Abstract
Understanding the signal transduction systems governing invasion is fundamental for the design of therapeutic strategies against metastasis. Na+/H+ exchanger regulatory factor (NHERF1) is a postsynaptic density 95/disc-large/zona occludens (PDZ) domain-containing protein that recruits membrane receptors/transporters and cytoplasmic signaling proteins into functional complexes. NHERF1 expression is altered in breast cancer, but its effective role in mammary carcinogenesis remains undefined. We report here that NHERF1 overexpression in human breast tumor biopsies is associated with metastatic progression, poor prognosis, and hypoxia-inducible factor-1α expression. In cultured tumor cells, hypoxia and serum deprivation increase NHERF1 expression, promote the formation of leading-edge pseudopodia, and redistribute NHERF1 to these pseudopodia. This pseudopodial localization of NHERF1 was verified in breast biopsies and in three-dimensional Matrigel culture. Furthermore, serum deprivation and hypoxia stimulate the Na+/H+ exchanger, invasion, and activate a protein kinase A (PKA)-gated RhoA/p38 invasion signal module. Significantly, NHERF1 overexpression was sufficient to induce these morphological and functional changes, and it potentiated their induction by serum deprivation. Functional experiments with truncated and binding groove-mutated PDZ domain constructs demonstrated that NHERF1 regulates these processes through its PDZ2 domain. We conclude that NHERF1 overexpression enhances the invasive phenotype in breast cancer cells, both alone and in synergy with exposure to the tumor microenvironment, via the coordination of PKA-gated RhoA/p38 signaling.
INTRODUCTION
Breast cancer is the most commonly diagnosed cancer among women (Jemal et al., 2005), and the dissemination of metastasis is the leading cause of breast cancer fatality (Weigelt et al., 2005), underlying the need for new therapeutic approaches specifically focusing on tumor cell spreading to distant sites. However, the complex molecular mechanisms governing tumor metastasis are still not very well understood (Mareel and Leroy, 2003), making elucidation of the basic mechanisms of tumor invasion one of the major challenges in tumor biology (Mareel and Leroy, 2003; Cardone et al., 2005b). In this regard, the identification of the still mostly undefined signal transduction systems governing metastatic progression is of particular importance (Christofori, 2006). The organization of discrete, functional macromolecular signaling modules/complexes constitutes a critical level of biological activity, and their construction is orchestrated by scaffolding proteins containing modular interaction domains that facilitate the association of multiple target proteins (Vondriska et al., 2004). One class of modular domains is the PDZ domain that associates with specific carboxyl-terminal motifs on target proteins and that can also oligomerize with other postsynaptic density 95/disc-large/zona occludens (PDZ) domains to enhance the scaffolding function (Nourry et al., 2003; Brone and Eggermont, 2005).
The Na+/H+ exchanger regulatory factor (NHERF1) is a PDZ domain-containing scaffolding protein that recruits membrane receptors/transporters and cytoplasmic signaling proteins into functional complexes (Shenolikar et al., 2004). This importance of NHERF1 in cell signaling and cytoskeleton remodeling has prompted the search for its role in cancer progression. NHERF1 is overexpressed in the tumor compared with the nontumor counterparts in hepatocellular carcinomas (Shibata et al., 2003), in schwannoma (Fraenzer et al., 2003), and in breast cancer (Stemmer-Rachamimov et al., 2001). However, information on the relationships between NHERF1 and clinical-pathological tumor characteristics are lacking. In breast cancer cell lines, NHERF1 mRNA expression was associated with a small percentage of loss of heterozygosis (Dai et al., 2004), and it increased upon estrogen receptor activation (Ediger et al., 1999). Although these studies suggest that NHERF1 plays a role in breast cancer progression, neither its specific mechanism(s) of action in driving cancer nor the signal transduction systems involved in its contribution to the metastatic spreading of breast tumors have been explored so far. Furthermore, although the signaling systems regulated by NHERF1 in normal cells are being defined, nothing is known concerning the signaling components downstream of NHERF1 in cancer cells. Here, we better define the specific role and mechanism(s) of NHERF1 action in driving breast cancer progression. We observe that NHERF1 is overexpressed in aggressive human breast tumors, and we report the ability of NHERF1 overexpression to enhance cell invasion and to induce an invasive phenotype in breast cancer cells in vitro, both alone and in synergy with exposure to the tumor metabolic microenvironment by coordinating a protein kinase A (PKA)-gated RhoA/p38 invasion signaling module which stimulates Na+/H+ exchanger (NHE1)-dependent invasion.
MATERIALS AND METHODS
Patients
Surgical specimens of breast cancer were obtained from 43 consecutive patients with a first diagnosis of primary breast cancer histologically confirmed at the Women's Department of the National Cancer Institute (Bari, Italy). Before undergoing routine surgery, all patients signed an informed consent authorizing the Institute to use their removed biological tissues for research purposes. Routine staging procedures were adopted for determination of stage disease extension according to Union Internationale Contre le Cancer criteria (Greene and Sobin, 2002). The patients underwent surgery before receiving any therapy. Just after surgical removal of biological tissues, the pathologist selected from the primary tumor and from contiguous macroscopically not involved breast tissues samples destined to routine diagnostic practice and to research activities. The characteristics of these tissue samples have been successively confirmed by hematoxylin and eosin histological analysis. The cytohistological tumor differentiation grade (G) was determined as reported previously (Elston and Ellis, 2002), and hormone receptors (estrogen receptor, ER; and progesterone receptor, PgR) expression were determined by immunohistochemical assays and categorized as positive or negative cases according to the cut-off value of 10% of positive-immunostained cells (Tommasi et al., 2005). Estrogen and progesterone and their receptors play important roles in the genesis and malignant progression of breast cancer; they are the prototype predictive markers in oncology. Both the ER and PgR are ligand-activated transcription factors belonging to the family of nuclear hormone receptors (Lefsky, 2001), and their principal application is for selecting patients with early breast cancer likely to respond to hormone therapy (Duffy, 2005). Tumor proliferative activity is one of the most crucial variables closely associated with the histopathological grade of malignancy (Offersen et al., 2003), and it was determined as the percentage of tumor cells expressing the growth-related MIB antigen by immunohistochemical assay (Tommasi et al., 2005). The Nottingham prognostic index (NPI) combining tumor size, lymph node stage, and histological grade information was used to score each patient in which patients with NPI values <2.5 are with an expected best prognosis, with NPI = 2.5–3.5 with an expected intermediate prognosis and with NPI > 4.5 associated with poorest prognosis (Sundquist et al., 1999).
Statistical Procedures
In the clinical measurements, Kruskal–Wallis nonparametric ANOVA test was applied to analyze NHERF1 expression between different grades and NPI stages, whereas the Mann–Whitney nonparametric test was applied to normal and tumor tissues, age, menopausal status, size, node status, and the positive versus negative classes of ER, PgR, and MIB. Correlation analysis for NHERF1 expression and ER, PgR, MIB, and hypoxia-inducible factor-1α (HIF-1α) expression were performed with the Spearman rank nonparametric test. In the in vitro experiments, Student's t test was applied to analyze the statistical significance between treatments in which p < 0.05 was considered as significant. All comparisons were performed with InStat (GraphPad Software, San Diego, CA).
Cell Culture and Transfection of Constructs
MCF-10A, MCF-7, and MDA-MB-435 cells were cultured as described previously (Reshkin et al., 2000). Cells were subjected to hypoxia by placing the culture dishes in a Modular Incubator Chamber (Billups-Rothenberg, Del Mar, CA) and flushing the chamber for 5 min with a 95% N2, 5% CO2 gas mixture. The box was closed and placed at 37°C for the remainder of the incubation. For three-dimensional (3D) colony growth on reconstituted 3D basement membrane cultures (Matrigel; Chemicon International, Temecula, CA), a stock solution of Matrigel (9.2 mg/ml; BD Matrigel MATRIX) containing growth factors was diluted to 4 mg/ml in serum-free DMEM and a 1-mm-thick layer was laid down on 25-mm glass coverslips. After a 30-min polymerization in the incubator, 60,000 cells were seeded on each layer in the presence of DMEM plus 10% serum, and after 2 d the cells formed small spheroid colonies. After 5–6 d of growth, some of these colonies started to spontaneously disrupt, and either single cells or small groups of cells emerged from the colony by protruding pseudopodia (30 ± 10% of colonies, n = 25 in 3 independent experiments). The colonies were prepared for immunofluorescence as described below, and they were selected at random for confocal analysis.
NHERF1 constructs truncated or mutated in the PDZ1 or PDZ2 domains were developed as described previously (Weinman et al., 2001; Weinman et al., 2003). A green fluorescent protein (GFP)-tagged NHERF1 construct was generously supplied by Prof. Stephen Lambert (University of Massachusetts Medical School, Worcester, MA). For transfection, 10 μg of plasmid cDNA or empty vector was incubated with 100 μl of LipoTaxi transfection reagent (Stratagene, La Jolla, CA) in 1 ml of simple DMEM growth medium for 30 min at room temperature (RT). Serum was added to 3%, and 200 μl of this mixture was pipetted onto confluent monolayers on glass coverslips and placed in an incubator at 5% CO2 and 37°C for 6 h. This mixture was then replaced with fresh complete medium (10% serum) for 24 h in normal growth conditions.
NHE1 Activity
Intracellular pH was determined spectrofluorometrically in cells loaded with the acetoxy-methyl ester derivative of the pH-sensitive dye 2,7-biscarboxyethyl-5(6)-carboxyfluorescein (Invitrogen, Carlsbad, CA). NHE1 activity was determined by measuring the rate of cytoplasmic pH (pHi) recovery from an acid load produced with the NH4Cl prepulse technique by evaluating the derivative of the slope of the time-dependent pHi recovery as described previously (Reshkin et al., 2000; Paradiso et al., 2004). After each experiment, trypan blue exclusion was also measured for each coverslip, and when it was >5%, the experiment was not used.
RhoA activity
For these experiments, RhoA activity was measured in fluorescence resonance energy transfer (FRET) microscopy by using the Raichu 1297 probe as described previously (Cardone et al., 2005a). In this sensor, the Rho binding domain (RBD) of the RhoA effector protein Rhotekin is sandwiched by VenusYFP and cyan fluorescent protein (CFP). The binding of endogenous GTP-RhoA to RBD generates a conformational change that displaces yellow fluorescent protein (YFP) and CFP, thereby decreasing FRET efficiency between the two fluorophores, and a reduction of intracellular active RhoA results in the opposite effect. To present ratio images in pseudocolor intensity-modulated display mode (IMD), the CFP channel images were divided by the YFP-FRET channel images. To eliminate the distracting data from regions outside of cells, the YFP channel is used as a saturation channel. FRET intensity is displayed between the low and high renormalization value, according to a temperature-based lookup table with blue (cold) indicating low values and red (hot) indicating high values. All calculations were performed using the MetaFluor Analyst module of the Meta Imaging Series 6.1 software (Molecular Devices, Sunnyvale, CA).
Western Blotting
Western blotting was performed as described previously (Reshkin et al., 2000). Samples were extracted in SDS sample buffer (6.25 mM Tris-HCl, pH 6.8, containing 10% [vol/vol] glycerol, 3 mM SDS, 1% [vol/vol] 2-mercaptoethanol, and 0.75 mM of bromphenol blue), separated by 4–12% SDS-polyacrylamide gel electrophoresis (PAGE), and blotted to Immobilon P. Western blotting was performed with monoclonal antibodies diluted 1:250 against NHERF1 (BD Biosciences Transduction Laboratories, Lexington, KY) and polyclonal antibodies diluted 1:1000 against RhoA (Santa Cruz Biotechnology, Santa Cruz, CA), produced against a peptide of RhoA phosphorylated at serine 188 by PRIMM (Milan, Italy) diluted 1:1000 or against total and phosphorylated p38 (Cell Signaling Technology, Beverly, MA) diluted at 1:1000. Molecular weight standards were Biotinylated Protein Ladder Detection Pack (Cell Signaling Technology).
Coimmunoprecipitation
After treatment monolayers were washed two times with ice-cold phosphate-buffered saline (PBS) and then lysed in ice-cold coimmunoprecipitation lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 100 μM Na3VO4, and 1 mM NaF, protease inhibitors) and homogenated by five passes through a 20-gauge needle to obtain the total cell homogenate. An aliquot was removed for the determination of total cellular protein. Approximately 150 μg of total cellular protein was incubated for 1 h at 4°C on a rotator with 1 μg of primary antibody. Five microliters of resuspended volume of protein A/G Plus-Agarose (Santa Cruz Biotechnology) was then incubated at 4°C on a rotator overnight. Immunoprecipitates were collected by centrifugation at 2500 rpm for 5 min at 4°C. Supernatant was carefully aspirated and discarded, and the pellet was washed four times with 1 ml of lysis buffer, each time repeating the centrifugation step above. After the final wash, the pellet was resuspended in 40 μl of SDS sample buffer (6.25 mM Tris-HCl, pH 6.8, containing 10% [vol/vol] glycerol, 3 mM SDS, 1% [vol/vol] 2-mercaptoethanol, and 0.75 mM bromophenol blue) and was run on 10% SDS-PAGE and analyzed by Western blotting with polyclonal antibodies diluted 1:250 against RhoA phosphorylated at serine 188 and produced by PRIMM, NHERF1 (BD Biosciences Transduction Laboratories), PKARIIβ (BD Biosciences Transduction Laboratories), RhoA (Santa Cruz Biotechnology) or NHE1 (Alpha Diagnostic, San Antonio, TX).
Cell Fractionation
After treatment, monolayers were washed two times with ice-cold PBS and then lysed in ice-cold buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM dithiothreitol, and 0.5 mM phenylmethylsulfonyl fluoride) and homogenated by five passes through a 20-gauge needle to obtain the total cell homogenate. An aliquot was removed for the determination of total cellular protein. The nuclear fraction was obtained by centrifuging the homogenate at 600 × g for 10 min. The resulting supernatant was centrifuged at 3500 × g for 10 min to obtain a pellet containing the endosomal fraction, and the supernatant was centrifuged again at 17,000 × g for 1 h to obtain a plasma membrane rich pellet. The supernatant is centrifuged again at 100,000 × g for 1.5 h, resulting in a microsomal pellet and the soluble cytoplasmic fraction in the supernatant. Thirty-five micrograms of each of the separated cellular fractions was extracted in SDS sample buffer and analyzed by Western blotting as described above. Equal loading was determined for each lane by staining the blot with Coomassie blue and analyzing the general density of each lane. Experiments with >10% differences between lanes were not used.
Invasion
A quantitative measure of the degree of in vitro invasion of MDA-MB-435 cells was measured as the ability to traverse an 8-μm polycarbonate membrane coated with Matrigel (Chemicon International) as described previously (Cardone et al., 2005a). MDA-MB-435 cells were transfected with the indicated construct DNA or empty vector 48 h before the experiment, and after 24 h they were serum deprived or not for a further 24 h. The fluorescent samples were read in a fluorescence plate reader at 480/520 nm (Cary Eclipse fluorescence spectrophotometer; Varian, Palo Alto, CA).
Immunofluorescence
Cells plated on coverslips or colonies on Matrigel were washed two times in sterile PBS at RT, fixed with 3.7% ice-cold paraformaldehyde/PBS for 20 min. The fixed coverslips were washed three times for 5 min each with ice-cold PBS, and then the colonies were permeabilized with 0.1% Triton X-100 for 10 min and saturated with 0.1% gelatin in PBS for 10 min. The coverslips were incubated with polyclonal anti-NHERF1 primary antibody (Affinity BioReagents, Golden, CO) diluted 1:300 in 0.1% gelatin in PBS at RT for 1 h, washed 3 times for 5 min each with 0.1% gelatin in PBS and incubated at RT for 1 h with the Alexa 488 goat anti-rabbit immunoglobulin G secondary antibody conjugate (Invitrogen). The coverslips were then washed three times with ice-cold PBS for 5 min each, rapidly rinsed in distilled H2O, and then mounted with Mowiol (Calbiochem, San Diego, CA). NHERF1 was detected with a Nikon TE 2000S epifluorescence microscope equipped with a MicroMax 512BFT charge-coupled device (CCD) camera (Princeton Scientific Instruments, Monmouth Junction, NJ) by using a Nikon lamp shutter with a mercury short arc photo optic HBO 103 W/2 lamp for excitation (Osram, Augsburg, Germany). In colocalization experiments, cells were observed at 600× magnification in oil immersion with a laser scanning confocal microscope (C1/TE2000-U; Nikon Instruments, Sesto Fiorentino, Italy) equipped with He/Ne 633 and argon 488 lasers with 495- to 519- (B2-A) and 642- to 660 (Cy5)-nm excitation filters. For each cell or tissue section, scanning was conducted with 25–30 optical series from the top to the bottom of the cell with a step size of 0.45 μm. Parameters related to fluorescence intensity were maintained at constant values in all measurements.
Image Analysis
For every image, a Z-stack was acquired using the MetaMorph software (Molecular Devices), and every two-color stack (red and green) is the sum of two stacks (one for each color) acquired separately in black and white (B/W). Before image analysis, each stack is deconvolved using the AutoDeblur 9.1 function of AutoQuant (AutoQuant Software, Troy, NY) and then merged by transforming the two channels corresponding to red (tetramethylrhodamine B isothiocyanate) and green (fluorescein isothiocyanate) into a single two-color stack by using the “RGB merge” command of ImageJ software (National Institutes of Health Bethesda, MD). To verify colocalization, the two separate B/W stacks were analyzed with the “colocalization” plugin of ImageJ with a ratio of 97, threshold of 50 for both channel 1 and 2. By then selecting the “colocalizated points (8-bit)” option, a new stack is obtained where the colocalized pixels look white on a black background. This stack is then converted into a voxel-gradient (VG) shading function of AutoVisualize (AutoQuant Software), which permits the observation of the 3D colocalization zones in a volume.
The random or codependent nature of the aforementioned calculated “apparent” dye-overlap colocalizations were tested using 1) intensity correlation analysis (ICA) in which the distribution of the intensity value (normalized to 1) for each pixel of a channel is plotted against the product of the difference of the mean of the two channels, and 2) the Li intensity correlation quotient (ICQ) is calculated (Li et al., 2004). Both ICA and ICQ were calculated using the JACoP image analysis package plugin in ImageJ. Apparent colocalization due to random staining or very high intensities in one window will have an ICQ near zero, whereas if the two intensities are interdependent (colocalized), the values will be positive with a maximum of 0.5. In addition to giving information on cellular localization of the proteins, the ICA-ICQ analysis is useful for identifying potential low-affinity interactions within protein complexes that may be missed in high-affinity coimmunoprecipitation or pull-down experiments.
Immunohistofluorescence
Immunohistofluorescence studies were performed on Formalin-fixed tissue sections embedded in paraffin wax. The breast cancer tissues, obtained from patient breast biopsy specimens, were fixed in 20% neutral buffered Formalin for 24 h and embedded in paraffin. Serial sections of 3 μm in thickness were obtained from tissue blocks, deparaffinized with xylene, and rehydrated in an ethanol series. Microwave pretreatment (slides were immersed in a 10 mM citrated buffer, pH 6.0, at 95°C for 15 min total) for antigen retrieval was carried out before incubation with primary antibody. After cooling, slides were washed in distilled water, treated 10 min with 0.2% bovine serum albumin (BSA) to block nonspecific protein binding, washed in water for 5 min, and then incubated with NHERF1 (1 μg/100 μl) and/or HIF-1 (monoclonal H1α67, 1:300; Abcam, Cambridge, United Kingdom) antibody overnight at 4°C in a humidified chamber. Positive tissue controls of the breast carcinoma as well as negative control slides that were run simultaneously were used to assess the quality of immunostaining. For negative controls, slide sections that were positive for staining were treated with 0.2% BSA instead of the primary antibody. No staining was observed in any of these controls. Images were obtained on a BX40 microscope (Olympus, Tokyo, Japan) with a SenSys 1401E-Photometrics CCD camera.
RESULTS
NHERF1 Expression Is Associated with More Aggressive Metastatic Progression, Poor Prognosis, and Tumor Hypoxia
We first asked whether NHERF1 is overexpressed in human breast tumors. Relative NHERF1 protein expression was measured in Western blots (Figure 1A) of tumor tissues and in their contiguous, noninvolved breast tissue from the same patient to account for intra- and interindividual variations. NHERF1 was overexpressed in 100% of the human tumor breast samples analyzed (0.38 ± 0.15 versus 2.28 ± 0.44 in contiguous noninvolved and tumor tissues, respectively; p = 0.0005, n = 25, ratio NHERF1/actin OD values normalized to MCF-7 expression). Figure 1B shows immunohistofluorescence of a nested series of images from 3-μm sections at the edge of a pT3N1aM0 (G3) mammary tumor in which image exposure is increased from left to right to bring out details where NHERF1 expression is lower. These images verify that NHERF1 expression was much higher in the tumor lobules than in normal lobules of the contiguous peritumoral breast tissues. Furthermore, the magnified view (Figure 1C) of the normal lobule, inside the box, in the right-most field of Figure 1B shows that NHERF1 expression is limited to the apical membrane region of the normal, primary lobules while being also intracellular in tumor lobular tissue (Figure 1B).
Figure 1.
Analysis of NHERF1 expression and localization in human normal and breast cancer. (A) Representative Western blot of NHERF1 protein expression in the patients breast tumor (T) and in its contiguous, nontumor breast tissue (NT) for four patients and for an aliquot of total cell extract from MCF-7 cells. Biopsies and cells were extracted for total protein and NHERF1 and actin expression analyzed by Western blot as described in Materials and Methods. (B) Immunolocalization of NHERF1 in a breast tumor section at increasing signal intensity from left to right panels, 40× objective. Bar, 10 μm. The increased NHERF1 expression in the large disorganized, breast tumor lobules (t) displays diffuse distribution in the cells of the lobule, whereas normal, organized breast lobules (n) have a strictly apical distribution. (C) That NHERF1 is limited to the apical membrane region of cells in a normal lobule can be better observed in the micrograph showing the zone enclosed in the white box with the 100× objective. Bar, 10 μm. Arrow indicates NHERF1 in the apical membrane.
We then correlated NHERF1 protein expression measured as in Figure 1A in primary breast tumors with respect to patient clinicopathological disease characteristics such as age, menopausal status, tumor size, proliferation index (MIB), presence of regional nodal metastases (node status), and estrogen and progesterone receptor status, tumor cytohistological dedifferentiation (grade), and NPI (Table 1). Overall, NHERF1 protein expression significantly increased with increasing tumor cytohistological dedifferentiation (grade) and with a poorer prognosis as indicated by an increasing Nottingham prognostic index (Sundquist et al., 1999). Correlation analysis further revealed that tumor NHERF1 protein expression levels were positively correlated with increasing ER levels in ER+ tumors (r = 0.49, p = 0.034 with Spearman rank, n = 18) and with increasing levels of the tumor microenvironment marker HIF-1α (Hopfl et al., 2004; r = 0.581, p = 0.027 with Spearman rank, n = 20).
Table 1.
Relationship between NHERF1 expression and clinicopathological parameters
| Frequency (%) | NHERF1 (mean ± SE) | NHERF1 (median) | p (median) | |
|---|---|---|---|---|
| Age | 0.285 | |||
| <55 | 22 (51.2) | 1.8 ± 0.53 | 0.56 | |
| >55 | 21 (51.2) | 2.77 ± 0.69 | 1.10 | |
| Menopausal status | 0.740 | |||
| Premenopause | 15 (23.03) | 2.2 ± 0.7 | 0.59 | |
| Postmenopause | 28 (65.1) | 2.3 ± 0.54 | 0.80 | |
| Tumor size | 0.625 | |||
| ≤2 cm | 10 (31.25) | 1.69 ± 0.51 | 0.87 | |
| ≥2 cm | 22 (68.75) | 2.04 ± 0.65 | 0.58 | |
| Node status | 0.078 | |||
| Negative (N0) | 23 (53.5) | 1.46 ± 0.39 | 0.51 | |
| Positive (N1-2) | 20 (46.5) | 3.02 ± 0.79 | 0.80 | |
| Grade | 0.028 | |||
| 1 | 6 (15.4) | 0.41 ± 0.13 | 0.41 | |
| 2 | 16 (41) | 1.51 ± 0.49 | 0.66 | |
| 3 | 17 (43.6) | 3.39 ± 0.72 | 3.14 | |
| NPI | 0.031 | |||
| <2.5 | 6 (14) | 0.40 ± 0.2 | 0.2 | |
| 2.5–3.5 | 19 (44.2) | 0.95 ± 0.3 | 0.71 | |
| >3.5 | 18 (41.9) | 4.62 ± 1.4 | 5.09 | |
| MIB | 0.920 | |||
| Positive | 28 (65.12) | 2.19 ± 0.48 | 0.79 | |
| Negative | 15 (34.88) | 2.46 ± 0.89 | 0.62 | |
| ER | 0.315 | |||
| Positive | 18 (50) | 1.85 ± 0.48 | 0.79 | |
| Negative | 18 (50) | 2.78 ± 0.78 | 0.95 | |
| PgR | 0.089 | |||
| Positive | 17 (46) | 1.44 ± 0.44 | 0.51 | |
| Negative | 24 (58.54) | 2.92 ± 0.65 | 1.15 |
NHERF1 expression measured in Western blot was relative to expression in identical extracts of MCF-7 cells. n = 43 invasive ductal breast carcinomas (median age 55). Significance between median NHERF1 expression values for grade and NPI were evaluated by the Kruskal–Wallis nonparametric analysis of variance test, whereas the Mann–Whitney nonparametric test was applied to age, menopausal status, size, MIB, node status, and receptor status.
This association with HIF-1α was also observed in confocal analysis of histological sections from the same patient in which an architecturally disorganized, tumor lobule (Figure 2B and Video 2) more strongly expressed HIF-1α and NHERF1 than in a nearby, normal lobule (Figure 2A and Video 1). Statistical analysis of the colocalization by cytofluorogram and by ICA of the variance in the distribution of the two proteins (Li et al., 2004; see Materials and Methods), demonstrated that HIF-1α and NHERF1 localization in the tumor lobule was highly codependent. The 3D distribution and colocalization of NHERF1 and HIF-1α in these two lobules can also be observed in Supplemental Videos 1 and 2, in which colocalization between the two proteins, denoted by yellow, was especially evident in the tumor lobule.
Figure 2.
Analysis of NHERF1 and HIF-1α coexpression in human normal and breast cancer. Single (x-y) plane views from confocal 3D analysis of NHERF1 and HIF-1α localization in a normal (A) and a tumor (B) breast lobule. Normal, organized breast lobules display an apical distribution of NHERF1 that, in a disorganized tumor lobule, becomes redistributed diffusely in the cells of the lobule. HIF-1α is distributed in the cytosol in both normal and tumor lobules but is much more expressed in the tumor lobule. The third panel shows their colocalization as analyzed in all the 3D planes and displayed in VG. Cytofluorogram and ICA plots of colocalization using the JACoP image analysis plugin in ImageJ demonstrate a low colocalization in the normal lobule (ICQ = 0.226) and highly significant colocalization in the tumor lobule (ICQ = 0.381). The relative intensity scale is identical for both lobules. Bar, 10 μm.
Hypoxia and Serum Deprivation Specifically Increase Tumor NHERF1 Protein Expression and Redistribute NHERF1 Expression to Newly Formed Leading Edge Pseudopodia
Although these clinical data indicate that NHERF1 is related overall to a more aggressive phenotype of breast cancer, the mechanism(s) involved in its role as a metastasis promoter are unknown. We were particularly intrigued by the strong correlation with HIF-1α, suggesting that NHERF1 expression could be related to the tumor metabolic microenvironment, which has been shown to play an important role in driving metastatic progression (Cardone et al., 2005b; Gillies et al., 2002). We further investigated this possible interaction between the metabolic microenvironment and NHERF1 by subjecting normal and tumor cells to hypoxia or serum deprivation, two components of the tumor microenvironment, in three cell lines that model breast cancer progression: the normal MCF-10A, the ER+ primary tumor MCF-7 and the highly metastatic MDA-MB-435 cell line (Reshkin et al., 2000; Cardone et al., 2005a). Western blot analysis revealed that both serum deprivation (Figure 3A) and hypoxia (Figure 3B) reduced NHERF1 protein expression in the normal cells (MCF-10A), but increased its expression in both cancer cell lines. Indeed, in an extended series of measurements in the metastatic MDA-MB-435 cells, serum deprivation and hypoxia increased NHERF1 protein expression compared with control cells by 186.6 ± 13.2% (n = 62) and 208 ± 22.7% (n = 35), p < 0.001, respectively. All three cell lines responded with this same pattern to chemical hypoxia (100 μM of either CoCl2 or deferoxamine for 2 h; data not shown). This up-regulation of NHERF1 expression was rapid for hypoxia (within 2 h) and slow for serum deprivation (1 d), suggesting that NHERF1 can serve as both a rapid and slow response element to the tumor metabolic microenvironment.
Figure 3.
Exposure to the tumor microenvironment increases NHERF1 protein expression in tumor cells and redistributes NHERF1 to the pseudopodial tip. Confluent cell monolayers of the human MCF-10A, MCF-7, or MDA-MB-435 breast cell lines were exposed to either serum deprivation (Dep, 24 h) (A) or hypoxia (Hyp, 2 h) (B), and NHERF1 expression was compared with nontreated monolayers (Con) by Western analysis. Histograms underneath each blot represent the trend of NHERF1 expression normalized to actin for five and four independent experiments for serum deprivation and hypoxia, respectively. (C) In MDA-MB-435 cells, NHERF1 changes localization upon stimulation with components of the tumor microenvironment. Cells were either not treated or subjected to serum deprivation for 24 h or hypoxia for 2 h and then probed for NHERF1 expression in 3D immunofluorescence as described in Materials and Methods. Top, typical 3D immunofluorescence views from above (x-y plane) and with the x-z and y-z plane profiles shown in the panels on the side. Hypoxia and serum deprivation elongated the cells to form a leading edge pseudopodia and NHERF1 is strongly redistributed to the pseudodial tip (arrowheads). Bar, 10 μm. Bottom, map plots of the relative intensity of NHERF1 expression in the representative x-y plane of the immunofluorescence image above. To analyze both this localization in the pseudopodial tip and to determine the relative pixel “concentration” of NHERF1 in the various parts of the cell we analyzed the pixel density at planes 1–30 starting from the bottom of the cell. We then took the “high map” feature of the AutoVisualize software (AutoQuant Software) to show the relative pixel density throughout the cell in which white is the highest pixel density. The relative intensity scale is identical for all three cells and increases from blue to white as indicated in the scale to the right. (D) Elongation factor of cells subjected to these treatments defined as the greatest length divided by the greatest width of each cell. Error bars are mean + SE, n = 100.
In normal epithelial cells, NHERF1 is thought to be expressed primarily subapically in association with apical membrane proteins (Mery et al., 2002) as we observed in normal mammary lobules (Figures 1C and 2A). However, NHERF1 has also been reported to be redistributed to the distal tip of long growth cones in “aggressively motile,” myelinating Schwann cells when stimulated (Gatto et al., 2003). We therefore examined the pattern of NHERF1 localization in the highly invasive MDA-MB-435 cells before and after stimulation by serum deprivation or hypoxia with 3D epifluorescence microscopy (Figure 3C). Both serum deprivation and hypoxia induced a cellular shape change with the production of long, leading-edge pseudopodia (Figure 3, C and D) and NHERF1 expression was particularly strong at the tip of the leading edge pseudopodia (Figure 3C, arrowheads) in 86 ± 2.4% of 100 cells measured in serum deprivation and 81 ± 4.7% of 100 cells in hypoxia. The strong nuclear staining induced us to isolate nuclei and determine the effect of serum deprivation and hypoxia on nuclear NHERF1 expression. We found that although NHERF1 is highly expressed in the nucleus, neither serum deprivation nor hypoxia had any effect on its nuclear expression (Supplemental Figure 1).
To verify whether similar patterns of NHERF1 localization are present in in vivo tumor tissues, indirect immunohistofluorescence images were obtained in 3-μm sections of a pT2N3aM0 (G2) mammary tumor. Figure 4A shows that tumor cells that are escaping from a laterally disrupted lobule in the process of malignant progression have a NHERF1 expression in the tip of the leading-edge pseudopodia. Furthermore, a collective group of tumor cells that have escaped from a tumor lobule (Friedl et al., 2004; Sahai, 2005) and are invading have a mesenchymal shape with a strong NHERF1 expression in the tip of the leading edge pseudopodia of the leading cell (Supplemental Figure 2).
Figure 4.
NHERF1 is localized preferentially in the tip of invading cells and is colocalized with NHE1. (A) NHERF1 immunohistofluorescence in escaping cells of a tumor lobule of a patient. In the bottom panel, the entire lobule is shown in which NHERF1 staining is particularly strong in a group of cells at one border of the lobule. This section of the lobule is shown magnified in the top panel where a small group of cells can be seen to be escaping from the tumor lobule. Interestingly, a number of small pseudopodia extend from these cells and NHERF1 is strongly expressed along these pseudopodia and at their tips. Bar, 8 μm. (B) NHERF1 (red) and NHE1 (green) expression in a representative MDA-MB-435 tumor mass grown in 3D Matrigel culture in which a cell on one side of the mass is escaping and sending a pseudopodia that is invading the matrigel (top). Higher magnifications of the pseudopodia for NHERF1, NHE1 and their merge are shown in the bottom panels. Bars, 10 μm. (C) NHERF1 (red) and NHE1 (green) expression in a representative single MDA-MB-435 cell in Matrigel 3D culture, which has escaped from a mass and is actively invading the matrigel. Top, view from above the cell (x-y plane). Bottom, pseudopodia seen in the x-z plane. Higher magnification of the pseudopodial tip enclosed in the box is shown for both panels. Bar, 10 μm. The ICA graphs for each pseudopodial tip are shown next to the confocal expression images.
To further test this hypothesis that NHERF1 redistributes to the leading edge pseudopodia and especially at its tip during invasion and to determine the relationship between NHERF1 and NHE1 localization, we took advantage of a powerful technique to grow the cancer cells in reconstituted basement membrane (Matrigel) as 3D structures similar to the tumor lobule, and we recapitulate the process of tumor cell invasive escape from the tumor mass (Debnath and Brugge, 2005; Moyano et al., 2006.; Shackleton et al., 2006). As can be seen in the sample colony observed in 3D epifluorescence (Figure 4B), in 3D Matrigel culture the tumor cells formed colonies with a highly disorganized architecture and a general, diffuse NHERF1 expression (red) that resembled in vivo tumor lobules (Figure 1B). Inspection of colonies revealed that some cells formed protrusions that penetrated the Matrigel with high levels of NHERF1 (red) and NHE1 (green) along the pseudopodia, and, particularly, at the tip (arrowheads) in 100% of 25 colonies inspected in three independent experiments. A similar pattern with high NHERF1 and NHE1 expression at the pseudopodial tip also was observed in a single cell that has fully escaped a colony and is invading through the Matrigel (Figure 4C). In both cases, intensity correlation analysis (ICA, panels to right of images) and Li's ICQ revealed a high codependence between the distribution of NHERF1 and NHE1 in the pseudopodia (ICQ = 0.310 and 0.403 for the colony and single cell, respectively). This colocalization suggested the possibility that NHERF1 could be involved in the regulation of NHE1 activity in the tumor cells.
NHERF1 Overexpression Activates Invasion through Stimulation of NHE1 via a PKA–RhoA–p38 Signal Cascade
Although much support now exists for the hypothesis that the Na+/H+-exchanger isoform 1, NHE1, plays a prominent role in tumor invasion, the driving forces and regulatory processes underlying it are still not well known (for reviews, see Cardone et al., 2005b; Harguindey et al., 2005). We first approached the question of the role of increased endogenous NHERF1 expression in microenvironment-driven regulation of tumor cell NHE1 and invasive activity by overexpressing exogenous wild-type (wt) NHERF1 in both control and serum deprivation conditions. We asked whether an increase of exogenous NHERF1 protein expression in MDA-MB-435 cells could 1) per se duplicate the effects of serum deprivation and hypoxia, i.e., confer increased basal NHE1 activity, alter cell shape, and stimulate invasive capacity; and 2) also potentiate the previously described serum deprivation-dependent increase in all these activities (Reshkin et al., 2000; Cardone et al., 2005a).
In serum replete conditions transfection of the wtNHERF1 construct stimulated Na+/H+ exchange activity to the level induced by serum deprivation and potentiated this serum deprivation-dependent stimulation of Na+/H+ exchanger activity (Figure 5A) without an increase in NHE1 expression (data not shown). This NHERF1-stimulated Na+/H+ exchange activity (Figure 5A) was inhibited by greater than 90% by 2 μM of the specific NHE1 isoform inhibitor cariporide. If NHERF1 regulates invasive activity through its modulation of NHE1 activity, then we can hypothesize that we will find the same regulatory pattern of directed NHERF1 overexpression on invasion and invasive morphology as was observed for NHE1 activity. Indeed, increased exogenous NHERF1 overexpression stimulated control levels of invasion and potentiated serum deprivation-induced invasion of MDA-MB-435 cells (Figure 5B) and was sufficient to increase pseudopodial extension and redistribute NHERF1 to the tip of the pseudopodia (Figure 5C) in 89 ± 8.3% of wtNHERF1 and 79 ± 11.3% in GFP-NHERF1–transfected cells. These data demonstrate that, indeed, exogenous overexpression of NHERF1 is sufficient to duplicate the effects of direct exposure to either hypoxia or serum deprivation.
Figure 5.
Effect of exogenous NHERF1 overexpression on NHE1 activity and NHE1-dependent invasion, cellular morphology, and pseudopodial protrusion. (A) Transfection of wtNHERF1 (wt) in the pcDNA3.1hygro expression vector increased Na+/H+ exchanger activity in nonserum-deprived (solid bars) conditions and potentiated the serum deprivation-dependent stimulation (striped bars) of Na+/H+ exchanger activity. Both basal and NHERF1 overexpressed-dependent activity were >90% inhibited by the benzoylguanidine derivative cariporide (HOE642; 2 μM). (B) Transfection of wtNHERF1 increased invasive activity in nonserum-deprived conditions (solid bars) and potentiated the serum deprivation-dependent stimulation of invasion (striped bars). Both basal and NHERF1 overexpressed-dependent invasion were >90% inhibited by 2 μM cariporide, demonstrating that NHE1 is the principle mechanism underlying invasion. Error bars are mean ± SE from five independent experiments performed in quintuplicate. (C) Transfection of either wtNHERF1 or wtGFP-tagged NHERF1 elongated the cells to form a leading edge pseudopodia and redistributed NHERF1 expression to the tip of these pseudopodia. NHERF1 was visualized by immunofluorescence microscopy with anti-NHERF1 or by GFP fluorescence microscopy. Bars, 10 μm. (D) Light transmission images of wtNHERF1-transfected cells treated with 2 μM cariporide at time zero (t0, top) and after 60 min at 100×. Bar, 10 μm.
Importantly, invasion (Figure 5B) and pseudopodial extension (Figure 5D and Supplemental Video 3) were reversed by inhibition of NHE1 by cariporide. Furthermore, transfection with small interfering RNA targeting NHE1 into either control cells or cells overexpressing wtNHERF1 reduced both NHE1 expression and invasion by ∼40%, supporting the cariporide data that NHE1 is indeed involved also in NHERF1-dependent induction of invasion (Supplemental Figure 3). Together, these data show that, in the cancer cells, serum deprivation drives invasion through NHERF1 overexpression-dependent stimulation of NHE1 and that NHERF1 overexpression is both necessary and sufficient to drive these processes. Potential interaction of NHERF1 with the NHE3 isoform rather than with NHE1 in the pHi response was ruled out because 1) neither NHE3 mRNA nor protein is expressed in the cancer cells and 2) the selective NHE3 inhibitor S-3226 had no effect on pHi recovery after acidification in either control or wtNHERF1-transfected cells (Supplemental Figure 4).
We have previously reported that serum deprivation confers increased invasive ability in human breast cancer cells by a stimulation of the NHE1 via the PKA-mediated phosphorylation of RhoA at serine 188 and the subsequent inhibition of RhoA and p38 activities (Cardone et al., 2005a). Figure 6A shows that both serum deprivation and hypoxia increase RhoA phosphorylation at serine 188 with a time course consistent with their stimulation of NHERF1 expression. We hypothesize that the altered expression/distribution of the scaffolding protein NHERF1 plays a fundamental role in coordinating the action of this signaling module by redirecting PKA to RhoA. Cell fractionation measurements before and after serum deprivation (Figure 6B) and NHERF1 coimmunoprecipitation experiments (Figure 6C) support this hypothesis. As described previously (Cardone et al., 2005a), serum deprivation increased the phosphorylation state of RhoA without altering total expression (see homogenate) and produced a redistribution of total RhoA from the plasma membrane to the endosomal fraction. This phosphorylation and removal of RhoA from the plasma membrane corresponded directly with the increase in NHERF1 expression at the plasma membrane and suggested that increased NHERF1 expression at the plasma membrane shifts PKA phosphorylation to RhoA, removing it from the plasma membrane and turning on the previously described neoplastic signal transduction module (Cardone et al., 2005a; Jia et al., 2006). This hypothesis was supported by coimmunoprecipitation experiments in which serum deprivation, hypoxia, or transfection with wtNHERF1 increased the amount of the RIIβ subunit of PKA and phospho-RhoA that coimmunoprecipitated with NHERF1 and that serum deprivation and hypoxia further increased the coimmunoprecipitation of these proteins observed with the transfection of wtNHERF1 (Figure 6C). In the homogenates tested for input, there was no change in either PKA RIIβ or total RhoA expression, suggesting that the treatments do not alter their total cellular expression and that the differences observed in the immunoprecipitates is due to changes in the interaction with NHERF1. That the pattern for total RhoA in the NHERF1 immunoprecipitate was so similar to that for phospho-RhoA suggests that only phospho-RhoA is actually bound to NHERF1.
Figure 6.
Hypoxia- and serum deprivation-induced NHERF1 overexpression at the pseudopodial tip stimulates RhoA phosphorylation, which is necessary for increased invasion. (A) MDA-MB-435 cells were treated for 24 h with serum deprivation (D, 24 h) or increasing times of hypoxia; and the expression of NHERF1, RhoA, and phospho-RhoA was determined by Western blot. (B) NHERF1, total RhoA, and phospho-RhoA expression in 35 μg of various cellular fractions of nondeprived (ND) or 24-h-serum–deprived (D) MDA-MB-435 cell monolayers. Control or 24-h-serum–deprived cell monolayers were fractionated and separated as described in Materials and Methods. (C) Cells were exposed to serum deprivation (SD), hypoxia (hyp), or transfected with wtNHERF1 and then exposed to these conditions. The homogenate was then immunoprecipitated with anti-NHERF1 and precipitated RIIβ subunit of PKA, phospho-RhoA, total RhoA, and NHERF1 were analyzed by Western blot. Protein input for each protein was measured by Western blot of the homogenate with the same antibodies and total input anti-β-actin antibody. Typical experiment of five replicates. (D) Confocal 3D immunofluorescence x-y plane image of NHERF1 (green) and phospho-RhoA (red) of a MDA-MB-435 cell in control conditions or treated with 2-h hypoxia. Inset in the hypoxic cell shows enlargement of NHERF1 and phospho-RhoA localization in the pseudopodial tip. Right, VG rendering of NHERF1 and phospho-RhoA colocalization. Bars, 5 μm. (E) NHERF1-dependent stimulation of invasion is reversed by treatment with the PKA inhibitor H89 or by transfection with RhoA mutated to alanine at serine 188 such that it can no longer be phosphorylated by PKA (pdRhoA). Mean ± SE, n = 5, ***p < 0.001 and n.s., not significant with respect to pcDNA3.1-transfected cells; ††p < 0.01 and †††p < 0.001 with respect to wtNHERF1-transfected cells.
To further analyze this interaction between NHERF1 and phospho-RhoA, we analyzed their relative localization in confocal immunofluorescence in control and serum deprivation or hypoxia conditions (Figure 6D). Colocalization analysis showed that in untreated cells, NHERF1 and phospho-RhoA staining were localized along the plasmalemma, but they were only slightly coexpressed in any part of the cell consistent with a lack of interaction. Hypoxia produced a strong apparent colocalization of NHERF1 and phospho-RhoA expression in the nucleus and, primarily, at the tip of the long, leading edge pseudopodia. Statistical analysis of the colocalization in these two subcellular regions by intensity correlation analysis (Li et al., 2004) showed that the nuclear colocalization was random due probably to heavy staining in the NHERF1 channel, whereas the colocalization in the pseudopodial tip was strongly codependent (Supplemental Figure 5). Last, transfection of wtNHERF1-expressing cells with the phospho-dead RhoA mutant construct (pdRhoA) or treatment with the PKA inhibitor H89 reversed the NHERF1 overexpression-dependent increase in invasive capacity (Figure 6E).
Together, these data demonstrate that NHERF1 expression is involved in the regulation of tumor cell NHE1 activity and invasive capacity and regulates the phosphorylation of RhoA by PKA. The increase in NHERF1 protein expression activates a rapid stimulation of NHE1, pseudopodial formation, cell movement, and invasion, lending further support for the notion that changes in the expression of a scaffolding protein alters its function (Burack and Shaw, 2000).
NHERF1 Regulates NHE1-dependent Invasion through a PDZ2-dependent Activation of the PKA-gated RhoA/p38 Signal Module Located in the Leading Edge Pseudopodia
NHERF1 scaffolding function is mediated by its binding to other proteins via one of its PDZ domains. To determine which binding domain of NHERF1 is necessary for its mediation of the microenvironmental activation of the NHE1 and invasion, we overexpressed NHERF1 constructs that had been either PDZ domain truncated (ΔPDZ1 or ΔPDZ2) or mutated in the binding groove consensus sequence from GYGF to GAGA of either of the domains (PDZ1, HRF1 or PDZ2, HRF2). Overexpression of either the ΔPDZ1 or HRF1 construct had the same effect on the activity of the NHE1 (Figure 7A) as overexpression of the wild-type (wt) construct (Figure 5A) in both serum-replete and serum-deprived conditions. However, overexpression of either the ΔPDZ2 or HRF2 construct did not produce this increase in NHE1 activity in nondeprived conditions and greatly reduced, instead of potentiating, the serum deprivation-dependent stimulation of NHE1 activity (Figure 7B). This same regulatory pattern was observed on invasive capacity and pseudopodial extension after transfection of these mutated constructs: overexpression of only HRF2 abrogated the NHERF1-dependent increase in basal invasion and blocked the serum deprivation-dependent stimulation of invasion (Figure 7C). Transfection with the HRF1 construct increased pseudopodial extension and translocation of NHERF1 to the pseudopodial tip similarly to that produced by the wtNHERF1 (Figure 5C), whereas transfection with HRF2 blocked these processes (Figure 7D).
Figure 7.
Inhibition of PDZ2 binding blocks the NHERF1-dependent increase of NHE1 activity and invasion. To determine whether PDZ1 or PDZ2 are implicated in the NHERF1-dependent stimulation of NHE1 and invasion, the binding ability of these domains was blocked by either their truncation or mutation (HRF1 or HRF2), and the NHE1 activity (A and B), invasive ability (C), and cell morphology/NHERF1 distribution (D) were measured. NHE1 activity and invasive ability were measured as in Figure 5. Error bars are mean ± SE of 15–20 measurements for NHE1, ***p < 0.001 and **p < 0.01 in comparison with nondeprived empty pcDNA3.1 vector; †††p < 0.001 and ††p < 0.01 in comparison with deprived empty pcDNA3.1 vector. Error bars are mean ± SE of 20–25 measurements for invasive activity, **p < 0.001 and **p < 0.01 in comparison with nondeprived empty pcDNA3.1 vector, and the asterisks outside the lines refer to nondeprived empty pcDNA3.1 vector. Transfected NHERF1 expression was visualized by immunofluorescence microscopy with an anti-6His antibody. Bar in large field, 10 μm, and in the inset, 5 μm.
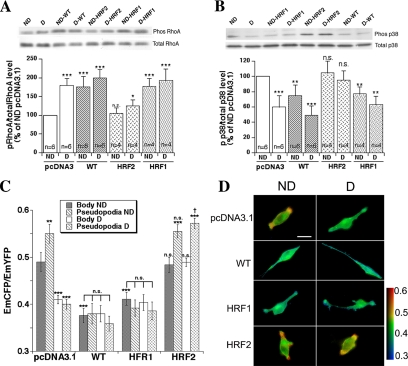
In line with these results, transfection with either wtNHERF1 or its PDZ1-mutated (HRF1) constructs stimulated PKA phosphorylation of RhoA (Figure 8A) and inhibited p38 phosphorylation (Figure 8B) similarly to that observed by serum deprivation, whereas the PDZ2 mutated construct (HRF2) inhibited the activation of these processes by serum deprivation. These last data were further supported by simultaneous live cell measurements of cell body and pseudopodial RhoA activity in FRET microscopy (Figure 8, C and D). In the control cells, serum deprivation preferentially reduced RhoA activity especially in the pseudopodia as described previously (Cardone et al., 2005a). Transfection of either wt or HRF1 constructs was sufficient to mimic the effect of serum deprivation on RhoA activity levels in both the cell body and pseudopodia, whereas transfection with the HRF2 construct had no effect on RhoA in serum replete conditions and abrogated the inhibition of RhoA activity by serum deprivation.
Figure 8.
Inhibition of PDZ2 binding blocks the NHERF1-dependent signal transduction. To determine which PDZ domain is implicated in the NHERF1-dependent regulation of RhoA and p38 phosphorylation/activity, the binding ability of these domains was blocked by mutation. The phosphorylation state of RhoA (A) and p38 (B) were then measured in Western blot, and the histograms underneath each blot represent the value for the number of experiments indicated in each bar. (C) RhoA activity in the cell body and in the pseudopodia was measured by FRET analysis. In FRET measurements, a relative increase in the CFP emission compared with the YFP emission (EmCFP/EmYFP) denotes an increase in active RhoA. Error bars are mean ± SE of 30–45 measurements for RhoA activity, ***p < 0.001 and **p < 0.01 in comparison with nondeprived empty pcDNA3.1 vector. (D) Pseudocolor images of representative cells from each of the treatments in the FRET measurements. FRET images, obtained as described in Materials and Methods, are presented in pseudocolor IMD mode such that red indicated the highest EmCFP/EmYFP ratio (highest RhoA activity), and blue reflected the lowest EmCFP/EmYFP ratio (lowest RhoA activity). Bar, 10 μm.
DISCUSSION
Although a role for NHERF1 in breast tumor progression has been suggested in a immunohistochemical study that observed an elevated NHERF1 expression in breast tumors compared with adjacent normal breast tissue (Stemmer-Rachamimov et al., 2001), to date there has not been an evaluation of the clinicopathological parameters associated with its overexpression. Here, we demonstrate, in a series of tumor and contiguous, noninvolved breast tissues from the same patient (Table 1 and Figure 1, A and B), that NHERF1 protein is highly overexpressed in tumors compared with their contiguous, noninvolved tissue. Furthermore, this overexpression is associated with increasingly aggressive clinical characteristics, with poor prognosis, and with increasing HIF-1α expression, a marker of the tumor microenvironment (Zhong et al., 1999; Covello and Simon, 2004). In line with previous reports (Ediger et al., 1999; Stemmer-Rachamimov et al., 2001), we did observe a strong association with ER expression in ER-positive tumors.
The tumor metabolic microenvironment arises from the abnormal process of neoplastic growth with the resultant reduction of circulatory supplied oxygen and nutrients from the host (Gillies et al., 2002; Axelson et al., 2005). This metabolic microenvironment up-regulates important molecules that mediate neoplastic processes that are under the control of HIF-1α, such as vascular endothelial growth factor and angiopoietin-2 (Pouyssegur et al., 2006). In the present report, we add NHERF1 to the list of proteins up-regulated by hypoxia; furthermore, we present data that another of the tumor metabolic microenvironmental conditions, serum deprivation, also contributes to its protein overexpression (Figure 3A). In both conditions, the up-regulation of NHERF1 expression was limited to tumor cells but occurred with different time scales: hypoxia-dependent up-regulation reached a maximum within hours, whereas the up-regulation dependent on serum deprivation needed a day. A study of a panel of human cancer cell lines has demonstrated that combined hypoxia and glucose deprivation or acidosis are necessary for the complete up-regulation of p53 protein expression (Pan et al., 2004).
As recently suggested (Cardone et al., 2005b; Pouyssegur et al., 2006), cellular pH regulatory mechanisms are targeted by hypoxia and serum deprivation, and, in turn, they could be important targets for antimetastasis therapy. We, indeed, demonstrate here that regulated NHERF1 overexpression leads to the increased activity of the membrane ion transporter NHE1 and to characteristics that contribute to basement membrane invasion, one of the defining features of metastatic cells (Mareel and Leroy, 2003). In analogy to HIF-1α where its genetically targeted overexpression is sufficient to mimic hypoxia (Krishnamachary et al., 2003), we find here that genetically targeted overexpression of exogenous NHERF1 in tumor cells mimics the action of the tumor microenvironment on NHE1 activity (Figure 5A), invasive potential (Figure 5B), and NHERF1 localization/cell shape (Figure 5C) such that its overexpression is sufficient to govern an increased NHE1 activity and invasive potential. The underlying mechanism for NHERF1-mediated tumor invasiveness was related to an increase in the activity of the NHE1, because both NHERF1-dependent NHE1 activity and invasion were abrogated by the specific NHE1 inhibitor cariporide (Figure 5, A and B). Furthermore, NHERF1 overexpression was also sufficient to mimic the activation of the pseudopodial located RhoA–p38–NHE1 signal transduction cascade by the tumor microenvironment (Figures 6 and 8). The hypothesis that NHERF1 overexpression influences invasive potential in breast cancer cells, at least in part, through the regulation of the activity of the NHE1 is further supported by the data showing that NHE1 activity, pseudopodial extension, invasive capacity, and the PKA-gated RhoA–p38 signal cascade were all regulated by only the PDZ2 domain of NHERF1 (Figures 7 and 8). We suggest that NHERF1 overexpression enables tumors to take advantage of the physiological response mechanism to hypoxia and serum deprivation to improve their own survival; furthermore, these data, together with the in vivo data (Figure 1 and Table 1), could mean that tumors exist in vivo almost completely in a state influenced by their self-generated, physiological microenvironment.
It is now well established that tumor cells have acquired altered morphological characteristics to facilitate their increased chemotaxic and invasive ability: leading edge pseudopodia (Cardone et al., 2005b; Jia et al., 2006). Establishment of the directed cell polarity involved in invasion requires reorganization of the cytoskeleton and sorting of proteins to this pseudopodial compartment. The NHERF1 scaffolding protein organizes transmembrane and cytoplasmic proteins to integrate both structure and signaling at the apical junctional complex of polarized epithelial cells. In MDA-MB-435 cells, the increase in NHERF1 expression in either serum-deprived (Figure 3A) and hypoxic conditions (Figure 3B) or by targeted exogenous NHERF1 overexpression (Figure 5C) induced leading edge pseudopodia and a corresponding redistribution of NHERF1 to the pseudopodial tip. Furthermore, MDA-MB-435 cells escaping from tumor lobules developed in 3D Matrigel culture had NHERF1 preferentially localized to the pseudopodial tip of the escaping and invading tumor cells (Figure 4, B and C). This localization of NHERF1 to the pseudopodia was also observed in vivo in cancer cells escaping from a tumor lobule (Figure 4A) and in a group of migrating cancer cells that had already escaped (Supplemental Figure 2). We also demonstrate with confocal microscopy that the colocalization of NHERF1 with phospho-RhoA, and with FRET microscopy analysis, we demonstrate that NHERF1-dependent inactivation of RhoA occurs preferentially at the pseudopodia. We hypothesize that the protein reorganization in the leading edge pseudopodia occurs, in part, through binding of the NHERF1 PDZ2 domain with still unidentified submembrane proteins. Intriguingly, a similar polarized distribution of NHERF1 to the distal tip of long growth cones in myelinating, “aggressively motile” Schwann cells (Gatto et al., 2003) and a similar inhibition of the RhoA/ROCK pathway has been reported in controlling the process of neurite outgrowth (Busca et al., 1998; Luo, 2000; Scaife et al., 2003; Yuan et al., 2003; Kishida et al., 2004; Wu et al., 2005). These data suggest that neurite outgrowth has profound similarities to the tumor-invasive process.
The mechanism by which NHERF1 stimulates the activity of NHE1 is unknown at present. NHERF1 binds to the NHE3 isoform of the cation exchanger family of proteins but there is no evidence, direct or based on the amino acid sequence of NHE1, to predict that NHE1 and NHERF1 would interact directly with each other. A growing body of evidence now indicates that a localized decrease in RhoA expression or activity controls pseudopodial extension and motility/invasion in a variety of cell types (Wang et al., 2003; Wittchen et al., 2003; De Wever et al., 2004; McHardy et al., 2004; Paradiso et al., 2004; Cardone et al., 2005a; Wicki et al., 2006). Accordingly, we favor the view that NHERF1 facilitates the assembly of a protein complex that affects the targeting of PKA to RhoA during exposure to the microenvironment (Figure 6, B and C) that results in RhoA inactivation (Figure 8C) and removal from the plasma membrane (Figure 6B). We have previously described a novel signal transduction module in breast cancer cells localized to the dominant leading edge pseudopodia that, during serum deprivation, increases invasive ability in human breast cancer cells by a stimulation of the NHE1 via the PKA-mediated phosphorylation of RhoA at serine 188 and the subsequent inhibition of RhoA and p38 activities (Cardone et al., 2005a). We hypothesize that the altered expression and distribution of NHERF1 during exposure to the tumor metabolic microenvironment promotes invasion in that its altered expression and distribution redirects PKA to RhoA, which inhibits a RhoA–ROCK–p38 signal pathway that normally represses NHE1. The release of this repression results in an increase in NHE1 activity and in subsequent invasion.
The mechanism by which NHERF1 activates the PKA–RhoA–p38 regulation of NHE1 activity is still unknown. Ezrin is a member of the ezrin–radixin–moesin family of cytoskeleton-associated proteins that binds to NHE1 (Denker et al., 2000) to link it with the cytoskeleton and to NHERF1 to determine the trafficking and PKA-dependent phosphorylation of specific target proteins (Shenolikar et al., 2004). Ezrin has been shown to have key roles in the coordination of signal complexes required for metastasis (Hunter, 2004). Our identification of NHERF1 as a crucial molecule in breast cancer metastasis suggests that the roles of ezrin (Hunter, 2004) and NHE1 (Cardone et al., 2005b) in directing metastasis may well be governed by NHERF1. An important potential confirmation of this hypothesis comes in a recent study showing that transfection of human gastric cancer cells with Ht31, a peptide that blocks PKA binding to anchoring proteins such as ezrin, disrupts the phosphorylation and inhibition of RhoA by PKA (Wang et al., 2006b). Another intriguing possibility, linked to RhoA-dependent ezrin function, could be the involvement of cofilin, which was recently shown to be important in regulating leading edge protrusion in metastatic breast tumor cells (Wang et al., 2006a).
The present study adds NHERF1 as an important player in the race for understanding the molecular mechanisms behind the breast cancer invasive process. We think that NHERF1 could serve as a marker potentially applicable to the detection and identification of presymptomatic cancers and, second, could be exploited as a therapeutic target in those cancers, probably by targeting the binding of downstream partner protein(s) to the PDZ2 domain of NHERF1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Prof. Stephen Lambert for the GFP-tagged NHERF1 construct, Prof. Antonio Frigeri (University of Bari, Bari, Italy) for the use of the hypoxia chamber, and Nicola Paciolla (Laboratory of Clinical and Experimental Oncology, National Cancer Institute Giovanni Paolo II, Bari, Italy) for help in the management of the tumor tissues. We thank Dr. Simona Granata (Division of Nephrology, Department of Emergency and Transplantation, University of Bari) for the RPTEC cell line. We thank Drs. Gianni M. Simone and Stella Petroni (Laboratory of Cytopathology, National Cancer Institute Giovanni Paolo II) for the clinical marker measurements and for discussions on the interpretation of clinical parameters in relation to NHERF1 expression. This work was supported by Progetti Strategici Oncologia Consiglio Nazionale delle Ricerche-Ministero dell'Istruzione, dell'Università e della Ricerca grant CU03.00304 and Fondo per gli Investimenti della Ricerca di Base grant RBAU01B3A3-001.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-07-0617) on March 1, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Axelson H., Fredlund E., Ovenberger M., Landberg G., Pahlman S. Hypoxia-induced dedifferentiation of tumor cells–a mechanism behind heterogeneity and aggressiveness of solid tumors. Semin. Cell Dev. Biol. 2005;16:554–563. doi: 10.1016/j.semcdb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Brone B., Eggermont J. PDZ proteins retain and regulate membrane transporters in polarized epithelial cell membranes. Am. J. Physiol. 2005;288:C20–C29. doi: 10.1152/ajpcell.00368.2004. [DOI] [PubMed] [Google Scholar]
- Burack W. R., Shaw A. S. Signal transduction: hanging on a scaffold. Curr. Opin. Cell Biol. 2000;12:211–216. doi: 10.1016/s0955-0674(99)00078-2. [DOI] [PubMed] [Google Scholar]
- Busca R., Bertolotto C., Abbe P., Englaro W., Ishizaki T., Narumiya S., Boquet P., Ortonne J. P., Ballotti R. Inhibition of Rho is required for cAMP-induced melanoma cell differentiation. Mol. Biol. Cell. 1998;9:1367–1378. doi: 10.1091/mbc.9.6.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone R. A., Bagorda A., Bellizzi A., Busco G., Guerra L., Paradiso A., Casavola V., Zaccolo M., Reshkin S. J. Protein kinase A gating of a pseudopodial-located RhoA/ROCK/p38/NHE1 signal module regulates invasion in breast cancer cell lines. Mol. Biol. Cell. 2005a;16:3117–3127. doi: 10.1091/mbc.E04-10-0945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone R. A., Casavola V., Reshkin S. J. The role of disturbed pH dynamics and the Na+/H+ exchanger in metastasis. Nat. Rev. Cancer. 2005b;5:786–795. doi: 10.1038/nrc1713. [DOI] [PubMed] [Google Scholar]
- Christofori G. New signals from the invasive front. Nature. 2006;441:444–450. doi: 10.1038/nature04872. [DOI] [PubMed] [Google Scholar]
- Covello K. L., Simon M. C. HIFs, hypoxia, and vascular development. Curr. Top. Dev. Biol. 2004;62:37–54. doi: 10.1016/S0070-2153(04)62002-3. [DOI] [PubMed] [Google Scholar]
- Dai J. L., Wang L., Sahin A. A., Broemeling L. D., Schutte M., Pan Y. NHERF (Na+/H+ exchanger regulatory factor) gene mutations in human breast cancer. Oncogene. 2004;23:8681–8687. doi: 10.1038/sj.onc.1207962. [DOI] [PubMed] [Google Scholar]
- Debnath J., Brugge J. S. Modelling glandular epithelial cancers in three-dimensional cultures. Nat. Rev. Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- Denker S. P., Huang D. C., Orlowski J., Furthmayr H., Barber D. L. Direct binding of the Na-H exchanger NHE1 to ERM proteins regulates the cortical cytoskeleton and cell shape independently of H(+) translocation. Mol. Cell. 2000;6:1425–1436. doi: 10.1016/s1097-2765(00)00139-8. [DOI] [PubMed] [Google Scholar]
- De Wever O., Nguyen Q. D., Van Hoorde L., Bracke M., Bruyneel E., Gespach C., Mareel M. Tenascin-C and SF/HGF produced by myofibroblasts in vitro provide convergent pro-invasive signals to human colon cancer cells through RhoA and Rac. FASEB J. 2004;18:1016–1018. doi: 10.1096/fj.03-1110fje. [DOI] [PubMed] [Google Scholar]
- Duffy M. J. Predictive markers in breast and other cancers: a review. Clin. Chem. 2005;51:494–503. doi: 10.1373/clinchem.2004.046227. [DOI] [PubMed] [Google Scholar]
- Ediger T. R., Kraus W. L., Weinman E. J., Katzenellenbogen B. S. Estrogen receptor regulation of the Na+/H+ exchange regulatory factor. Endocrinology. 1999;140:2976–2982. doi: 10.1210/endo.140.7.6885. [DOI] [PubMed] [Google Scholar]
- Elston C. W., Ellis I. O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. C. W. Elston & I. O. Ellis. Histopathology 1991; 19, 403–410. Histopathology. 2002;41:151. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- Fraenzer J. T., Pan H., Minimo L., Jr, Smith G. M., Knauer D., Hung G. Overexpression of the NF2 gene inhibits schwannoma cell proliferation through promoting PDGFR degradation. Int. J. Oncol. 2003;23:1493–1500. [PubMed] [Google Scholar]
- Friedl P., Hegerfeldt Y., Tusch M. Collective cell migration in morphogenesis and cancer. Int. J. Dev. Biol. 2004;48:441–449. doi: 10.1387/ijdb.041821pf. [DOI] [PubMed] [Google Scholar]
- Gatto C. L., Walker B. J., Lambert S. Local ERM activation and dynamic growth cones at Schwann cell tips implicated in efficient formation of nodes of Ranvier. J. Cell Biol. 2003;162:489–498. doi: 10.1083/jcb.200303039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies R. J., Raghunand N., Karczmar G. S., Bhujwalla Z. M. MRI of the tumor microenvironment. J. Magn. Reson. Imaging. 2002;16:430–450. doi: 10.1002/jmri.10181. [DOI] [PubMed] [Google Scholar]
- Greene F. L., Sobin L. H. The TNM system: our language for cancer care. J. Surg. Oncol. 2002;80:119–120. doi: 10.1002/jso.10114. [DOI] [PubMed] [Google Scholar]
- Harguindey S., Orive G., Luis Pedraz J., Paradiso A., Reshkin S. J. The role of pH dynamics and the Na+/H+ antiporter in the etiopathogenesis and treatment of cancer. Two faces of the same coin–one single nature. Biochim. Biophys. Acta. 2005;1756:1–24. doi: 10.1016/j.bbcan.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Hopfl G., Ogunshola O., Gassmann M. HIFs and tumors–causes and consequences. Am. J. Physiol. 2004;286:R608–R623. doi: 10.1152/ajpregu.00538.2003. [DOI] [PubMed] [Google Scholar]
- Hunter K. W. Ezrin, a key component in tumor metastasis. Trends Mol. Med. 2004;10:201–204. doi: 10.1016/j.molmed.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Jemal A., Murray T., Ward E., Samuels A., Tiwari R. C., Ghafoor A., Feuer E. J., Thun M. J. Cancer statistics, 2005. CA Cancer J. Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- Jia Z., Vadnais J., Lu M. L., Noel J., Nabi I. R. Rho/ROCK-dependent pseudopodial protrusion and cellular blebbing are regulated by p38 MAPK in tumour cells exhibiting autocrine c-Met activation. Biol. Cell. 2006;98:337–351. doi: 10.1042/BC20050088. [DOI] [PubMed] [Google Scholar]
- Kishida S., Yamamoto H., Kikuchi A. Wnt-3a and Dvl induce neurite retraction by activating Rho-associated kinase. Mol. Cell Biol. 2004;24:4487–4501. doi: 10.1128/MCB.24.10.4487-4501.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamachary B., Berg-Dixon S., Kelly B., Agani F., Feldser D., Ferreira G., Iyer N., LaRusch J., Pak B., Taghavi P., Semenza G. L. Regulation of colon carcinoma cell invasion by hypoxia-inducible factor 1. Cancer Res. 2003;63:1138–1143. [PubMed] [Google Scholar]
- Lefsky J. M. Nuclear receptor minireview series. J. Biol. Chem. 2001;276:36863–36864. doi: 10.1074/jbc.R100047200. [DOI] [PubMed] [Google Scholar]
- Li Q., Lau A., Morris T. J., Guo L., Fordyce C. B., Stanley E. F. A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J. Neurosci. 2004;24:4070–4081. doi: 10.1523/JNEUROSCI.0346-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L. Rho GTPases in neuronal morphogenesis. Nat. Rev. Neurosci. 2000;1:173–180. doi: 10.1038/35044547. [DOI] [PubMed] [Google Scholar]
- Mareel M., Leroy A. Clinical, cellular, and molecular aspects of cancer invasion. Physiol. Rev. 2003;83:337–376. doi: 10.1152/physrev.00024.2002. [DOI] [PubMed] [Google Scholar]
- McHardy L. M., Sinotte R., Troussard A., Sheldon C., Church J., Williams D. E., Andersen R. J., Dedhar S., Roberge M., Roskelley C. D. The tumor invasion inhibitor dihydromotuporamine C activates RHO, remodels stress fibers and focal adhesions, and stimulates sodium-proton exchange. Cancer Res. 2004;64:1468–1474. doi: 10.1158/0008-5472.can-03-2733. [DOI] [PubMed] [Google Scholar]
- Mery L., Strauss B., Dufour J. F., Krause K. H., Hoth M. The PDZ-interacting domain of TRPC4 controls its localization and surface expression in HEK293 cells. J. Cell Sci. 2002;115:3497–3508. doi: 10.1242/jcs.115.17.3497. [DOI] [PubMed] [Google Scholar]
- Moyano J. V., et al. AlphaB-crystallin is a novel oncoprotein that predicts poor clinical outcome in breast cancer. J. Clin. Investig. 2006;116:261–270. doi: 10.1172/JCI25888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourry C., Grant S. G., Borg J. P. PDZ domain proteins: plug and play! Sci. STKE. 2003;2003:RE7. doi: 10.1126/stke.2003.179.re7. [DOI] [PubMed] [Google Scholar]
- Offersen B. V., Sørensen F. B., Knoop J. The prognostic relevance of estimates of proliferative activity in early breast cancer. Overgaard. Histopathol. 2003;43:573. doi: 10.1111/j.1365-2559.2003.01745.x. [DOI] [PubMed] [Google Scholar]
- Pan Y., Oprysko P. R., Asham A. M., Koch C. J., Simon M. C. p53 cannot be induced by hypoxia alone but responds to the hypoxic microenvironment. Oncogene. 2004;23:4975–4983. doi: 10.1038/sj.onc.1207657. [DOI] [PubMed] [Google Scholar]
- Paradiso A., Cardone R. A., Bellizzi A., Bagorda A., Guerra L., Tommasino M., Casavola V., Reshkin S. J. The Na+-H+ exchanger-1 induces cytoskeletal changes involving reciprocal RhoA and Rac1 signaling, resulting in motility and invasion in MDA-MB-435 cells. Breast Cancer Res. 2004;6:R616–R628. doi: 10.1186/bcr922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouyssegur J., Dayan F., Mazure N. M. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006;441:437–443. doi: 10.1038/nature04871. [DOI] [PubMed] [Google Scholar]
- Reshkin S. J., Bellizzi A., Albarani V., Guerra L., Tommasino M., Paradiso A., Casavola V. Phosphoinositide 3-kinase is involved in the tumor-specific activation of human breast cancer cell Na(+)/H(+) exchange, motility, and invasion induced by serum deprivation. J. Biol. Chem. 2000;275:5361–5369. doi: 10.1074/jbc.275.8.5361. [DOI] [PubMed] [Google Scholar]
- Sahai E. Mechanisms of cancer cell invasion. Curr. Opin. Genet. Dev. 2005;15:87–96. doi: 10.1016/j.gde.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Scaife R. M., Job D., Langdon W. Y. Rapid microtubule-dependent induction of neurite-like extensions in NIH 3T3 fibroblasts by inhibition of ROCK and Cbl. Mol. Biol. Cell. 2003;14:4605–4617. doi: 10.1091/mbc.E02-11-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton M., Vaillant F., Simpson K. J., Stingl J., Smyth G. K., Asselin-Labat M. L., Wu L., Lindeman G. J., Visvader J. E. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Shenolikar S., Voltz J. W., Cunningham R., Weinman E. J. Regulation of ion transport by the NHERF family of PDZ proteins. Physiology. 2004;19:362–369. doi: 10.1152/physiol.00020.2004. [DOI] [PubMed] [Google Scholar]
- Shibata T., Chuma M., Kokubu A., Sakamoto M., Hirohashi S. EBP50, a beta-catenin-associating protein, enhances Wnt signaling and is over-expressed in hepatocellular carcinoma. Hepatology. 2003;38:178–186. doi: 10.1053/jhep.2003.50270. [DOI] [PubMed] [Google Scholar]
- Stemmer-Rachamimov A. O., Wiederhold T., Nielsen G. P., James M., Pinney-Michalowski D., Roy J. E., Cohen W. A., Ramesh V., Louis D. N. NHE-RF, a merlin-interacting protein, is primarily expressed in luminal epithelia, proliferative endometrium, and estrogen receptor-positive breast carcinomas. Am. J. Pathol. 2001;158:57–62. doi: 10.1016/S0002-9440(10)63944-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist M., Thorstenson S., Brudin L., Nordenskjold B. Applying the Nottingham Prognostic Index to a Swedish breast cancer population. South East Swedish Breast Cancer Study Group. Breast Cancer Res. Treat. 1999;53:1–8. doi: 10.1023/a:1006052115874. [DOI] [PubMed] [Google Scholar]
- Tommasi S., et al. BRCA1 mutations and polymorphisms in a hospital-based consecutive series of breast cancer patients from Apulia, Italy. Mutat. Res. 2005;578:395–405. doi: 10.1016/j.mrfmmm.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Vondriska T. M., Pass J. M., Ping P. Scaffold proteins and assembly of multiprotein signaling complexes. J. Mol. Cell Cardiol. 2004;37:391–397. doi: 10.1016/j.yjmcc.2004.04.021. [DOI] [PubMed] [Google Scholar]
- Wang H. R., Zhang Y., Ozdamar B., Ogunjimi A. A., Alexandrova E., Thomsen G. H., Wrana J. L. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science. 2003;302:1775–1779. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]
- Wang W., Mouneimne G., Sidani M., Wyckoff J., Chen X., Makris A., Goswami S., Bresnick A. R., Condeelis J. S. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J. Cell Biol. 2006a;173:395–404. doi: 10.1083/jcb.200510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chen Y., Chen M., Xu W. AKAPs competing peptide HT31 disrupts the inhibitory effect of PKA on RhoA activity. Oncol. Rep. 2006b;16:755–761. [PubMed] [Google Scholar]
- Weigelt B., Peterse J. L., van 't Veer L. J. Breast cancer metastasis: markers and models. Nat. Rev. Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- Weinman E. J., Steplock D., Wade J. B., Shenolikar S. Ezrin binding domain-deficient NHERF attenuates cAMP-mediated inhibition of Na(+)/H(+) exchange in OK cells. Am. J. Physiol. 2001;281:F374–F380. doi: 10.1152/ajprenal.2001.281.2.F374. [DOI] [PubMed] [Google Scholar]
- Weinman E. J., Wang Y., Wang F., Greer C., Steplock D., Shenolikar S. A C-terminal PDZ motif in NHE3 binds NHERF-1 and enhances cAMP inhibition of sodium-hydrogen exchange. Biochemistry. 2003;42:12662–12668. doi: 10.1021/bi035244l. [DOI] [PubMed] [Google Scholar]
- Wicki A., Lehembre F., Wick N., Hantusch B., Kerjaschki D., Christofori G. Tumor invasion in the absence of epithelial-mesenchymal transition: podoplanin-mediated remodeling of the actin cytoskeleton. Cancer Cell. 2006;9:261–272. doi: 10.1016/j.ccr.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Wittchen E. S., Haskins J., Stevenson B. R. NZO-3 expression causes global changes to actin cytoskeleton in Madin-Darby canine kidney cells: linking a tight junction protein to Rho GTPases. Mol. Biol. Cell. 2003;14:1757–1768. doi: 10.1091/mbc.E02-08-0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K. Y., Hengst U., Cox L. J., Macosko E. Z., Jeromin A., Urquhart E. R., Jaffrey S. R. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436:1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X. B., Jin M., Xu X., Song Y. Q., Wu C. P., Poo M. M., Duan S. Signalling and crosstalk of Rho GTPases in mediating axon guidance. Nat. Cell Biol. 2003;5:38–45. doi: 10.1038/ncb895. [DOI] [PubMed] [Google Scholar]
- Zhong H., De Marzo A. M., Laughner E., Lim M., Hilton D. A., Zagzag D., Buechler P., Isaacs W. B., Semenza G. L., Simons J. W. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59:5830–5835. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.