Abstract
In this report, an antisense RNA strategy has allowed us to show that disruption of ALP expression affects the expression of the muscle transcription factors myogenin and MyoD, resulting in the inhibition of muscle differentiation. Introduction of a MyoD expression construct into ALP-antisense cells is sufficient to restore the capacity of the cells to differentiate, illustrating that ALP function occurs upstream of MyoD. It is known that MyoD is under the control of serum response factor (SRF), a transcriptional regulator whose activity is modulated by actin dynamics. A dramatic reduction of actin filament bundles is observed in ALP-antisense cells and treatment of these cells with the actin-stabilizing drug jasplakinolide stimulates SRF activity and restores the capacity of the cells to differentiate. Furthermore, we show that modulation of ALP expression influences SRF activity, the level of its coactivator, MAL, and muscle differentiation. Collectively, these results suggest a critical role of ALP on muscle differentiation, likely via cytoskeletal regulation of SRF.
INTRODUCTION
Development of skeletal muscle cells is characterized by a succession of events starting with the determination of mesodermal progenitor cells into muscle precursor cells called myoblasts. Later, upon induction of muscle differentiation, proliferating myoblasts withdraw from the cell cycle, align, and fuse to become multinucleate myotubes that will mature into functional myofibers (for review, see Olson, 1992). These morphological changes are accompanied by the up-regulation of muscle-specific genes, including those encoding members of the myogenic regulatory factor (MRF) family of basic helix-loop-helix transcription factors and the myocyte enhancer factor-2 (MEF-2). The MRF family, comprised of MyoD, Myf5, myogenin, and myogenic regulatory factor 4 (MRF4), regulates the transcription of genes necessary for the specification of myoblasts from somatic mesoderm and subsequently for their differentiation into contractile muscle cells. MEF-2 collaborates with MRFs to regulate muscle-specific gene expression required for terminal differentiation (for review, see Ludolph and Konieczny, 1995).
The earliest steps in skeletal muscle differentiation involve the formation and maintenance of myoblasts and are controlled by MyoD and Myf5 (for review, see Weintraub, 1993). Knowledge of how these key muscle determination factors are controlled is thus pivotal for understanding how muscle is formed during development. The demonstration that Myf5 and MyoD modulate different pathways of muscle development (Megeney et al., 1996; Tajbakhsh et al., 1996) and display distinct spatiotemporal expression patterns (Cossu et al., 1996) suggested that these two myogenic factors might be activated by different upstream signaling pathways. Indeed, it is now recognized that various hormones and growth factors differentially influence MyoD and Myf5 expression (Carnac et al., 1992; Cossu et al., 1996; Montarras et al., 1996). Moreover, it has been demonstrated that serum response factor (SRF), a MADS box transcription factor essential for muscle differentiation (Arsenian et al., 1998), regulates MyoD, but not Myf5, gene expression in myoblasts (Carnac et al., 1998) through a functional SRF-binding CarG element present in the distal regulatory region of the MyoD gene (L'Honore et al., 2003), providing molecular insight into one mechanism by which MRFs are differentially controlled.
SRF regulates both cellular immediate-early genes such as c-fos as well as a variety of muscle-specific genes (Johansen and Prywes, 1995; Treisman, 1995; Arsenian et al., 1998). As occurs with many transcriptional regulators, SRF interacts with a variety of other factors to control tissue-specific gene expression (Treisman, 1994; Wang and Olson, 2004). SRF and its partners are themselves influenced by a variety of intracellular signals. In the case of SRF-dependent transcription of the c-fos gene, the critical SRF partner, TCF (ternary complex factor), is regulated by MAP kinase–dependent phosphorylation (Treisman, 1994). Alternatively, SRF-dependent regulation of some genes can be influenced by changes in actin dynamics, independent of known MAP kinase signaling pathways. In this case, which has been well-documented in fibroblasts, SRF-dependent transcriptional activity is positively regulated through Rho-stimulated increases in actin polymerization and stabilization (Sotiropoulos et al., 1999), which induce the cytoplasmic-to-nuclear redistribution of MAL, a recently identified SRF coactivator (Miralles et al., 2003). It is postulated that MAL's accumulation in the nucleus is inhibited by direct binding of G-actin that occurs when monomeric actin levels are elevated. Furthermore, the two actin-depolymerizing agents, latrunculin B and cytochalasin D, which have opposing effects on SRF activity (Geneste et al., 2002), also exhibit differential effects on MyoD expression (Dhawan and Helfman, 2004) in accordance with the view that actin dynamics affect MyoD expression through SRF activity. RhoA can also stimulate SRF-dependent expression of muscle-specific genes (Carnac et al., 1998; Wei et al., 1998); thus this, or a similar, mechanism may be relevant in muscle cells as well.
The link between actin dynamics and transcriptional control illustrates an exciting point of convergence between cytoarchitecture and nuclear function. Recently, certain proteins that regulate actin assembly state have been shown to influence SRF-dependent transcription, providing a more detailed picture of how this interplay might be achieved in vivo. For example, overexpression of LIM kinase, which phosphorylates and inactivates the actin disassembly factor, cofilin, enhances actin stability and activates SRF (Sotiropoulos et al., 1999; Geneste et al., 2002). Likewise, expression of inactive VASP mutants disturbs both actin assembly within cells and abrogates SRF activation (Grosse et al., 2003). The exquisite sensitivity of SRF activation state to the dynamics and organization of the actin cytoskeleton highlights the potential importance of cytoskeletal factors in the regulation of gene expression.
We have recently purified and characterized a PDZ-LIM protein called the α-actinin associated LIM protein (ALP), which displays dramatically upregulation during muscle differentiation (Pomies et al., 1999). ALP is expressed in smooth, cardiac, and skeletal muscle cells (Pomies et al., 1999; Xia et al., 1997). Two ALP isoforms which exhibit identical N-terminal PDZ domains and C-terminal LIM domains with a variable central core are produced as a result of alternative splicing (Pomies et al., 1999). As its name implies, ALP interacts directly with the actin-binding protein α-actinin and is colocalized with α-actinin at the Z-discs of striated muscle cells (Xia et al., 1997; Pomies et al., 1999). The α-actinin binding capacity of ALP maps to the N-terminal PDZ domain and both ALP isoforms interact with α-actinin (Xia et al., 1997; Pomies et al., 1999). Biochemical studies revealed that purified ALP enhances α-actinin's capacity to cross-link actin filaments, raising the possibility that ALP plays a role in actin organization and anchorage within muscle cells (Pashmforoush et al., 2001). Indeed, overexpression of ALP induces the assembly of highly organized, robust actin arrays (Pashmforoush et al., 2001). The regulated expression of ALP during muscle differentiation coupled with its capacity to enhance cytoskeletal organization led us to explore its role during differentiation of cultured C2C12 myoblasts.
Here we report that differentiation of C2C12 myoblasts is accompanied by a dramatic increase in expression and switching of ALP isoforms. Antisense-mediated down-regulation of ALP expression in C2C12 myoblasts interferes with the differentiation response. Knock-down of ALP expression results in failure in the expression of the critical myogenic transcription factors, myogenin and MyoD, but not Myf5. Reintroduction of MyoD restores the capacity of the cells to differentiate, indicating that ALP functions upstream of MyoD. As discussed above, MyoD expression is regulated by SRF, a transcription factor whose activity can depend on actin filament organization. Loss of ALP function leads to a dramatic reduction in actin filament arrays in C2C12 myoblasts, a decrease in levels of the SRF coactivator, MAL, and a concomitant reduction in SRF activity. Induction of actin filament assembly and stabilization by jasplakinolide restores the capacity of C2C12 cells to fully differentiate in absence of ALP. Finally, we show that overexpression of ALP in C2C12 cells grown in proliferation medium induces differentiation of C2C12 cells, accompanied by an increase of SRF activity. Our results thus suggest a critical role for ALP in the integration of cytoskeletal architecture and transcriptional regulation that is essential for myogenic differentiation of C2C12 myoblasts.
MATERIALS AND METHODS
Gel Electrophoresis and Western Immunoblotting
Protein fractions were separated by SDS-PAGE according to the method of Laemmli (1970) except with a bisacrylamide concentration of 0.13%. 12.5% or 8% polyacrylamide gels were used in this work.
For Western immunoblotting, proteins were resolved by SDS-PAGE and transferred to nitrocellulose. Rabbit polyclonal antibodies raised against chicken ALP isoforms (K55; Pomies et al., 1999), Myf5 (Santa Cruz Biotechnology, Santa Cruz, CA), MyoD (Santa Cruz Biotechnology), SRF (Santa Cruz Biotechnology), MRTF-A/MAL (Santa Cruz Biotechnology), GFP (Torrey Pines Biolabs, Houston, TX), and actin (Sigma, Saint Louis, MO) were used, followed by horseradish peroxidase linked to protein A (Amersham Life Science, Cleveland, OH). A goat polyclonal antibody directed against MRTF-A/MAL (Santa Cruz Biotechnology) was also used, followed by horseradish peroxidase linked to rabbit antibodies anti-goat immunoglobulins (Dako, Copenhagen, Denmark). Monoclonal antibodies specific for skeletal myosin heavy chain (Sigma), nonmuscle α-actinin (ICN Biomedicals, Aurora, OH), sarcomeric α-actinin (Sigma), myogenin (PharMingen, San Diego, CA), and α-tubulin (Monosan, Uden, The Netherlands) were used, followed by horseradish peroxidase linked to whole sheep antibody directed against mouse immunoglobulins (Amersham Life Science, Piscataway, NJ). Immunodetection was enhanced using chemiluminescent techniques (ECL, Amersham Life Science).
Cell Culture and Microscopy
The C2C12 myogenic cell line was grown in DMEM supplemented with 10% fetal bovine serum (growth medium). C2C12 differentiation was induced by transferring the cells into DMEM supplemented with 2% horse serum (differentiation medium).
Phase contrast images of living cells were collected on an inverted Nikon Eclipse TE300 microscope (Melville, NY) and analyzed using the QED imaging software (QED Imaging, Pittsburgh, PA).
Indirect immunofluorescence of C2C12 cells was performed as described previously by Arber et al. (1994). C2C12 cells were cultured on glass coverslips in growth medium and were induced to differentiate by transferring the cells in differentiation medium. For immunostaining, the polyclonal antibody raised against ALP (K55) and the mAb specific for sarcomeric α-actinin (Sigma) were used. They were followed by a FITC-conjugated goat anti-rabbit secondary antibody (Molecular Probes, Eugene, OR) and a Texas Red–conjugated goat anti-mouse secondary antibody (Molecular Probes). Staining of the nuclei was obtained using Hoechst 33258 (Sigma), and staining of the actin bundles was obtained using phalloidin-TRITC (Sigma). Images of the cells were collected on a Zeiss Axiophot microscope (Thornwood, NY) and analyzed using the Openlab software (Improvision, Lexington, MA).
Construction of the ALP-Antisense Expression Vector and Establishment of Stable C2C12 Cells Lines Harboring ALP-Antisense Transcripts
The full-length rat skALP was cloned in the antisense orientation into the pcDNA3.1 expression vector (Invitrogen, Carlsbad, CA) via the EcoRV and NotI sites.
C2C12 cells were transfected with the ALP-antisense vector or the empty pcDNA3.1 expression vector using the transfection reagent Lipofectamine (GIBCO BRL, Grand Island, NY). In brief, C2C12 cells were maintained in growth medium until they reached 70% confluence. For transfection, 6 μg of either ALP-antisense vector or empty pcDNA3.1 vector were mixed with Lipofectamine in serum-free DMEM for 45 min and added to the C2C12 cells placed in serum-free DMEM. Transfection was carried out for 12 h at 37°C. Forty-eight hours after transfection the C2C12 cells were grown in growth medium containing the neomycin analogue G418 (GIBCO BRL) to enable selection of stably transfected cells. Individual clones were isolated using cloning rings and were further grown in growth medium supplemented with G418.
RT-PCR Amplification of Mouse cDNA Fragments
Total RNA was isolated from ALP-antisense C2C12 cells (clone Anti 14I) and control C2C12 cells (clone Vector 13G) cultured in growth medium using the Mini RNA Isolation II kit (Zymo Research, Orange, CA). First-strand cDNA was synthesized with 0.5 μg of total RNA from ALP-antisense cells and control cells in a final volume of 30 μl, and PCR was then used to assess expression of mouse Myf5, MyoD, and smooth muscle ALP (smALP). The following sequences of forward and reverse primers were used: Myf5, 5′-CACCATGCGCGAGCGTAGA-3′ and 5′-TCTGTCCCGGCAGGCTGTAA-3′; MyoD, 5′-GGAAGAGTGCGGCTGTGT-3′ and 5′-CTGTTCTGTGTCGCTTAGGG-3′; smALP, 5′-ATGCCCCAGAACGTAGTTCTC-3′ and 5′-AGCTTTGGGGTACAGAGTGAC-3′. As an internal control, PCR was also performed in parallel using primers for mouse tubulin.
Transfection of ALP-Antisense C2C12 Cells with a pcDNA3/MyoD Construct
ALP-antisense C2C12 cells (clone Anti 14I) were transiently transfected with 3 μg of either the pcDNA3 expression vector alone or a pcDNA3/MyoD construct using Lipofectamine PLUS reagent (GIBCO BRL). Transfection was carried out for 6 h at 37°C. The cells were then grown in proliferation medium for 36 h before induction of differentiation.
Generation of ALP-Antisense Adenoviral Vectors and Adenoviral Infection of C2C12 Cells
Adenoviral vectors were generated according to protocols previously described (Wang et al., 1996). In summary, mouse smALP cDNA was cloned downstream of the cytomegalovirus (CMV) promoter of the pXCJL.1 shuttle plasmid in the antisense orientation. A full-length smALP cDNA, a 262-base pair fragment of the 5′ region of ALP containing the PDZ domain, and a 284-base pair fragment of the 3′ region containing the LIM domain sequences were used to generate the antisense shuttle plasmids. The adenoviral vectors were then generated through homologous recombination between pJM17 plasmid and the shuttle plasmids in 293 cells as previously described. The orientation of the ALP sequences with respect to the CMV promoter was confirmed by PCR analysis of the viral DNA. The viral vector Ad/ALP/as contains the full-length smALP, Ad/ALP-PDZ/as contains the PDZ domain sequences, and Ad/ALP-LIM/as contains the LIM domain sequences in an antisense orientation with respect to the CMV promoter.
C2C12 cells were infected with 20–30 PFUs (plaque forming units) for 12 h while growing in growth medium and then transferred to differentiation medium to induce myogenic differentiation. After 5 d in differentiation medium, cells were assayed for differentiation of myoblasts to myotubes by Western immunoblot and indirect immunofluorescence.
Jasplakinolide Treatment
ALP-antisense C2C12 cells were placed in differentiation medium in absence or in presence of 0.16 μM jasplakinolide (Calbiochem, Darmstadt, Germany) for 1 h. After treatment, the cells were washed once and then grown in differentiation-promoting conditions for 7 d.
SRF Reporter Assay
For luciferase assays, proliferating ALP-antisense C2C12 cells (clone Anti 14I), control C2C12 cells (clone Mock 13G) or C2C12 cells were cotransfected with 2 μg of the SRF reporter plasmid pSRE3-Luc, which contains a luciferase cDNA controlled by 3 SRF binding sites, and 40 ng of a Renilla standardization reporter plasmid, used as an internal transfection control to normalize luciferase reporter activity. In some experiments, C2C12 cells were cotransfected with 2 μg of either the expression vector pQE-TriSystem (QIAGEN, Valencia, CA) encoding mouse smALP or the empty pQE-TriSystem vector. Cells were then placed in differentiation-promoting conditions for 2 d before quantitation of reporter activity. In other experiments, C2C12 cells were cotransfected with 2 μg of either the expression vector pEGFP-N1 (Clontech, Mountain View, CA) encoding mouse smALP or the empty pEGFP-N1 vector. Cells were then placed in growth medium for 5 d before analysis.
RESULTS
Induction of ALP Expression during Myogenic Differentiation
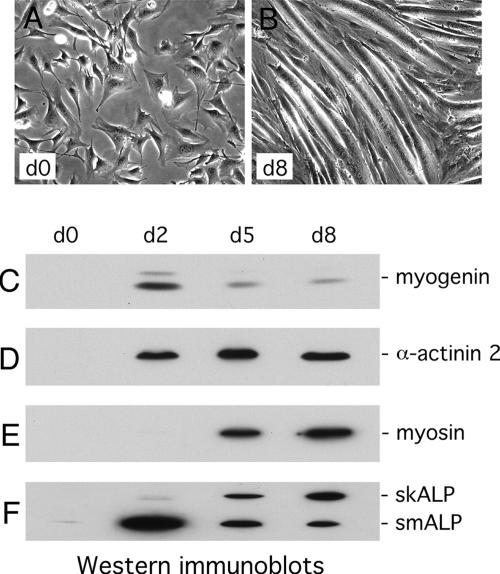
To examine the potential role of ALP in muscle cytoarchitecture and function, we have used the myogenic C2C12 cell line, a well-established model for testing the role of muscle proteins in differentiation and contractile function (Arber et al., 1994; Ornatsky et al., 1997). C2C12 myoblasts proliferate in presence of high concentration of serum and are induced to differentiate into functional myotubes upon removal of growth factors. As differentiation proceeds, the cells elongate, fuse, and develop contractile properties (Figure 1, A and B). The induction of differentiation can also be monitored biochemically (Figure 1, C–F). Within 2 d of growth factor removal, expression of the myogenic transcription factor, myogenin, is evident (Figure 1C). The increase in myogenin is transient and by day 5, the protein levels have already declined significantly relative to the earlier peak expression. Muscle-specific markers of the contractile machinery such as α-actinin-2 and muscle myosin heavy chain are absent in undifferentiated cells, but are induced during the differentiation process and remain at high levels (Figure 1, D and E).
Figure 1.
Expression of muscle-specific markers during differentiation of C2C12 cells. Phase contrast images of C2C12 cells grown in proliferation medium (A) and placed in differentiation-promoting conditions for 8 d (B). Different muscle markers were detected by Western immunoblotting in proliferating (d0) as well as in differentiating C2C12 cells, 2 d (d2), 5 d (d5), and 8 d (d8) after inducing differentiation: (C) myogenin; (D) α-actinin 2; (E) myosin; and (F) the 2 ALP isoforms, skALP and smALP.
The expression of ALP isoforms is also regulated in response to differentiation cues. In proliferating C2C12 cells, the lower molecular mass isoform, referred to as smALP based on its prominence in cardiac and smooth muscle cells (Pomies et al., 1999), is expressed at a low level, whereas the higher molecular mass isoform, referred to as skeletal muscle ALP (skALP), based on its strong expression in differentiated skeletal muscle, is not detected (Figure 1F). The expression of smALP in proliferating myoblasts highlights its potential role before induction of terminal differentiation. Similar to what is observed for myogenin, the expression of smALP increases dramatically 2 d after induction of differentiation and then declines, a pattern that suggests a role for smALP during the early stages of myogenesis (Figure 1, C and F). In contrast, skALP expression increases, progressively mirroring what occurs for the late myogenic differentiation marker, myosin (Figure 1, E and F).
Disruption of ALP Expression Inhibits Muscle Cell Differentiation
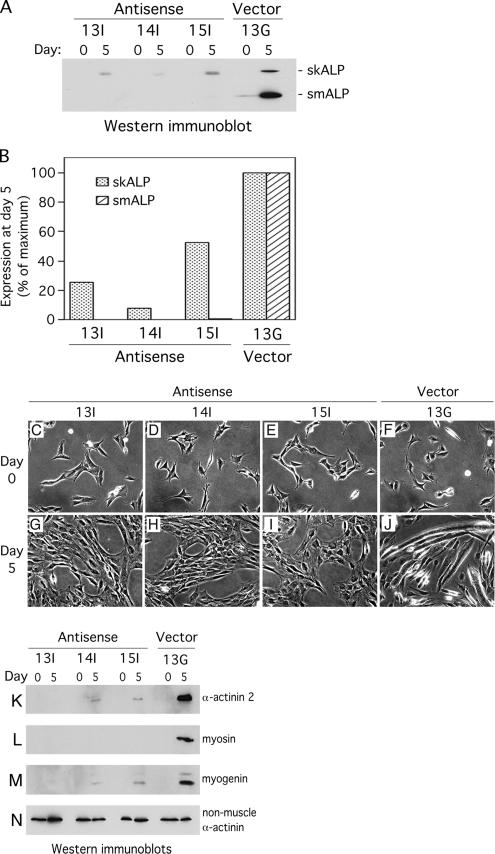
The finding that smALP expression is up-regulated upon induction of differentiation raised the possibility that smALP plays an important role during the early stages of skeletal muscle differentiation. To examine this possibility, we have transfected the C2C12 cell line with a mammalian expression vector containing the full-length skALP cDNA in the antisense orientation. This construct would be expected to lead to reduction in both smALP and skALP expressions because these are encoded by alternatively spliced transcripts that have much sequence in common (Pomies et al., 1999). Stably transfected C2C12 clones were screened by examining the levels of smALP and skALP expression relative to C2C12 clones transfected with an empty vector. Three antisense clones and one empty vector clone were selected for further characterization. These C2C12 clones were maintained in growth medium and then induced to differentiate by transfer into differentiation medium. Examination of antisense cells maintained in growth medium revealed elimination of smALP expression compared with vector controls (Figure 2A). After 5 d in differentiation-promoting conditions, the three antisense clones display dramatically decreased smALP and skALP expression relative to the control vector clone, which shows the typical increase in both smALP and skALP levels (Figure 2A). By densitometric analysis of the bands detected by Western immunoblot, smALP is undetectable in all three antisense clones. skALP expression is also consistently reduced although we observed variable extents of remaining skALP, from 8 to 52%, depending on the clone analyzed (Figure 2B).
Figure 2.
ALP-antisense C2C12 clones fail to differentiate when placed in differentiation-promoting conditions. Expression levels of smALP and skALP were analyzed in 3 ALP-antisense clones (Antisense 13I, 14I and 15I) and in 1 empty vector clone (Vector 13G). (A) smALP and skALP were detected by Western immunoblotting in proliferating cells (day 0) and in cells placed in differentiation medium for 5 d (day 5). (B) Densitometric analysis of the bands detected by Western immunoblotting at day 5 in differentiation medium. The data are expressed as a percentage of the maximum protein expression detected for the Vector 13G clone. This graph represent the densitometric analysis of the autoradiography shown in A, representative itself of five different autoradiographies. (C–J) Phase contrast pictures of the 3 ALP antisense clones (Antisense 13I, 14I and 15I) and the empty vector clone (Vector 13G). The cells were grown in proliferation medium (day 0) then placed in differentiation medium for 5 d (day 5). (K–N) Cell lysates were prepared from the 3 antisense clones (Antisense 13I, 14I and 15I) and the empty vector clone (Vector 13G) grown in proliferation medium (day 0) or placed in differentiation medium for 5 d (day 5). Expression of 3 different muscle-specific proteins was detected by Western immunoblotting: (K) α-actinin 2; (L) myosin; (M) myogenin. As a control, the nonmuscle isoform of α-actinin was detected using the same technique (N).
Antisense inhibition of ALP expression does not affect cell viability, and no gross morphological differences are detected between control transfectants and the three antisense cell lines (Figure 2, C–F). However, a striking difference in cell behavior is observed when a muscle differentiation signal is applied. In contrast with vector-transfected cells, which establish elongated large myotubes after 5 d of induction of differentiation, ALP-antisense C2C12 cells retain a fibroblast-like shape typical of proliferating myoblasts (Figure 2, G–J).
To confirm that the muscle differentiation program is inhibited in ALP-antisense C2C12 cells, we evaluated the expression levels of three muscle differentiation markers, α-actinin 2, myosin, and myogenin, in the three ALP-antisense clones. In contrast with the control transfectant in which all three differentiation markers display elevated expression after 5 d in differentiation medium, only a very minimal induction of differentiation markers is observed in the antisense cell lines (Figure 2, K–N). Importantly, the expression of the nonmuscle isoform of α-actinin (Figure 2N) and α-tubulin (data not shown) is unaffected in antisense clones, suggesting that targeting ALP expression specifically affects the production of muscle-specific proteins. Consistent with the biochemical analysis, neither myogenin nor sarcomeric α-actinin are detected by indirect immunofluorescence of the ALP-antisense 14I cells grown in differentiation medium for 5 d (data not shown). Taken together, these results demonstrate that the differentiation process is severely affected in C2C12 myoblasts in which ALP expression is perturbed. The loss of myogenin expression in ALP-antisense C2C12 cells placed in differentiation-promoting conditions illustrates that the impact of loss of ALP on myogenesis occurs at an early stage in the muscle differentiation process.
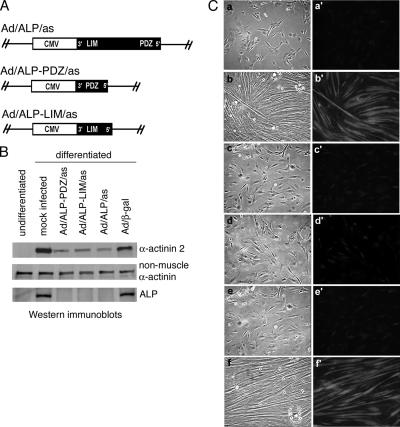
Several PDZ-LIM proteins related to ALP, including CLP-36/hCLIM1, Enigma, ENH, and Cypher/ZASP, are also expressed in striated muscle cells (Kuroda et al., 1996; Guy et al., 1999; Kotaka et al., 1999; Zhou et al., 1999). Therefore, to confirm the specificity of the ALP-antisense targeting construct, we have first determined the expression levels of two ALP-family members, Cypher 1 and CLP-36, in ALP-antisense C2C12 cells. We have observed that Cypher 1 is only expressed in differentiated C2C12 cells (data not shown). In accordance with its exclusive expression in differentiated C2C12 cells and with the fact that ALP-antisense C2C12 cells are unable to enter the differentiation program, we did not detect Cypher 1 expression in ALP-antisense cells (data not shown). We have also observed that CLP-36 expression is not significantly affected in ALP-antisense cells compared with control C2C12 cells (data not shown). Furthermore, we have generated two more specific antisense constructs corresponding to the PDZ domain and the LIM domain of ALP (Figure 3A). As we observed for full-length ALP-antisense, the expression of ALP-PDZ- and ALP-LIM-antisense RNAs totally abolishes the expression of ALP in C2C12 cells grown for 5 d in differentiation medium (Figure 3B). Furthermore, α-actinin-2 expression is reduced in both PDZ-ALP- and LIM-ALP-antisense cells, suggesting that cell differentiation is affected (Figure 3B). Examination of cell morphology after stimulation of differentiation confirmed the effect of ALP antisense constructs on muscle differentiation. Control cells infected with an empty vector or with a vector containing the β-galactosidase gene form myotubes and express α-actinin 2 when they are placed in differentiation-promoting conditions for 5 d. Unlike control cells, the PDZ-ALP- and LIM-ALP-antisense C2C12 cells do not form myotubes and express very low levels of the muscle-specific protein, α-actinin 2 (Figure 3C). Taken together, these results demonstrate the specificity of our ALP-antisense construct.
Figure 3.
Expression of ALP-PDZ- and ALP-LIM-antisense transcripts inhibits differentiation of C2C12 cells. (A) Generating ALP-antisense transcripts. A full-length smALP cDNA was cloned in the antisense orientation with respect to the CMV promoter to generate the Ad/ALP/as virus. The Ad/ALP-PDZ/as and Ad/ALP-LIM/as adenoviruses contain the PDZ and LIM domain, respectively, in the antisense orientation. (B) C2C12 cells were infected with ALP-antisense, ALP-PDZ-antisense and ALP-LIM-antisense adenoviruses and then allowed to differentiate in differentiation medium for 5 d. Expression of the three antisense transcripts results in loss of skALP expression (ALP) and in marked reduction of sarcomeric α-actinin expression (α-actinin 2) whereas the nonmuscle isoform remained unchanged (nonmuscle α-actinin). The control adenovirus Ad/β-gal expressing the β-galactosidase gene had no effect on α-actinin 2 expression. (C) Panels a–f show light microscopy and a′–f′ show the corresponding α-actinin 2 immunostaining of the infected cells. Panels a and a′, undifferentiated mock infected cells; b and b′, differentiated mock infected cells; c and c′, Ad/ALP-PDZ/as infected cells; d and d′, Ad/ALP-LIM/as infected cells; e and e′, Ad/ALP/as infected cells; f and f′, Ad/β-gal infected cells. All adenoviral-infected cells were maintained in differentiation medium for 5 d.
The MyoD Signaling Pathway Is Affected in ALP-Antisense C2C12 Cells
Myf5, MyoD, myogenin, and MRF4 are MRFs belonging to the basic helix-loop-helix protein family (Rudnicki and Jaenisch, 1995). Myogenin and MRF4 expression are required for fusion and terminal differentiation (Hinterberger et al., 1991; Venuti et al., 1995). Myf5 and MyoD are expressed in proliferating C2C12 myoblasts and play critical roles in early muscle determination events, with Myf5 acting upstream of MyoD (Delfini et al., 2000; Hadchouel et al., 2003).
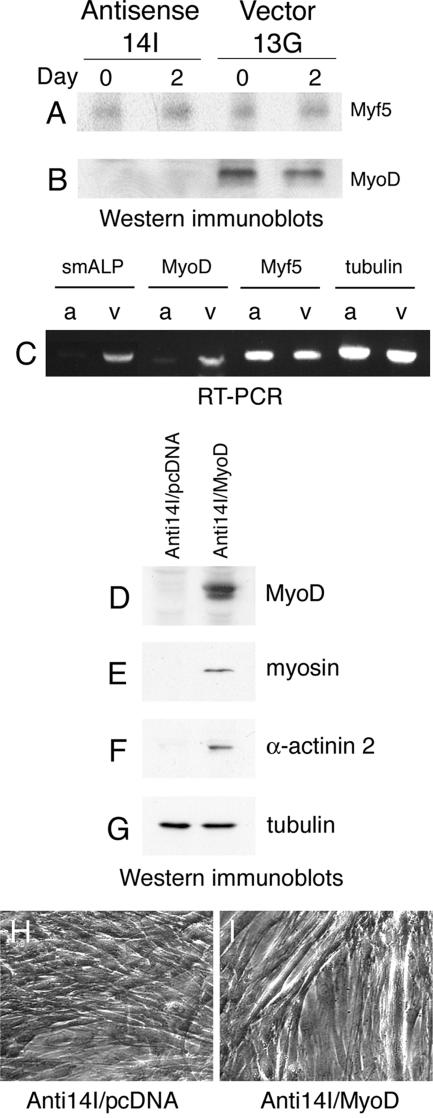
To determine whether the signaling pathways involving Myf5 or MyoD are affected in C2C12 cells lacking ALP expression, we evaluated the expression levels of these factors in ALP-antisense C2C12 cells. As shown in Figure 4A, Myf5 expression levels are not disrupted in the ALP-antisense cells. In contrast, MyoD expression is barely detectable in ALP-antisense C2C12 cells grown in proliferation medium or placed in differentiation-promoting conditions for 2 d (Figure 4B), suggesting that MyoD expression or stability depends, either directly or indirectly, on the presence of ALP. Furthermore, analysis of transcript levels corresponding to smALP, MyoD and Myf5 revealed that in ALP-antisense C2C12 cells, a decrease in the expression of ALP transcripts is accompanied with a reduction of MyoD, but not Myf5, transcripts (Figure 4C). These observations provide evidence that ALP acts either downstream of Myf5 or in a parallel pathway that regulates the transcription of MyoD.
Figure 4.
Alteration of the MyoD signaling pathway in ALP-antisense C2C12 cells. Myf 5 (A) and MyoD (B) expression levels were detected by Western immunoblotting in ALP-antisense C2C12 cells (Antisense 14I) and control C2C12 cells (Vector 13G) grown in proliferation medium (Day 0) or placed in differentiation-promoting conditions for 2 d (Day 2). Unlike Myf5, MyoD expression is severely affected in ALP-antisense C2C12 cells. (C), RT-PCR analysis of RNA from ALP-antisense C2C12 cells (a) and control C2C12 cells (v) cultured in growth medium was performed using primers corresponding to smALP, MyoD and Myf5. Tubulin served as a control. Representative expression patterns are shown. In ALP-antisense C2C12 cells the level of MyoD transcripts is severely reduced. In a rescue experiment ALP-antisense C2C12 cells were transfected with a MyoD construct (Anti14I/MyoD) or with an empty vector (Anti14I/pcDNA). Expression of muscle-specific proteins was detected by Western immunoblotting: MyoD (D) in cells grown in proliferation medium for 36 h following transfection; myosin (E) and α-actinin 2 (F) in cells placed in differentiation-promoting conditions for 5 d. As a control, tubulin was detected in these cells using the same technique (G). Phase contrast images of the ALP-antisense C2C12 cells transfected with the vector alone (H) or with the MyoD construct (I) and grown in differentiation-promoting conditions for 5 d.
To test whether the absence of differentiation observed in ALP-antisense C2C12 cells can be attributed to a decrease in MyoD activity, we introduced a MyoD expression construct into the ALP-antisense cells and tested their ability to undergo differentiation in response to serum removal. In contrast with what we observe in ALP-antisense C2C12 cells, MyoD is detected in ALP-antisense cells transfected with the pcDNA3/MyoD construct (Figure 4D). Moreover, in ALP-antisense C2C12 cells programmed to express MyoD, expression of the muscle differentiation markers, myosin and α-actinin 2, is restored (Figure 4, E and F). Phase-contrast images show the presence of large myotubes in the ALP-antisense C2C12 cells transiently transfected with the MyoD construct (Figure 4, H and I). Thus, the reexpression of MyoD in the ALP-antisense cells is sufficient to restore the capacity of the cells to differentiate. This illustrates that the essential role of ALP in C2C12 muscle differentiation occurs upstream of MyoD.
Decreased Filamentous Actin in ALP-Antisense C2C12 Cells
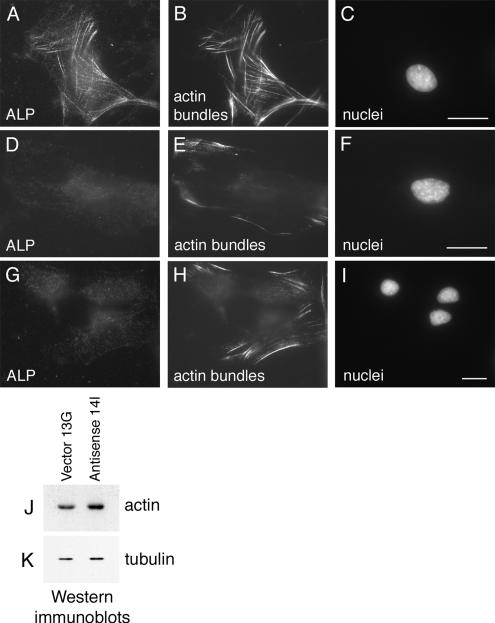
MyoD expression is controlled by SRF (Gauthier-Rouviere et al., 1996), a transcriptional regulator whose activity is modulated by actin dynamics (Sotiropoulos et al., 1999). Stabilization of filamentous actin is sufficient to promote the activation of SRF, leading to the expression of certain target genes (Sotiropoulos et al., 1999), thus regulators of the actin cytoskeleton may impact SRF function. Recently, it has been shown that the F-actin–bundling protein Scp1 reduces the rate of actin disassembly illustrating that actin cross-linking proteins can affect actin assembly dynamics in addition to affecting actin filament organization (Goodman et al., 2003). We demonstrated previously that ALP facilitates α-actinin–dependent bundling of actin filaments in vitro and therefore could participate in the stabilization of cellular actin filaments (Pashmforoush et al., 2001). To examine whether actin filament arrays are affected in ALP-antisense cells, a fluorescent derivative of the F-actin–binding drug, phalloidin, was used to stain cells. As shown in Figure 5, A–C, a meshwork of actin bundles is present in control C2C12 myoblasts expressing ALP. In contrast, in C2C12 myoblasts lacking ALP expression, only few short bundles of actin filaments appear at the periphery of the cells (Figure 5, D–I), illustrating that loss of ALP expression leads to a reduction in actin bundles. This diminution of actin bundles in ALP-antisense C2C12 cells is not due to a down-regulation of actin expression because the amount of total actin in the ALP-antisense cells are at least as high as that detected in control C2C12 cells (Figure 5, J and K). Rather, the reduced phalloidin labeling in the ALP-antisense cells appears to reflect a shift in the organization or stability of cellular actin filaments when ALP is absent.
Figure 5.
Loss of actin bundles in ALP-antisense C2C12 cells. Immunofluorescence microscopy of control Vector 13G C2C12 cells (A–C) and Antisense 14I C2C12 cells (D–F and G–I) grown in proliferation-promoting conditions. Cells were triple-labeled for ALP (A, D, and G), actin bundles (B, E, and H), and nuclei (C, F, and I). Only few short actin bundles are detected in ALP-antisense C2C12 cells. Total actin and tubulin were detected by Western immunoblotting in control C2C12 cells (Vector 13G) and ALP-antisense C2C12 cells (Antisense 14I) grown in proliferation medium. Bar, 30 μm.
Differentiation of the ALP-Antisense C2C12 Cells Is Induced by F-Actin Stabilization
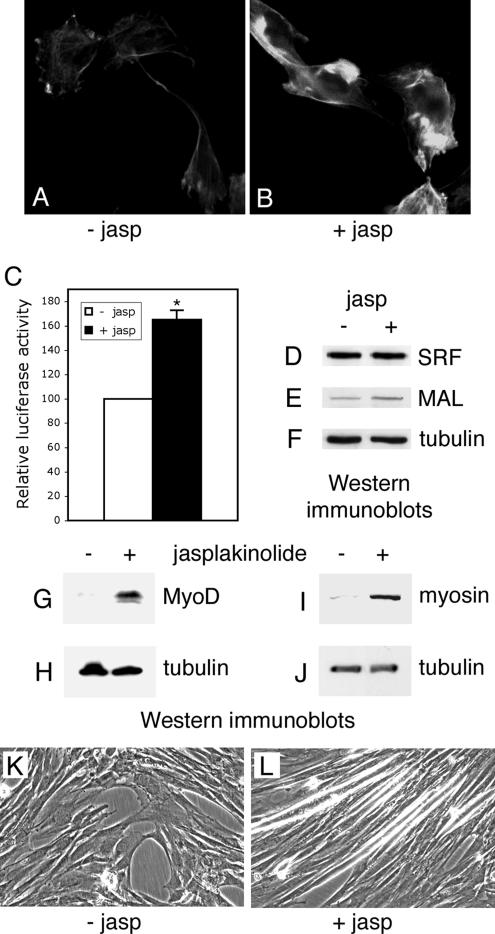
It has been demonstrated that agents promoting F-actin accumulation and stabilization induce SRF activity. In particular, exposure of cells to the F-actin enhancing drug, jasplakinolide, has been shown to strongly activate SRF (Sotiropoulos et al., 1999). To test whether actin stabilization could restore the differentiation capacity of the ALP-antisense cells, we have treated these cells with jasplakinolide. ALP-antisense C2C12 cells were placed in differentiation medium in presence of jasplakinolide for 1 h. This treatment was able to induce the formation of large F-actin aggregates in ALP-antisense C2C12 cells (Figure 6, A and B) as previously observed in REF52 cells (Bubb et al., 2000). Using a SRF reporter assay described in Materials and Methods, we observed that SRF activity is increased in the jasplakinolide-treated ALP-antisense cells (Figure 6C). Furthermore, although the expression level of SRF did not vary, the expression level of the SRF cofactor, MAL, slightly increased after treatment (Figure 6, D–F). It has been demonstrated previously that MAL is a SRF coactivator making a link between actin dynamics and SRF transcriptional activity (Miralles et al., 2003). The jasplakinolide treatment was also able to induce MyoD expression when the ALP-antisense C2C12 cells were subsequently grown in differentiation-promoting conditions for 3 d (Figure 6, G and H). Furthermore, myosin expression (Figure 6, I and J) accompanied with myotube formation (Figure 6, K and L) was also induced in ALP-antisense C2C12 cells placed in differentiation medium for 7 d after jasplakinolide treatment. These results demonstrated that jasplakinolide-induced stabilization of actin filaments is able to activate myogenic differentiation of the C2C12 cells in absence of ALP. Therefore, stabilization of actin filaments by the jasplakinolide drug is able to bypass the ALP requirement in the MyoD-dependent activation of myogenesis.
Figure 6.
Differentiation of the ALP-antisense C2C12 cells after jasplakinolide treatment. ALP-antisense C2C12 cells were placed in differentiation-promoting conditions in absence or in presence of 0.16 μM jasplakinolide for 1 h and then grown in differentiation medium for 1 d (A–F), 3 d (G and H) or 7 d (I–L). Fluorescence microscopy was performed to visualize F-actin in ALP-antisense C2C12 cells treated (B) or not (A) with jasplakinolide. SRF activity was determined in a similar experiment where the jasplakinolide-treated cells were previously cotransfected with the SRF reporter plasmid pSRE3-Luc and a Renilla standardization reporter plasmid (C). Luciferase activities are expressed after correction for Renilla reporter activities. Results are the mean ± SEM of three independent experiments. Asterisks indicate statistical significance at p < 0.003 according to the unpaired Student's t test. Cell lysates were analyzed by immunoblotting using SRF (D), MAL (E), MyoD (G), myosin (I) and tubulin (F, H, and J) antibodies. Phase contrast images of ALP-antisense C2C12 cells treated (L) or not (K) with jasplakinolide.
Activation of SRF Is Modulated by ALP Expression
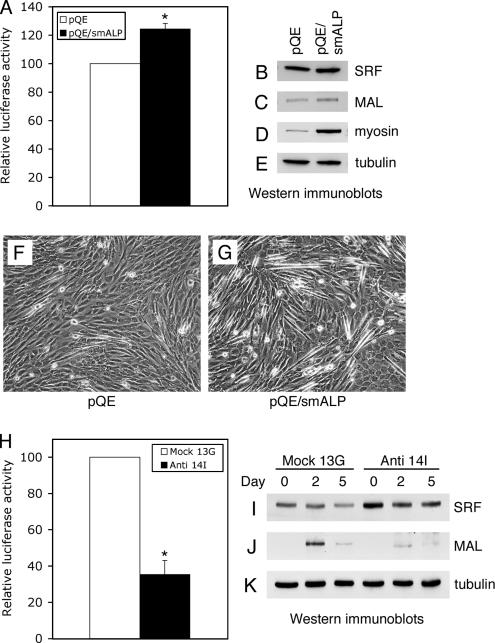
Finally, in order to test whether ALP could play a role in MyoD regulation via cytoskeletal regulation of SRF activity, we used a SRF reporter assay in C2C12 cells overexpressing or lacking smALP. We first studied the activity of SRF in C2C12 cells transiently transfected with a vector encoding mouse smALP. Overexpression of smALP induced a 25% increase in SRF activity when the C2C12 cells were placed in differentiation medium for 2 d (Figure 7A). Under these conditions, SRF expression levels remained unchanged, whereas MAL expression increased somewhat (Figure 7, B and C). Furthermore, smALP overexpression stimulated differentiation of the C2C12 cells, as can be seen by the enhanced myosin expression level (Figure 7D) and the enhanced formation of elongated C2C12 cells (Figure 6, F and G). We next tested the activity of SRF in ALP-antisense C2C12 cells grown in differentiation-promoting conditions for 2 d. As shown in Figure 7H, activation of SRF was strongly reduced in ALP-antisense C2C12 cells compared with control C2C12 cells. Nevertheless, this diminution in SRF activity is not due to a loss of SRF expression because the amount of SRF is even slightly increased in ALP-antisense cells compared with control C2C12 cells (Figure 7I). In contrast with SRF, MAL expression levels are drastically reduced in the ALP-antisense cells (Figure 7J).
Figure 7.
SRF activities and MAL expression levels are dependent on ALP expression. C2C12 cells were cotransfected with a mouse smALP expression vector (pQE/smALP) or an empty vector (pQE), and the SRF reporter plasmid pSRE3-Luc associated with a Renilla standardization reporter plasmid, and were subsequently placed in differentiation medium for 2 d. SRF activity was determined in (A). In a similar experiment, expression levels of SRF (B), MAL (C), myosin (D), and tubulin (E) were analyzed by Western immunoblotting, while phase contrast images show the aspect of the C2C12 cells transfected with the vector alone (F) or the smALP construct (G) after 2 d in differentiation medium. (H) SRF activity was also determined in ALP-antisense C2C12 cells (Anti 14I) or control C2C12 cells (Mock 13G) that were cotransfected with the SRF reporter plasmid pSRE3-Luc and a Renilla standardization reporter plasmid, and subsequently grown for 2 d in differentiation-promoting conditions. Expression levels of SRF (I) and MAL (J) were detected by Western immunoblotting in ALP-antisense C2C12 cells (Anti 14I) and control C2C12 cells (Mock 13G) grown in proliferation medium (Day 0) or placed in differentiation-promoting conditions for 2 d (Day 2) and 5 d (Day 5). As a control, tubulin was detected under the same conditions (K). Luciferase activities are expressed after correction for Renilla reporter activities. Results are the mean ± SEM of three independent experiments. Asterisks indicate statistical significance at p < 0.003 according to the unpaired Student's t test.
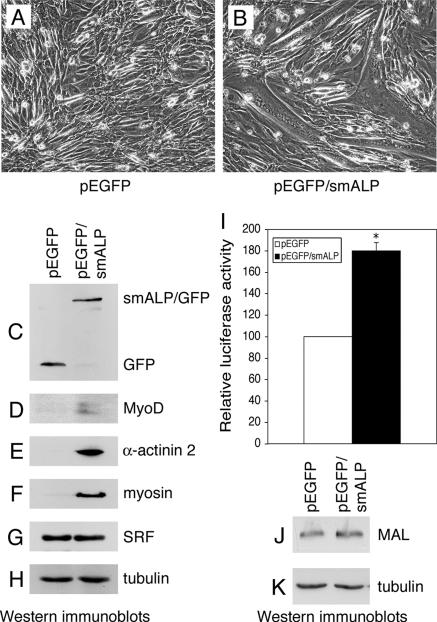
Because smALP overexpression was able to increase SRF activity in C2C12 cells placed in differentiation-promoting conditions (Figure 7A), we examined the effects of a similar overexpression on C2C12 cells grown in proliferation medium. Unlike C2C12 cells transfected with the control vector, C2C12 cells expressing exogenous smALP (Figure 8C) form myotubes when grown under proliferation conditions for 5 d (Figure 8, A and B). Furthermore, all the myotubes formed under these conditions expressed the recombinant smALP/GFP protein as observed by direct fluorescence microscopy (data not shown). Differentiation is fully achieved in these cells since the muscle-specific markers MyoD, myosin and α-actinin-2 are expressed under these conditions (Figure 8, D–F). Although SRF expression levels are similar in control and in smALP-overexpressing C2C12 cells (Figure 8G), overexpression of smALP induces an increase of SRF activity accompanied by a small but reproducible increase in MAL expression when the cells were grown in proliferation medium (Figure 8, I–K). These results demonstrated that overexpression of smALP stimulates SRF activity and induces differentiation of the C2C12 cell line even when the cells are grown under proliferation-promoting conditions.
Figure 8.
SmALP overexpression induces differentiation of C2C12 cells grown in proliferation conditions. C2C12 cells were cotransfected with a smALP construct (pEGFP/smALP) or with an empty vector (pEGFP), and the SRF reporter plasmid pSRE3-Luc and a Renilla standardization reporter plasmid, and were subsequently grown under proliferation conditions for 5 d. Phase contrast images of the C2C12 cells transfected with the vector alone (A) or the smALP construct (B). Corresponding cell lysates were analyzed by immunoblotting using GFP (C), MyoD (D), α-actinin 2 (E), myosin (F), SRF (G), and tubulin (H) antibodies. (I) SRF activity was determined in these cells. Luciferase activities are expressed after correction for Renilla reporter activities. Results are the mean ± SEM of three independent experiments. Asterisks indicate statistical significance at p < 0.0005 according to the unpaired Student's t test. MAL expression levels (J) were also detected in the C2C12 cells transfected with the vector alone (pEGFP) or the smALP construct (pEGFP/smALP) grown in proliferation medium for 3 d. As a control, tubulin was detected under the same conditions (K).
DISCUSSION
Regulation of ALP Expression and Isoform Diversity during Muscle Development
To gain insight into the role of ALP in muscle differentiation and organization, we studied ALP expression during the differentiation process of the myogenic C2C12 cell line. We observed a striking regulation of ALP expression and isoform switching during muscle cell differentiation. We show that the lower molecular mass protein, smALP, is the only isoform expressed in proliferating C2C12 myoblasts, suggesting that smALP plays a role before the induction of differentiation.
A striking up-regulation of smALP expression occurs in C2C12 cells placed in differentiation medium. At later stages in the differentiation program, smALP expression declines and there is a corresponding increase in the expression of skALP. SkALP is the sole isoform present in mature myotubes where it colocalizes at the Z-discs with its binding partner, α-actinin (Xia et al., 1997; Pomies et al., 1999). The dramatic shift in ALP isoform expression during muscle differentiation suggests that smALP and skALP have some unique properties that are functionally relevant at different stages of muscle development. SmALP and skALP are encoded by a single gene and are generated as a result of alternative splicing. The murine ALP isoforms display identical N- and C-termini, including PDZ and LIM domains, and vary only in their central regions where 63 amino acids in smALP are replaced by 111 amino acids in skALP. Thus far, no specific function has been assigned to this variable domain, which must be responsible for any nonoverlapping roles of smALP and skALP. Determination of the unique properties, and possibly protein partners, of smALP and skALP will likely provide valuable insight into the specific actions of these proteins at different stages in muscle differentiation.
ALP-dependent Regulation of the Myogenic Transcription Factor, MyoD
An antisense mRNA strategy has been used previously to assess the roles of a variety of proteins during C2C12 differentiation. The approach has enabled the identification of proteins involved at multiple stages in the differentiation process (Galbiati et al., 1999; Montanaro et al., 1999; Chen et al., 2000). Using an antisense RNA approach, we have generated C2C12 cell clones in which smALP and skALP expression has been severely reduced. We find that ALP-antisense C2C12 cells are unable to differentiate when they are placed in differentiation-promoting conditions. This differentiation block is attributed to a specific effect on ALP transcripts because multiple antisense constructs that target different ALP sequences have comparable effects.
Compromised expression of ALP in C2C12 cells eliminates the accumulation of the myogenic transcription factors MyoD and myogenin, as well as the expression of terminal differentiation markers such as skeletal muscle myosin heavy chain, without affecting Myf5 levels. MyoD is normally expressed in both determined, proliferating myoblasts as well as differentiating muscle cells (Montarras et al., 1991), and MyoD levels are suppressed in both populations of cells when antisense-ALP is expressed. In myoblasts, MyoD plays an important role in controlling the proliferation-to-differentiation switch and at later stages in the differentiation process, MyoD regulates the expression of the essential terminal differentiation factor, myogenin (for review, see Weintraub, 1993). Thus, loss of MyoD expression is sufficient to explain the failure of antisense-ALP C2C12 cells to differentiate. Indeed, restoration of MyoD expression is sufficient to bypass the requirement for ALP illustrating that ALP is an essential regulator of myogenesis that functions upstream of MyoD in the differentiation pathway.
The C2C12 system provides an excellent model for assessing the role of particular proteins in muscle differentiation but does not provide a high-resolution view of functional attributes of mature myotubes. Consequently, it is possible that ALP also functions later in mature myotubes, perhaps in muscle homeostasis or stress-induced hypertrophy. In this regard, it is interesting that MyoD has been implicated in the regulation of gene expression programs necessary for muscle maintenance and repair (Lowe and Alway, 1999; Parker et al., 2003).
Role of ALP in Muscle Development In Vivo
In mouse embryos, ALP is expressed in precardial mesoderm and later is found in cardiac, skeletal, and smooth muscle (Pashmforoush et al., 2001). Given the profound effects of abrogating ALP expression for the differentiation of C2C12 cells, one would anticipate that ALP deficiency would have serious deleterious consequences for muscle development. Surprisingly, mice that lack ALP did not display a failure of skeletal muscle differentiation (Jo et al., 2001). However, homozygous mutation of the ALP gene resulted in moderately penetrant (15%) embryonic lethality that appears to result from heart failure (Pashmforoush et al., 2001). This phenotype depends on the genetic background, which may account for the lack of phenotype detected in skeletal muscle in the parallel study by Jo et al. (2001).
Analysis of mice in which the gene encoding ALP was disrupted by homologous recombination revealed a role for ALP in the development of cardiac muscle, in particular the right ventricular chamber. Loss of ALP resulted in embryonic chamber dysmorphogenesis, a failure of trabeculation, and chamber dilation (Pashmforoush et al., 2001). In surviving adults, cardiomyopathy resulted. Thus, ALP clearly contributes to normal cardiac development and function. The existence of multiple PDZ-LIM proteins closely related to ALP that are expressed in overlapping patterns with ALP (Kuroda et al., 1996; Guy et al., 1999; Kotaka et al., 1999; Zhou et al., 1999) may have provided some compensation for the lack of ALP and contributed to the muscle function in the ALP-deficient animals. Similarly, in skeletal muscle, it is well established that there are numerous signaling inputs that contribute to the upregulation of MyoD expression during embryogenesis, including some that come from mesoderm-adjacent structures like dorsal ectoderm (Parker et al., 2003). The complexity and plasticity of a three-dimensional developing embryo is much greater than can be achieved in cell culture models of differentiation. Thus, mechanisms for compensatory signaling to abrogate the loss of ALP may occur within a developing organism that would not be possible in cell culture. Indeed, it is well established that elimination of MyoD function in C2C12 cells blocks myogenic differentiation when this is not sufficient in the mouse (Rudnicki et al., 1992; Dedieu et al., 2002).
Mechanism of ALP Influence on SRF Activity
We have shown that suppression of ALP levels leads to the loss of MyoD protein in C2C12 cells, thus providing evidence that ALP influences the cellular levels of a master regulatory protein for muscle differentiation. This regulation is likely to occur at the level of transcription because MyoD transcripts are also reduced in the ALP-antisense cells. ALP may contribute to control MyoD expression by indirect effects on SRF activation state. It is now well established that F-actin stabilization positively regulates SRF-dependent gene transcription, and depletion of the G-actin pool in the cells seems to be responsible for the activation of SRF (Sotiropoulos et al., 1999). Thus, proteins that regulate the assembly state of actin can influence SRF signaling (Sotiropoulos et al., 1999; Copeland and Treisman, 2002). ALP is localized along the actin filaments in proliferating C2C12 myoblasts, and we showed previously that it enhances the ability of α-actinin to cross-link actin filaments in vitro (Pashmforoush et al., 2001). These observations suggest that ALP might participate in the stabilization of F-actin in muscle cells, and we report here that suppression of ALP expression gives rise to a reduction in F-actin arrays. Interestingly, the RIL protein that is closely related to ALP has been reported to modulate the turnover of actin stress fibers (Vallenius et al., 2004), consistent with the idea that ALP-related proteins have shared roles in regulation of actin dynamics. The properties of ALP suggest that it could impact SRF-dependent muscle gene transcription by virtue of its ability to stabilize F-actin. Several factors that influence the actin cytoskeleton and SRF activity, including LIM-kinase and VASP, have been identified by overexpression studies (Sotiropoulos et al., 1999). The fact that we observed deficits in actin organization and MyoD expression when ALP is compromised using a loss-of-function approach provides powerful indication for a specific role of ALP in the cytoskeletal regulation of SRF. This hypothesis is strengthened by experiments showing that overexpression of ALP stimulates SRF activity, but not SRF expression, and that SRF activity, but not SRF expression level, is strongly reduced in ALP-antisense C2C12 cells. This variation of activity may result, at least in part, from a modulation in the level of the SRF coactivator, MAL. SRF is postranslationally down-regulated by proteolytic degradation in NIH3T3 fibroblasts (Shin et al., 2006), and it is possible that MAL stability may be influenced by actin monomer or polymer. Importantly, our ability to rescue muscle differentiation in the ALP-antisense cells by simple application of the actin stabilizing drug, jasplakinolide, illustrates that it is possible to bypass the requirement for ALP by manipulating actin dynamics. Actin dynamics is known to influence SRF via regulation of the nuclear accumulation of MAL (Miralles et al., 2003). Our work raises the possibility that actin dynamics may also contribute to SRF activity by influencing MAL stability or expression. Future work will be required to define the detailed molecular mechanism by which ALP-induced changes in the actin cytoskeleton influence SRF function.
ACKNOWLEDGMENTS
We thank Dr. D. S. Bredt for the rat skALP vector, Dr. S. Ait-Si-Ali for the pcDNA3/MyoD expression vector, Dr. R. A. Hipskind for the pSRE3-Luc vector, and Dr. T. P. Makela and Dr. T. Vallenius for the CLP-36 antibody. Support was provided by grants from the Association Française contre les Myopathies and the Fondation pour la Recherche Médicale to P.P. and from the Muscular Dystrophy Association and the National Institutes of Health Grant HL60591 to M.C.B. Support of the University of Utah Shared Resources (Cancer Center Support Grant P30CA42014) is also acknowledged.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-09-0815) on March 1, 2007.
REFERENCES
- Arber S., Halder G., Caroni P. Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell. 1994;79:221–231. doi: 10.1016/0092-8674(94)90192-9. [DOI] [PubMed] [Google Scholar]
- Arsenian S., Weinhold B., Oelgeschlager M., Ruther U., Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 1998;17:6289–6299. doi: 10.1093/emboj/17.21.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubb M. R., Spector I., Beyer B. B., Fosen K. M. Effects of jasplakinolide on the kinetics of actin polymerization. An explanation for certain in vivo observations. J. Biol. Chem. 2000;275:5163–5170. doi: 10.1074/jbc.275.7.5163. [DOI] [PubMed] [Google Scholar]
- Carnac G., Albagli-Curiel O., Vandromme M., Pinset C., Montarras D., Laudet V., Bonnieu A. 3,5,3′-Triiodothyronine positively regulates both MyoD1 gene transcription and terminal differentiation in C2 myoblasts. Mol. Endocrinol. 1992;6:1185–1194. doi: 10.1210/mend.6.8.1406697. [DOI] [PubMed] [Google Scholar]
- Carnac G., Primig M., Kitzmann M., Chafey P., Tuil D., Lamb N., Fernandez A. RhoA GTPase and serum response factor control selectively the expression of MyoD without affecting Myf5 in mouse myoblasts. Mol. Biol. Cell. 1998;9:1891–1902. doi: 10.1091/mbc.9.7.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. L., Dowhan D. H., Hosking B. M., Muscat G. E. The steroid receptor coactivator, GRIP-1, is necessary for MEF-2C-dependent gene expression and skeletal muscle differentiation. Genes Dev. 2000;14:1209–1228. [PMC free article] [PubMed] [Google Scholar]
- Copeland J. W., Treisman R. The diaphanous-related formin mDia1 controls serum response factor activity through its effects on actin polymerization. Mol. Biol. Cell. 2002;13:4088–4099. doi: 10.1091/mbc.02-06-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossu G., Kelly R., Tajbakhsh S., Di Donna S., Vivarelli E., Buckingham M. Activation of different myogenic pathways: myf-5 is induced by the neural tube and MyoD by the dorsal ectoderm in mouse paraxial mesoderm. Development. 1996;122:429–437. doi: 10.1242/dev.122.2.429. [DOI] [PubMed] [Google Scholar]
- Dedieu S., Mazeres G., Cottin P., Brustis J. J. Involvement of myogenic regulator factors during fusion in the cell line C2C12. Int. J. Dev. Biol. 2002;46:235–241. [PubMed] [Google Scholar]
- Delfini M. C., Hirsinger E., Pourquie O., Duprez D. Delta 1-activated notch inhibits muscle differentiation without affecting Myf5 and Pax3 expression in chick limb myogenesis. Development. 2000;127:5213–5224. doi: 10.1242/dev.127.23.5213. [DOI] [PubMed] [Google Scholar]
- Dhawan J., Helfman D. M. Modulation of acto-myosin contractility in skeletal muscle myoblasts uncouples growth arrest from differentiation. J. Cell Sci. 2004;117:3735–3748. doi: 10.1242/jcs.01197. [DOI] [PubMed] [Google Scholar]
- Galbiati F., Volonte D., Engelman J. A., Scherer P. E., Lisanti M. P. Targeted down-regulation of caveolin-3 is sufficient to inhibit myotube formation in differentiating C2C12 myoblasts. Transient activation of p38 mitogen-activated protein kinase is required for induction of caveolin-3 expression and subsequent myotube formation. J. Biol. Chem. 1999;274:30315–30321. doi: 10.1074/jbc.274.42.30315. [DOI] [PubMed] [Google Scholar]
- Gauthier-Rouviere C., Vandromme M., Tuil D., Lautredou N., Morris M., Soulez M., Kahn A., Fernandez A., Lamb N. Expression and activity of serum response factor is required for expression of the muscle-determining factor MyoD in both dividing and differentiating mouse C2C12 myoblasts. Mol. Biol. Cell. 1996;7:719–729. doi: 10.1091/mbc.7.5.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geneste O., Copeland J. W., Treisman R. LIM kinase and Diaphanous cooperate to regulate serum response factor and actin dynamics. J. Cell Biol. 2002;157:831–838. doi: 10.1083/jcb.200203126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman A., Goode B. L., Matsudaira P., Fink G. R. The Saccharomyces cerevisiae calponin/transgelin homolog Scp1 functions with fimbrin to regulate stability and organization of the actin cytoskeleton. Mol. Biol. Cell. 2003;14:2617–2629. doi: 10.1091/mbc.E03-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse R., Copeland J. W., Newsome T. P., Way M., Treisman R. A role for VASP in RhoA-Diaphanous signalling to actin dynamics and SRF activity. EMBO J. 2003;22:3050–3061. doi: 10.1093/emboj/cdg287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy P. M., Kenny D. A., Gill G. N. The PDZ domain of the LIM protein enigma binds to beta-tropomyosin. Mol. Biol. Cell. 1999;10:1973–1984. doi: 10.1091/mbc.10.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadchouel J. Analysis of a key regulatory region upstream of the Myf5 gene reveals multiple phases of myogenesis, orchestrated at each site by a combination of elements dispersed throughout the locus. Development. 2003;130:3415–3426. doi: 10.1242/dev.00552. [DOI] [PubMed] [Google Scholar]
- Hinterberger T. J., Sassoon D. A., Rhodes S. J., Konieczny S. F. Expression of the muscle regulatory factor MRF4 during somite and skeletal myofiber development. Dev. Biol. 1991;147:144–156. doi: 10.1016/s0012-1606(05)80014-4. [DOI] [PubMed] [Google Scholar]
- Jo K., Rutten B., Bunn R. C., Bredt D. S. Actinin-associated LIM protein-deficient mice maintain normal development and structure of skeletal muscle. Mol. Cell. Biol. 2001;21:1682–1687. doi: 10.1128/MCB.21.5.1682-1687.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen F. E., Prywes R. Serum response factor: transcriptional regulation of genes induced by growth factors and differentiation. Biochim. Biophys. Acta. 1995;1242:1–10. doi: 10.1016/0304-419x(94)00014-s. [DOI] [PubMed] [Google Scholar]
- Kotaka M., Ngai S. M., Garcia-Barcelo M., Tsui S. K., Fung K. P., Lee C. Y., Waye M. M. Characterization of the human 36-kDa carboxyl terminal LIM domain protein (hCLIM1) J. Cell. Biochem. 1999;72:279–285. doi: 10.1002/(sici)1097-4644(19990201)72:2<279::aid-jcb12>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Kuroda S., Tokunaga C., Kiyohara Y., Higuchi O., Konishi H., Mizuno K., Gill G. N., Kikkawa U. Protein-protein interaction of zinc finger LIM domains with protein kinase C. J. Biol. Chem. 1996;271:31029–31032. doi: 10.1074/jbc.271.49.31029. [DOI] [PubMed] [Google Scholar]
- L'Honore A., Lamb N. J., Vandromme M., Turowski P., Carnac G., Fernandez A. MyoD distal regulatory region contains an SRF binding CArG element required for MyoD expression in skeletal myoblasts and during muscle regeneration. Mol. Biol. Cell. 2003;14:2151–2162. doi: 10.1091/mbc.E02-07-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lowe D. A., Alway S. E. Stretch-induced myogenin, MyoD, and MRF4 expression and acute hypertrophy in quail slow-tonic muscle are not dependent upon satellite cell proliferation. Cell Tissue Res. 1999;296:531–539. doi: 10.1007/s004410051314. [DOI] [PubMed] [Google Scholar]
- Ludolph D. C., Konieczny S. F. Transcription factor families: muscling in on the myogenic program. FASEB J. 1995;9:1595–1604. doi: 10.1096/fasebj.9.15.8529839. [DOI] [PubMed] [Google Scholar]
- Megeney L. A., Kablar B., Garrett K., Anderson J. E., Rudnicki M. A. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- Miralles F., Posern G., Zaromytidou A. I., Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- Montanaro F., Lindenbaum M., Carbonetto S. alpha-Dystroglycan is a laminin receptor involved in extracellular matrix assembly on myotubes and muscle cell viability. J. Cell Biol. 1999;145:1325–1340. doi: 10.1083/jcb.145.6.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montarras D., Aurade F., Johnson T., Ilan J., Gros F., Pinset C. Autonomous differentiation in the mouse myogenic cell line, C2, involves a mutual positive control between insulin-like growth factor II and MyoD, operating as early as at the myoblast stage. J. Cell Sci. 1996;109(Pt 3):551–560. doi: 10.1242/jcs.109.3.551. [DOI] [PubMed] [Google Scholar]
- Montarras D., Chelly J., Bober E., Arnold H., Ott M. O., Gros F., Pinset C. Developmental patterns in the expression of Myf5, MyoD, myogenin, and MRF4 during myogenesis. New Biol. 1991;3:592–600. [PubMed] [Google Scholar]
- Olson E. N. Interplay between proliferation and differentiation within the myogenic lineage. Dev. Biol. 1992;154:261–272. doi: 10.1016/0012-1606(92)90066-p. [DOI] [PubMed] [Google Scholar]
- Ornatsky O. I., Andreucci J. J., McDermott J. C. A dominant-negative form of transcription factor MEF2 inhibits myogenesis. J. Biol. Chem. 1997;272:33271–33278. doi: 10.1074/jbc.272.52.33271. [DOI] [PubMed] [Google Scholar]
- Parker M. H., Seale P., Rudnicki M. A. Looking back to the embryo: defining transcriptional networks in adult myogenesis. Nat. Rev. Genet. 2003;4:497–507. doi: 10.1038/nrg1109. [DOI] [PubMed] [Google Scholar]
- Pashmforoush M., Pomies P., Peterson K. L., Kubalak S., Ross J., Jr, Hefti A., Aebi U., Beckerle M. C., Chien K. R. Adult mice deficient in actinin-associated LIM-domain protein reveal a developmental pathway for right ventricular cardiomyopathy. Nat. Med. 2001;7:591–597. doi: 10.1038/87920. [DOI] [PubMed] [Google Scholar]
- Pomies P., Macalma T., Beckerle M. C. Purification and characterization of an alpha-actinin-binding PDZ-LIM protein that is up-regulated during muscle differentiation. J. Biol. Chem. 1999;274:29242–29250. doi: 10.1074/jbc.274.41.29242. [DOI] [PubMed] [Google Scholar]
- Rudnicki M. A., Braun T., Hinuma S., Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- Rudnicki M. A., Jaenisch R. The MyoD family of transcription factors and skeletal myogenesis. Bioessays. 1995;17:203–209. doi: 10.1002/bies.950170306. [DOI] [PubMed] [Google Scholar]
- Shin S. Y. Suppression of Egr-1 transcription through targeting of the serum response factor by oncogenic H-Ras. EMBO J. 2006;25:1093–1103. doi: 10.1038/sj.emboj.7600987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiropoulos A., Gineitis D., Copeland J., Treisman R. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell. 1999;98:159–169. doi: 10.1016/s0092-8674(00)81011-9. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S., Rocancourt D., Buckingham M. Muscle progenitor cells failing to respond to positional cues adopt non-myogenic fates in myf-5 null mice. Nature. 1996;384:266–270. doi: 10.1038/384266a0. [DOI] [PubMed] [Google Scholar]
- Treisman R. Ternary complex factors: growth factor regulated transcriptional activators. Curr. Opin. Genet. Dev. 1994;4:96–101. doi: 10.1016/0959-437x(94)90097-3. [DOI] [PubMed] [Google Scholar]
- Treisman R. Journey to the surface of the cell: Fos regulation and the SRE. EMBO J. 1995;14:4905–4913. doi: 10.1002/j.1460-2075.1995.tb00173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallenius T., Scharm B., Vesikansa A., Luukko K., Schafer R., Makela T. P. The PDZ-LIM protein RIL modulates actin stress fiber turnover and enhances the association of alpha-actinin with F-actin. Exp. Cell Res. 2004;293:117–128. doi: 10.1016/j.yexcr.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Venuti J. M., Morris J. H., Vivian J. L., Olson E. N., Klein W. H. Myogenin is required for late but not early aspects of myogenesis during mouse development. J. Cell Biol. 1995;128:563–576. doi: 10.1083/jcb.128.4.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. Z., Olson E. N. Control of smooth muscle development by the myocardin family of transcriptional coactivators. Curr. Opin. Genet. Dev. 2004;14:558–566. doi: 10.1016/j.gde.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Wang Y., Krushel L. A., Edelman G. M. Targeted DNA recombination in vivo using an adenovirus carrying the cre recombinase gene. Proc. Natl. Acad. Sci. USA. 1996;93:3932–3936. doi: 10.1073/pnas.93.9.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Zhou W., Croissant J. D., Johansen F. E., Prywes R., Balasubramanyam A., Schwartz R. J. RhoA signaling via serum response factor plays an obligatory role in myogenic differentiation. J. Biol. Chem. 1998;273:30287–30294. doi: 10.1074/jbc.273.46.30287. [DOI] [PubMed] [Google Scholar]
- Weintraub H. The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- Xia H., Winokur S. T., Kuo W. L., Altherr M. R., Bredt D. S. Actinin-associated LIM protein: identification of a domain interaction between PDZ and spectrin-like repeat motifs. J. Cell Biol. 1997;139:507–515. doi: 10.1083/jcb.139.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Ruiz-Lozano P., Martone M. E., Chen J. Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to alpha-actinin-2 and protein kinase C. J. Biol. Chem. 1999;274:19807–19813. doi: 10.1074/jbc.274.28.19807. [DOI] [PubMed] [Google Scholar]