Abstract
Leading edge protrusion in the amoeboid sperm of Ascaris suum is driven by the localized assembly of the major sperm protein (MSP) cytoskeleton in the same way that actin assembly powers protrusion in other types of crawling cell. Reconstitution of this process in vitro led to the identification of two accessory proteins required for MSP polymerization: an integral membrane phosphoprotein, MSP polymerization–organizing protein (MPOP), and a cytosolic component, MSP fiber protein 2 (MFP2). Here, we identify and characterize a 34-kDa cytosolic protein, MSP polymerization–activating kinase (MPAK) that links the activities of MPOP and MFP2. Depletion/add-back assays of sperm extracts showed that MPAK, which is a member of the casein kinase 1 family of Ser/Thr protein kinases, is required for motility. MPOP and MPAK comigrated by native gel electrophoresis, coimmunoprecipitated, and colocalized by immunofluorescence, indicating that MPOP binds to and recruits MPAK to the membrane surface. MPAK, in turn, phosphorylated MFP2 on threonine residues, resulting in incorporation of MFP2 into the cytoskeleton. Beads coated with MPAK assembled a surrounding cloud of MSP filaments when incubated in MPAK-depleted sperm extract, but only when supplemented with detergent-solubilized MPOP. Our results suggest that interactions involving MPOP, MPAK, and MFP2 focus MSP polymerization to the plasma membrane at the leading edge of the cell thereby generating protrusion and minimizing nonproductive filament formation elsewhere.
INTRODUCTION
Amoeboid cell motility, characteristic of many eukaryotic cells, involves protrusion of the leading edge coordinated with attachment to the substrate and retraction of the trailing cell body (reviewed in Mitchison and Cramer, 1996; Rafelski and Theriot, 2004). There is now general agreement that localized assembly of actin filaments drives leading edge protrusion (reviewed in Pollard and Borisy, 2003). Many of the components of the protrusion machinery have been identified and in a closely related motile system, the actin comet tails that propel intracellular pathogens such as Listeria, movement has been reconstituted from purified proteins (Loisel et al., 1999). However, the mechanism by which filament assembly pushes the leading edge membrane of a crawling cell forward is still under debate (Mogilner, 2006).
Sperm from nematodes such as Ascaris offer distinct advantages for investigating the mechanism of protrusion. These remarkably simple cells exhibit the same pattern of movement as other crawling cells although their locomotion is based on a system of filaments comprised of major sperm protein (MSP) in place of the more typical actin cytoskeleton (Sepsenwol et al., 1989; reviewed in Roberts and Stewart, 2000; Stewart and Roberts, 2005). The MSP motility apparatus is dedicated to locomotion and its simplicity is illustrated by the ease with which leading edge protrusion can be reconstituted in vitro. Addition of ATP to cell-free extracts of sperm triggers the assembly of a column of MSP filaments, called a fiber, behind vesicles derived from the plasma membrane at the leading edge of the sperm lamellipod (Italiano et al., 1996). As the fiber grows it pushes the vesicle forward in the same way that localized assembly of the MSP cytoskeleton powers protrusion of the leading edge in intact sperm (reviewed in Roberts and Stewart, 2000; Italiano et al., 2001).
Analysis of the MSP in vitro motility system has generated important insights into the mechanism of amoeboid locomotion that also apply to actin-based cells (Roberts and Stewart, 2000; Miao et al., 2003). To exploit fully the advantages of this simple experimental system and its faithful reconstitution of leading edge protrusion requires identification of the complete set of proteins that comprise the motility apparatus. Previous work identified two sperm proteins, in addition to MSP, that are required for fiber assembly and vesicle movement. One, called MSP polymerization–organizing protein (MPOP), is an integral membrane phosphoprotein that is the only vesicle component needed to build fibers. Tyrosine phosphorylation of MPOP directs MSP polymerization to the membrane surface (LeClaire et al., 2003). The other component is a cytosolic protein, MSP fiber protein 2 (MFP2), expressed as three closely related isoforms all of which incorporate into fibers (Buttery et al., 2003; Grant et al., 2005). Although the exact function of MFP2 is unclear, it is required for fiber assembly and its concentration in sperm extracts modulates the rate of fiber growth.
Here we describe the identification and characterization a third essential component of the MSP motility apparatus, MSP polymerization–activating kinase (MPAK), a 34-kDa putative Ser/Thr kinase. MPAK, which exhibits sequence homology to several members of the casein kinase 1 (CK1) family, binds to MPOP and phosphorylates MFP2. Both interactions are required for fiber formation. Moreover, we show that beads coated with MPAK trigger the assembly of a surrounding cloud of MSP filaments when incubated in a cell-free sperm extract, but only when supplemented with detergent-solubilized MPOP. On the basis of these data, we propose a model for how MPOP, MPAK, and MFP2 interact to induce membrane-associated polymerization and discuss how closely this process parallels localized assembly of the actin machinery even though none of the proteins in the two motility systems are shared.
MATERIALS AND METHODS
Sperm Extracts and Fiber Assembly In Vitro
Methods for collecting males of Ascaris suum, harvesting sperm, preparing cell-free extracts (S100), and assembling fibers in those preparations have been described in detail previously (Italiano et al., 1996; Buttery et al., 2003; LeClaire et al., 2003).
Production of Anti-MPAK Antibody
To generate polyclonal antibodies to MPAK, protein was electroeluted from the Mr 34,000 band in SDS-PAGE gels of 10–25% cut of ammonium sulfate precipitation, dialyzed into PBS, combined with 1 ml of RIBI adjuvant (Corixia, Hamilton, MT), and injected into New Zealand white rabbits using a total of 1 mg of antigen. Antibodies were purified from immune serum using Pierce Immunopure plus Immobilized Protein A/IgG purification kits (Rockford, IL).
Purification of MPAK from S100
S100 was diluted 1:4 with KPM buffer (10 mM potassium phosphate, 0.5 mM MgCl2, pH 6.8) (LeClaire et al., 2003) and centrifuged at 100,000 × g for 1 h to separate the sedimentable vesicle fraction from the soluble cytosolic proteins. The cytosol was fractionated by ammonium sulfate precipitation as described (Buttery et al., 2003). The 10–20% cut, which was enriched in an Mr 34,000 band, was dialyzed against 10 mM sodium phosphate (NaP) buffer, pH 7.0, and injected onto a 5-ml ceramic hydroxyapatite (HA) high-performance liquid chromatography (HPLC) column (Bio-Rad, Hercules, CA), equilibrated in the same buffer. Protein was eluted with a 10–250 mM gradient of NaP, pH 7.0. The 34-kDa–enriched fractions, identified by SDS-PAGE, were pooled and dialyzed into immunoaffinity binding buffer (50 mM PBS, 100 mM NaCl, pH 7.2) and applied at 4°C to a column comprised of 1.6 mg of anti-MPAK antibody coupled to Pierce AminoLink Plus gel, as described in the manufacturer's protocol. The column was washed with KPM buffer and then eluted with discontinuous gradient of 100–500 mM NaCl. The eluted fraction was dialyzed against KPM buffer and used immediately or aliquoted for storage at −70°C.
Peptide Sequencing and cDNA Cloning of MPAK
We used methods described for other sperm motility proteins (Buttery et al., 2003) to obtain and sequence four peptides of MPAK by digestion of the Mr 34,000 band from SDS-PAGE gels of a 10–25% cut of ammonium sulfate precipitation of S100 and to obtain a full-length cDNA encoding the protein. The cDNA obtained was inserted into a pCR 2.1 TOPO vector (Invitrogen, Carlsbad, CA) for sequencing of both strands. The full-length cDNA sequence (NCBI accession No. AY326289) was translated by ExPASy translate program. Proteins sequences were aligned using online ClustalW (http://www.ch.embnet.org/software/ClustalW.html) and exported with ESPript (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi).
Immunofluorescence Microscopy
Fibers grown in vitro were prepared for immunolabeling as described (Buttery et al., 2003; LeClaire et al., 2003). We used anti-MPAK, anti-MPOP (LeClaire et al., 2003), anti-MFP2 (Buttery et al., 2003), and mouse monoclonal anti-phosphotyrosine (anti-pY) and anti-phosphothreonine (anti-pT; Cell Signaling Technology, Beverly, MA) as primary antibodies and AlexaFluor 568– or AlexaFluor 488–conjugated goat anti-mouse or goat anti-rabbit (Molecular Probes, Eugene, OR), as appropriate, as secondary antibodies. All antibodies were used at 5 μg/ml for 3 h at 25°C. For controls, primary antibodies were omitted. Immunolabeled cells and fibers were imaged with a Zeiss 510 (Carl Zeiss, Thornwood, NY) laser scanning confocal microscope equipped with dual HeNe laser with appropriate filters or with a Zeiss Axioskop2 microscope equipped with a 40× acroplan/phase objective with appropriate filters and imaged with an Orca 12-bit digital camera (Hamamatsu, Bridgewater, NJ). Images were captured and processed with MetaMorph (Molecular Devices, Sunnyvale, CA) software.
Immunodepletion and Immunoprecipitation
MPAK-depleted extracts were obtained by passing cytosol three times over a column of anti-MPAK antibody coupled to agarose beads (Pierce). Columns constructed with beads bound to IgG from preimmunization serum served as a control. We used SDS-PAGE and Western blotting to monitor the depletion efficiency of the column. Purified MPAK used to replenish these depleted extracts was obtained as described above.
Samples of S100 for immunoprecipitation were incubated in KPM with 0.5% Triton X-100 for 30 min at 25°C and then mixed with equal volume of 2× immunoprecipitation (IP) buffer (2% Triton X-100, 300 mM NaCl, 20 mM Tris, pH 7.4, 2 mM EGTA) followed by immunoprecipitation with appropriate antibody and protein G–coupled agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA), which were prewashed three times with IP buffer. The mixture was incubated at 4°C for 3 h. The beads were harvested by centrifugation at 1000 × g for 3 min and then washed five times with IP buffer. The protein composition of immunoprecipitates was analyzed by SDS-PAGE and Western blots.
Additional Protein–Protein Binding Assays
To assay binding of sperm cytosolic proteins to vesicles, S100 was incubated at 4°C with 1 mM ATP for 10 min, diluted 1:5 into KPM buffer, and centrifuged at 100,000 × g for 1 h in a TLA100.3 rotor (Beckman Coulter, Fullerton, CA). The vesicle pellet was resuspended in KPM containing 1.5 M KCl for 1 h to remove peripheral proteins and then washed three times in KPM. The salt-extracted vesicles were incubated in MPAK in KPM for 2 h and then centrifuged at 100,000 g for 1 h to repellet the vesicles. The protein composition of the vesicle pellets was examined by SDS-PAGE gel and Western blots.
To assess further the interaction between MPAK and MPOP, we ran blue native (BN) gels as described by Schagger and von Jagow (1991). To remove low-molecular-weight material from the samples, S100 was spun three times on a 50-kDa molecular weight cutoff filter at 15,000 × g for 30 min at 4°C. After each centrifugation, we added 10 volumes of cold BN cathode buffer (50 mM Tricine, 15 mM Bis-Tris, pH 7.0). After the third centrifugation, samples were brought to 0.5% of TX-100, incubated for 2 h on ice, and then separated on 4–16% BN gels run at 4°C at 100 V constant voltage until the blue dye front reached the bottom. BSA was used as molecular weight marker. The strips were cut from the BN gel, incubated in SDS-PAGE loading buffer for 2 h at 22°C, and placed individually into the wells of 15% SDS-PAGE gels for electrophoresis in the second dimension. SDS-PAGE and immunoblotting were performed according to standard protocols. Membranes were probed first with anti-MPOP antibody, rinsed with TBST overnight, blocked with 0.5% BSA in TBST, and blotted again with anti-MPAK antibody.
Pharmacological Studies
We tested the effects of five protein kinase inhibitors, including bisindolylmaleimide-1 (Bisin-1) for protein kinase C, H89 for protein kinase A, IC 261 for casein kinase 1, KN93 for calmodulin kinase, and protein kinase G inhibitor (PKG-I) for PKG. S100 was diluted 1:10 with KPM in the presence of each reagent individually. Fiber assembly was triggered by addition of 1 mM ATP. The concentration used for each inhibitor was 10- to 100- fold greater than its Ki (Bisin-1, 1 μM; H89, 1 μM; IC 261, 200 μM; KN93, 300 μM; and PKG-I, 10 mM).
In Vitro Kinase Assays
We used in vitro assays to test phosphorylation of two proteins, casein and MFP2, by MPAK. Casein was obtained from Sigma-Aldrich (St. Louis, MO). Partially purified MFP2 was obtained from the cytosolic fraction of S100 by sequential molecular size exclusion chromatography and anion exchange chromatography (Buttery et al., 2003). The assays were performed in a total volume of 30 μl containing of 1.5 μM purified MPAK, ∼5 μM casein or MFP2, and 4 μCi of [γ-32P]ATP in kinase reaction buffer (20 mM Tris·Cl, pH 7.5, 5 mM MgCl2, 150 mM KCl). In control assays either MPAK or the target protein was omitted from the reaction. The reaction mixtures were incubated for 30 min at 23oC and stopped by the addition of 5× loading buffer. After heating to 95°C for 5 min, the mixtures were separated on 15% gels by SDS-PAGE. The gels were dried, exposed to phosphorscreen, and analyzed on a STORM phosphoimager (Amersham Biosciences, Piscataway, NJ).
To complement these tests of the enzymatic activity of MPAK, we also used immunolabeling to assess the kinase activity of the protein. These assays were performed in a total volume of 30 μl comprised of 1.5 μM purified MPAK, 5 μM MFP2, and 2 mM ATP. The reactions were incubated for 3 h at room temperature and then stopped by addition of same amount of 2× SDS-PAGE buffer. The phosphorylation state of protein was examined with SDS-PAGE and Western blot using anti-phosphothreonine (Cell Signaling Technology), anti-phosphoserine (Chemicon, Temecula, CA), and anti-phosphotyrosine antibodies. In control assays we omitted either MPAK or MFP2 from the reaction mixture.
Filament Assembly on MPAK-coated Beads
Carboxylated polystyrene beads (2-μm diameter) were coated with a fraction enriched in MPAK (∼3 μM), obtained by passing cytosol over an anti-MPAK immunoaffinity column. Bead were incubated for 1 h at room temperature, followed by BSA (10 mg/ml) for another 15 min to block remaining free adsorption sites, and then sedimented, washed with KPM buffer, and resuspended in the same buffer. To determine if the MPAK-coated beads triggered MSP polymerization, aliquots were added to S100 that had been treated with 0.5% Triton X-100 on ice for 1 h to lyse membrane vesicles and then diluted 1:5 with KPM. BSA-coated beads were used as controls. In some assays we added 3 μM Alexa-488–labeled MSP to enable detection of bead-associated polymerization by fluorescence. ATP was added to initiate filament assembly and the samples were incubated for 30 min. Samples containing Alexa 488-MSP were examined by fluorescence microscopy. Digital images were captured and processed using MetaMorph software as described above. Samples for electron microscopy were incubated on a 7 × 10-mm glass coverslips for 45 min, fixed for 20 min in 2.5% glutaraldehyde, washed with KPM, dehydrated in ethanol, and critical point dried as described previously (Ris, 1985). Platinum replicas were prepared according to method described previously (LeClaire et al., 2003) and examined on a Philips (Wilmington, DE) CM 120 electron microscope operated at 80 kV.
RESULTS
A 34-kDa Protein Is a Required Component of the MSP Motility Apparatus
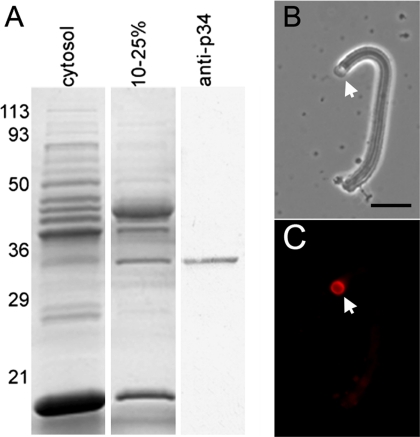
We identified a 34-kDa protein as a potential component of MSP cytoskeleton based on its presence and abundance in the 10–25% ammonium sulfate cut of sperm cytosol (S100 with vesicles removed by centrifugation; Figure 1A). This fraction contains all of the components required to build fibers when supplemented with vesicles, MSP and ATP (Buttery et al., 2003). To determine whether this protein was a fiber component, we generated a polyclonal antibody to the protein extracted from the Mr ∼34-kDa band on SDS-PAGE gels. This antibody labeled only a single Mr ∼34-kDa band on Western blots of cytosol (Figure 1A) and also labeled fibers by indirect immunofluorescence with the signal localized at the vesicle-bearing end of the fiber (Figure 1, B and C).
Figure 1.
Identification of a 34-kDa protein as a component of fibers. (A) Coomassie-stained gel of cytosol (obtained by removing the vesicles from S100 by centrifugation) and the fraction of cytosol obtained by precipitation in 10–25% ammonium sulfate. A corresponding Western blot of the 10–25% ammonium sulfate fraction probed with an antibody to the 34-kDa protein (anti-MPAK) is shown at the right. Numerals indicate the masses of molecular weight markers (in kDa). (B and C) Paired phase-contrast and fluorescence images of a fiber probed with anti-MPAK antibody by indirect immunofluorescence. Labeling is primarily at the vesicle-bearing end of the fiber (arrow) with the heaviest staining at the vesicle–fiber interface. Bar, 5 μm.
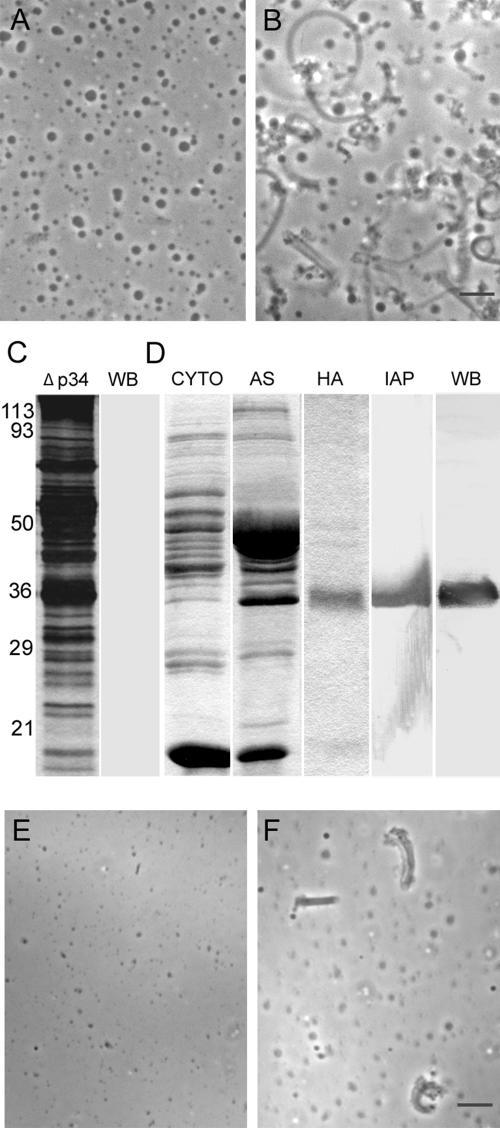
Addition of the antibody generated against the 34-kDa protein at 1 mg/ml to 1:10-diluted S100 completely inhibited fiber formation (Figure 2, A and B). By contrast, anti-MFP3 antibody, directed against a protein that is found in fibers but not required for their assembly (our unpublished observations), at the same concentration had no effect on fiber formation (data not shown), suggesting that the 34-kDa protein may be required for fiber growth. To explore this hypothesis, we used the antibody to immunodeplete the 34-kDa protein from S100. After passing the S100 over the column three times, the 34-kDa protein was no longer detectable by Western blot analysis (Figure 2C), and this 34-kDa-depleted material, after concentration to its original volume, was not able to support fiber growth (Figure 2E). Fiber assembly in the depleted S100 could be rescued by addition of the 34-kDa protein that had been purified from S100 by sequential centrifugation, ammonium sulfate precipitation, HA chromatography, and immunoaffinity chromatography (Figure 2D). This procedure yielded a single 34-kDa protein band on a heavily loaded, silver-stained gel. The purified 34-kDa protein obtained in this way restored fiber assembly when added back to immunodepleted S100 (Figure 2F).
Figure 2.
A 34-kDa protein is required for fiber assembly. (A and B) Phase-contrast micrographs of 1:10 diluted S100 plus ATP with (A) and without (B) 1 mg/ml anti-p34 antibody. No fibers were detected in the presence of the antibody. (C) SDS-PAGE and Western blot analysis of the fractions used for depletion/add-back assays. The two lanes show a silver-stained lane (Δp34) of cytosol after three passes over an anti-p34 immunoaffinity column and a Western blot of the same material probed with anti-p34 antibody. The 34-kDa protein could not be detected after immunodepletion. (D) Sequential purification of the 34-kDa protein from Ascaris sperm extracts. CYTO, cytosol fraction obtained by centrifugation of S100; AS, 10–20% cut obtained by ammonium sulfate precipitation of the CYTO fraction; HA, 34-kDa protein-enriched fraction separated from the 10–20% cut by hydroxyapatite chromatography (the CYTO, AS, and HA lanes were Coomassie-stained); IAP: silver-stained lane of the protein obtained from the 34-kDa protein-enriched HA fraction by immunoaffinity purification by passage over an anti-p34 affinity column. WB, Western blot of the material shown in the IAP lane probed with anti-p34 antibody. Numerals indicate the position of molecular weight markers in kDa for both C and D. (E and F) Fiber assembly in depletion/add-back assays. Addition of vesicles, MSP, and ATP to the 34-kDa protein-depleted fraction shown in C failed to trigger fiber assembly (E). However, addition of the 34-kDa protein purified as shown in D to the depleted material along with vesicles, MSP, and ATP resulted in the formation of numerous fibers (F). Bars, 5 μm in B and F.
The 34-kDa Protein Is a Member of the CK1 Family of Protein Kinases
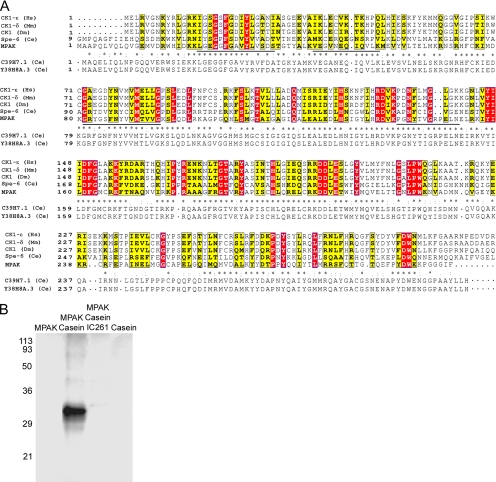
To determine the sequence of the Mr ∼34-kDa band, we used the sequences of four peptides (Figure 3A) obtained by endo-LysC digestion of the 34-kDa band eluted from SDS-PAGE gels to prepare degenerate oligonucleotide primers for RT-PCR against total polyA+ RNA obtained from Ascaris testis. These probes identified a single full-length cDNA the sequence of which predicted a 34-kDa polypeptide that matched exactly the sequences of all four peptides from the 34-kDa band and exhibited 67% amino acid identity (205/307) to two predicted proteins, C39H7.1 and Y38H8A.3, from Caenorhabditis elegans (Figure 3A), both of which exhibit spermatogenesis-enriched expression in microarray analyses (Reinke et al., 2000). Searches of sequence databases indicated that the 34-kDa protein from Ascaris sperm and its homologues in C. elegans are putative members of the CK1 family of protein serine/threonine kinases. The kinase domain of known CK1s is highly conserved. Within this region, which comprises residues 14–298 of the 34-kDa polypeptide, the sperm protein sequence shares 15% amino acid identity and 54% similarity with other members of the CK1 family (Figure 3A). We found that the 34-kDa protein, purified from S100 (Figure 2D), phosphorylated purified casein in the presence of γ-32P-ATP (Figure 3B), which was consistent with its sequence similarity to CK1. Moreover, IC261, a CK1 inhibitor, blocked phosphorylation of casein in this assay. Thus, the 34-kDa protein appears to be a member of the CK1 family on the basis of both sequence and enzymatic activity. On the basis of these data and studies of the function of the 34-kDa protein in assembly of the MSP cytoskeleton (see below), we refer to the protein hereafter as MPAK.
Figure 3.
Amino acid sequence homology and enzymatic activity indicate that the 34-kDa protein is a member of the casein kinase 1 family of Ser/Thr kinases. (A) Alignment of the predicted sequence of the 34-kDa protein (now designated MPAK) with four representative sequences from the casein kinase 1 family: CK1-ε from humans (Hs, NP_001885), CK1-δ from mouse (Mm, NP_620690), CK1 from Drosophila melanogaster (Dm, AAC39134), and spe-6 from C. elegans (Ce, NP_499482). Completely conserved residues are highlighted in red and nearly conserved ones in yellow. The sequence of MPAK (AY326289) is also aligned with that of its C. elegans homolog, C39H7.1; asterisks represent identical residues, colons indicate highly conservative substitutions, and periods denote less conservative differences. The sequence of Y38H8A.3, a closely related predicted protein from C. elegans, is shown below that of C39H7.1. The four peptide sequences obtained via proteolysis of MPAK and used to generate oligonucleotides for RT-PCR are underlined in the MPAK sequence. The sequence of each peptide is identical to the predicted amino acid sequence from the MPAK cDNA. (B) Autoradiogram of an SDS-PAGE gel of kinase assays of purified MPAK. Components incubated with 32P-ATP in the kinase assays are indicated above the gel lanes. Casein was phosphorylated by MPAK and the casein 1 kinase inhibitor, IC 261, blocked this activity. No phosphorylation was detected with MPAK or casein alone. Numerals indicate the positions of molecular weight markers in kDa.
Efforts to obtain soluble recombinant MPAK by bacterial expression as either a native or a GST- or His-tagged fusion protein were unsuccessful. The cDNA obtained by RT-PCR could be expressed in bacteria using a variety of vectors and several growth temperatures. The expressed protein labeled with anti-p34 antibody on Western blots, confirming that it corresponded to the protein purified from S100. However, all of the expressed protein was found in inclusion bodies. We were unable to obtain any of this protein in soluble form, even after expression at low temperatures or, in some cases, subjecting the insoluble material to high pressure. Reagents that dissolve inclusion bodies did solubilize the recombinant MPAK, but the protein reprecipitated when returned to the benign buffers used to assay enzyme activity and fiber assembly. Therefore, because we were unable to obtain soluble bacterially expressed protein, we used native MPAK purified from S100 (Figure 2D) for further analyses of its role in MSP-based motility.
MPAK Is Recruited to the Membrane by Interaction with Phosphorylated MPOP
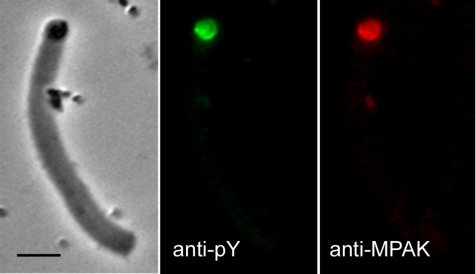
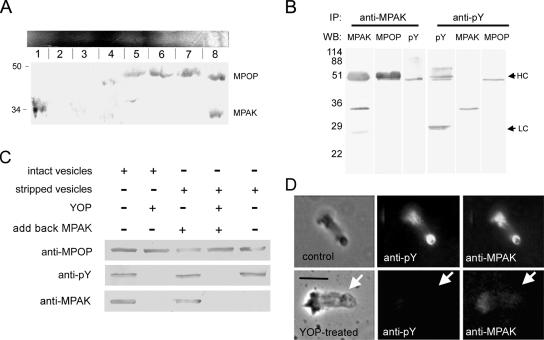
Previous work showed that MPOP, the integral membrane phosphoprotein that determines the site of MSP polymerization, is located in the lamellipodial plasma membrane of Ascaris sperm and in the vesicles that build fibers in S100 (LeClaire et al., 2003). Immunofluorescence labeling showed that MPAK colocalized with phospho-MPOP (labeled with anti-pY antibody; see LeClaire et al., 2003) at the vesicle at the growing end of the fiber (Figure 4) Both proteins were localized to the vesicle surface and, in most cases, the labeling was particularly pronounced at the vesicle-fiber interface.
Figure 4.
Colocalization of MPAK and phospho-MPOP in fibers. Phase-contrast and corresponding fluorescence images of a fiber labeled with anti-pY (green) and anti-MPAK (red) antibodies by indirect immunofluorescence. Bar, 5 μm.
Four lines of evidence indicated that the colocalization of MPAK and MPOP detected by immunofluorescence is due to interaction of MPAK with the phosphorylated form of the membrane protein. First, when we separated the proteins in S100 on BN gels and then analyzed the protein components in slices of the native gel by SDS-PAGE, in addition to the slices that contained MPAK or MPOP alone, we also identified a slice from the native gel that contained both MPAK and MPOP, detectable on Western blots probed with antibodies specific to each protein (Figure 5A). MPOP was present in a broad band, encompassing slices 5–8 of the native gel, as is often seen with membrane proteins. Some of this heterogeneity may also reflect interactions of MPOP with other components of S100. However, the presence of both MPAK and MPOP in band 8, but not in bands 5–7, was clearly consistent with some of each protein existing in a complex in S100. Therefore, this analysis suggests that at least part of the MPAK and MPOP in S100 bind with sufficient affinity to comigrate by native gel electrophoresis.
Figure 5.
MPAK binds to phosphorylated MPOP. (A) Two-dimensional native/SDS-PAGE gel electrophoresis of S100. A Coomassie-stained native gel is shown horizontally at the top. An identical gel was cut into eight slices as indicated, and each slice was loaded into a well of a 15% SDS-PAGE gel, blotted, and probed with both anti-MPOP and anti-MPAK. Slice 8 from the native gel contained both proteins. Slice 1 contained MPAK but no MPOP; slices 5–7 contained MPOP but no MPAK. (B) Western blots of the proteins obtained by immunoprecipitation (IP) with anti-MPAK and anti-pY antibodies. WB denotes the antibody used for Western blots of the gel lanes indicated. In addition to its target antigen, anti-MPAK also pulled down tyrosine phosphorylated MPOP, detectable by both anti-MPOP and anti-pY on Western blots. HC and LC indicate the positions of rabbit and mouse Ig heavy and light chains, respectively. Note that because anti-MPAK and anti-MPOP were produced in rabbits the secondary anti-rabbit IgG used for detection labeled the immunoglobulin used for immunoprecipitation. Ig heavy chain migrates slightly slower than the 48-kDa MPOP, the position of which can be seen in the lane probed with anti-pY, a mouse monoclonal that does not react with rabbit IgG. Anti-pY coimmunoprecipitated MPAK and MPOP detected on Western blots probed with anti-MPAK and anti-MPOP antibodies. Numerals indicate positions of molecular-weight markers (in kDa). (C) Vesicle pulldown assay. Selected portions of Western blots of material pelleted after incubation of vesicles (intact or stripped with 1.5M KCl) with or without MPAK. Incubation conditions are shown above the Western blot panels. In preparations treated with YOP (second and fourth lanes), the vesicles were incubated with the tyrosine phosphatase, washed in assay buffer, and then pelleted. Antibodies used for Western blots are shown at the left. (D) Effect of YOP on the presence of MPAK in fibers. The top series of panels shows a control fiber, washed with KPM assembly buffer, fixed, and labeled with anti-pY and anti-MPAK antibodies by indirect immunofluorescence. The bottom panels show a fiber treated with buffer containing YOP before fixation and labeling. YOP treatment abolished labeling of both phosphorylated MPOP and MPAK at the vesicle-bearing end of the fiber. Arrows indicate the position of the vesicle on each fiber. Bar, 5 μm.
Second, we found that MPAK and MPOP coimmunoprecipitated from S100. For example, addition of anti-MPAK antibody conjugated to beads to S100 that had been treated with Triton X-100 to lyse the vesicles, pulled down MPAK as well as a Mr ∼48-kDa band that labeled with both anti-MPOP and anti-pY antibodies on Western blots (Figure 5B). Therefore, at least part of the MPOP that coimmunoprecipitated by anti-MPAK antibody was phosphorylated. In the reciprocal assay, immunoprecipitation of Triton-treated S100 with anti-pY brought down phosphorylated MPOP and MPAK (Figure 5B).
Third, we used vesicles harvested from S100 to assess further the interaction between MPAK and MPOP. When these vesicles were isolated by centrifugation, they contained associated MPAK, as judged by SDS-PAGE and Western blots (Figure 5C). Treatment of the isolated vesicles with 1.5 M KCl removed all detectable MPAK (Figure 5C). When these salt-extracted vesicles were washed into KPM buffer, incubated with purified MPAK in the presence of ATP, and then pelleted by centrifugation, MPAK cosedimented with the vesicles. However, when the salt-extracted vesicles were incubated with MPAK and ATP in the presence of YOP, a protein tyrosine phosphatase from Yersinia enterocolytica shown previously to dephosphorylate MPOP (LeClaire et al., 2003), the MPOP in the vesicles no longer labeled with anti-pY, and no detectable MPAK cosedimented with the vesicles (Figure 5C). These data suggest that MPAK associated with the vesicle by binding to phosphorylated MPOP.
Fourth, we used YOP to ask if dephosphorylation of MPOP on vesicles that had assembled fibers affected the localization of MPAK at the vesicle–fiber interface. As shown in Figure 5D, when growing fibers were washed with KPM buffer and then fixed for immunofluorescence, they retained both anti-pY and anti-MPAK labeling. By contrast, fibers treated before fixation with KPM containing YOP failed to label with either anti-pY or anti-MPAK. Thus, treatment of fiber-associated vesicles with YOP not only dephosphorylated MPOP but also released bound MPAK from the vesicle-fiber interface.
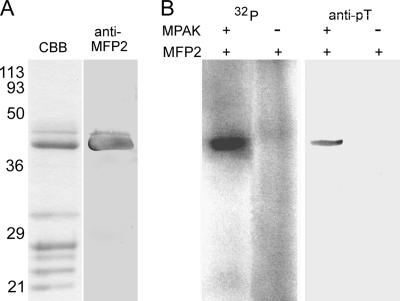
MFP2 Is Phosphorylated by MPAK
Fiber elongation is driven by polymerization at the vesicle-fiber interface (Roberts et al., 1998), where MPOP and MPAK are located, and the rate of fiber growth is modulated by MFP2 (Buttery et al., 2003), probably operating at the site of filament assembly. Therefore, we reasoned that MFP2 may be a target of the kinase activity of MPAK. To test this hypothesis, we obtained an MFP2-enriched fraction from S100 by sequential centrifugation, molecular exclusion chromatography, and anion exchange chromatography (Figure 6A; see also Buttery et al., 2003) and combined this material with purified MPAK in the presence of γ-32P-ATP. These conditions resulted in prominent γ32P-labeling of MFP2, the most abundant protein in the immunoprecipitate, but none of the other proteins in the MFP2-enriched fraction was labeled (Figure 6B). Moreover, we detected no labeling of MFP2 when MPAK was omitted. To assess further the phosphorylation of MFP2 by MPAK, we repeated the incubation of the MFP2-enriched fraction with purified MPAK, in this case in the presence of unlabeled ATP. After incubation of in the presence of the kinase, MFP2 labeled with anti-phosphothreonine (anti-pT) antibody on Western blots (Figure 6B). By contrast, when MFP2 was incubated with ATP alone, we were unable to detect phosphorylation of the protein on anti-pT Western blots (Figure 6B). Addition of anti-MPAK antibody or the CK1 inhibitor, IC 261, blocked the phosphorylation of MFP2 by MPAK (data not shown). MFP2 incubated with MPAK did not label with either anti-pY or anti-phosphoserine on Western blots, and neither of these antibodies immunoprecipitated MFP2 from S100. Thus, MPAK appears to phosphorylate MFP2 exclusively on threonine residues.
Figure 6.
MPAK phosphorylates MFP2. (A) SDS-PAGE analysis of MFP2 partially purified from the cytosolic fraction of S100 by sequential molecular size exclusion and anion exchange chromatography. The most prominent band in a Coomassie-stained lane (CBB) in this material was labeled in a duplicate lane (anti-MFP2) blotted and probed with polyclonal anti-MFP2 antibody. (B) Phosphorylation of MFP2 by purified MPAK. The left two lanes (32P) show autoradiograms obtained after incubation of 32P-ATP with components indicated above the gel lanes. No phosphorylation was detected when MPAK was omitted from the reaction. By contrast, addition of MPAK resulted in heavy labeling of a band at Mr ∼38 kDa, which comigrated with MFP2. The right two lanes (anti-pT) show Western blot analysis of MFP2 threonine phosphorylation after incubation in ATP in the presence and absence of MPAK. Anti-phosphothreonine (anti-pT) antibody labeled MFP2 only after the protein was incubated with MPAK. Numerals at the left indicate the positions of molecular weight markers in kDa in all lanes.
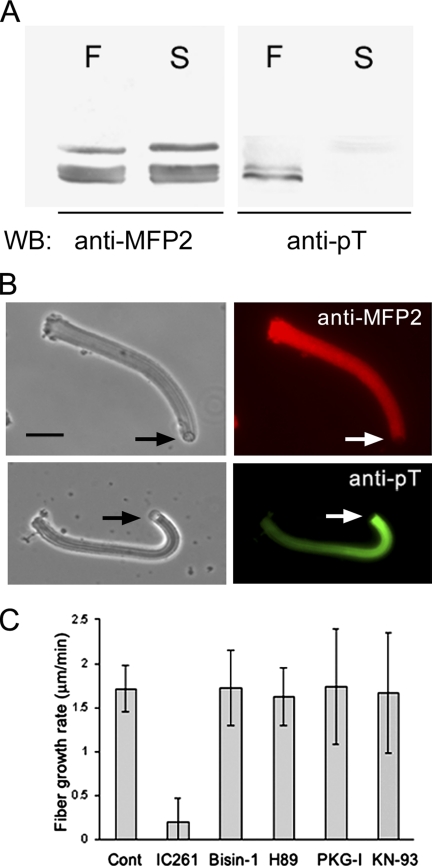
To determine if the phosphorylation of MFP2 is associated with fiber assembly, we grew fibers in S100 and then separated them from the remaining soluble proteins by centrifugation. Comparison of the fiber pellet and the supernatant by SDS-PAGE and Western blot analysis (Figure 7A) showed that both fractions contained MFP2. However, when we blotted the gel with anti-pT, we detected labeling of the MFP2 that had been incorporated into fibers but did not detect phosphorylation of the MFP2 remaining in the supernatant. Consistent with the presence of threonine-phosphorylated MFP2 in the fiber, anti-pT–labeled fibers in indirect immunofluorescence assays (Figure 7B). However, the labeling pattern did not coincide exactly with that obtained with anti-MFP2 antibody. Anti-MFP2 labeled the fiber uniformly. By contrast, anti-pT produced a gradient of labeling intensity along the length of the fiber. The fluorescent signal was greatest at the newly assembled vesicle-bearing end of the fiber and decreased in intensity with distance from the vesicle. Thus, the fraction of phosphorylated MFP2 in the fiber may be highest near the vesicle where the protein is phosphorylated by MPAK anchored to MPOP and first incorporated into the fiber.
Figure 7.
Phosphorylated MFP2 in fibers. (A) Western blot of the MFP2-containing region of SDS-PAGE gels of fibers (F) and the supernatant (S) remaining after separating fibers from S100 by centrifugation. Both fractions contain MFP2 detectable by Western blots with anti-MFP2 antibody but only the MFP2 in fibers labels with anti-pT on Western blots. (B) Paired phase-contrast and immunofluorescence micrographs comparing the labeling pattern of anti-MFP2 (top panels) and anti-pT (bottom panels) in fibers. MFP2 is distributed nearly uniformly along the length of the fiber, whereas anti-pT exhibits a gradient of decreased staining from the vesicle-associated end rearward. Bar, 5 μm. (C) IC261, an inhibitor of casein kinase 1, significantly reduced the rate of fiber growth in S100 compared with that of untreated control fibers. The fiber growth rate in the presence of bisindolylmaleimide-1 (Bisin-1), H89, PKG-I, or KN93, which inhibit protein kinase C, A, G, and calmodulin kinase, respectively, was not significantly different from that of untreated control fibers. Bars, mean ± SE of three trials, each including 5–10 fibers.
In keeping with its inhibition of MPAK-catalyzed phosphorylation of MFP2, we found that addition of 200 μM IC261 to S100 diluted 1:10 reduced the rate of fiber growth by 90% compared with that of fibers grown in the absence of the inhibitor (Figure 7C). By contrast, inhibitors of other Ser/Thr protein kinases, including PKA (H89), PKC (bisindolylmaleimide-1), PKG (PKG-I), and Cam kinase (KN93) had no effect on fiber elongation when added to S100 at concentrations up to 100-fold greater than their reported Kis.
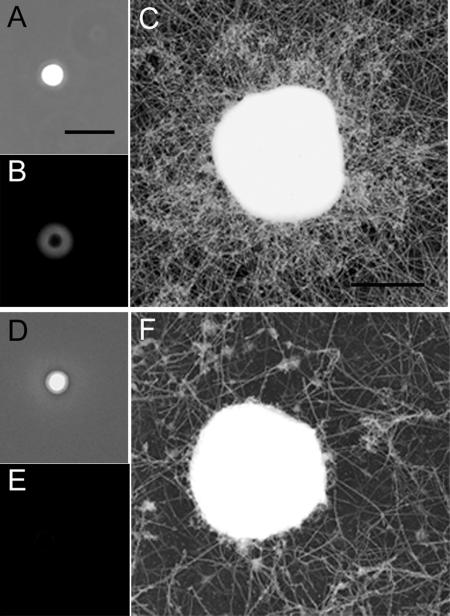
MPAK-coated Beads Trigger MSP Polymerization
To assess further the role of MPAK in MSP filament assembly, we asked if plastic beads coated with the kinase could trigger MSP polymerization. Because of the high concentration of protein required for these assays, we obtained MPAK directly from cytosol by anti-MPAK immunoaffinity chromatography. The fraction obtained in this way was enriched in MPAK but also contained small amount of MFP2 and MSP. When beads coated with this MPAK-enriched fraction were incubated in Triton-treated S100 supplemented with Alexa 488-MSP, a cloud of fluorescence formed around the beads (Figure 8, A and B). Examination of platinum replicas of similar beads by EM showed that the cloud of fluorescent MSP corresponded to a dense meshwork of filaments surrounding the beads (Figure 8C). These preparations also contained a looser meshwork of filaments in the background. When we used control beads, coated with BSA in place of MPAK, in these assays, we still observed filaments in the background but there was no accumulation of filaments on the bead itself (Figure 8, D and F). Moreover, neither MSP nor MFP2, the other proteins in the MPAK-enriched fraction used to coat the beads, stimulated MSP filament assembly alone or in combination (data not shown). MPAK-coated beads did not form filament clouds when they were incubated in S100 that had not been treated with detergent and, thus, contained intact vesicles. Likewise, no filament clouds formed when coated beads were incubated in S100 depleted of vesicles by centrifugation. Thus, the polymerization of MSP on MPAK-coated beads appears to depend on the presence of solubilized vesicle components, presumably MPOP.
Figure 8.
Assembly of filaments on MPAK-coated beads. (A and B) Phase-contrast and fluorescence micrographs of 2-μm polystyrene beads coated with MPAK obtained by immunoaffinity purification from cytosol and incubated in S100 containing 0.5% Triton X-100 and supplemented with 3 μM Alexa488-MSP. A cloud of material containing fluorescent labeled MSP formed around the beads during a 30-min incubation. (C) Electron micrograph of a platinum replica of a MPAK-coated bead from preparations like those in A and B showing that the cloud of material detected by fluorescence is comprised of filaments surrounding the bead. Note that there are also filaments in the background that assemble under these conditions. (D–F) Control assay in which BSA-coated beads were incubated in Triton-treated S100 plus ATP and Alexa488-MSP. These beads lack the cloud of filaments detectable by either fluorescence (E) or electron microscopy (F), although filaments are still detectable in the background. Bars, (A) 5 μm; (C) 1 μm.
We also used an in vitro assembly assay to determine if MPAK, MPOP, and MFP2 are sufficient to trigger MSP polymerization. However, we were unable to detect filaments by EM after incubation of 3 μM MPAK, 20 μM MFP2, and 0.8 mM MSP (their approximate concentrations in 1:5 diluted S100), 2 mM ATP, and a Triton extract of vesicles as a source of MPOP.
DISCUSSION
Reconstitution of MSP-based motility in vitro has facilitated investigation of the biophysical basis of both lamellipodial protrusion (Italiano et al., 1996) and cell body retraction (Miao et al., 2003) in amoeboid cell motility. To capitalize fully on this powerful experimental system, it is necessary identify the full roster of the components required for motility. Previous work identified MPOP and MFP2 as essential components of the motility apparatus, but these proteins alone are not sufficient to build fibers when added to MSP and ATP (Buttery et al., 2003; LeClaire et al., 2003). We have now identified a third accessory protein, MPAK, required to generate protrusion by membrane-associated MSP polymerization. Further, we have shown that MPAK is a serine/threonine kinase that couples the activities of MPOP and MFP2 by binding to phospho-MPOP and by phosphorylating MFP2.
Some caution is appropriate in assigning a specific cellular function to a protein isolated from a cellular extract, especially considering that, despite extensive effort, we have not able to obtain active MPAK by bacterial expression. However, several lines of evidence suggest that MPAK is responsible for the activities described in this study. First, both the casein kinase activity of the native protein and its effect on fiber growth are inhibited by the CK1 inhibitor, IC 261, and not by inhibitors of other Ser/Thr kinases. Second, the four peptides obtained from native material matched exactly the sequence predicted from the MPAK cDNA. Third, an antibody raised against the native protein recognized the insoluble bacterially expressed protein encoded by the MPAK cDNA. Finally, the same antibody blocked the phosphorylation of MFP2 by MPAK and inhibited fiber assembly in S100. These results are all consistent with the 34-kDa protein isolated from S100 being responsible for the activities we have identified for MPAK.
All of the accessory proteins in the Ascaris MSP motility apparatus that have been sequenced have homologues in C. elegans, but in all previous cases these homologues are predicted proteins with no assigned function and are encoded in genes that have not been defined by mutation (Buttery et al., 2003; LeClaire et al., 2003). Thus, MPAK is the first MSP motility component that has significant sequence homology to a protein with a common fold and a well-defined activity. The CK1 family to which MPAK belongs is a subset of the serine/threonine kinases that is expressed ubiquitously in eukaryotes and is involved in a broad range of cellular activities (Knippschild et al., 2005). Like other CK1s, the predicted kinase domain of MPAK is located near in the N-terminal region of the sequence and is flanked by a C-terminal noncatalytic domain. The length and sequence of these C-terminal domains differ substantially among the CK1s. The C-terminal region of MPAK is particularly short (8 residues), thus accounting for the lower molecular mass of this protein compared with the 37–51 kDa range that is more typical for the members of the CK1 family.
Our data suggest that the interactions among MPOP, MPAK, and MFP2 direct MSP polymerization to the membrane at the leading edge of the lamellipod. Previous work (LeClaire et al., 2003) established that MPOP is the only membrane protein required for fiber assembly. Our antibody inhibition tests and immunodepletion/add-back assays established that MPAK is also necessary for fiber assembly. Moreover, comigration of MPAK and MPOP on native gels together with reciprocal immunoprecipitation of the two proteins with antibodies to MPAK and tyrosine phosphorylated MPOP showed that these proteins interact in a complex mixture consistent with the interaction being highly specific. The disruption of this interaction by dephosphorylation of MPOP, both on the vesicles used in pulldown assays and also on the vesicles attached to growing fibers indicates that MPAK is recruited to the membrane surface by binding to phosphorylated MPOP. These results are consistent with previous studies that showed that addition of YOP to S100 blocks fiber elongation (Italiano et al., 1996; Miao et al., 2003) and indicate that this effect of YOP is due, at least in part, to the loss of MPAK from the site of filament assembly upon dephosphorylation of MPOP. The sensitivity of the interaction of the two proteins to tyrosine phosphorylation implies that MPAK may bind to phosphotyrosine residues on MPOP. However, the sequence of MPAK does not include an SH2 domain by which many effector molecules bind to phosphorylated tyrosine residues on the cytoplasmic domain of membrane proteins (Felder et al., 1993). Thus, the binding of these two proteins appears to involve a different phosphorylation-dependent molecular interaction.
Our data also indicate that the presence of both MPOP and MPAK are required to trigger MSP polymerization at the membrane. For example, we found that the assembly of filaments around MPAK-coated beads did not occur unless extracts were supplemented with solubilized MPOP. This observation suggests that MPOP may turn on the catalytic activity of MPAK. However, purified MPAK phosphorylated both casein and MFP2 in the absence of MPOP. Therefore, MPOP is not required to activate MPAK and it may be that each protein plays an essential but independent role in assembling the MSP motility apparatus.
Although there may be additional substrates for MPAK, we have shown that MFP2 is a primary target of the enzyme and that the phosphorylation of MFP2 is associated with its incorporation into fibers. MFP2 is required for fiber assembly and, although its exact function is not known, the protein enhances the rate of MSP polymerization thereby increasing the rate of fiber elongation (Buttery et al., 2003). Thus, the protein likely exerts its effect at the growing end of the fiber where filaments are assembled. However, unlike its partner proteins, MPOP and MPAK, MFP2 is not concentrated at the vesicle-fiber interface and, instead, is distributed throughout the fiber (Buttery et al., 2003). Therefore, MFP2 may be activated by MPAK-catalyzed phosphorylation at the growing end of a fiber where it modulates the rate of polymerization and then remains bound to the MSP filament system as fiber elongation continues and the vesicle moves away.
Like the MSP-based motility of nematode sperm, the localized polymerization of actin filaments is used to move membranes, either in the form of protrusion of the leading edge of crawling cells (Borisy and Svitkina, 2000) or the intracellular movement of membrane vesicles, bacteria, and other pathogens by actin comet tails (Cameron et al., 2000). Although the identities of the molecular players in the MSP and actin systems differ, the overall scheme by which they operate to direct filament assembly to the membrane surface has some intriguing parallels. For example, in actin-based cells a number of different membrane proteins, depending on cell type, define sites of filament assembly by recruiting and activating a member of the WASp/Scar family of proteins to the membrane surface, an interaction that is analogous to that of MPOP-MPAK. Activated WASp, in turn activates the Arp2/3 complex resulting in the nucleation and elongation of actin filaments near the membrane (Rohatgi et al., 1999; reviewed in Pollard and Borisy, 2003) to form a branched filament network. The role of MPAK appears generally analogous to that of WASp as it phosphorylates MFP2 and perhaps other components needed to assemble fibers. Thus, although the MSP cytoskeleton consists of a meshwork of individual filaments and not the branched network characteristic of Arp2/3-nucleated actin filament array, the two systems both use a series of protein interactions to direct assembly of the force-generating machinery to the site of membrane protrusion. The identification of MPAK and its essential role in linking MSP filament assembly to the plasma membrane lays the groundwork for a deeper understanding of the biochemistry of MSP-based motility and should enable a more detailed comparison of MSP- and actin-based motility to define the shared features of the two systems that power amoeboid cell motility.
ACKNOWLEDGMENTS
We thank Margaret Seavy, Gail Ekman, Lori McFadden, Kim Riddle, and Tom Fellers for expert technical assistance in protein biochemistry and microscopy. This work was supported by National Institutes of Health Grant GM-29994.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-08-0741) on March 7, 2007.
REFERENCES
- Borisy G. G., Svitkina T. M. Actin machinery: pushing the envelope. Curr. Opin. Cell Biol. 2000;12:104–112. doi: 10.1016/s0955-0674(99)00063-0. [DOI] [PubMed] [Google Scholar]
- Buttery S. M., Ekman G. C., Seavy M., Stewart M., Roberts T. M. Dissection of the Ascaris sperm motility machinery identifies key proteins involved in major sperm protein-based amoeboid locomotion. Mol. Biol. Cell. 2003;14:5082–5088. doi: 10.1091/mbc.E03-04-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron L. A., Giardini P. A., Soo F. S., Theriot J. A. Secrets of actin-based motility revealed by a bacterial pathogen. Nat. Rev. Mol. Cell Biol. 2000;1:110–119. doi: 10.1038/35040061. [DOI] [PubMed] [Google Scholar]
- Felder S., Zhou M., Hu P., Urena J., Ullrich A., Chaudhuri M., White M., Shoelson S. E., Schlessinger J. SH2 domains exhibit high-affinity binding to tyrosine-phosphorylated peptides yet also exhibit rapid dissociation and exchange. Mol. Cell. Biol. 1993;13:1449–1455. doi: 10.1128/mcb.13.3.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant R. P., Buttery S. M., Ekman G. C., Roberts T. M., Stewart M. Structure of MFP2 and its function in enhancing MSP polymerization in Ascaris sperm amoeboid motility. J. Mol. Biol. 2005;347:583–595. doi: 10.1016/j.jmb.2005.01.054. [DOI] [PubMed] [Google Scholar]
- Italiano J. E., Jr, Roberts T. M., Stewart M., Fontana C. A. Reconstitution in vitro of the motile apparatus from the amoeboid sperm of Ascaris shows that filament assembly and bundling move membranes. Cell. 1996;84:105–114. doi: 10.1016/s0092-8674(00)80997-6. [DOI] [PubMed] [Google Scholar]
- Italiano J. E., Jr, Stewart M., Roberts T. M. How the assembly dynamics of the nematode major sperm protein generate amoeboid cell motility. Int. Rev. Cytol. 2001;202:1–34. doi: 10.1016/s0074-7696(01)02002-2. [DOI] [PubMed] [Google Scholar]
- Knippschild U., Wolff S., Giamas G., Brockschmidt C., Wittau M., Wurl P. U., Eismann T., Stoter M. The role of the casein kinase 1 (CK1) family in different signaling pathways linked to cancer development. Onkologie. 2005;28:508–514. doi: 10.1159/000087137. [DOI] [PubMed] [Google Scholar]
- LeClaire L. L., III, Stewart M., Roberts T. M. A 48 kDa integral membrane phosphoprotein orchestrates the cytoskeletal dynamics that generate amoeboid cell motility in Ascaris sperm. J. Cell Sci. 2003;116:2655–2663. doi: 10.1242/jcs.00469. [DOI] [PubMed] [Google Scholar]
- Loisel T. P., Boujemaa R., Pantaloni D., Carlier M. F. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- Miao L., Vanderlinde O., Stewart M., Roberts T. M. Retraction in amoeboid cell motility powered by cytoskeletal dynamics. Science. 2003;302:1405–1407. doi: 10.1126/science.1089129. [DOI] [PubMed] [Google Scholar]
- Mitchison T. J., Cramer L. P. Actin-based cell motility and cell locomotion. Cell. 1996;84:371–379. doi: 10.1016/s0092-8674(00)81281-7. [DOI] [PubMed] [Google Scholar]
- Mogilner A. On the edge: modeling protrusion. Curr. Opin. Cell Biol. 2006;18:32–39. doi: 10.1016/j.ceb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Pollard T. D., Borisy G. G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Rafelski S. M., Theriot J. A. Crawling toward a unified model of cell motility: spatial and temporal regulation of actin dynamics. Annu. Rev. Biochem. 2004;73:209–239. doi: 10.1146/annurev.biochem.73.011303.073844. [DOI] [PubMed] [Google Scholar]
- Reinke V., et al. A. global profile of germline gene expression in C. elegans. Mol. Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Ris H. The cytoplasmic filament system in critical point-dried whole mounts and plastic-embedded sections. J. Cell Biol. 1985;100:1474–1487. doi: 10.1083/jcb.100.5.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts T. M., Salmon E. D., Stewart M. Hydrostatic pressure shows that lamellipodial motility in Ascaris sperm requires membrane-associated major sperm protein filament nucleation and elongation. J. Cell Biol. 1998;140:367–375. doi: 10.1083/jcb.140.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts T. M., Stewart M. Acting like actin. The dynamics of the nematode major sperm protein (msp) cytoskeleton indicate a push-pull mechanism for amoeboid cell motility. J. Cell Biol. 2000;149:7–12. doi: 10.1083/jcb.149.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R., Ma L., Miki H., Lopez M., Kirchhausen T., Takenawa T., Kirschner M. W. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- Schagger H., von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Sepsenwol S., Ris H., Roberts T. M. A unique cytoskeleton associated with crawling in the amoeboid sperm of the nematode, Ascaris suum. J. Cell Biol. 1989;108:55–66. doi: 10.1083/jcb.108.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M., Roberts T. M. Cytoskeletal dynamics powers nematode sperm motility. Adv. Prot. Chem. 2005;71:384–399. doi: 10.1016/S0065-3233(04)71010-4. [DOI] [PubMed] [Google Scholar]