant (data not shown). Therefore, these results negated the possibility that the high FRET signal observed on the endosomes of Raichu-R-Ras–expressing cells was caused by an accumulation of the probe. Interestingly, we could not observe remarkable difference in the localization of the wild-type and mutant Raichu-Ras proteins. This is probably because the effector domain of R-Ras-GTP is masked by the Ras-binding domain of the probe and suggests that the localization of the probe was determined primarily by the carboxy terminus of R-Ras.
Endosomal Localization of Rgl2/Rlf, an R-Ras Effector
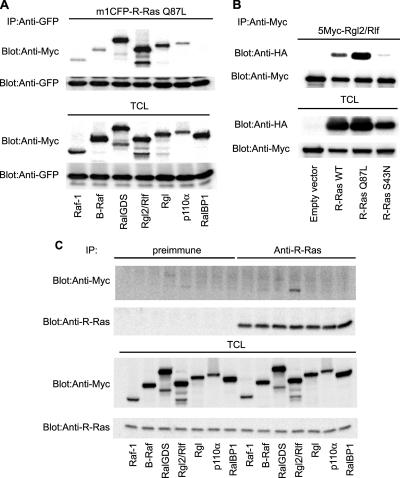
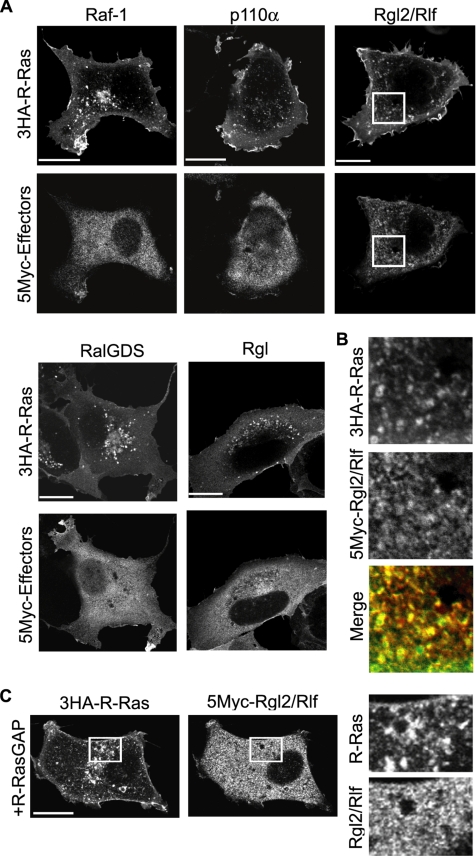
Next, we attempted to identify the signaling molecules downstream of R-Ras on the endosomes. To this end, we first compared the affinity of R-Ras for Raf-1, B-Raf, RalGDS, Rgl2/Rlf, Rgl, and the p110α subunit of PI3K, all of which are known to bind to a wide range of Ras-family GTPases (Figure 4A). K-Ras and Rap1A were used as controls for the GTPases (Supplemental Figure S5A). In a coimmunoprecipitation assay, a constitutively active mutant of R-Ras (R-Ras Q87L) was found to interact most strongly with three GEFs for Ral, i.e., RalGDS, Rgl, and Rgl2/Rlf, and less strongly with Raf-1, B-Raf, and p110α. These interactions were shown to depend on GTP loading, because the R-Ras Q87L mutant showed a markedly higher affinity for Rgl2/Rlf than did the wild-type protein and the nucleotide-free mutant, R-Ras S43N (Figure 4B). However, it should be noted that the high-affinity binding detected by coimmunoprecipitation does not necessarily indicate the signaling strength between the two associated proteins. Thus, we examined the effect of the activated R-Ras mutant, R-Ras Q87L, on the activity of downstream effectors. In agreement with the coimmunoprecipitation experiments, RalA, RalB, and Akt1 (downstream of p110 PI3K), but not MEK1 (downstream of the Raf proteins), were activated by R-Ras Q87L (Supplemental Figure S5). Next, we used R-Ras antiserum to examine whether the endogenous R-Ras protein is also associated with Ras effectors. Among the effector proteins tested, Rgl2/Rlf exhibited the strongest affinity for endogenous R-Ras (Figure 4C). Finally, we confirmed that Rgl2/Rlf, but not Raf-1 or p110α, colocalized efficiently with R-Ras on the endosomes (Figure 5, A and B). This endosomal colocalization of Rgl2/Rlf with R-Ras was abrogated by the expression of R-RasGAP (Figure 5C). These results strongly suggested that R-Ras is bound to Rgl2/Rlf on endosomes in a GTP-dependent manner.
Figure 4.
Binding of R-Ras to Rgl2/Rlf. (A) Cos7 cells expressing mCFP-tagged R-Ras-Q87L and the Myc-tagged effector proteins indicated at the bottom of the panel were used for the analysis. In p110α expression, p85α was used for coexpression to stabilize the PI3K heterodimer complex. From the cell lysates, GFP-tagged proteins were immunoprecipitated, and bound proteins were analyzed by immunoblotting with anti-Myc mAb or anti-GFP mAb. (B) Cos7 cells expressing HA-tagged R-Ras mutants and Myc-tagged Rgl2/Rlf were lysed and analyzed as described in A. (C) Endogenous R-Ras protein was immunoprecipitated with anti-R-Ras serum from Cos7 cells expressing the Myc-tagged effector proteins. The immunoprecipitates were analyzed as described in A. Preimmune serum was used as a control.
Figure 5.
Colocalization of R-Ras with Rgl2/Rlf. (A) Cos7 cells expressing HA-tagged R-Ras and Myc-tagged R-Ras effector proteins were stained with anti-HA and anti-Myc antibodies, and the cells were observed by confocal microscopy. Bar, 10 μm. (B) Enlarged images of the outlined regions in A. In the merged image, green and red indicate 3HA-R-Ras and 5Myc-Rgl2/Rlf, respectively. (C) Cos7 cells expressing HA-tagged R-Ras, Myc-tagged Rgl2/Rlf, and R-RasGAP were stained with anti-HA and anti-Myc antibodies. Right panels are enlarged images of the outlined regions in the left panels.
Regulation of Vesicular RalA Activity by R-Ras
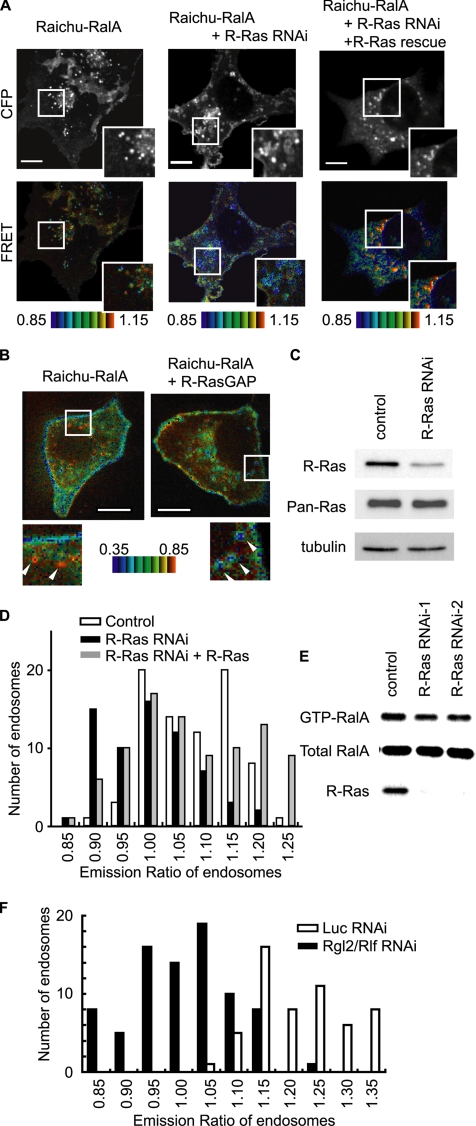
We next addressed the question of whether R-Ras regulates the activities of Ral proteins on the endosomes. In results that were consistent with those of a previous report (Shipitsin and Feig, 2004), RalA and RalB were detected both on the plasma membrane and on endosomes (Supplemental Figure S4C). Because colocalization with R-Ras was clearer in the case of RalA than RalB, we focused on RalA and examined the RalA activity on endosomes by use of Raichu-RalA, a marker for RalA activity (Takaya et al., 2004) (Figure 6). In contrast to the FRET images obtained with Raichu-R-Ras, the FRET level varied significantly among endosomes, suggesting that RalA activity varied among different types of endosomes (Figure 6A). We considered it likely that such differences in RalA activity among endosomes might have depended on the presence of active R-Ras; therefore, we examined RalA activity in cells expressing R-RasGAP or shRNA for R-Ras. Under both conditions, the number of endosomes showing high RalA activity was reduced significantly (Figure 6, B–D). This shRNA-mediated decrease in RalA activity was recovered by the coexpression of exogenous R-Ras, the cDNA of which harbors mutations in the shRNA-binding sequence. The dependence of RalA activity on R-Ras was confirmed by a pull-down assay with the RalA-binding region of RalBP1; the net amount of GTP-RalA was thus shown to have decreased by 20% (Figure 6E). It should be noted that the basal GTP level of RalA is ∼7% of total guanine nucleotides bound to RalA (Takaya et al., 2004). Therefore, if we assume based on the immunofluorescence data that the proportion of RalA on the endosomes is significantly smaller than that at the plasma membrane, a 20% decrease in the net amount of GTP-RalA seemed to be consistent with the significant reduction in the number of endosomes showing a high GTP-RalA level, as demonstrated by Raichu-RalA. Finally, we found that knockdown of Rgl2/Rlf profoundly decreased the RalA activity on the endosomes, suggesting that the R-Ras–Rgl2/Rlf complex is the principal activator of RalA on the endosomes (Figure 6F).
Figure 6.
Effect of R-Ras knockdown on RalA activity on the endosomes. (A) Cos7 cells were transfected with expression vectors as indicated at the top of the panel. For knockdown, we used pSuper-R-Ras, an shRNA vector. For the rescue from knockdown, an R-Ras mutant resistant to the shRNA vector was expressed. Sixty hours after transfection, CFP and FRET images were obtained with a spinning confocal microscope. (B) MDCK cells were transfected with expression vectors as indicated at the top of the panel and imaged 20 h after transfection as in A. Outlined regions were enlarged and are shown in the insets. Arrowheads indicate representative vesicles. Bar, 10 μm. (C) Cells transfected with an empty pSuper vector and pSuper-R-Ras were selected as described in A, and the proteins were analyzed by immunoblotting with the antibodies shown on the left. (D) Histogram of the FRET level of Raichu-RalA on the endosomes. The histograms were drawn from the data obtained from 79 endosomes in four Cos7 cells, those obtained from 66 endosomes in six pSuper-R-Ras–expressing Cos7 cells and those obtained from 89 endosomes in nine Cos7 cells expressing both pSuper-R-Ras and pCXN2-5Myc-R-Ras-rRNAi. (E) HeLa cells were transfected with control siRNA or two different siRNAs for R-Ras. After 72 h, GTP-RalA levels in the cells were analyzed by Bos' pull-down method with GST-RalBP1-RBD. The knockdown of R-Ras was also confirmed by immunoblotting. (F) The histograms were drawn from the data obtained from 55 endosomes in two Raichu-RalA-expressing Cos7 cells transfected with siRNA for luciferase and those obtained from 83 endosomes in three Raichu-RalA–expressing Cos7 cells transfected with siRNA for Rgl2/Rlf.
Requirement of R-Ras and RalA for Calcium-dependent Exocytosis
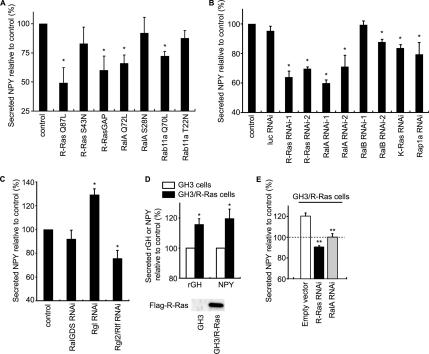
It has been reported that RalA is engaged in calcium-triggered exocytosis (Moskalenko et al., 2002; Vitale et al., 2005), which prompted us to examine the role played by R-Ras in the same process. YFP-tagged NPY and rat growth hormone (rGH) were used as markers of exocytosis (Nagai et al., 2002; Matsuno et al., 2005) (Figure 7 and Supplemental Figure S4C). In PC12 cells, depolarization-induced NPY secretion was as significantly inhibited by the expression of active R-Ras (Q87L) and R-RasGAP as by the expression of active RalA (G23V) and Rab11a (Q70L). The expression levels of recombinant proteins were examined by a quantitative immunoblotting analysis (Supplemental Figure S6). Because the expression of Rab11a T22N was less than R-Ras or RalA mutants, the ineffectiveness of this particular mutant might be ascribable to its low expression level. The role played by R-Ras and RalA was further confirmed with small interfering RNA (siRNA) (Figure 7B). The reduction of R-Ras and RalA inhibited depolarization-induced NPY secretion to a similar extent. Among the three Ral GEFs knocked down by siRNA, only the reduction of Rgl2/Rlf had an inhibitory effect on NPY secretion (Figure 7C). In GH3 pituitary adenoma cells, in which the level of expression of endogenous R-Ras was found to be low, the exogenous expression of R-Ras significantly enhanced the depolarization-induced exocytosis of both rGH and NPY (Figure 7D). The enhancement by R-Ras was abrogated by the knockdown of either R-Ras or RalA (Figure 7E). These results demonstrated that R-Ras is involved in depolarization-induced exocytosis, most likely due to the activation of Ral proteins. Furthermore, the finding that not only inhibition but also constitutive activation of R-Ras inhibits exocytosis suggests that the on-off cycle of R-Ras is required for this process.
Figure 7.
Requirement of both R-Ras and RalA for depolarization-induced exocytosis. (A) PC12 cells were transfected with pVenus-NPY together with the indicated constructs. Sixty hours after transfection, the cells were stimulated with high-potassium saline for 20 min. The efficiency of NPY-Venus secretion was determined as the ratio of NPY-Venus present in the medium versus that remaining in the cell lysates. The values were normalized to a vector control. Error bars indicate the SD from at least three experiments. The symbols indicate the results of t test analysis; *p < 0.002 compared with the control. (B and C) PC12 cells were transfected with pVenus-NPY and siRNA for luciferase, R-Ras, RalA, RalB, RalGDS, Rgl, or Rgl2/Rlf. NPY secretion upon depolarization was examined as described in A. Error bars indicate the SD from at least three experiments. The symbols indicate the results of t test analysis; *p < 0.001 compared with the control. (D) GH3 cells and GH3/ R-Ras cells were transfected with expression vectors for NPY-Venus or EYFP-GH1. Depolarization-induced secretion was monitored as described in A. The symbols indicate the results of t test analysis; *p < 0.001 compared with the control. (E) GH3/R-Ras cells were transfected with expression vectors for NPY-Venus and an shRNA vector for R-Ras, or RalA. Depolarization-induced secretion of NPY was examined as described in A. The symbols indicate the results of t test analysis; **p < 0.001 compared with the control (GH3/R-Ras cells).
DISCUSSION
We propose the following scenario for the role of R-Ras on endosomes: 1) R-Ras is activated on the surface of recycling and early endosomes, and it remains active on the vesicles derived from these endosomes. 2) The active R-Ras on these vesicles recruits Ral GEF(s). Among the candidate proteins, Rgl2/Rlf is the most plausible, because R-Ras activates both Rgl and Rgl2/Rlf more efficiently than it does RalGDS (Rodriguez-Viciana et al., 2004), and also because the endogenous R-Ras was found to colocalize with Rgl2/Rlf (Figure 5). However, other Ral GEFs that are also known to bind to R-Ras (Nancy et al., 1999; Shao and Andres, 2000; Rodriguez-Viciana et al., 2004) may be recruited to R-Ras on endosomes in other cell types. 3) These R-Ras-recruited Ral GEFs on the endosomes activate RalA, followed by the recruitment of RalA effectors to the endosomes. Among these effectors is Exo84, a component of the exocyst complex, which marks a microdomain at the plasma membrane as a delivery site for exocytotic vesicles (Grindstaff et al., 1998; Yeaman et al., 2001; Inoue et al., 2003). 4) The R-Ras-loaded vesicles from endosomes are tethered to the plasma membrane via the exocyst complex. It has been demonstrated that the interaction between RalA and the exocyst complex is essential for exocytosis (Moskalenko et al., 2002; Polzin et al., 2002; Shipitsin and Feig, 2004), and the inhibition of the exocyst complex has been shown to impair calcium-triggered exocytosis (Tsuboi et al., 2005). Therefore, our observations that R-Ras was required for the calcium-induced secretion of NPY and rGH (Figure 7) are suggestive of the positive role played by R-Ras in the formation of the exocyst complex.
Another prediction contained in our model, but not directly assessed, is that RalA regulates the assembly of the exocyst complex, both on the vesicles and on the plasma membrane, by means of binding to different components of the exocyst complex: RalA interacts not only with Exo84 but also with Sec5, another component of the exocyst complex (Feig, 2003). Sec5 is primarily present on the plasma membrane, whereas Exo84 is localized primarily on the vesicles (Moskalenko et al., 2003). In this context, it should be noted that RalA is activated on the plasma membrane either by Ras-dependent or by calcium-dependent pathways (Hofer et al., 1998; Wolthuis and Bos, 1999). Hence, the tethering of vesicles may be promoted by the two portions of the exocyst complex, both of which are anchored to the lipid membranes via RalA. One portion consists of subunits containing Exo84 and is anchored to the endosomes and/or the vesicles and endosomes by R-Ras-activated RalA, whereas the other portion consists of subunits containing Sec5, and it is anchored to the plasma membrane by RalA activated by either Ras or calcium. The results have thus far suggested that the high activity of R-Ras and RalA on the surface of vesicles is constitutive rather than stimulation regulated. Therefore, RalA activity at the plasma membrane, but not that on endosomes, may play a regulatory role in the assembly of the complete exocyst complex. In agreement with this view, we have shown that RalA is locally activated in the nascent lamellipodia of epidermal growth factor (EGF)-stimulated or migrating cells (Takaya et al., 2004). Because exocytosis plays critical roles in EGF-induced membrane ruffling and cell migration (Bretscher and Aguado-Velasco, 1998; Schmoranzer et al., 2003; Proux-Gillardeaux et al., 2005; Tayeb et al., 2005), RalA activation at the site of the membrane protrusion may indicate its role in the transport of lipid bilayer and/or integral proteins via exocytosis.
In agreement with the results of a previous report showing that RalA but not RalB regulates the delivery of E-cadherin in MDCK cells (Shipitsin and Feig, 2004), we found that only RalA was involved in calcium-triggered exocytosis (Figure 7). This difference between the two Ral proteins with respect to their involvement in exocytosis may be ascribable not only to the low binding affinity of RalB to Sec5 (Shipitsin and Feig, 2004) but also to the predominant localization of RalB on the plasma membrane (Shipitsin and Feig, 2004; Lim et al., 2005).
Currently, the mechanism underlying the high R-Ras activity on the endosomes remains unknown. A dominant-negative mutant of R-Ras has been shown to be more enriched on endosomes than is the wild-type protein (Furuhjelm and Peranen, 2003). Because a dominant-negative mutant of Ras family GTPases sequesters GEFs (Feig, 1999), these observations strongly suggest that the GEFs for R-Ras are enriched on endosomes. In contrast to RalA on the vesicles, the activity of R-Ras on the vesicles seems constant (Figures 3C and 6A). Thus, R-Ras may be activated as soon as nascent R-Ras is recruited to the vesicles and inactivated when the vesicles are fused to the plasma membrane. Although none of the GEFs for R-Ras (i.e., RasGRF1, CalDAG-GEF-I/RasGRP2, CalDAG-GEF-II/RasGRP1, CalDAG-GEF-III/RasGRP3, and C3G) have been shown to localize on the endosomes (Ohba et al., 2000), this failure to detect GEFs on the endosomes may simply reflect a lack of high-affinity antibodies that could be applied for immunostaining. Interestingly, GAPs for R-Ras seem to localize primarily on the plasma membrane (Anderson et al., 1990; Margolis et al., 1990; Cozier et al., 2003; Oinuma et al., 2004). Thus, R-Ras would be expected to be inactivated when it is transported from the endosomes to the plasma membrane. This inactivation of R-Ras might serve to liberate the components of the exocyst complex and send them back into the cytoplasm or to send R-Ras back to the endosomes from the plasma membrane.
Previous studies have implicated R-Ras in the activation of integrin (Zhang et al., 1996; Keely et al., 1999; Berrier et al., 2000; Self et al., 2001; Oinuma et al., 2006) and also in cell migration and adhesion (Nakada et al., 2005; Wozniak et al., 2005); however, the molecular mechanisms underlying these phenomena remain elusive. Our finding that the R-Ras-Rgl2/Rlf-RalA pathway regulates exocytosis may account for some of these biological activities of R-Ras. It is already known that the inhibition of exocytosis impairs integrin recycling and thereby also cell migration and cell adhesion (Proux-Gillardeaux et al., 2005; Tayeb et al., 2005). Hence, the inhibition of integrin by the suppression of R-Ras might initially be caused by the disruption of exocytosis. Although no direct evidence supporting the involvement of RalA in the recycling of integrin has yet been reported, Ral proteins have been shown to be implicated in cell migration (Gildea et al., 2002; Takaya et al., 2004; Oxford et al., 2005), a process in which integrin is thought to be coordinately activated and inactivated. Therefore, it is reasonable to speculate that the R-Ras-Rgl2/Rlf-RalA pathway is involved in the recycling of integrin and thereby also in the regulation of integrin activity.
In conclusion, we observed high R-Ras activity on early and recycling endosomes. This high R-Ras activity recruits Rlf/Rgl2 and thereby activates RalA, followed by the assembly of a portion of the exocyst complex. Importantly, in addition to RalA, other low-molecular-weight GTPases belonging to different families (i.e., TC10, Arf6, and Rab11) are also known to interact with the exocyst complex (Prigent et al., 2003; Inoue et al., 2003; Zhang et al., 2004; Wu et al., 2005). Further study will be needed to determine whether these GTPases of different families coordinately regulate the exocyst complex on the same endosomes, or whether there are distinct classes of endosomes containing only some of these GTPases. It would be of particular importance to examine the dynamic activity changes of these GTPases during the vesicular transport, which would be best examined by FRET-based imaging techniques used here.
Supplementary Material
ACKNOWLEDGMENTS
We thank L. A. Feig, R. Y. Tsien, Y. Fukui, K. Kaibuchi, A. Kikuchi, S. Kuroda, B. J. Mayer, N. Minato, A. Miyawaki, and Y. Takai for the provision of reagents and N. Yoshida, N. Fujimoto, and K. Fukuhara for technical assistance. This work was supported by grants-in-aid for scientific research and for cancer research from the Ministry of Education, Science, Sports and Culture of Japan, and by a grant from the Health Science Foundation of Japan. A.T. was supported by Research Fellowships from the Japan Society for the Promotion of Science for Young Scientists.
Abbreviations used:
- EGF
epidermal growth factor
- FRET
fluorescence (Förster's) resonance energy transfer
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchange factor
- NPY
neuropeptide Y
- PI3K
p110α subunit of phosphoinositide-3-kinase.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-08-0765) on March 7, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Akagi T., Sasai K., Hanafusa H. Refractory nature of normal human diploid fibroblasts with respect to oncogene-mediated transformation. Proc. Natl. Acad. Sci. USA. 2003;100:13567–13572. doi: 10.1073/pnas.1834876100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D., Koch C. A., Grey L., Ellis C., Moran M. F., Pawson T. Binding of SH2 domains of phospholipase C gamma 1, GAP, and Src to activated growth factor receptors. Science. 1990;250:979–982. doi: 10.1126/science.2173144. [DOI] [PubMed] [Google Scholar]
- Aoki K., Nakamura T., Fujikawa K., Matsuda M. Local phosphatidylinositol 3,4,5-trisphosphate accumulation recruits Vav2 and Vav3 to activate Rac1/Cdc42 and initiate neurite outgrowth in nerve growth factor-stimulated PC12 cells. Mol. Biol. Cell. 2005;16:2207–2217. doi: 10.1091/mbc.E04-10-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrier A. L., Mastrangelo A. M., Downward J., Ginsberg M., LaFlamme S. E. Activated R-ras, Rac1, PI 3-kinase and PKCepsilon can each restore cell spreading inhibited by isolated integrin beta1 cytoplasmic domains. J. Cell Biol. 2000;151:1549–1560. doi: 10.1083/jcb.151.7.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher M. S., Aguado-Velasco C. EGF induces recycling membrane to form ruffles. Curr. Biol. 1998;8:721–724. doi: 10.1016/s0960-9822(98)70281-7. [DOI] [PubMed] [Google Scholar]
- Cox A. D., Brtva T. R., Lowe D. G., Der C. J. R-Ras induces malignant, but not morphologic, transformation of NIH3T3 cells. Oncogene. 1994;9:3281–3288. [PubMed] [Google Scholar]
- Cozier G. E., Bouyoucef D., Cullen P. J. Engineering the phosphoinositide-binding profile of a class I pleckstrin homology domain. J. Biol. Chem. 2003;278:39489–39496. doi: 10.1074/jbc.M307785200. [DOI] [PubMed] [Google Scholar]
- Feig L. A. Tools of the trade: use of dominant–inhibitory mutants of Ras-family GTPases. Nat. Cell Biol. 1999;1:E25–E27. doi: 10.1038/10018. [DOI] [PubMed] [Google Scholar]
- Feig L. A. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 2003;13:419–425. doi: 10.1016/s0962-8924(03)00152-1. [DOI] [PubMed] [Google Scholar]
- Finger F. P., Hughes T. E., Novick P. Sec3p is a spatial landmark for polarized secretion in budding yeast. Cell. 1998;92:559–571. doi: 10.1016/s0092-8674(00)80948-4. [DOI] [PubMed] [Google Scholar]
- Friedrich G. A., Hildebrand J. D., Soriano P. The secretory protein Sec8 is required for paraxial mesoderm formation in the mouse. Dev. Biol. 1997;192:364–374. doi: 10.1006/dbio.1997.8727. [DOI] [PubMed] [Google Scholar]
- Furuhjelm J., Peranen J. The C-terminal end of R-Ras contains a focal adhesion targeting signal. J. Cell Sci. 2003;116:3729–3738. doi: 10.1242/jcs.00689. [DOI] [PubMed] [Google Scholar]
- Gildea J. J., Harding M. A., Seraj M. J., Gulding K. M., Theodorescu D. The role of Ral A in epidermal growth factor receptor-regulated cell motility. Cancer Res. 2002;62:982–985. [PubMed] [Google Scholar]
- Gonzalez-Garcia A., Pritchard C. A., Paterson H. F., Mavria G., Stamp G., Marshall C. J. RalGDS is required for tumor formation in a model of skin carcinogenesis. Cancer Cell. 2005;7:219–226. doi: 10.1016/j.ccr.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Gotoh T., Niino Y., Tokuda M., Hatase O., Nakamura S., Matsuda M., Hattori S. Activation of R-Ras by Ras-guanine nucleotide-releasing factor. J. Biol. Chem. 1997;272:18602–18607. doi: 10.1074/jbc.272.30.18602. [DOI] [PubMed] [Google Scholar]
- Grindstaff K. K., Yeaman C., Anandasabapathy N., Hsu S. C., Rodriguez-Boulan E., Scheller R. H., Nelson W. J. Sec6/8 complex is recruited to cell-cell contacts and specifies transport vesicle delivery to the basal-lateral membrane in epithelial cells. Cell. 1998;93:731–740. doi: 10.1016/s0092-8674(00)81435-x. [DOI] [PubMed] [Google Scholar]
- Gromley A., Yeaman C., Rosa J., Redick S., Chen C. T., Mirabelle S., Guha M., Sillibourne J., Doxsey S. J. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell. 2005;123:75–87. doi: 10.1016/j.cell.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Hamad N. M., Elconin J. H., Karnoub A. E., Bai W., Rich J. N., Abraham R. T., Der C. J., Counter C. M. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 2002;16:2045–2057. doi: 10.1101/gad.993902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer F., Berdeaux R., Martin G. S. Ras-independent activation of Ral by a Ca(2+)-dependent pathway. Curr. Biol. 1998;8:839–842. doi: 10.1016/s0960-9822(98)70327-6. [DOI] [PubMed] [Google Scholar]
- Huff S. Y., Quilliam L. A., Cox A. D., Der C. J. R-Ras is regulated by activators and effectors distinct from those that control Ras function. Oncogene. 1997;14:133–143. doi: 10.1038/sj.onc.1200815. [DOI] [PubMed] [Google Scholar]
- Inoue M., Chang L., Hwang J., Chiang S. H., Saltiel A. R. The exocyst complex is required for targeting of Glut4 to the plasma membrane by insulin. Nature. 2003;422:629–633. doi: 10.1038/nature01533. [DOI] [PubMed] [Google Scholar]
- Keely P. J., Rusyn E. V., Cox A. D., Parise L. V. R-Ras signals through specific integrin alpha cytoplasmic domains to promote migration and invasion of breast epithelial cells. J. Cell Biol. 1999;145:1077–1088. doi: 10.1083/jcb.145.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Ruoslahti E. R-Ras is a global regulator of vascular regeneration that suppresses intimal hyperplasia and tumor angiogenesis. Nat. Med. 2005;11:1346–1350. doi: 10.1038/nm1324. [DOI] [PubMed] [Google Scholar]
- Kurokawa K., Itoh R. E., Yoshizaki H., Nakamura T., Matsuda M. Coactivation of Rac1 and Cdc42 at lamellipodia and membrane ruffles induced by epidermal growth factor. Mol. Biol. Cell. 2004a;15:1003–1010. doi: 10.1091/mbc.E03-08-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K., Takaya A., Terai K., Fujioka A., Matsuda M. Visulalizing the signal transduction pathways in living cells with GFP-based FRET probes. Acta Histochem. Cytochem. 2004b;37:347–355. [Google Scholar]
- Lim K. H., Baines A. T., Fiordalisi J. J., Shipitsin M., Feig L. A., Cox A. D., Der C. J., Counter C. M. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell. 2005;7:533–545. doi: 10.1016/j.ccr.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Lipschutz J. H., Mostov K. E. Exocytosis: the many masters of the exocyst. Curr. Biol. 2002;12:R212–R214. doi: 10.1016/s0960-9822(02)00753-4. [DOI] [PubMed] [Google Scholar]
- Lowe D. G., Capon D. J., Delwart E., Sakaguchi A. Y., Naylor S. L., Goeddel D. V. Structure of the human and murine R-ras genes, novel genes closely related to ras proto-oncogenes. Cell. 1987;48:137–146. doi: 10.1016/0092-8674(87)90364-3. [DOI] [PubMed] [Google Scholar]
- Margolis B., Li N., Koch A., Mohammadi M., Hurwitz D. R., Zilberstein A., Ullrich A., Pawson T., Schlessinger J. The tyrosine phosphorylated carboxy terminus of the EGF receptor is a binding site for GAP and PLC-gamma. EMBO J. 1990;9:4375–4380. doi: 10.1002/j.1460-2075.1990.tb07887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marte B. M., Rodriguez-Viciana P., Wennstrom S., Warne P. H., Downward J. R-Ras can activate the phosphoinositide 3-kinase but not the MAP kinase arm of the Ras effector pathways. Curr. Biol. 1997;7:63–70. doi: 10.1016/s0960-9822(06)00028-5. [DOI] [PubMed] [Google Scholar]
- Matsuno A., Mizutani A., Itoh J., Takekoshi S., Nagashima T., Okinaga H., Takano K., Osamura R. Y. Establishment of stable GH3 cell line expressing enhanced yellow fluorescein protein-growth hormone fusion protein. J. Histochem. Cytochem. 2005;53:1177–1180. doi: 10.1369/jhc.5B6708.2005. [DOI] [PubMed] [Google Scholar]
- Mochizuki N., Yamashita S., Kurokawa K., Ohba Y., Nagai T., Miyawaki A., Matsuda M. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 2001;411:1065–1068. doi: 10.1038/35082594. [DOI] [PubMed] [Google Scholar]
- Moskalenko S., Henry D. O., Rosse C., Mirey G., Camonis J. H., White M. A. The exocyst is a Ral effector complex. Nat. Cell Biol. 2002;4:66–72. doi: 10.1038/ncb728. [DOI] [PubMed] [Google Scholar]
- Moskalenko S., Tong C., Rosse C., Mirey G., Formstecher E., Daviet L., Camonis J., White M. A. Ral GTPases regulate exocyst assembly through dual subunit interactions. J. Biol. Chem. 2003;278:51743–51748. doi: 10.1074/jbc.M308702200. [DOI] [PubMed] [Google Scholar]
- Murthy M., Garza D., Scheller R. H., Schwarz T. L. Mutations in the exocyst component Sec5 disrupt neuronal membrane traffic, but neurotransmitter release persists. Neuron. 2003;37:433–447. doi: 10.1016/s0896-6273(03)00031-x. [DOI] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Nakada M., Niska J. A., Tran N. L., McDonough W. S., Berens M. E. EphB2/R-Ras signaling regulates glioma cell adhesion, growth, and invasion. Am. J. Pathol. 2005;167:565–576. doi: 10.1016/S0002-9440(10)62998-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima S., Morinaka K., Koyama S., Ikeda M., Kishida M., Okawa K., Iwamatsu A., Kishida S., Kikuchi A. Small G protein Ral and its downstream molecules regulate endocytosis of EGF and insulin receptors. EMBO J. 1999;18:3629–3642. doi: 10.1093/emboj/18.13.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nancy V., Wolthuis R. M., de Tand M. F., Janoueix-Lerosey I., Bos J. L., de Gunzburg J. Identification and characterization of potential effector molecules of the Ras-related GTPase Rap2. J. Biol. Chem. 1999;274:8737–8745. doi: 10.1074/jbc.274.13.8737. [DOI] [PubMed] [Google Scholar]
- Nishigaki M., Aoyagi K., Danjoh I., Fukaya M., Yanagihara K., Sakamoto H., Yoshida T., Sasaki H. Discovery of aberrant expression of R-RAS by cancer-linked DNA hypomethylation in gastric cancer using microarrays. Cancer Res. 2005;65:2115–2124. doi: 10.1158/0008-5472.CAN-04-3340. [DOI] [PubMed] [Google Scholar]
- Ohba Y., Mochizuki N., Yamashita S., Chan A. M., Schrader J. W., Hattori S., Nagashima K., Matsuda M. Regulatory proteins of R-Ras, TC21/R-Ras2, and M-Ras/R-Ras3. J. Biol. Chem. 2000;275:20020–20026. doi: 10.1074/jbc.M000981200. [DOI] [PubMed] [Google Scholar]
- Oinuma I., Ishikawa Y., Katoh H., Negishi M. The Semaphorin 4D receptor Plexin-B1 is a GTPase activating protein for R-Ras. Science. 2004;305:862–865. doi: 10.1126/science.1097545. [DOI] [PubMed] [Google Scholar]
- Oinuma I., Katoh H., Negishi M. Semaphorin 4D/Plexin-B1-mediated R-Ras GAP activity inhibits cell migration by regulating beta(1) integrin activity(2) J. Cell Biol. 2006;173:601–613. doi: 10.1083/jcb.200508204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxford G., Owens C. R., Titus B. J., Foreman T. L., Herlevsen M. C., Smith S. C., Theodorescu D. RalA and RalB: antagonistic relatives in cancer cell migration. Cancer Res. 2005;65:7111–7120. doi: 10.1158/0008-5472.CAN-04-1957. [DOI] [PubMed] [Google Scholar]
- Polzin A., Shipitsin M., Goi T., Feig L. A., Turner T. J. Ral-GTPase influences the regulation of the readily releasable pool of synaptic vesicles. Mol. Cell. Biol. 2002;22:1714–1722. doi: 10.1128/MCB.22.6.1714-1722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent M., Dubois T., Raposo G., Derrien V., Tenza D., Rosse C., Camonis J., Chavrier P. ARF6 controls post-endocytic recycling through its downstream exocyst complex effector. J. Cell Biol. 2003;163:1111–1121. doi: 10.1083/jcb.200305029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proux-Gillardeaux V., Gavard J., Irinopoulou T., Mege R. M., Galli T. Tetanus neurotoxin-mediated cleavage of cellubrevin impairs epithelial cell migration and integrin-dependent cell adhesion. Proc. Natl. Acad. Sci. USA. 2005;102:6362–6367. doi: 10.1073/pnas.0409613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilliam L.A., Rebhun J. F., Castro A. F. A growing family of guanine nucleotide exchange factors is responsible for activation of Ras-family GTPases. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:391–444. doi: 10.1016/s0079-6603(02)71047-7. [DOI] [PubMed] [Google Scholar]
- Rangarajan A., Hong S. J., Gifford A., Weinberg R. A. Species- and cell type-specific requirements for cellular transformation. Cancer Cell. 2004;6:171–183. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Rey I., Taylor-Harris P., van Erp H., Hall A. R-ras interacts with rasGAP, neurofibromin and c-raf but does not regulate cell growth or differentiation. Oncogene. 1994;9:685–692. [PubMed] [Google Scholar]
- Rodriguez-Viciana P., Sabatier C., McCormick F. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol. Cell. Biol. 2004;24:4943–4954. doi: 10.1128/MCB.24.11.4943-4954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoranzer J., Kreitzer G., Simon S. M. Migrating fibroblasts perform polarized, microtubule-dependent exocytosis towards the leading edge. J. Cell Sci. 2003;116:4513–4519. doi: 10.1242/jcs.00748. [DOI] [PubMed] [Google Scholar]
- Self A. J., Caron E., Paterson H. F., Hall A. Analysis of R-Ras signalling pathways. J. Cell Sci. 2001;114:1357–1366. doi: 10.1242/jcs.114.7.1357. [DOI] [PubMed] [Google Scholar]
- Shao H., Andres D. A. A novel RalGEF-like protein, RGL3, as a candidate effector for rit and Ras. J. Biol. Chem. 2000;275:26914–26924. doi: 10.1074/jbc.M002241200. [DOI] [PubMed] [Google Scholar]
- Shipitsin M., Feig L. A. RalA but not RalB enhances polarized delivery of membrane proteins to the basolateral surface of epithelial cells. Mol. Cell. Biol. 2004;24:5746–5756. doi: 10.1128/MCB.24.13.5746-5756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaargaren M., Bischoff J. R. Identification of the guanine nucleotide dissociation stimulator for Ral as a putative effector molecule of R-ras, H-ras, K-ras, and Rap. Proc. Natl. Acad. Sci. USA. 1994;91:12609–12613. doi: 10.1073/pnas.91.26.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaargaren M., Martin G. A., McCormick F., Fernandez-Sarabia M. J., Bischoff J. R. The Ras-related protein R-ras interacts directly with Raf-1 in a GTP-dependent manner. Biochem. J. 1994;300:303–307. doi: 10.1042/bj3000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaya A., Ohba Y., Kurokawa K., Matsuda M. RalA activation at nascent lamellipodia of epidermal growth factor-stimulated Cos7 cells and migrating Madin-Darby canine kidney cells. Mol. Biol. Cell. 2004;15:2549–2557. doi: 10.1091/mbc.E03-11-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayeb M. A., Skalski M., Cha M. C., Kean M. J., Scaife M., Coppolino M. G. Inhibition of SNARE-mediated membrane traffic impairs cell migration. Exp. Cell Res. 2005;305:63–73. doi: 10.1016/j.yexcr.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Tsuboi T., Ravier M. A., Xie H., Ewart M. A., Gould G. W., Baldwin S. A., Rutter G. A. Mammalian exocyst complex is required for the docking step of insulin vesicle exocytosis. J. Biol. Chem. 2005;280:25565–25570. doi: 10.1074/jbc.M501674200. [DOI] [PubMed] [Google Scholar]
- Vega I. E., Hsu S. C. The exocyst complex associates with microtubules to mediate vesicle targeting and neurite outgrowth. J. Neurosci. 2001;21:3839–3848. doi: 10.1523/JNEUROSCI.21-11-03839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale N., Mawet J., Camonis J., Regazzi R., Bader M. F., Chasserot-Golaz S. The Small GTPase RalA controls exocytosis of large dense core secretory granules by interacting with ARF6-dependent phospholipase D1. J. Biol. Chem. 2005;280:29921–29928. doi: 10.1074/jbc.M413748200. [DOI] [PubMed] [Google Scholar]
- Wolthuis R. M., Bos J. L. Ras caught in another affair: the exchange factors for Ral. Curr. Opin. Genet. Dev. 1999;9:112–117. doi: 10.1016/s0959-437x(99)80016-1. [DOI] [PubMed] [Google Scholar]
- Wozniak M. A., Kwong L., Chodniewicz D., Klemke R. L., Keely P. J. R-Ras controls membrane protrusion and cell migration through the spatial regulation of Rac and Rho. Mol. Biol. Cell. 2005;16:84–96. doi: 10.1091/mbc.E04-04-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Mehta S. Q., Pichaud F., Bellen H. J., Quiocho F. A. Sec15 interacts with Rab11 via a novel domain and affects Rab11 localization in vivo. Nat. Struct. Mol. Biol. 2005;12:879–885. doi: 10.1038/nsmb987. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Matsui T., Nakafuku M., Iwamatsu A., Kaibuchi K. A novel GTPase-activating protein for R-Ras. J. Biol. Chem. 1995;270:30557–30561. doi: 10.1074/jbc.270.51.30557. [DOI] [PubMed] [Google Scholar]
- Yeaman C., Grindstaff K. K., Wright J. R., Nelson W. J. Sec6/8 complexes on trans-Golgi network and plasma membrane regulate late stages of exocytosis in mammalian cells. J. Cell Biol. 2001;155:593–604. doi: 10.1083/jcb.200107088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- Zhang X. M., Ellis S., Sriratana A., Mitchell C. A., Rowe T. Sec15 is an effector for the Rab11 GTPase in mammalian cells. J. Biol. Chem. 2004;279:43027–43034. doi: 10.1074/jbc.M402264200. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Vuori K., Wang H., Reed J. C., Ruoslahti E. Integrin activation by R-ras. Cell. 1996;85:61–69. doi: 10.1016/s0092-8674(00)81082-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.