Abstract
Scar/WAVE proteins, members of the conserved Wiskott-Aldrich syndrome (WAS) family, promote actin polymerization by activating the Arp2/3 complex. A number of proteins, including a complex containing Nap1, PIR121, Abi1/2, and HSPC300, interact with Scar/WAVE, though the role of this complex in regulating Scar function remains unclear. Here we identify a short N-terminal region of Dictyostelium Scar that is necessary and sufficient for interaction with HSPC300 and Abi in vitro. Cells expressing Scar lacking this N-terminal region show abnormalities in F-actin distribution, cell morphology, movement, and cytokinesis. This is true even in the presence of wild-type Scar. The data suggest that the first 96 amino acids of Scar are necessary for participation in a large-molecular-weight protein complex, and that this Scar-containing complex is responsible for the proper localization and regulation of Scar. The presence of mis-regulated or unregulated Scar has significant deleterious effects on cells and may explain the need to keep Scar activity tightly controlled in vivo either by assembly in a complex or by rapid degradation.
INTRODUCTION
Changes in the actin cytoskeleton are necessary for a variety of eukaryotic cellular functions such as cell migration, chemotaxis, morphogenesis, endocytosis, and cytokinesis. Expansion and protrusion of a cell's leading edge is driven by a force generated by cycles of actin polymerization and depolymerization (reviewed in (Lauffenburger and Horwitz, 1996; Borisy and Svitkina, 2000; Pollard and Borisy, 2003). A major component of the molecular machinery involved in actin polymerization is the Arp2/3 complex, a highly conserved set of proteins (Machesky et al., 1994) that catalyze the nucleation of actin filaments (Welch et al., 1997; Mullins et al., 1998). Purified Arp2/3 complex itself is essentially inactive in vitro and requires extrinsic activators. The best studied activators are Wiskott-Aldrich syndrome protein (WASp) family members, which bind and activate the complex via a conserved carboxyl terminal region (Machesky and Insall, 1998; Rohatgi et al., 1999; Winter et al., 1999; Yarar et al., 1999; Marchand et al., 2001; Zalevsky et al., 2001).
The WASp protein family has two subgroups, the WASps and the Scar/WAVEs. The protein family is highly conserved; every eukaryote examined has at least one ortholog. The proteins share a high degree of sequence similarity at the carboxy terminus that include conserved regions that interact with actin and the Arp2/3 complex. The amino terminal region of WASp family proteins is, however, highly divergent (Bear et al., 1998; Saxe, 2003; Bompard and Caron, 2004). WASp has a unique GTPase binding region (GBD) and WH1 domain, whereas SCAR has a unique Scar homology domain (SHD) and basic region. Vertebrates have multiple isoforms of each subgroup: WASp, N-WASp, Scar1-3 (also called WAVE1-3), whereas animals such as C. elegans, Drosophila melanogaster, and the amoeba Dictyostelium discoideum have single WASp and Scar genes. Dictyostelium Scar null cells are reduced in size, have aberrant actin distribution and impaired motility, and proceed more quickly through development culminating in smaller multicellular fruiting bodies compared with wild-type cells (Bear, 1998; Bear et al., 1998; Blagg et al., 2003b). Subsequent studies showed Scar to be crucial for the integrity of actin dependent structures in Drosophila development (Zallen et al., 2002), as well as murine and Arabidopsis development (Dahl et al., 2003; Soderling et al., 2003; Yamazaki et al., 2003; Yan et al., 2003; Frank et al., 2004).
WASp family proteins are downstream effectors of Rho family GTPases (Miki et al., 1998; Bishop and Hall, 2000; Mullins, 2000; Hufner et al., 2002; Vartiainen and Machesky, 2004). WASp binds directly to Cdc42 via a conserved 21-amino acid GTPase binding domain (Symons et al., 1996), but there is no obvious GTPase binding domain present in Scar, nor any reported evidence for Scar binding directly to small G-proteins. Studies of WASp and N-WASp reported the proteins to exist in an autoinhibited conformation in vitro until activated (Miki and Takenawa, 1998; Kim et al., 2000). Recent data, however, suggest that WASp may also be physiologically regulated by a multiprotein complex (Ho et al., 2004). Scar also appears to be regulated by a large-molecular-weight protein complex. Mammalian Scar1/WAVE1 (and more recently Scar2/WAVE2) was copurified with four other proteins: HSPC300, PIR121 (a Rac-interacting protein), Nap1 (a Nck-associated protein), and Abi1/2 (abelson tyrosine kinase–interacting protein). Data indicate this protein complex regulates Scar (Eden et al., 2002; Blagg et al., 2003a; Kunda et al., 2003; Rogers et al., 2003; Innocenti et al., 2004), but the precise nature of molecular mechanisms underlying this regulation or the biological significance of the complex is not understood.
Here we show that D. discoideum Scar does exist in a large protein complex in vivo and binds to Abi and HSPC300 in vitro and that the binding site is contained within the extreme N-terminus of Scar. We show that cells expressing Scar lacking this binding region produce a stable protein that is not properly localized or degraded and results in cells with motility defects and are unable to chemotax in a spatial gradient of the chemoattractant cAMP. The cells have poorly defined leading edges, lack normal F-actin distribution and cell polarity, are unusually adherent to the underlying substrate, and become multinucleate, all even in the presence of endogenous Scar.
MATERIALS AND METHODS
Cells and Reagents
Cells of the Ax3-derived parental HPS400 strain were cultured in HL-5 medium either on 100 × 15-mm Petri dishes or in suspension. Mutant strains were maintained in HL-5 medium supplemented with 10 μg of G418/ml.
PCR
All PCR reactions (100 μl volume) contained 50 ng template DNA, 10 nM primers (Scar) or 250 nM primers (HSPC300 and Abi), 1.0 U VentR polymerase (New England Biolabs, Beverly, MA) for Scar or 1 U Recombinant Taq polymerase (Invitrogen, Carlsbad, CA) for HSPC300 and Abi, MgCl2-containing buffer, and 250 μM dNTPs. Reactions were 30 cycles of denaturing for 45 s at 94°C, annealing for 60 s at 50°C (Scar and HSPC300) or 55°C (Abi), 2-min extension at 72°C (Scar) or 65°C (Abi) or 60-s extension at 65°C (HSPC300). All templates were full-length cDNAs including pScar9 (Scar), SSL226 (HSPC300), and FC-AN22 (Abi). Primers are listed in the Supplementary Materials.
In Vitro Transcription and Translation
In vitro translation reactions were performed using TNT T7 Quick for PCR (Promega, Madison, WI), following the manufacturer's instructions. Briefly, ∼1 μg of PCR-generated DNA, 10–20 μCi [35S]methionine (Amersham Biosciences, Piscataway, NJ) and 1× Complete protease inhibitors (Roche, Indianapolis, IN) were added to each reaction. KCl and MgCl2 were added to a final concentration for 30 and 35 mM, respectively. The reactions were incubated at 30°C for 1.5 h and then placed on ice until used for immunoprecipitation reactions.
Binding Assay
Anti-T7 agarose-coupled beads were purchased from Bethyl Laboratories (Montgomery, TX), and used according to the manufacturer's instructions. Protein G Plus Agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) were coupled to either anti-hemagglutinin (HA) high-affinity rat monoclonal antibody 3F10 (Roche) or anti-Myc monoclonal antibody 9B11 (Cell Signaling, Beverly, MA) by end-over-end mixing overnight at 4°C. Antibody, 0.5 μg, was added to 500 μl 50% bead slurry. Unbound antibody was removed by washing beads three times with buffer 1 (PBS with 0.01% Triton X-100 and Complete protease inhibitors; Roche), with gentle shaking for 10 min. For binding assay, 300 μl buffer 1, 50 μl antibody coupled beads, and a 10-μl aliquot of appropriate TNT reaction mixture, representing 20% of the total reaction, were added to a 1.5-ml tube, sealed with parafilm, and mixed end-over-end overnight at 4°C. Beads were collected by centrifugation, 1500 rpm for 1 min at 4°C. Beads were washed three times with 500 μl buffer 1, 15 min per wash with gentle shaking at 4°C. Beads were then resuspended in 50 μl 1× solubilization buffer, placed at 65°C for 15 min, and then loaded 20 μl/lane onto 4–20% SDS–PAGE gel (Bio-Rad, Hercules, CA). Electrophoresis was performed at 150 V. For autoradiography, gel was soaked in 16% sialicylic acid for 20 min, fixed with a 7% acetic acid, 7% methanol, 2% glycerol solution, and dried. Signal was detected using Hyperfilm ECL (Amersham Biosciences). For Western analysis, proteins were transferred to nitrocellulose, blocked for nonspecific binding overnight with nonfat milk (5%) in PBS/Tween (0.05%). After incubation with antibody, the blots were washed three times with PBS/Tween and further incubated with horseradish peroxidase–conjugated secondary antibody. After washing, ECL Western Detection Reagent (Amersham Biosciences) was used to develop the chemiluminescent signal, which was detected using Hyperfilm ECL (Amersham Biosciences).
Chemotaxis Assays
Cells were polarized by shaking a 50-ml flask with ∼1 × 107 cells/ml at 225 rpm while pulsing cells with cAMP at 6-min intervals to a final concentration of 10 nM for 4–6 h. After cells were polarized, they were diluted and placed on a no. 1.5 glass-bottomed microscope slide (Lab-Tek II/Nalge Nunc, Naperville, IL) and allowed to adhere to the slide for ∼10 min. A micropipette (Femtotips, Eppendorf, Fremont, CA) containing 100 μM cAMP solution was brought into the field of view and introduced to cells using a micromanipulator (Eppendorf), and a gradient was created using an Eppendorf FemtoJet Microinjector. Cells were exposed to the cAMP gradient, and cellular responses were recorded by time-lapse videomicroscopy. Cells were imaged every 12 s for 20 min.
Actin Assay
To assess the basal level of F-actin within cells, a modified assay based on the cAMP-stimulated actin polymerization assay (Condeelis et al., 1990) was performed. Vegetatively growing cells were collected into 15-ml conical tubes (Corning Glass Works, Corning, NY), counted, and then pelleted in a clinical centrifuge (International Equipment, Needham Heights, MA) for 3 min. Cells were washed twice with DB (5 mM Na2PO4, 5 mM KH2PO4, 2 mM MgSO4, 0.2 mM CaCl2), resuspended in 5 ml DB, and counted again to correct for losses during washes. Volumes were adjusted so that cells were at a density of 2 × 107 cells/ml. Aliquots of cells, 100 μl, were pipetted into 400 μl of fixative (0.1% Triton X-100, 10 mM K2PO4, 10 mM Pipes, 5 mM EGTA, 2 mM MgSO4, 0.1% saponin, 3% formaldehyde) containing 5 μM tetramethylrhodamine B isothiocyanate (TRITC)-phalloidin. Samples were wrapped in foil to protect from light and placed on a platform shaker for 1 h at room temperature to allow the TRITC-phalloidin to bind to the cellular actin. Samples were centrifuged in an Eppendorf microfuge for 2 min at 14K rpm. Pellets were washed twice with 200 μl of saponin buffer (10 mM K2PO4, 10 mM KH2PO4, 10 mM Pipes, 5 mM EGTA, 2 mM MgSO4, 0.1% saponin), resuspended in 500 μl 100% methanol, and placed on a platform shaker overnight at 4°C. Cellular material was pelleted by centrifugation in an Eppendorf microfuge at 14K rpm for 2 min. The supernatant containing the TRITC-phalloidin–bound F-actin was collected, placed in cuvettes (Bio-Rad), and the fluorescence was measured using a fluorimeter set at an excitation of 554 nm and emission of 573 nm. Relative F-actin was calculated as a ratio of measured fluorescence of mutant strain to measured fluorescence of the HPS400 strain.
Imaging
Axenically grown parental (HPS400) and scar− cells expressing the empty vector, parental, and scar− cells expressing ScarΔ96-GFP (green fluorescent protein) were allowed to adhere to glass coverslips and were observed with Nomarski differential interference contrast (DIC). For live cell imaging, a Nikon Eclipse TE2000 with a 60× objective was used (Melville, NY). Images were recorded using IPLab3 software. Movies are 10 min in length, with 12-s intervals. For fixed cells analysis, cells were fixed in 2% formaldehyde/DB for 20 min, washed twice with DB, permeabilized with 0.02% Triton X-100 (Sigma, St. Louis, MO)/DB for 5 min, and washed twice with DB. Cell were blocked overnight at 4°C with 3% BSA/DB. F-actin was visualized with 50 nM TRITC-phalloidin and imaged using a Zeiss LSM510/Axiovert 100M laser scanning confocal microscope.
Cell Extract
Cells, 5 × 107, were collected and resuspended in 750 μl buffer 2 (50 mM Tris, 20 mM KCl, 1 mM MgCl2, 1 mM DTT, pH 7.4) containing 0.5 mM ATP and Complete protease inhibitors (Roche). All manipulations were done on ice or at 4°6 C. Cells were mechanically lysed using polycarbonate, 5-μm, 25-mm membranes (Osmonics, Minnetonka, MN) The extract was cleared by centrifugation (14,000 rpm for 30 min).
Sucrose Gradient
Eleven-milliliter 7–20% sucrose gradients were generated in buffer 2 (50 mM Tris, pH 7.4, 20 mM KCl, 1 mM MgCl2, 1 mM DTT) containing 0.5 mM ATP and Complete protease inhibitors (Roche). Markers of 1 μg/ml (thyroglobin 669 kDa, apoferritin 443 kDa, bovine serum albumin 66 kDa, and carbonic Anhydrase 29 kDa; Sigma) or 400–500 μl clarified cell extract was loaded on to the top of a sucrose gradient. The gradients were run for 16 h at 36,000 rpm in a SW41 rotor in a Beckman Optima LE-80K Ultracentifuge (Fullerton, CA). Fractions of 0.5-ml were collected from the bottom of the gradient. A 15-μl fraction was mixed with 5 μl 4× sample buffer and loaded onto a 10% SDS-PAGE gel, and electrophoresis, protein transfer, and immunoblotting were carried out as described above.
RESULTS
Dictyostelium Scar Is Present in a Large-Molecular-Weight Complex
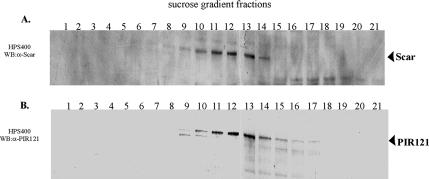
Mammalian Scar/WAVE proteins are reported to exist in a large-molecular-weight complex containing the proteins Abi, HSPC300, PIR121, and Nap1 (Eden et al., 2002; Innocenti et al., 2004). To determine whether Dictyostelium Scar is found in a similar multiprotein complex, we fractionated clarified Dictyostelium cell lysates on 7–20% sucrose gradients and analyzed the fractions via Western immunoblots using anti-Scar and anti-PIR121 antibodies. We found the majority of endogenous Scar and PIR121 in fractions 9–14, corresponding to protein complexes of 400–600 kDa (Figure 1, A and B). These results suggest that both endogenous Scar and PIR121 reside in large protein complexes of similar molecular weights, although it does not directly prove that Scar and PIR121 exist in the same complex.
Figure 1.
Endogenous Scar is present in a large-molecular-weight complex. Clarified cell lysate from growing HPS400 cells was loaded onto a 7–20% sucrose gradient. Fractions of 0.5 ml were collected and immunoblotted for Scar (A) or PIR121 (B). Numbers above blots indicate fraction number with the bottom of the gradient on the left. The majority of endogenous Scar is present in fractions 9–14 (A), which contain protein complexes of ∼400–600 kDa. The majority of PIR121 protein was detected in the same fractions (B).
Scar Binds to HSPC300 and Abi In Vitro
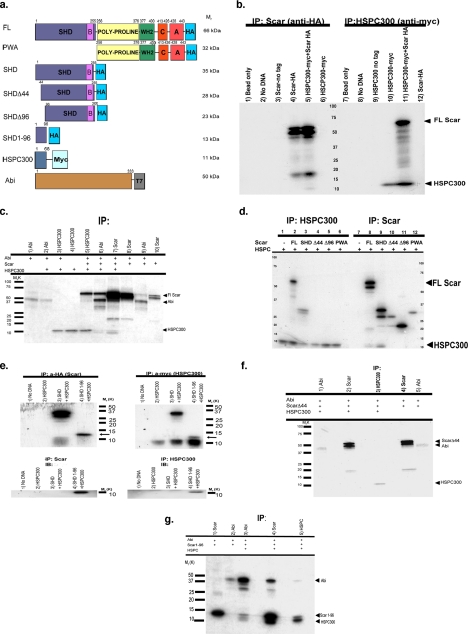
Having found Scar in a large-molecular-weight complex, we sought to determine if any of the Dictyostelium orthologues of the mammalian complex associate with Scar. In mammalian cells two members of the complex, HSPC300 and Abi, bind directly to Scar (Eden et al., 2002; Gautreau et al., 2004). To determine if similar interactions exist in Dictyostelium, Scar and the Dictyostelium genes encoding HSPC300 and Abi were transcribed and translated in vitro with C-terminal peptide tags added (Figure 2A), and their interactions were assessed by coimmunoprecipitation assays. The in vitro translation of Scar-HA routinely generated three polypeptide products ranging in size from ∼66 to 48 kDa (Figure 2B, lane 4). As confirmed below, these products represent full-length Scar (66 kDa) as well as two truncated forms, initiated from internal methionines at amino acid 45 (56 kDa) and amino acid 97 (48 kDa). In vitro interactions were found to exist between HA-tagged full-length Scar and myc-tagged HSPC300 (11 kDa, Figure 2B, lane 10). The α-HA antibody immunoprecipitated HSPC300-myc only when Scar-HA was present (Figure 2B, compare lanes 5 and 6). Conversely, full-length Scar-HA was immunoprecipitated with the α-myc antibody only when HSPC300-myc was present in the reaction (Figure 2B, compare lanes 11 and 12).
Figure 2.
The N-terminus of Scar binds HSPC300 and Abi in vitro. (a) Constructs used in these experiments. Numbers represent position of amino acids relative to the N-terminus. Relative molecular weights are indicated on the right. All Scar constructs contain C-terminal HA tag. FL, full-length Scar; SHD, construct containing the Scar homology domain (SHD) and Basic (B) regions; PWA, construct containing the poly-proline region, WH2, connecting (C), and acidic (A) regions; SHDΔ44, SHD construct lacking the first 44 amino acids; SHDΔ96, SHD construct lacking the first 96 amino acids. SHD1-96, Scar construct containing only amino acids 1–96; HSPC300, HSPC300 protein with C-terminal myc tag; Abi protein, amino acids 1–333 with C-terminal T7 peptide tag. (b) SDS-PAGE of in vitro binding and coimmunoprecipitation assays. Scar constructs with a C-terminal HA tag and HSPC300 with C-terminal myc tag were separately transcribed and translated in vitro, mixed, and then immunoprecipitated with protein G agarose beads conjugated to anti-HA (lanes 1–6) or anti-myc antibodies (lanes 7–12). Bead only controls representing lysates mixed with agarose beads are shown in lanes 1 and 7. Immunoprecipitates of lysate containing no added DNA are shown in lanes 2 and 8. Lysates containing PCR-generated DNAs of untagged Scar (lane 3) or untagged HSPC300 (lane 9) immunoprecipitated with anti-HA or anti-myc–conjugated beads, respectively, are shown. Transcription and translation of untagged proteins was verified (data not shown). Lysates containing both Scar-HA and HSPC300-myc immunoprecipitated with anti-HA (lane 5) or anti-myc (lane 11) are shown. FL Scar-HA migrates at ∼66 kDa; HSPC300 migrates at ∼11 kDa. Controls for anti-HA bead specificity are shown in lanes 4 and 6. Controls for anti-myc bead specificity are shown in lanes 10 and 12. (c) Scar-HA, Abi-T7, and HSPC300-myc were cotranslated and immunoprecipitated in various combinations. Lysates immunoprecipitated with anti-T7 (Abi), anti-HA (Scar), or anti-myc–conjugated agarose beads are shown. Assays were done as described in b. Molecular-weight marker sizes are indicated on the left. (d) Amino acids 1–44 of Scar are necessary to bind HSPC300 in vitro. Assay was done as described in b. Proteins added in binding assay are indicated for each lane. Lysates immunoprecipitated for HSPC300 using anti-myc–conjugated agarose beads (lanes 1–6) or for Scar using anti-HA–conjugated agarose beads (lanes 7–12) are shown. (e) Amino acids 1–96 of Scar are sufficient to bind HSPC300 in vitro. Assays were as in b. Lysates were immunoprecipitated for Scar using anti-HA bound agarose beads. The gels were either detected for 35S-Met (top panels) or immunoblotted with anti-HA antibody (bottom panels). Control lysate containing no added DNA is shown in lane 1. HSCP300 control is shown in lane 2. Lane 3 represents lysate containing both SHD-HA and HSPC300-myc proteins. Lane 4 represents lysate containing 1–96 fragment of SHD domain and HSPC300. (f) Amino acids 1–44 of Scar are necessary to bind to Abi. ScarΔ44-HA (full-length Scar minus the first 44 amino acids), Abi-T7, and HSPC300-myc were cotranslated and immunoprecipitated with anti-T7– conjugated agarose beads (lane 1), anti-HA–conjugated agarose beads (lane 2), or anti-myc–conjugated agarose beads (lane 3). ScarΔ44-HA (full-length Scar minus the first 44 amino acids) and Abi-T7 were cotranslated and immunoprecipitated with anti-HA–conjugated agarose beads (lane 4) or anti-T–conjugated agarose beads (lane 5). (g) Amino acids 1–96 of Scar are sufficient to bind Abi in the presence of HSPC300. Scar SHD1-96-HA and Abi-T7 were cotranslated and immunoprecipitated with anti-HA–conjugated agarose beads (lane 1) or anti-T7–conjugated agarose beads (lane 2). Scar SHD1-96-HA, Abi-T7, and HSPC300-myc were cotranslated and immunoprecipitated with anti-T7–conjugated agarose beads (lane 3), anti-HA–conjugated agarose beads (lane 4), or anti-myc–conjugated agarose beads (lane 5).
In Dictyostelium, an Abi ortholog exists that has significant homology to the N-terminal region of mammalian Abi1/2 proteins, but does not contain the C-terminal poly-proline or SH3 domains (our observations; Blagg et al., 2003a). We found Scar-HA bound T7-tagged Abi when both proteins were present in the in vitro reaction (Figure 2C, lanes 8 and 9), and full-length Scar appears to be the predominant Scar protein binding to Abi. Adding HSPC300 to the reaction cocktail may have slightly enhanced the binding of Scar to Abi (Figure 2C, compare lanes 5, and 7–9), although the effect, if real, is small. Although every attempt was made to add equivalent amounts of DNA to each reaction, the differences seen whether HSPC300 was added may reflect the nonquantitative nature of both the TNT reactions and the immunoprecipitations. We found no evidence for Abi and HSPC300 binding directly to one another (Figure 2C, lanes 2 and 3).
No interactions were detected between Scar and either Nap1 or PIR121 (data not shown).
Amino Acids 1–44 of Scar Are Necessary and Amino Acids 1–96 Are Sufficient To Bind to HSPC300
To further test the requirement of the N-terminus of Scar for these interactions, epitope-tagged, truncated Scar proteins were generated utilizing the internal methionine residues at amino acid (aa) 45 (Scar-SHDΔ44 HA; 28 kDa) and aa97 (Scar-SHDΔ96 HA; 23 kDa). As controls, PCR oligos were used to generate in vitro truncated proteins encompassing the entire SHD and basic domain, referred to as Scar-SHD HA (35 kDa), or only the C-terminus of Scar beginning at the poly-proline region, referred to as Scar-PWA HA (32 kDa; Figure 2A). When the different Scar truncations were combined in vitro with HSPC300 and immunoprecipitated using the α-HA antibody, only proteins containing aa1-44 of Scar bound to HSPC300 (Figure 2D, lanes 8 and 9); truncations lacking this region were unable to bind HSPC300 (Figure 2D, lanes 10–12). The same binding specificity was found when the reaction was immunoprecipitated for HSPC300 (Figure 2D, lanes 1–6), demonstrating that aa1-44 of Scar are necessary to bind HSPC300.
Because it proved beyond the technical limits of the in vitro system to stably produce the Scar 1-44 peptide, a Scar peptide encompassing only the first 96 amino acids of Scar tagged with HA (Scar 1-96 HA, 13 kDa, Figure 2E) was generated. When incubated with HSPC300-myc and immunoprecipitated using the α-HA antibody, a small amount of HSPC300-myc (11 kDa) coprecipitated (Figure 2E, lane 4, left). When Scar 1-96 HA and HSPC300-myc are immunoprecipitated with α-myc, a more extensive interaction between Scar 1-96 HA and HSPC300 myc was detected (Figure 2E, lane 4, right). The 13 kDa band was confirmed to be Scar 1-96 HA via immunoblotting with the monoclonal α-HA antibody (Figure 2E, bottom panels). These data indicate that the first 96 amino acids of Scar alone are sufficient to bind to HSPC300.
The Amino Terminus of Scar Interacts with Abi
Data presented in Figure 2F show that the first 44 amino acids of Scar are also necessary to bind Abi in vitro. Abi does not bind to Scar missing the first 44 amino acids, whether or not HSPC300 is present in the reaction (compare lanes 1 and 2 with lanes 4 and 5). In the presence of HSPC300, in vitro generated Scar 1-96-HA detectably binds Abi (Figure 2G, lanes 3–5); however, there is little apparent interaction between Abi and Scar 1-96 in the absence of HSPC300 (Figure 2G, lanes 1 and 2). This suggests that Abi may interact with a region between amino acids 44–97 of Scar and that HSPC300 may facilitate efficient binding.
Absence of Amino Acids 1–96 of Scar Has In Vivo Consequences
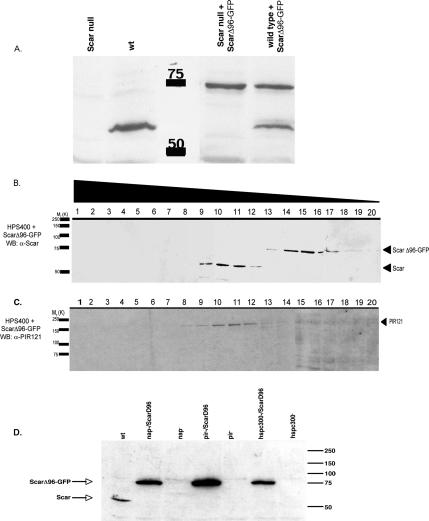
To determine if deleting the first 96 amino acids of Scar affected the protein's ability to interact in vivo with the large multiprotein complex, we expressed an inducible ScarΔ96-GFP construct in both parental and Scar null cells. Expression was confirmed by Western blot analysis using both an anti-Scar antibody (Figure 3A) and a monoclonal GFP antibody (data not shown). ScarΔ96-GFP–expressing cells were subsequently fractionated on 7–20% sucrose gradients. In parental cells expressing ScarΔ96-GFP, the endogenous, full-length, Scar fractionation profile on sucrose gradients remained unchanged (Figure 3B). However, the majority of truncated ScarΔ96-GFP was shifted to later fractions (fractions 14–18), representing smaller protein complexes of ∼150–250 kDa. Notably, little or no ScarΔ96-GFP is found in the same sucrose gradient fractions as endogenous full-length Scar (Figure 3B) and also does not migrate as a monomer. The fractionation profile for PIR121 remained unaffected in parental cells expressing ScarΔ96-GFP (compare 1B to 3C), consistent with PIR121 being unable to interact with Scar in the absence of abi and/or HSPC300 binding to Scar. Therefore the first 96 amino acids of Scar appears to be essential for Scar's ability to interact with the large-molecular-weight complex that is presumed to also contain PIR121 (and Nap1). It should be noted that despite being absent from the pentameric complex, ScarΔ96-GFP was stably expressed in both parental and scar null genetic backgrounds (Figure 3A). In other systems full-length Scar is unstable when any single member of the complex is missing (Rogers et al., 2003). We expressed ScarΔ96-GFP in genetic backgrounds missing PIR121, Nap1, or HSPC300 and found the truncated, but not full-length Scar protein to be stable in each case (Figure 3D). The N-terminus and/or its interactions with HSPC300 and abi, therefore, are critical for regulating Scar stability.
Figure 3.
ScarΔ96-GFP is stably expressed and complexes differently than wild-type Scar. (A) Growing HPS400 cells expressing Scar (∼66 kDa) and/or ScarΔ96-GFP (∼75 kDa) were collected and Western blotted for Scar. ScarΔ96-GFP is stably expressed in both backgrounds. Control cells are HPS400 and Scar null cells expressing empty vector. Clarified cell lysate from parental cells expressing ScarΔ96-GFP was loaded onto a 7–20% sucrose gradient. Fractions of 0.5 ml, were collected and immunoblotted for Scar (B) or PIR121 (C). Numbers above blots indicate fraction number. The majority of endogenous Scar is present in fractions 9–12 as in Figure 1. ScarΔ96-GFP is present in fractions 14–18 (B), which contain protein complexes of ∼150–300 kDa. PIR121 protein is not detected in the fractions containing ScarΔ96-GFP but continues to track with wild-type Scar (C). (D) Cells null for each of the Scar complex members with or without expressed ScarΔ96-GFP were collected and Western blotted as in A. ScarΔ96-GFP is stably expressed in each background.
Expression of ScarΔ96-GFP Results in an Abnormal Cellular Morphology
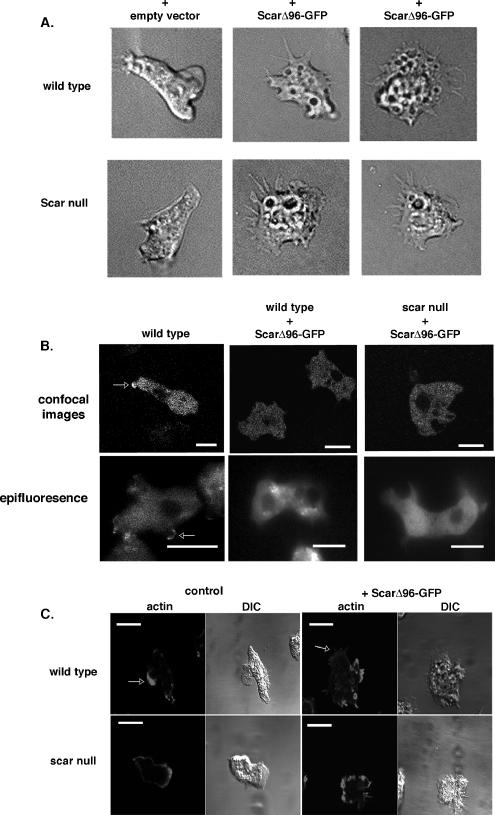
Vegetatively growing cells expressing ScarΔ96-GFP have an unusually flattened appearance, adhere more tightly to plastic and glass surfaces, have an increased number of visible vesicles, and extend long, thin protrusions when compared with parental controls (Figure 4, Supplementary Figure S1, Supplementary Movies 1–4). This abnormal phenotype is dominant being equally evident both in parental and Scar null cells (Figure 4A). The morphological phenotype, and the rest of the characteristics described below, was not attributed to the presence of the GFP tag because cells expressing ScarΔ96 without the tag exhibited identical alterations (data not shown). Expression of full-length Scar or the C-terminal, potentially Arp2/3-activating, PWA fragment also did not recreate the ScarΔ96-GFP phenotype (Supplementary Figure S1).
Figure 4.
Expression of ScarΔ96-GFP results in an abnormal morphology. (A) Axenically grown parental (HPS400) and scar null cells expressing empty vector or ScarΔ96-GFP were allowed to adhere to glass coverslips and observed with Nomarski differential interference contrast. Movies of cells are available in the Supplementary Material. (B) ScarΔ96-GFP does not localize to leading edges of newly forming pseudopods. Full-length Scar-GFP was expressed in scar null cells developed for 6 h (leftmost panels) and ScarΔ96-GFP was expressed in similarly developed parental (middle panels), and scar null backgrounds (right panels). The GFP in the cells was visualized using confocal microscopy (top panels) and epifluorescence microscopy (bottom panels). Scale bar, 10 μm. (C) Cells expressing ScarΔ96-GFP have aberrant actin staining. Six-hour developed parental and scar− cells expressing ScarΔ96-GFP were stained with TRITC-phalloidin. DIC images of cells are also shown. Bar, 10 μm.
In both parental and scar null backgrounds ScarΔ96-GFP affected cell size and the number of pseudopods extended over time. Cells expressing ScarΔ96-GFP had a significantly larger cell area when compared with their parent and extended fewer pseudopods per minute (Table 1). Parental and scar null cells growing vegetatively extend an average of 6–7 psedopods/10 min, which is in agreement with previous studies (Wessels et al., 2000). Parental cells expressing ScarΔ96-GFP extend fewer pseudopods, 3.1 pseudopods/10 min (Table 1). Scar null cells expressing ScarΔ96-GFP exhibit a significantly increased cell area, and although there was a trend toward fewer pseudopods, the reduction was not statistically significant (Table 1). Expressing full-length Scar in a scar null background affected neither psuedopod number nor cell area (Table 1).
Table 1.
Morphometrics
| Cell type | Average no. of pseudopods extended/10 min | Average cell area |
|---|---|---|
| Parental (HPS400) + empty vector | 6.9 | 71.2 |
| scar− + empty vector | 6.4 | 78.2 |
| HPS400 + ScarΔ96-GFP | 3.1* | 118* |
| scar− + ScarΔ96-GFP | 5.2 | 103* |
| scar− + full length Scar | 6.5 | 76.6 |
Wild-type (HPS400), scar null, Parental cells expressing ScarΔ96-GFP, and scar null cells expressing ScarΔ96-GFP or full-length Scar were filmed under Nomarski differential interference contrast, and the number of pseudopods extended over a 10-min period was scored. Values represent averages of ≥50 cells. The number of pseudopods extended by parental cells expressing ScarΔ96-GFP over a 10-min period was found to be significantly lower (p < 0.01) than in wild-type cells, scar null cells, and scar null cells expressing ScarΔ96-GFP or full-length Scar. Average cell area was measured using NIH ImageJ Software. Values represent average of ≥40 cells. Cells expressing ScarΔ96-GFP were found to have a significantly larger cell area (p < 0.01) than the parental counterparts. Asterisks indicate values statistically different than control.
Amino Acids 1–96 of Scar Are Necessary for Proper Localization
The aberrant morphology of cells expressing ScarΔ96-GFP suggested there might be a misregulation and/or mislocalization of the truncated Scar protein. We found full-length Scar-GFP expressed in Scar null cells transiently localize to the leading edges of newly forming pseudopods (Figure 4B, white arrow; Steiner, 2000) which is corroborated in other systems (Nakagawa et al., 2001; Kunda et al., 2003; Echarri et al., 2004; Leng et al., 2005). In contrast, ScarΔ96-GFP protein appears to be ubiquitously expressed within both parental and Scar null cells, with no visible enrichment to newly forming pseudopods or membranes whether observed by confocal or epifluorescence microscopy (Figure 4B). Thus, maintenance of Scar in the complex appears to play an important role in the proper localization of Scar.
Cells Expressing ScarΔ96-GFP Have Aberrant F-Actin Distribution
Because of the known relationship between Scar, Arp2/3, and actin polymerization, we examined the integrity of the actin cytoskeleton in cells expressing ScarΔ96-GFP by staining filamentous actin (F-actin) with TRITC-phalloidin. Wild-type Dictyostelium cells are enriched for F-actin in lamellae and pseudopods as well as at the uropod (Fukui and Inoue, 1997; Noegel and Schleicher, 2000). Developing scar null cells, however, have a diminished actin meshwork at the front and rear of the cells and display an increased cortical staining when compared with parental cells (Bear et al., 1998). In similar cells expressing ScarΔ96-GFP, regardless of background, there is no enrichment of actin that would distinguish the front or rear of the cell (Figure 4C, arrows). Instead, there appears to be sporadic enrichments of actin around the cell periphery at areas where there are irregular shaped protrusions (but not defined lamellae) and around crown structures.
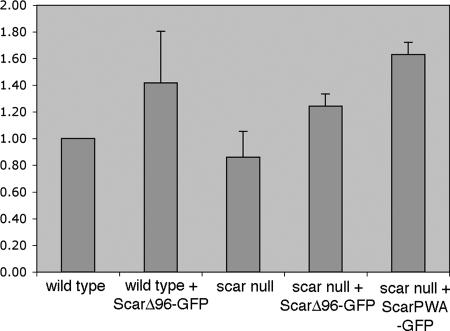
The overall F-actin levels of growing cells expressing ScarΔ96-GFP were measured and compared with levels in parental cells with no statistical difference found in any of the strains tested. In contrast, the Scar PWA-GFP construct that contains only the polyproline, actin binding, and Arp2/3-activating regions does show an increase in basal F-actin content (Figure 5). Minimal differences from controls were also seen when developmentally competent cells were stimulated with cAMP (data not shown). Therefore, the differences seen in the F-actin–staining patterns of cells expressing ScarΔ96-GFP and their parental counterparts cannot be attributed to global changes in F-actin levels. Rather the data suggest that expression of Scar missing the first 96 amino acids has a disruptive effect on the proper regulation of F-actin assembly and the establishment of cell polarity but does not lead to constitutive activation of Arp2/3 via unregulated Scar activity.
Figure 5.
Cells expressing ScarΔ96 show no dramatic increases in F-actin levels. Cellular F-actin levels were measured as described in Materials and Methods. Basal F-actin levels in all ScarΔ96-GFP–expressing cells measured were not statistically different from levels in wild-type cells. The difference between scar null and scar null/Scar PWA-GFP was statistically significant. The Y-axis is fold change in F-actin polymerization relative to parental control. Data represent results from three separate experiments.
Expressing Truncated Scar Affects Motility and Chemotaxis
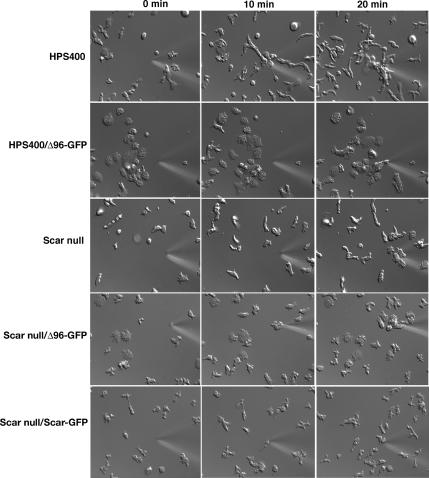
WASp family proteins are known to be required for specific actin-based cellular responses to chemoattractants (Ochs et al., 1980; Zicha et al., 1998; Baba et al., 1999; Haddad et al., 2001; Jones et al., 2002; Myers et al., 2005). Dictyostelium cells chemotax toward a point source of cAMP via intracellular signaling (see Figure 6 and Supplementary Movies 5–9; reviewed in van Haastert and Devreotes, 2004). Scar is necessary for proper cellular response to cyclic AMP in D. discoideum (Steiner, 2000; Blagg et al., 2003b); scar null cells fail to chemotax as efficiently as parental cells in response to a cAMP spatial gradient (Figure 6, compare Supplementary Movies 5 and 6; Steiner, 2000; unpublished results; Blagg et al., 2003). Expressing full-length Scar-GFP in a Scar null background fully restores the ability of cells to chemotax (Supplementary Movie 7; Steiner, 2000). To determine if the interaction of Scar with HSPC300 and/or Abi is critical for an appropriate cellular response to cAMP, parental and Scar null cells expressing ScarΔ96-GFP were challenged with a spatial gradient of cAMP (Figure 6, Supplementary Movies 8 and 9). Unlike results for full-length Scar, expressing ScarΔ96-GFP in scar null cells does not restore wild-type chemotaxis (Figure 6, Supplementary Movies 6 and 9). Many of the cells are flattened, tightly adherent and move only sporadically toward the chemoattractant source. Significantly, chemotaxis and basic motility are similarly impaired when ScarΔ96-GFP is expressed in parental cells (Figure 6, compare Supplementary Movies 5 and 8). Individual cells expressing ScarΔ96-GFP, whether in the wild-type or scar null backgrounds, displayed chemotactic responses ranging from a slight delay to having movement completely blocked (Figure 6, Supplementary Movie 8). Therefore, ScarΔ96-GFP could not rescue chemotaxis in Scar null cells and has a dominant inhibitory effect on normal cell movement and chemotaxis in parental cells.
Figure 6.
Cells expressing ScarΔ96-GFP have motility and chemotaxis defects. Chemotaxis assays were performed on 8-h developed HPS400, Scar null, and HPS400 and Scar null cells expressing ScarΔ96-GFP and Scar null cells expressing full-length Scar-GFP. Times of exposure to a cAMP gradient are shown at the top of the figure.
Expressing Truncated Scar Affects Cytokinesis
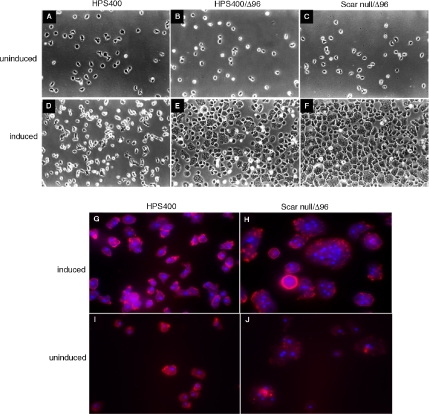
With extended periods of induction, growing cells expressing ScarΔ96-GFP became enlarged and adherent (Figure 7, A–F) and were multinucleate (Figure 7, G–J). Cells with 4 to more than 20 nuclei were observed, with no evidence of cleavage furrows. Multinucleate cells were observed after 10–12 h of induction and by ∼72 h virtually every cell was converted to the large, multinucleate phenotype. There was no significant change in ScarΔ96-GFP protein levels during that time. The phenotype was roughly equivalent in both parental and Scar null backgrounds and appears to be a dominant consequence of expressing ScarΔ96-GFP. There was no similar cytokinesis effect when full-length Scar was expressed (data not shown).
Figure 7.
Induction of cytokinesis defect by extended induction of Δ96 constructs. (A–C, I, and J) Axenically growing strains with the ScarΔ96-GFP construct uninduced; (D–H) each strain after 72 h of ScarΔ96 induction. A–F, DIC images; G–J, epifluorescent images with nuclei stained with DAPI and cells outlined with TRITC-phalloidin.
DISCUSSION
The data presented in this article show that Dictyostelium Scar is present in a large-molecular-weight complex and interacts with Abi and HSPC300 and that the N-terminal 96 amino acids are necessary and sufficient for this interaction. The physiological consequences of removing the Abi and HSPC300 binding site(s) of Scar leads to disruption of the Scar containing complex, loss of Scar localization, increased stability of the truncated protein, abnormal cellular actin dynamics, aberrant cell adhesion and motility, and failure of normal cytokinesis.
We determined that the first 44 amino acids of Scar are necessary for binding to both HSPC300 and Abi, and the first 96 amino acids were sufficient for this binding in vitro. It is quite possible that the first 44 amino acids of Scar are also sufficient for binding to Abi and HSPC300, but for technical reasons we were unable to establish that unequivocally. Consistent with that idea, the phenotype of cells expressing ScarΔ44-GFP is virtually identical to that of ScarΔ96-GFP–expressing cells (data not shown). We observed that the in vitro binding of Scar to Abi may be subtly enhanced in the presence of HSPC300. Although our data are only suggestive, others have also reported an enhanced recruitment of Scar to the complex when HSPC300 and Abi are present (Gautreau et al., 2004). In contrast, the presence or absence of Abi had no effect on the binding between Scar and HSPC300.
The first 96 amino acids of Scar are contained within the SHD (Bear et al., 1998). The SHD region has been implicated in binding to Abi (Echarri et al., 2004; Innocenti et al., 2004; Leng et al., 2005), and overexpression studies in Cos-7 cells indicate that amino acids 32–66 of Scar2/WAVE2 are necessary for Abi binding (Leng et al., 2005). In addition, expressing Scar/WAVE in cultured cells depleted for Abi result in loss of Scar/WAVE localization (Kunda et al., 2003; Rogers et al., 2003). Our data show that the first 96 amino acids of Scar are not only necessary to bind Abi, consistent with Leng et al. (2005), but are sufficient for that binding. The SHD region also appears to play a role in the localization of Scar2/WAVE2 to lamellipodia and filopodia (Nakagawa et al., 2001, 2003; Nozumi et al., 2003; Leng et al., 2005; Mitsushima et al., 2006). When overexpressed in NG108 neuroblastoma cells Scar2/WAVE2 fragments containing only amino acids 1–83 localize to filopodial tips, but fragments containing amino acids 1–54 do not (Nozumi et al., 2003). We report here that Scar missing the first 96 amino is unable to localize properly. This is consistent if Abi binding is required for proper Scar localization (Leng et al., 2005). An iso/leucine-rich region is present in the conserved SHD region of Scar family members (Bear et al., 1998) and may play a role in protein-protein interactions (Miki et al., 1998; Tu et al., 2004). In Dictyostelium the iso/leucine-rich region spans approximately amino acids 28–90, overlapping the region that we find critical for binding to HSPC300 and Abi. There is no direct evidence linking any of the hydrophobic residues to Scar interactions and/or localization, but it is quite possible. More specific mutagenesis studies are needed to establish involvement of these residues in interactions between Scar, Abi, and HSPC300.
Endogenous Dictyostelium Scar and PIR121 are found in complexes of ∼400–600 kDa, consistent with the existing model of Scar/WAVE regulation and with the Insall laboratory's demonstration of coimmunoprecipitation of endogenous Scar and PIR121 (Ibarra et al., 2006; Stradal and Scita, 2006). ScarΔ96-GFP is found in a smaller sized complex(es). Neither full-length Scar nor PIR121 comigrate with ScarΔ96-GFP in the smaller complex(es), arguing that there is no interaction in vivo. The other components of the ScarΔ96-GFP–containing complex(es) are unknown and are presently being pursued. Suetsugu et al. (2006) reported that overexpression of full-length tagged WAVE2, in the presence of endogenous WAVE2, remains largely as monomers, though they did not directly establish that the WAVE in lower fractions was monomeric (Suetsugu et al., 2006). The same, tagged, WAVE2 is found in the high-molecular-weight complex when expressed in cells devoid of endogenous WAVE2. Consistent with their observations, and with our Scar null rescue data, we find full-length Scar-GFP, expressed in Scar null cells, present in fractions representing the high-molecular-weight complex (not shown). We have been unable to stably express full-length Scar-GFP in cells expressing endogenous Scar. Transient expression has been detected, but only at very low levels and for short periods of time. This is consistent with the idea that this protein is more unstable that ScarΔ96. The presence in cells of ScarΔ96-GFP does not interfere with the ability of endogenous Scar or PIR121 to participate in a high-molecular-weight complex. This is consistent with the in vitro data that amino acids 1–96 of Scar are needed to bind Abi and HSPC300 and that Abi is necessary for the assembly of the Scar-containing macromolecular protein complex (Gautreau et al., 2004; Innocenti et al., 2004).
Expression of ScarΔ96-GFP has a dramatic effect on cell morphology, cytoskeletal organization, substrate adhesion, motility, and cytokinesis. This is true whether it is expressed in the presence (HPS400) or absence (scar−) of the normal Scar-containing complex and is likely to reflect inability of ScarΔ96-GFP to respond to normal Scar regulation. Although ScarΔ96-GFP does not associate with the pentapeptide complex, it is expected to still bind actin and Arp2/3 and stimulate F-actin production, potentially with little or no regulation. It was, therefore, somewhat surprising to see the overall level of F-actin in ScarΔ96-GFP–expressing cells to be the same as in control cells. This is in contrast to expression of a construct, Scar PWA-GFP, which does result in cells having an increased basal level of F-actin. Those same Scar PWA-GFP cells do not produce a phenotype at all like ScarΔ96-GFP (Figure 5 and Supplementary Figure S1). The data suggest that ScarΔ96-GFP does not act like a constitutively active Arp2/3 activator. It may be that there is transient, localized activation of Arp2/3 and F-actin production but not enough to be detected on a cell-wide basis. Consistent with that is the abnormal distribution of F-actin into patches around the periphery of the cell, a different pattern from that seen in either parental or scar null cells. Whether the control of ScarΔ96-GFP is through the protein complex(es) seen in the sucrose gradients or by some other means awaits further characterization of ScarΔ96-GFP. There is also an increase in the number of vesicles present in ScarΔ96-GFP cells. Dictyostelium scar null cells are known to have defects in vesicle maturation and export (Seastone et al., 2001) and the phenotype of ScarΔ96-GFP–expressing cells may reflect a misregulation of a normal role for Scar in vesicle morphogenesis or trafficking. Excessive formation of long, thin protrusions is seen in Drosophila S2 cells when Scar, Abi, Nap1, or PIR121 are RNA interference depleted (Kunda et al., 2003; Rogers et al., 2003). A similar increase is seen when ScarΔ96-GFP is expressed. In both systems this increase occurs when one or more of the complex members is absent. In the absence of PIR121, Abi, or Nap1 there is a decrease in the level of wild-type Scar within the cells (Blagg et al., 2003a; Kunda et al., 2003; Rogers et al., 2003; Ibarra et al., 2006). This is likely to be, at least in part, proteasome-mediated degradation (Rogers et al., 2003; Mitsushima et al., 2006). In contrast to wild-type Scar, ScarΔ96-GFP is not rapidly degraded. This is true in parental as well as in null backgrounds for Scar, PIR121, Nap1, and HSPC300. This strongly suggests that in addition to ScarΔ96-GFP being resistant to regulation by the pentapeptide complex, it is resistant to degradation. It is possible that the N-terminal 96 amino acids of Scar contain a targeting site for degradation or is important for masking such a site. It was recently reported that an Arabidopsis ortholog of HSPC300, BRICK1, plays a role in stabilizing SCAR2 in that organism (Le et al., 2006). They suggest that BRICK1 binds Scar and contributes to the masking of a degradation sequence. It is also possible that regulated degradation of Scar is dependent on proper localization. It may be that the presence of Scar in the peptapeptide complex not only targets it to sites of new F-actin assembly, but is responsible for presenting Scar to the degradation machinery. It is also possible that the deletion of the first 44 or 96 amino acids of Scar changes the conformation of the protein such that abi and/or HSPC300 no longer have an intact recognition site. Although the experiments showing amino acids 1–96 of Scar are sufficient to bind HSPC300, we cannot rule out that possibility for abi.
Expression of ScarΔ96-GFP has a dominant, gain-of-function effect on cell motility. Chemotaxis is not rescued in Scar null cells by expression of ScarΔ96-GFP, and chemotaxis of parental cells is severely compromised. The effects do not seem to be on the ability of the cells to detect the chemoattractant, because movies show that cells are able to identify the direction of the source. Rather the defect in motility may be an indirect consequence of enhanced substrate adhesion and an inability of cells to polarize properly. Movies show many cells expressing ScarΔ96-GFP adhering to the substrate, flattening out, and moving very little. These cells can convert back to less adherent cells but still extend multiple, poorly directed protrusion resulting in inefficient directed motion. The lack of chemotactic response appears to reflect more on the cell's general inability to organize its actin cytoskeleton, as seen by phalloidin F-actin staining, rather than on its ability to orient correctly. It is possible that ScarΔ96-GFP is affecting the Phg2-Adrm pathway and/or SadA-mediated adhesion as both play prominent roles in regulating F-actin organization, adhesion, and motility (Fey et al., 2002; Gebbie et al., 2004; Cherix et al., 2006). Further studies are underway to investigate these possibilities.
Perhaps the most surprising effect of expressing ScarΔ96-GFP was on cytokinesis. Unlike myosin II null mutants that are multinucleate in suspension but can recover a form of cell division when returned to a solid surface (De Lozanne and Spudich, 1987; Knecht and Loomis, 1987; Fukui et al., 1990), ScarΔ96-GFP–expressing cells become highly multinucleate on solid support. In that sense the ScarΔ96-GFP–expressing cells are more reminiscent of null mutants in coronin (de Hostos et al., 1993), SAPKα (Sun et al., 2003), or the multiple mutants in PI3-kinase and PTEN involved in the phosphatidyl 4,5-bisphosphate synthetic pathway (Janetopoulos et al., 2005). The latter connection is particularly interesting given the proposed relationship between phophoinositide signaling and members of the Scar/WASp family of proteins (e.g., Oikawa et al., 2004; Weiner et al., 2006). Janetopoulos et al. (2005) and others have suggested a connection between G-protein–coupled receptor (GPCR) signaling, PI3K/PTEN and cytokinesis. Because Scar was originally identified as a suppressor of GPCR signaling, it is possible that expression of the truncated Scar is interfering with that normal regulation. It is also possible that the cytokinesis defect is due to the abnormal substrate adhesion seen in ScarΔ96-GFP– expressing cells. This could result in new cells being unable to effectively make a division furrow and move apart. SadA mutants also show a cytokinesis defect, again suggesting a possible connection between the abnormal actin regulation of ScarΔ96-GFP–expressing cells and SadA-mediated adhesion.
Expression of N-terminally truncated Scar results in a wide array of deleterious consequences including effects on cell morphology, substrate adhesion, motility, and cytokinesis. This protein is defective in at least two different ways: it is not regulated by the normal involvement in a large multiprotein complex and it is unusually stable. These two things may be related as targeting to the plasma membrane by the complex may be required for both activation and degradation of Scar. The findings reported here underline the importance of maintaining strict regulation over Arp2/3 activators like Scar and WASp. Localized activation and rapid degradation of Scar/WASp proteins provide mechanisms for both spatial and temporal regulation of Arp2/3-mediated actin polymerization. Our data suggest that necessary and perhaps sufficient components of that regulation are contained within the first 100 amino acids of Scar. The ability to perform site-directed mutagenesis on the N-terminus of Scar and return it to an in vivo context provides us with a means to more precisely define the role of the N-terminus of Scar provide insight into its regulation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Daniel Kalman and Win Sale for discussion and helpful suggestions on the manuscript, Robert Insall for the PIR121 antibody, and the Japanese cDNA projects for providing clones and information. D.C. and C.L.S. were supported in part by National Institutes of Health Grant GM45705 and by a grant from the University Research Committee of Emory.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-06-0518) on February 21, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Baba Y., et al. Involvement of wiskott-aldrich syndrome protein in B-cell cytoplasmic tyrosine kinase pathway. Blood. 1999;93:2003–2012. [PubMed] [Google Scholar]
- Bear J. E. Atlanta, GA: Emory University; 1998. SCAR Defines a New Family of WASP-related Actin Cytoskeleton Regulators. [Google Scholar]
- Bear J. E., Rawls J. F., Saxe C. L., III SCAR, a WASP-related protein, isolated as a suppressor of receptor defects in late Dictyostelium development. J. Cell Biol. 1998;142:1325–1335. doi: 10.1083/jcb.142.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A. L., Hall A. Rho GTPases and their effector proteins. Biochem. J. 2000;348(Pt 2):241–255. [PMC free article] [PubMed] [Google Scholar]
- Blagg S. L., Stewart M., Sambles C., Insall R. H. PIR121 regulates pseudopod dynamics and SCAR activity in Dictyostelium. Curr. Biol. 2003a;13:1480–1487. doi: 10.1016/s0960-9822(03)00580-3. [DOI] [PubMed] [Google Scholar]
- Blagg S. L., Stewart M., Sambles C., Insall R. H. PIR121 regulates pseudopod dynamics and SCAR activity in Dictyostelium. Curr. Biol. 2003b;13:1480–1487. doi: 10.1016/s0960-9822(03)00580-3. [DOI] [PubMed] [Google Scholar]
- Bompard G., Caron E. Regulation of WASP/WAVE proteins: making a long story short. J. Cell Biol. 2004;166:957–962. doi: 10.1083/jcb.200403127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisy G. G., Svitkina T. M. Actin machinery: pushing the envelope. Curr. Opin. Cell Biol. 2000;12:104–112. doi: 10.1016/s0955-0674(99)00063-0. [DOI] [PubMed] [Google Scholar]
- Cherix N., Froquet R., Charette S. J., Blanc C., Letourneur F., Cosson P. A Phg2-Adrm1 pathway participates in the nutrient-controlled developmental response in Dictyostelium. Mol. Biol. Cell. 2006;17:4982–4987. doi: 10.1091/mbc.E06-07-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis J., Bresnick A., Demma M., Dharmawardhane S., Eddy R., Hall A. L., Sauterer R., Warren V. Mechanisms of amoeboid chemotaxis: an evaluation of the cortical expansion model. Dev. Genet. 1990;11:333–340. doi: 10.1002/dvg.1020110504. [DOI] [PubMed] [Google Scholar]
- Dahl J. P., Wang-Dunlop J., Gonzales C., Goad M.E.P., Mark R. J., Kwak S. P. Characterization of the WAVE1 knock-out mouse: implications for CNS development. J. Neurosci. 2003;23:3343–3352. doi: 10.1523/JNEUROSCI.23-08-03343.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hostos E. L., Rehfuess C., Bradtke B., Waddell D. R., Albrecht R., Murphy J., Gerisch G. Dictyostelium mutants lacking the cytoskeletal protein coronin are defective in cytokinesis and cell motility. J. Cell Biol. 1993;120:163–173. doi: 10.1083/jcb.120.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lozanne A., Spudich J. A. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 1987;236:1086–1091. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- Echarri A., Lai M. J., Robinson M. R., Pendergast A. M. Abl interactor 1 (Abi-1) wave-binding and SNARE domains regulate its nucleocytoplasmic shuttling, lamellipodium localization, and wave-1 levels. Mol. Cell. Biol. 2004;24:4979–4993. doi: 10.1128/MCB.24.11.4979-4993.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden S., Rohatgi R., Podtelejnikov A. V., Mann M., Kirschner M. W. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- Fey P., Stephens S., Titus M. A., Chisholm R. L. SadA, a novel adhesion receptor in Dictyostelium. J. Cell Biol. 2002;159:1109–1119. doi: 10.1083/jcb.200206067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M., Egile C., Dyachok J., Djakovic S., Nolasco M., Li R., Smith L. G. Activation of Arp2/3 complex-dependent actin polymerization by plant proteins distantly related to Scar/WAVE. Proc. Natl. Acad. Sci. USA. 2004;101:16379–16384. doi: 10.1073/pnas.0407392101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y., De Lozanne A., Spudich J. A. Structure and function of the cytoskeleton of a Dictyostelium myosin-defective mutant. J. Cell Biol. 1990;110:367–378. doi: 10.1083/jcb.110.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y., Inoue S. Amoeboid movement anchored by eupodia, new actin-rich knobby feet in Dictyostelium. Cell Motil. Cytoskelet. 1997;36:339–354. doi: 10.1002/(SICI)1097-0169(1997)36:4<339::AID-CM4>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Gautreau A., Ho H. Y., Li J., Steen H., Gygi S. P., Kirschner M. W. Purification and architecture of the ubiquitous Wave complex. Proc. Natl. Acad. Sci. USA. 2004;101:4379–4383. doi: 10.1073/pnas.0400628101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebbie L., et al. Phg2, a kinase involved in adhesion and focal site modeling in Dictyostelium. Mol. Biol. Cell. 2004;15:3915–3925. doi: 10.1091/mbc.E03-12-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad E., Zugaza J. L., Louache F., Debili N., Crouin C., Schwarz K., Fischer A., Vainchenker W., Bertoglio J. The interaction between Cdc42 and WASP is required for SDF-1-induced T-lymphocyte chemotaxis. Blood. 2001;97:33–38. doi: 10.1182/blood.v97.1.33. [DOI] [PubMed] [Google Scholar]
- Ho H. Y., Rohatgi R., Lebensohn A. M., Le M., Li J., Gygi S. P., Kirschner M. W. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell. 2004;118:203–216. doi: 10.1016/j.cell.2004.06.027. [DOI] [PubMed] [Google Scholar]
- Hufner K., Schell B., Aepfelbacher M., Linder S. The acidic regions of WASp and N-WASP can synergize with CDC42Hs and Rac1 to induce filopodia and lamellipodia. FEBS Lett. 2002;514:168–174. doi: 10.1016/s0014-5793(02)02358-x. [DOI] [PubMed] [Google Scholar]
- Ibarra N., Blagg S. L., Vazquez F., Insall R. H. Nap1 regulates Dictyostelium cell motility and adhesion through SCAR-dependent and -independent pathways. Curr. Biol. 2006;16:717–722. doi: 10.1016/j.cub.2006.02.068. [DOI] [PubMed] [Google Scholar]
- Innocenti M., Zucconi A., Disanza A., Frittoli E., Areces L. B., Steffen A., Stradal T. E., Di Fiore P. P., Carlier M. F., Scita G. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat. Cell Biol. 2004;6:319–327. doi: 10.1038/ncb1105. [DOI] [PubMed] [Google Scholar]
- Janetopoulos C., Borleis J., Vazquez F., Iijima M., Devreotes P. Temporal and spatial regulation of phosphoinositide signaling mediates cytokinesis. Dev. Cell. 2005;8:467–477. doi: 10.1016/j.devcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Jones G. E., Zicha D., Dunn G. A., Blundell M., Thrasher A. Restoration of podosomes and chemotaxis in Wiskott-Aldrich syndrome macrophages following induced expression of WASp. Int. J. Biochem. Cell Biol. 2002;34:806–815. doi: 10.1016/s1357-2725(01)00162-5. [DOI] [PubMed] [Google Scholar]
- Kim A. S., Kakalis L. T., Abdul-Manan N., Liu G. A., Rosen M. K. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 2000;404:151–158. doi: 10.1038/35004513. [DOI] [PubMed] [Google Scholar]
- Knecht D. A., Loomis W. F. Antisense RNA inactivation of myosin heavy chain gene expression in Dictyostelium discoideum. Science. 1987;236:1081–1085. doi: 10.1126/science.3576221. [DOI] [PubMed] [Google Scholar]
- Kunda P., Craig G., Dominguez V., Baum B. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr. Biol. 2003;13:1867–1875. doi: 10.1016/j.cub.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Lauffenburger D. A., Horwitz A. F. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- Le J., Mallery E. L., Zhang C., Brankle S., Szymanski D. B. Arabidopsis BRICK1/HSPC300 is an essential WAVE-complex subunit that selectively stabilizes the Arp2/3 activator SCAR2. Curr. Biol. 2006;16:895–901. doi: 10.1016/j.cub.2006.03.061. [DOI] [PubMed] [Google Scholar]
- Leng Y., Zhang J., Badour K., Arpaia E., Freeman S., Cheung P., Siu M., Siminovitch K. Abelson-interactor-1 promotes WAVE2 membrane translocation and Abelson-mediated tyrosine phosphorylation required for WAVE2 activation. Proc. Natl. Acad. Sci. USA. 2005;102:1098–1103. doi: 10.1073/pnas.0409120102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky L. M., Atkinson S. J., Ampe C., Vandekerckhove J., Pollard T. D. Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J. Cell Biol. 1994;127:107–115. doi: 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky L. M., Insall R. H. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- Marchand J. B., Kaiser D. A., Pollard T. D., Higgs H. N. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat. Cell Biol. 2001;3:76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- Miki H., Suetsugu S., Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 1998;17:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H., Takenawa T. Direct binding of the verprolin-homology domain in N-WASP to actin is essential for cytoskeletal reorganization. Biochem. Biophys. Res. Commun. 1998;243:73–78. doi: 10.1006/bbrc.1997.8064. [DOI] [PubMed] [Google Scholar]
- Mitsushima M., Sezaki T., Akahane R., Ueda K., Suetsugu S., Takenawa T., Kioka N. Protein kinase A-dependent increase in WAVE2 expression induced by the focal adhesion protein vinexin. Genes Cells. 2006;11:281–292. doi: 10.1111/j.1365-2443.2006.00932.x. [DOI] [PubMed] [Google Scholar]
- Morio T., Maeda M. The Dictyostelium developmental cDNA project. Tanpakushitsu Kakusan Koso. 2001;46:2419–2424. [PubMed] [Google Scholar]
- Mullins R. D. How WASP-family proteins and the Arp2/3 complex convert intracellular signals into cytoskeletal structures. Curr. Opin. Cell Biol. 2000;12:91–96. doi: 10.1016/s0955-0674(99)00061-7. [DOI] [PubMed] [Google Scholar]
- Mullins R. D., Heuser J. A., Pollard T. D. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S. A., Han J. W., Lee Y., Firtel R. A., Chung C. Y. A Dictyostelium homologue of WASP is required for polarized F-actin assembly during chemotaxis. Mol. Biol. Cell. 2005;16:2191–2206. doi: 10.1091/mbc.E04-09-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa H., Miki H., Ito M., Ohashi K., Takenawa T., Miyamoto S. N-WASP, WAVE and Mena play different roles in the organization of actin cytoskeleton in lamellipodia. J. Cell Sci. 2001;114:1555–1565. doi: 10.1242/jcs.114.8.1555. [DOI] [PubMed] [Google Scholar]
- Nakagawa H., Miki H., Nozumi M., Takenawa T., Miyamoto S., Wehland J., Small J. V. IRSp53 is colocalised with WAVE2 at the tips of protruding lamellipodia and filopodia independently of Mena. J. Cell Sci. 2003;116:2577–2583. doi: 10.1242/jcs.00462. [DOI] [PubMed] [Google Scholar]
- Noegel A. A., Schleicher M. The actin cytoskeleton of Dictyostelium: a story told by mutants. J. Cell Sci. 2000;113:759–766. doi: 10.1242/jcs.113.5.759. [DOI] [PubMed] [Google Scholar]
- Nozumi M., Nakagawa H., Miki H., Takenawa T., Miyamoto S. Differential localization of WAVE isoforms in filopodia and lamellipodia of the neuronal growth cone. J. Cell Sci. 2003;116:239–246. doi: 10.1242/jcs.00233. [DOI] [PubMed] [Google Scholar]
- Ochs H. D., Slichter S. J., Harker L. A., Von Behrens W. E., Clark R. A., Wedgwood R. J. The Wiskott-Aldrich syndrome: studies of lymphocytes, granulocytes, and platelets. Blood. 1980;55:243–252. [PubMed] [Google Scholar]
- Oikawa T., Yamaguchi H., Itoh T., Kato M., Ijuin T., Yamazaki D., Suetsugu S., Takenawa T. PtdIns(3,4,5)P3 binding is necessary for WAVE2-induced formation of lamellipodia. Nat. Cell Biol. 2004;6:420–426. doi: 10.1038/ncb1125. [DOI] [PubMed] [Google Scholar]
- Pollard T. D., Borisy G. G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Rogers S. L., Wiedemann U., Stuurman N., Vale R. D. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 2003;162:1079–1088. doi: 10.1083/jcb.200303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R., Ma L., Miki H., Lopez M., Kirchhausen T., Takenawa T., Kirschner M. W. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- Saxe C. L. Handbook of Cell Signaling. Vol. 2. New York: Elsevier Science; 2003. WASp/Scar/WAVE. [Google Scholar]
- Seastone D. J., Harris E., Temesvari L. A., Bear J. E., Saxe C. L., Cardelli J. The WASp-like protein scar regulates macropinocytosis, phagocytosis and endosomal membrane flow in Dictyostelium. J. Cell Sci. 2001;114:2673–2683. doi: 10.1242/jcs.114.14.2673. [DOI] [PubMed] [Google Scholar]
- Soderling S. H., Langeberg L. K., Soderling J. A., Davee S. M., Simerly R., Raber J., Scott J. D. Loss of WAVE-1 causes sensorimotor retardation and reduced learning and memory in mice. Proc. Natl. Acad. Sci. USA. 2003;100:1723–1728. doi: 10.1073/pnas.0438033100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner J. S. Atlanta, GA: Emory University; 2000. Scar, a WASp-like Protein, Is Required for Lateral Pseudopod Formation and Chemotaxis in Dictyostelium discoideum. Ph.D. Dissertation. [Google Scholar]
- Stradal T. E., Scita G. Protein complexes regulating Arp2/3-mediated actin assembly. Curr. Opin. Cell Biol. 2006;18:4–10. doi: 10.1016/j.ceb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Suetsugu S., Kurisu S., Oikawa T., Yamazaki D., Oda A., Takenawa T. Optimization of WAVE2 complex-induced actin polymerization by membrane-bound IRSp53, PIP3, and Rac. J. Cell Biol. 2006;173:571–585. doi: 10.1083/jcb.200509067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B. G., Ma H., Firtel R. A. Dictyostelium stress-activated protein kinase alpha, a novel stress-activated mitogen-activated protein kinase kinase kinase-like kinase, is important for the proper regulation of the cytoskeleton. Mol. Biol. Cell. 2003;14:4526–4540. doi: 10.1091/mbc.E03-01-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons M., Derry J. M., Karlak B., Jiang S., Lemahieu V., McCormick F., Francke U., Abo A. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- Tu H., Tang T. S., Wang Z., Bezprozvanny I. Association of type 1 inositol 1,4,5-trisphosphate receptor with AKAP9 (Yotiao) and protein kinase A. J. Biol. Chem. 2004;279:19375–19382. doi: 10.1074/jbc.M313476200. [DOI] [PubMed] [Google Scholar]
- van Haastert P.J.M., Devreotes P. N. Chemotaxis: signalling the way forward. Nat. Rev. Mol. Cell Biol. 2004;5:626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- Vartiainen M. K., Machesky L. M. The WASP-Arp2/3 pathway: genetic insights. Curr. Opin. Cell Biol. 2004;16:174–181. doi: 10.1016/j.ceb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Weiner O. D., Rentel M. C., Ott A., Brown G. E., Jedrychowski M., Yaffe M. B., Gygi S. P., Cantley L. C., Bourne H. R., Kirschner M. W. Hem-1 complexes are essential for Rac activation, actin polymerization, and myosin regulation during neutrophil chemotaxis. PLoS Biol. 2006;4:e38. doi: 10.1371/journal.pbio.0040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M. D., Depace A. H., Verma S., Iwamatsu A., Mitchison T. J. The human Arp2–3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J. Cell Biol. 1997;138:375–384. doi: 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels D. J., Zhang H., Reynolds J., Daniels K., Heid P., Lu S., Kuspa A., Shaulsky G., Loomis W. F., Soll D. R. The internal phosphodiesterase RegA. is essential for the suppression of lateral pseudopods during Dictyostelium chemotaxis. Mol. Biol. Cell. 2000;11:2803–2820. doi: 10.1091/mbc.11.8.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Lechler T., Li R. Activation of the yeast Arp2/3 complex by Bee1p, a WASP-family protein. Curr. Biol. 1999;9:501–504. doi: 10.1016/s0960-9822(99)80218-8. [DOI] [PubMed] [Google Scholar]
- Yamazaki D., Suetsugu S., Miki H., Kataoka Y., Nishikawa S., Fujiwara T., Yoshida N., Takenawa T. WAVE2 is required for directed cell migration and cardiovascular development. Nature. 2003;424:452–456. doi: 10.1038/nature01770. [DOI] [PubMed] [Google Scholar]
- Yan C., et al. WAVE2 deficiency reveals distinct roles in embryogenesis and Rac-mediated actin-based motility. EMBO J. 2003;22:3602–3612. doi: 10.1093/emboj/cdg350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarar D., To W., Abo A., Welch M. D. The Wiskott-Aldrich syndrome protein directs actin-based motility by stimulating actin nucleation with the Arp2/3 complex. Curr. Biol. 1999;9:555–558. doi: 10.1016/s0960-9822(99)80243-7. [DOI] [PubMed] [Google Scholar]
- Zalevsky J., Lempert L., Kranitz H., Mullins R. D. Different WASP family proteins stimulate different Arp2/3 complex-dependent actin-nucleating activities. Curr. Biol. 2001;11:1903–1913. doi: 10.1016/s0960-9822(01)00603-0. [DOI] [PubMed] [Google Scholar]
- Zallen J. A., Cohen Y., Hudson A. M., Cooley L., Wieschaus E., Schejter E. D. SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. J. Cell Biol. 2002;156:689–701. doi: 10.1083/jcb.200109057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zicha D., Allen W. E., Brickell P. M., Kinnon C., Dunn G. A., Jones G. E., Thrasher A. J. Chemotaxis of macrophages is abolished in the Wiskott-Aldrich syndrome. Br. J. Haematol. 1998;101:659–665. doi: 10.1046/j.1365-2141.1998.00767.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.