Abstract
In epithelial cells, p120 catenin (p120) localizes at cell–cell contacts and regulates adhesive function of the cadherin complex. In addition, p120 has been reported to localize in the nucleus, although the nuclear function of p120 is not fully understood. Here, we report the identification of Gli-similar 2 (Glis2) as a novel binding protein for p120. Glis2 is a Krüppel-like transcriptional repressor with homology to the Gli family, but its physiological function has not been well characterized. In this study, we show that coexpression of Glis2 and Src induces nuclear translocation of p120. Furthermore, p120 induces the C-terminal cleavage of Glis2, and this cleavage is further enhanced by Src. The cleaved form of Glis2 loses one of its five zinc finger domains, but it is still able to bind DNA. Functional studies in chick neural tube indicate that full-length Glis2 can affect neuronal differentiation, whereas the cleaved form requires coexpression of p120 to have a similar effect. These data indicate that p120 has additional novel functions in the nucleus together with Glis2.
INTRODUCTION
In multicellular organisms, individual cells often adhere to each other to form three-dimensionally structured tissues or organs. In addition to this structural role, cell–cell contacts can transduce various signaling pathways. Among cell–cell junctional proteins, cadherins, especially classical cadherins, have been shown to be involved in several signaling pathways, including cell proliferation, differentiation, and cell survival (for reviews, see Jamora and Fuchs, 2002; Wheelock and Johnson, 2003; Yap and Kovacs, 2003; Gumbiner, 2005). The cytoplasmic domain of cadherin interacts with several proteins, including β-catenin, p120 catenin (hereafter referred to as p120), C3G, and Hakai (McCrea et al., 1991; Reynolds et al., 1994; Fujita et al., 2002; Hogan et al., 2004). The role of β-catenin in the Wnt signaling pathway is well established (Hecht et al., 2000; Brembeck et al., 2006; Willert and Jones, 2006), whereas the involvement of other cadherin-binding proteins in signaling pathways is not yet fully understood. p120 was originally identified as a substrate for the Src tyrosine kinase (Reynolds et al., 1992). Subsequently, it was shown that p120 directly interacts with the juxtamembrane domain of classical cadherins (Reynolds et al., 1994) and that it regulates the adhesive function of cadherins (Aono et al., 1999; Thoreson et al., 2000; Ireton et al., 2002). Recently, p120 was reported to be involved in cadherin turnover and/or trafficking (Davis et al., 2003; Xiao et al., 2003). p120 is an Armadillo repeat domain protein that is structurally related to β-catenin. Despite this structural similarity, p120 binds to a different site on the cadherin tail and cannot function as a β-catenin substitute. In epithelial cells, p120 is predominantly localized at cadherin-based cell–cell contact sites, and it is also tethered to microtubules (Chen et al., 2003; Franz and Ridley, 2004; Yanagisawa et al., 2004). In addition, p120 is involved in dynamic regulation of the actin cytoskeleton by modulating activity of small GTPase Rho family members (Anastasiadis et al., 2000; Noren et al., 2000; Grosheva et al., 2001; Elia et al., 2006). However, under several experimental conditions, its nuclear localization has also been observed, suggesting a functional role of p120 in the nucleus (van Hengel et al., 1999; Roczniak-Ferguson and Reynolds, 2003; Kelly et al., 2004). Indeed, p120 interacts with Kaiso, a BTB/POZ domain zinc finger (ZnF) transcriptional repressor (Daniel and Reynolds, 1999; Prokhortchouk et al., 2001). It was also shown that the interaction with p120 prevents Kaiso from binding to DNA, thereby suppressing its repressor activity (Daniel et al., 2002; Kelly et al., 2004; Kim et al., 2004). In this study, we show that p120 has another nuclear binding partner, Gli-similar 2 (Glis2).
Glis2 was originally identified as a Gli-related Krüppel-like transcription factor that contains a tandem of five zinc finger domains (Zhang et al., 2002). By in situ hybridization, expression of Glis2 was detected in the neural tube, somites, and kidney (Lamar et al., 2001; Zhang et al., 2002). In vitro, Glis2 can act as a repressor of gene transcription (Zhang et al., 2002), and recently, it was reported to interact with CtBP1, a transcriptional corepressor (Kim et al., 2005). Glis2 was also shown to bind to the Gli binding site on DNA (Lamar et al., 2001), but its role in Gli signaling downstream of Sonic Hedgehog (Shh) has not been reported. Gli family proteins (Gli1, Gli2, and Gli3 in mammals and Cubitus interruptus [Ci] in fly) are the main transducers of Shh signaling, a pathway that plays a crucial role in several processes of embryonic development (Ingham, 1998). Shh signaling seems to regulate proteolytic processing of Gli proteins (for review, see Huangfu and Anderson, 2006). In Gli3 and Ci, when the soluble ligand Shh binds to its membrane-bound receptor Patched, the proteolytic cleavage of Gli is suppressed. The intact Gli then functions as a transcriptional activator. In the absence of Shh, Gli3 and Ci are cleaved to produce an N-terminal fragment. This cleaved N terminus of Gli retains its zinc finger DNA binding domains, but it lacks the coactivator binding domain; thus, it acts as a transcriptional repressor for target genes.
In this article, we report that p120 interacts with Glis2 and induces its proteolytic cleavage. Furthermore, the interaction with Glis2 increases the nuclear localization of p120. We also present evidence that indicates a novel functional role of p120 in regulating gene expression.
MATERIALS AND METHODS
Antibodies, Plasmids, and Materials
Anti-p120 antibody was purchased from BD Biosciences Transduction Laboratories (Lexington, KY). Anti-FLAG and peroxidase-conjugated anti-glutathione S-transferase (GST) antibodies were purchased from Sigma-Aldrich (St. Louis, MO). Anti-hemagglutinin (HA) and anti-phosphotyrosine antibodies were obtained from Roche Diagnostics (Mannheim, Germany) and Calbiochem (Darmstadt, Germany), respectively. Anti-GFP antibody is a rabbit serum from Invitrogen (Carlsbad, CA). Anti-mitogen-activated protein kinase (MAPK) antibody was purchased from QIAGEN (Crawley, United Kingdom). The rabbit polyclonal anti-Glis2 antibody was produced using GST-Glis2 (amino acids 35–174) as an antigen (Eurogentec, Seraing, Belgium). The Glis2 antibody can be used for Western blotting but not for immunoprecipitation. Anti-Lim1/2 4F2, anti-Pax6, and anti-Nkx2.2 antibodies were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by Department of Biological Sciences, The University of Iowa (Iowa City, IA).
To construct pGEX-Glis2, the partial Glis2 cDNA including the p120 binding site was excised from pVP16-Glis2 that was obtained by the yeast two-hybrid screen, and it was cloned into a NotI site of the pGEX-4T1 vector. The expressed sequence tag (EST) clone (IMAGE 4506253), which includes the open reading frame of mouse Glis2 was obtained from RZPD Deutsches Ressourcenzentrum für Genomforschung (Berlin, Germany). To construct FLAG-Glis2 and HA-Glis2, the cDNA of full-length Glis2 was amplified from the EST clone by polymerase chain reaction (PCR) and cloned into a NotI site of the pcDNA-FLAG and pcDNA-HA vectors, respectively. To produce various constructs expressing a series of Glis2 C-terminal deletion mutants (Glis2 ΔC), the cDNAs of Glis2 mutants were amplified by PCR and cloned into a NotI site of the pcDNA-FLAG vector. The amino acid numbers for the deletions were as follows: Glis2-ΔC1 (1-257), -ΔC2 (1-290), -ΔC3 (1-307), -ΔC4 (1-317), -ΔC5 (1-324), and -ΔC6 (1-357). To obtain pcAGGS-HA-p120, pcAGGS- FLAG-Glis2, and pcAGGS-FLAG-Glis2-ΔC, cDNAs of HA-p120, FLAG-Glis2 and FLAG-Glis2 ΔC were excised from the pcDNA vector, and they were cloned into a NotI site in the pcAGGS vector. pcAGGS-IRES-GFP was constructed by cloning the IRES-GFP construct into XhoI and BglII sites of the pcAGGS vector. To construct pcDNA-HA-E-cadherin, the cDNA of the whole cytoplasmic domain of mouse E-cadherin was amplified by PCR from pBAT-E-cadherin and cloned into a NotI site of the pcDNA-HA vector. To construct pcDNA-FLAG-p120, the cDNA of p120 was excised from pcDNA-HA-p120 and cloned into a NotI site of the pcDNA-FLAG vector. To construct pcDNA-HA-N-terminus and C terminus of p120, the cDNA of p120 was cleaved from pcDNA-HA-p120 by NotI and XhoI and PvuII and NotI, respectively, and cloned into the pcDNA-HA vector. pBSSR-HA-Ubiquitin, pcDNA-HA-p120 isoform 1B, pcDNA-myc-β-catenin, pSG-v-Src, and pRK5-myc-RacQ12V were described previously (Fujita et al., 2002; Hogan et al., 2004). pCMV5-FLAG-δ-catenin and pRK5-myc-Rho T19N were kindly provided by T. Okada (Kobe University, Kobe, Japan) and A. Hall (Memorial Sloan-Kettering Cancer Center, New York, NY), respectively.
Lipofectamine Plus reagent was obtained from Invitrogen. Hi-PerFect reagent was purchased from QIAGEN. Site-directed mutagenesis was performed using QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, CA).
Yeast Two-Hybrid Screen
The full length of p120 was used as bait. The cDNA of p120 was amplified from pBS-p120 by PCR and cloned into a SalI site of pBTM. The yeast two-hybrid screen was performed as described previously (Fujita et al., 2002).
Cell Culture, Immunofluorescence, Immunoprecipitation, GST-Glis2 Pulldown Assay, and Overlay-blotting Assay
Human embryonic kidney (HEK)293 cells were cultured and transfected as described previously (Schaeper et al., 2000). COS-1 and L fibroblast cells were cultured in DMEM supplemented with 10% fetal calf serum and 1% penicillin/streptomycin at 37°C and ambient air supplemented with 5% CO2, and transfected with Lipofectamine Plus reagent. Immunofluorescence was performed as described previously (Hogan et al., 2004). To measure the ratio of nucleus/cytoplasm p120 staining intensity, confocal images were analyzed by ImageJ software (National Institutes of Health, Bethesda, MD). After the location of the nucleus was confirmed by Hoechst staining, immunofluorescent intensity within a defined region (diameter 1.5 μm) in the nucleus (excluding nucleolus) or cytosol was quantified. Student's t tests assuming paired variances were performed for statistical analysis. To obtain epifluorescence and confocal images, we used an Axioskop 1 (Carl Zeiss, Jena, Germany) with a CoolSNAP camera (Roper Scientific, Trenton, NJ) and a Bio-Rad 1024 (Bio-Rad, Hercules, CA) mounted on an Optiphot 2 microscope (Nikon, Tokyo, Japan). Images were captured and analyzed using Openlab software (Improvision, Lexington, MA).
Immunoprecipitation and Western blotting were performed as described previously (Hogan et al., 2004). Expression of Src sometimes enhances the expression of cotransfected exogenous proteins by enhancing their transcription in cultured cells. Because of the detergent-insoluble nature of Glis2 ΔC, total cell lysates were used to examine Glis2 ΔC, except for in Figure 2C. To obtain total cell lysate, cells were washed once with phosphate-buffered saline (PBS) and suspended in Triton X-100 lysis buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, and 1% Triton X-100) containing 5 μg ml−1 leupeptin, 50 mM phenylmethylsulfonylfluoride (PMSF), and 7.2 trypsin inhibitor units ml−1 aprotinin. The suspended cells were directly boiled in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer. Densitometric analysis was performed with the image acquisition system Gel Doc 170 (Bio-Rad). For the GST-Glis2 pulldown assay, 10 μl of glutathione-Sepharose beads (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) attached to 10 μg of GST or GST-Glis2 protein was incubated with HEK293 cell lysates, followed by the same procedures described above for immunoprecipitation.
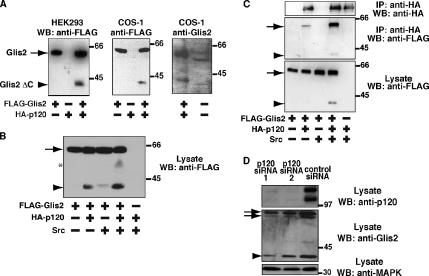
Figure 2.
Cleavage of Glis2 induced by coexpression of p120. (A) Cleavage of Glis2. Left and middle, FLAG-Glis2 was expressed in HEK293 and COS-1 cells with or without coexpression of HA-p120. Lysates were examined by Western blotting with anti-FLAG antibody. Right, COS-1 cells expressing FLAG-Glis2 and HA-p120, or untransfected, were examined by Western blotting with anti-Glis2 antibody. The minor size difference of the bands between two lanes is due to FLAG-tag of exogenous Glis2 and/or different species (monkey and mouse) of Glis2. It should be noted that the smaller amount of lysate was loaded in the left lane (1/20 of the right lane) because of much higher expression level of exogenous Glis2 than that of endogenous Glis2. (B) Expression of Src increases the p120-induced cleavage of Glis2. Lysates of HEK293 cells expressing FLAG-Glis2, HA-p120, and Src were examined by Western blotting with anti-FLAG antibody. Asterisk indicates the position of a potential intermediate proteolytic product. (C) p120 binds to Glis2 ΔC. Lysates of HEK293 cells expressing FLAG-Glis2, HA-p120, and Src were immunoprecipitated using anti-HA antibody, followed by Western blotting with anti-FLAG and anti-HA antibodies. (A–C) The arrow and arrowhead indicate the positions of full-length Glis2 and its cleavage product Glis2 ΔC, respectively. (D) Effect of p120 RNAi on Glis2. COS-1 cells were transiently transfected with two different p120 RNAi oligos directed against sequences present in all splicing variants of simian p120 or a control nonsilencing oligo. After 96 h, cell lysates were examined by Western blotting using anti-p120, anti-Glis2, and anti-MAPK antibodies. The band intensities were analyzed by densitometry. The arrows and arrowhead indicate the positions of full-length Glis2 and the 40-kDa product, respectively. The double band for the endogenous full-length protein (arrows) may be due to alternative splicing or protein degradation, which is sometimes observed.
Overlay blotting assays were performed as described previously (Fujita et al., 1998). A membrane was incubated with 200 nM GST or GST-Glis2, and proteins bound to p120 on the membrane were detected by anti-GST antibody.
DNA-Cellulose Binding Assay
For DNA-cellulose binding assays, HEK293 cells expressing FLAG-Glis2 or -Glis2 ΔC with HA-p120 and Src were lysed in a buffer containing 10 mM HEPES-NaOH, pH 7.4, 0.1% Triton X-100, 100 mM NaCl, 10% glycerol, and 0.05 mM EDTA supplemented with 5 μg ml−1 leupeptin, 50 mM PMSF, and 7.2 trypsin inhibitor units ml−1 aprotinin, and clarified by centrifugation at 5000 × g for 20 min. Cell lysate (100 μg of protein) was incubated with 25 μl of DNA-cellulose (GE Healthcare) or cellulose (Sigmacell cellulose type 50; Sigma-Aldrich) in 100 μl of lysis buffer for 1 h at 4°C. For competition assays, lysates were preincubated with the stated amounts of DNA (sonicated, calf thymus; GE Healthcare) or RNA (calf-liver type IV; Sigma-Aldrich), rotated for 30 min at 4°C, and then added to 25 μl of DNA-cellulose beads. After incubation, the beads were washed four times with 1 ml of lysis buffer, and the amount of FLAG-Glis2 retained on the beads was determined by Western blotting with anti-FLAG antibody.
Nuclear Fractionation
Cells were washed twice with PBS and trypsinized thoroughly until well separated. Pelleted cells were resuspended in 150 μl of 2X lysis buffer (50 mM HEPES-NaOH, pH 7.4, 10 mM EGTA, 5 mM MgCl2, 20% glycerol, 2% NP-40, and 2 mM dithiothreitol) containing 5 μg ml−1 leupeptin, 50 mM PMSF, and 7.2 trypsin inhibitor units ml−1 aprotinin. Cells were then immediately triturated with a 25-gauge needle 12 times. After centrifugation at 110 × g at 4°C for 5 min, the supernatant was removed as the cytoplasmic fraction, and remaining nuclei were washed twice in 1X lysis buffer. The supernatant and nuclear fractions were then boiled for 10 min with SDS-PAGE sample buffer and examined by Western blotting. Purity of nuclear fractions was confirmed by Western blotting with antibodies against several compartment-specific markers.
RNA Interference (RNAi)
p120 oligonucleotides (oligos) were purchased from QIAGEN and transiently transfected into COS-1 cells by using Hi-PerFect reagent. Cells (5 × 104 cells/well) were plated in a 24-well dish. We used 1.5 μg of small interfering RNA (siRNA) with 9 μl of Hi-PerFect reagent per well. As a negative control, we used the nonsilencing control siRNA (AF 488) from QIAGEN. Ninety-six hours after transfection, cells were lysed in Triton X-100 lysis buffer (described above) and examined by Western blotting. To ensure equal loading, the protein concentration of lysates was quantified using the DC Protein Assay reagent from Bio-Rad and measured on a VERSAmax microplate reader (Molecular Devices, Sunnyvale, CA).
Chick In Ovo Electroporation and Immunohistochemistry
pcAGGS-HA-p120, pcAGGS-HA-p120-N, pcAGGS-FLAG-Glis2, and pcAGGS- FLAG-Glis2 ΔC were expressed in chick embryos with pcAGGS-IRES-GFP by electroporation. The indicated plasmids were injected into Hamburger and Hamilton (HH) stage 10–12 chick embryos. Electrodes were placed either side of the neural tube, and electroporation was carried out using a T820 electro-squareporator (BTX, San Diego, CA), delivering five 50-ms pulses of 30 V. Transfected embryos were incubated at 38°C for 48 h and fixed and processed at HH stage 20–24. Embryos were fixed for 1 h in 4% paraformaldehyde, washed in PBS with 0.1% Triton X-100 (PBST), cryoprotected in 30% sucrose in PBS, and embedded in OCT for sectioning at 15 μm on a cryostat. For antibody stainings, slides were air-dried, washed twice with PBST, incubated with PBST plus 1% bovine serum albumin (BSA) for 10 min, and incubated with the primary antibody in PBST with 1% BSA overnight at 4°C. Slides were then washed three times with PBST and incubated with fluorescein isothiocyanate- or Cy2-conjugated secondary antibodies in PBST with 1% BSA. Slides were then dehydrated and mounted with 4′,6-diamidino-2-phenylindole. Samples were imaged using an LSM510 confocal microscope (Carl Zeiss). Student's t tests assuming paired variances were performed for statistical analysis.
RESULTS
p120 Binds to Glis2 and Induces Its Cleavage
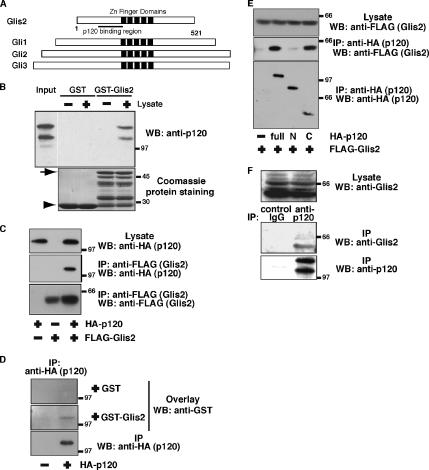
Using a yeast two-hybrid screen, we identified Glis2 as a novel binding protein for p120. Glis2 has a tandem of five Cys2-His2-type zinc finger domains in the central region. This zinc finger tandem shows high amino acid sequence identity with those of Gli family members (56–57%), but little homology was seen in the regions outside the zinc finger domains (Figure 1A) (Lamar et al., 2001; Zhang et al., 2002). The p120 binding site (amino acids 35–174), determined by yeast two-hybrid, is located just before the zinc finger domains. To confirm the interaction between p120 and Glis2, we performed a GST pulldown assay. Lysates from HEK293 cells were incubated with GST- or GST-Glis2 (amino acids 35–174)–coated beads. Endogenous p120 protein from HEK293 cell lysates interacted with GST-Glis2 beads but not with GST beads (Figure 1B). The interaction was further examined by immunoprecipitation by using HEK293 cell lysates. Exogenously expressed FLAG-Glis2 and HA-p120 were reciprocally coimmunoprecipitated (Figure 1, C and E). Next, we performed an overlay-blotting assay. HA-p120 on the membrane bound to GST-Glis2 but not to GST (Figure 1D), indicating that the interaction between p120 and Glis2 is direct. To study the Glis2 binding site, we produced truncation mutants of p120. The N terminus of p120 (amino acids 1–647), including the amino-terminal end and armadillo repeats 1–6, did not interact with Glis2, whereas the C terminus of p120 (amino acids 385–910), including armadillo repeats 2–10 and the carboxyl-terminal tail, bound to Glis2 (Figure 1E). Furthermore, endogenous Glis2 protein was coimmunoprecipitated using HEK293 cell lysates with anti-p120 antibody but not with control IgG (Figure 1F).
Figure 1.
Interaction between Glis2 and p120. (A) Molecular domain structure of Glis2. The clone isolated by the yeast two-hybrid screen encodes the N terminus of mouse Glis2 (amino acids 35–174). Glis2 is a transcription factor with five tandem zinc finger domains, with high homology to the Gli family in the zinc finger region. (B) Interaction between Glis2 and p120 by a GST pulldown assay. Endogenous p120 protein in HEK293 cells was pulled down by GST- or GST-Glis2 (amino acids 35–174)–coated beads and probed with anti-p120 antibody. Arrow and arrowhead in Coomassie protein staining indicate the position of GST-Glis2 and GST, respectively. Multiple bands below the GST–Glis2 band are its degradation products. (C) Interaction between Glis2 and p120 by immunoprecipitation. HA-p120 and FLAG-Glis2 were expressed in HEK293 cells, and cell lysates were immunoprecipitated with anti-FLAG antibody, followed by Western blotting with anti-FLAG and anti-HA antibodies. (D) Direct interaction between p120 and Glis2 by an overlay-blotting assay. HA-p120 expressed in HEK293 cells was immunoprecipitated with anti-HA antibody, followed by SDS-PAGE and an overlay-blotting assay with the purified recombinant GST or GST–Glis2 protein. Proteins bound to HA-p120 on the membrane were detected by Western blotting with anti-GST antibody. (E) Interaction between Glis2 and p120 truncation mutants by immunoprecipitation. FLAG-Glis2 was expressed with HA-p120 (full length), p120-N terminus (amino acids 1–647: designated as N), or p120-C terminus (amino acids 385–910: designated as C) in HEK293 cells. Cell lysates were immunoprecipitated with anti-HA antibody, followed by Western blotting with anti-FLAG and anti-HA antibodies. (F) Interaction between endogenous Glis2 and p120 by immunoprecipitation. HEK293 cell lysates were immunoprecipitated with control mouse IgG or anti-p120 antibody, and the precipitates were examined by Western blotting with anti-p120 and anti-Glis2 antibodies.
Because Glis2 is expressed in neurons, we also investigated its binding to δ-catenin, a p120-family member expressed at high levels in neurons. However, we did not see an interaction between Glis2 and δ-catenin (Supplemental Figure S1A). In addition, Gli1, one of Gli family members, was not coimmunoprecipitated with p120 (data not shown). Collectively, these data indicate the specificity of the interaction between p120 and Glis2.
In the process of coexpression of Glis2 and p120, we realized that expression of p120 induces the cleavage of Glis2 protein. When N-terminally FLAG-tagged full-length Glis2 was coexpressed with HA-p120 in HEK293 cells, cleavage of the Glis2 protein was observed, resulting in a product of ∼40 kDa lacking the C terminus, which we designated Glis2 ΔC (Figure 2A, left). The C terminus of p120 (amino acids 385–910) that can bind to Glis2 induced the cleavage of Glis2, whereas the N terminus of p120 (amino acids 1–647) did not (Supplemental Figure S1B), suggesting that the interaction between p120 and Glis2 is required for the cleavage of Glis2. A similar cleavage was also observed in COS-1 cells (Figure 2A, middle), indicating that this cleavage is not cell type specific. We then analyzed endogenous Glis2 protein with anti-Glis2 antibody in various cell lines. A 38- to 40-kDa product was detected in COS-1 cells (Figure 2A, right) and MDA-MB-231 cells (data not shown), whereas it was not detected in other cell lines, including HEK293, HeLa, and MCF-7.
p120 was originally identified as a Src substrate, but the functional significance of the tyrosine phosphorylation of p120 remains unclear (Alema and Salvatore, 2007). Thus, we examined the effect of overexpression of Src on the cleavage of Glis2. Expression of Src alone induced a small amount of Glis2 cleavage, but coexpression of Src and p120 induced tyrosine phosphorylation of p120 (Supplemental Figure S1C) and strongly enhanced the p120-induced cleavage of Glis2 (Figure 2B). The amount of p120-induced cleavage of Glis2 was variable under different experimental conditions; however, coexpression of p120 and Src always exhibited substantial Glis2 cleavage. To address whether the cleaved form of Glis2 (Glis2 ΔC) can still bind to p120, we performed immunoprecipitation using HEK293 cells expressing HA-p120, FLAG-Glis2 and Src. We found that both Glis2 and Glis2 ΔC were coimmunoprecipitated with p120 and that coexpression of Src enhanced the interaction between Glis2 and p120 (Figure 2C). In COS-1 cells, the Glis2 antibody detects a band at ∼40 kDa that has a similar molecular size as Glis2 ΔC (Figure 2A). To investigate whether this endogenous 40-kDa product is affected by p120, we transiently transfected two different siRNA oligos of p120 in COS-1 cells to knock down p120 expression (Figure 2D). RNAi treatment knocked down endogenous p120 protein by 90% and induced a marked reduction in the level of both endogenous full-length Glis2 and the 40-kDa product (Figure 2D). However, the level of the 40-kDa product was reduced to a greater extent than that of the full-length protein. These data suggest that p120 regulates the expression level of the Glis2 protein and that it is also involved in the processing of Glis2.
Characterization of p120-induced Cleavage of Glis2
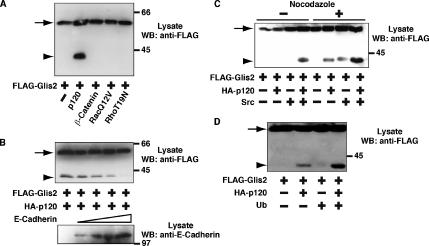
To assess whether the induction of cleavage of Glis2 is specific to p120, we coexpressed FLAG-Glis2 with β-catenin, which is structurally related to p120, but no cleavage of the Glis2 protein was observed (Figure 3A). p120 is known to up-regulate Rac and to down-regulate Rho activity (Anastasiadis et al., 2000; Noren et al., 2000). To mimic these downstream effects of p120, we coexpressed FLAG-Glis2 with constitutively active RacQ12V or inactive RhoT19N, but we did not observe cleavage of Glis2 under these conditions (Figure 3A). These data together with the coimmunoprecipitation result suggest that it is the direct interaction between Glis2 and p120 that induces Glis2 cleavage.
Figure 3.
Characterization of p120-induced cleavage of Glis2. (A) Specificity of the cleavage of Glis2. FLAG-Glis2 was expressed in HEK293 cells with coexpression of p120, β-catenin, constitutively active RacQ12V, or inactive RhoT19N, and the cleavage of Glis2 was examined by Western blotting with anti-FLAG antibody. (B) Effect of overexpression of E-cadherin on Glis2 cleavage. FLAG-Glis2 and HA-p120 were expressed in conjunction with increasing amounts of E-cadherin, and lysates were analyzed by Western blotting with anti-FLAG and anti-E-cadherin antibodies. One, 2, 4, and 8 μg of pcDNA-HA-E-cadherin was used for transfection. (C) Effect of nocodazole on Glis2 cleavage. FLAG-Glis2, HA-p120, and Src were expressed in HEK293 cells, followed by incubation with 5 μg/ml nocodazole for 4 h. Lysates were examined by Western blotting using anti-FLAG antibody. (D) Effect of ubiquitin on Glis2 cleavage. FLAG-Glis2, HA-p120, and HA-ubiquitin were expressed in HEK293 cells. Lysates were examined by Western blotting using anti-FLAG antibody. The arrow and arrowhead indicate the positions of full-length Glis2 and its cleavage product Glis2 ΔC, respectively.
p120 is bound to cadherin at cell–cell contact sites, and it can also interact with microtubules (Chen et al., 2003; Franz and Ridley, 2004; Yanagisawa et al., 2004). It has been shown that for p120 to localize in the nucleus, it must be free from sequestration by either cadherins or microtubules (Roczniak- Ferguson and Reynolds, 2003; Kelly et al., 2004). To investigate whether p120 sequestration affects the p120–Glis2 interaction, we examined the effect of coexpression of E-cadherin on the p120-induced cleavage of Glis2. As the expression level of E-cadherin increased, the cleavage of Glis2 decreased (Figure 3B), indicating that p120 complexed with E-cadherin is not able to induce cleavage of Glis2. We next examined the effect of nocodazole on Glis2 cleavage, which depolymerizes microtubules. In the presence of nocodazole, p120-induced cleavage of Glis2 was enhanced. Even in the absence of exogenous p120, nocodazole treatment together with Src induced the cleavage of Glis2 (Figure 3C). These data suggest that p120 must be free in the cytosol to interact with Glis2 and induce its cleavage.
The cleavage of Glis2 is reminiscent of the C-terminal cleavage of Gli-family transcription factors. It has been shown that the cleavage of Gli involves the ubiquitination system (Jiang and Struhl, 1998). To address the contribution of ubiquitination on Glis2 cleavage, we overexpressed ubiquitin with FLAG-Glis2 and HA-p120 in HEK293 cells. On overexpression of ubiquitin, p120-induced cleavage of Glis2 was enhanced (Figure 3D). This suggests that the mechanism of cleavage of Glis2 and Gli family members may be partly conserved.
Glis2 Directs Translocation of p120 into the Nucleus
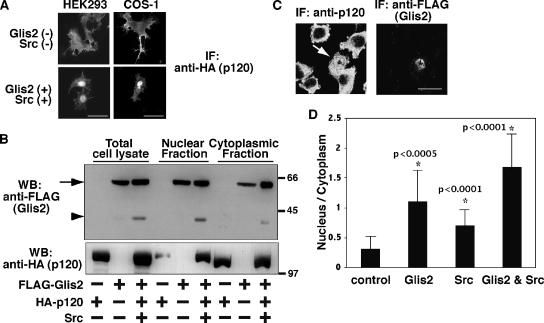
When p120 is overexpressed in HEK293 cells, it localizes to the plasma membrane, with diffuse cytoplasmic staining (Figure 4A, top left). However, when Glis2 and Src were coexpressed, we observed a redistribution of p120 into the nucleus (Figure 4A, bottom left). A similar effect was also observed in COS-1 cells (Figure 4A, right). When Glis2 or Src alone was expressed, the nuclear localization of p120 was not consistently observed (data not shown). We then looked at the nuclear localization of Glis2 by nuclear fractionation. Both Glis2 and Glis2 ΔC were able to translocate to the nucleus, although they were also found in the cytoplasm, even in the presence of p120 and Src (Figure 4B, top). Consistent with the immunostaining data, the nuclear localization of p120 was enhanced by coexpression of Glis2 and Src (Figure 4B, bottom). These data suggest that not only full-length Glis2 but also the cleavage product of Glis2 may have a function in the nucleus.
Figure 4.
Nuclear translocation of p120 by Glis2 and Src. (A) Immunofluorescence staining of exogenous p120. HA-p120 was transiently transfected in HEK293 and COS-1 cells with or without coexpression of FLAG-Glis2 and Src. The localization of p120 was examined by epifluorescent microscopy by using anti-HA antibody. Bars, 20 μm. (B) Both Glis2 and Glis2 ΔC localize in the nucleus. FLAG-Glis2 was coexpressed with HA-p120 and Src, and nuclear and cytoplasmic fractions were isolated and examined by Western blotting using anti-FLAG and anti-HA antibodies. The arrow and arrowhead indicate the positions of full-length Glis2 and its cleavage product Glis2 ΔC, respectively. (C) Immunofluorescence staining of endogenous p120. FLAG-Glis2 was transiently transfected in L fibroblast cells. The localization of endogenous p120 and exogenous Glis2 was examined by confocal microscopy by using anti-p120 and anti-FLAG antibodies, respectively. The arrow indicates the Glis2-positive cell, which contains nuclear p120. Bar, 20 μm. (D) Quantification of the nuclear translocation of endogenous p120. FLAG-Glis2 and/or Src were transiently transfected in L fibroblast cells. The empty pEGFP vector was cotransfected when FLAG-Glis2 was not transfected to identify transfected cells. The location of the nucleus was confirmed by Hoechst staining. Immunofluorescent staining intensity with anti-p120 antibody was measured in a defined region (diameter 1.5 μm) in the nucleus or cytosol, and the ratio of nucleus/cytoplasm staining intensity was analyzed in 15 cells per transfection. Asterisk (*) indicates significantly higher than control cells (p value is shown beside the asterisk).
To investigate whether the localization of endogenous p120 protein is also affected by Glis2 and Src, we used murine L fibroblast cells where no classical cadherins are expressed and p120 is predominantly localized in the cytosol (Thoreson et al., 2000). Overexpression of Glis2 significantly induced nuclear translocation of endogenous p120 (Figure 4, C and D). Expression of Src also showed a similar but weaker effect (Figure 4D). Coexpression of Glis2 and Src further augmented the nuclear translocation of endogenous p120 (Figure 4D) (p < 0.05; Glis2 vs. Glis2 and Src). Together, these data indicate that Glis2 promotes nuclear translocation of p120 and that Src enhances its effect.
Identification of the Cleavage Site of Glis2
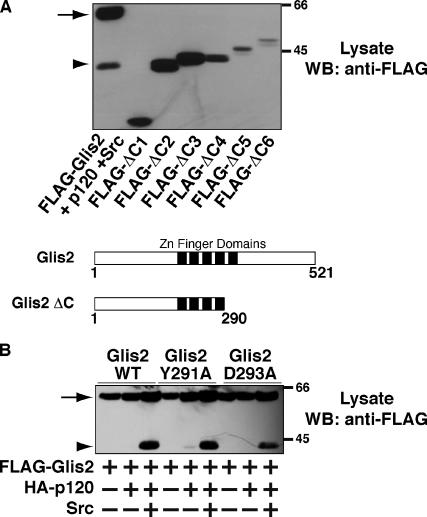
To analyze the role of the cleaved form of Glis2, we attempted to determine the C-terminal amino acid sequence of Glis2 ΔC by mass spectrometry. However, our attempts were unsuccessful, due to the low expression level of Glis2 and low detergent solubility of the cleaved form. As an alternative, we constructed a series of FLAG-tagged C-terminal truncation mutants of Glis2 by PCR that we named Glis2 ΔC1-6. For details of the mutants, see Materials and Methods. We expressed these truncation mutants in HEK293 cells, and the size of the mutants was compared with that of FLAG-Glis2 ΔC by Western blotting. As shown in Figure 5A, FLAG-Glis2 ΔC exhibited the same mobility in SDS-PAGE as FLAG-Glis2 ΔC2 (amino acids 1–290). The C terminus of Glis2 ΔC2 terminates at the second histidine residue of the fourth zinc finger domain. Glis2 ΔC therefore loses the last zinc finger domain of the full-length protein (Figure 5A, bottom schematic).
Figure 5.
Characterization of Glis2 ΔC. (A) Determination of the cleavage site of Glis2. A series of FLAG-tagged truncation mutants of Glis2 were constructed as described in Materials and Methods and expressed in HEK293 cells. Lysates were examined by Western blotting by using anti-FLAG antibody. The cleavage site is directly after the fourth zinc finger domain as shown in the diagram of Glis2 and Glis2 ΔC. (B) Effect of mutations between the fourth and fifth zinc finger domains on Glis2 cleavage. Lysates of HEK293 cells expressing either FLAG-Glis2, -Glis2 Y291A, or -Glis2 D293A, with HA-p120 and Src were examined by Western blotting by using anti-FLAG antibody. The band intensities were analyzed by densitometry. The arrow and arrowhead indicate the positions of full-length Glis2 and its cleavage product Glis2 ΔC, respectively.
Next, we studied the linker region between the fourth and fifth zinc finger domains of Glis2 to find candidate amino acids that may play a role in the cleavage of Glis2. Tyrosine(Y)291 and aspartic acid(D)293 are close to the putative cleavage site. Because it is possible that tyrosine phosphorylation or steric interaction of the charged side chain could affect the interaction of the proteolysis machinery with Glis2, we mutated each of these residues to alanine to examine the effect on p120-induced cleavage of Glis2. The Glis2 Y291A mutation did not affect the cleavage of Glis2, whereas the Glis2 D293A mutation exhibited a 30% reduction in cleavage induced by p120 and Src compared with wild-type Glis2 (Figure 5B). These data suggest that the sequence identity of the linker region between the fourth and fifth zinc finger domains is indeed important for the proteolytic cleavage.
Both Glis2 and Glis2 ΔC Have DNA Binding Ability
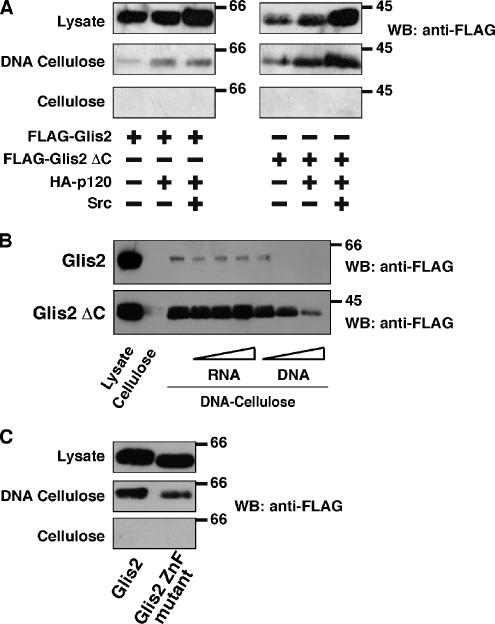
To examine whether Glis2 and Glis2 ΔC are both competent to bind DNA, we carried out DNA-cellulose binding experiments, by using sonicated calf thymus DNA conjugated to cellulose. This assay is widely used to examine the DNA binding ability of nuclear proteins when the DNA binding sites are not identified (Ritt et al., 1998; Surmacz et al., 2002; Johnson et al., 2004). Both Glis2 and Glis2 ΔC showed DNA binding ability in this assay, and they did not interact with cellulose alone (Figure 6A). Coexpression of p120 and Src did not significantly affect binding of either Glis2 or Glis2 ΔC to DNA (Figure 6A). This is consistent with the fact that the p120 binding region of Glis2 is outside the zinc finger domains. To confirm the specificity of the interaction with DNA, cell lysates were preincubated with increasing amounts of DNA or RNA. As expected, RNA was unable to affect binding of Glis2 or Glis2 ΔC with DNA-cellulose, whereas preincubation with DNA successfully inhibited the binding of Glis2 and Glis2 ΔC to the DNA-cellulose (Figure 6B). This indicates a specific interaction between DNA and Glis2 or Glis2 ΔC.
Figure 6.
In vitro DNA binding of Glis2 and Glis2 ΔC. (A) Both Glis2 and Glis2 ΔC can interact with DNA. Lysates of HEK293 cells expressing either FLAG-Glis2 or -Glis2 ΔC with HA-p120 and Src were incubated with DNA-cellulose or cellulose beads, followed by extensive washing. The amount of bound Glis2 was examined by Western blotting by using anti-FLAG antibody. (B) DNA, but not RNA, inhibits the interaction of Glis2 and Glis2 ΔC with DNA-cellulose beads. Lysates of HEK293 cells expressing FLAG-Glis2 or -Glis2 ΔC were preincubated for 30 min with 1, 5, or 20 μg of DNA or RNA, followed by incubation with DNA-cellulose beads. The amount of bound Glis2 was examined by Western blotting by using anti-FLAG antibody. (C) A zinc finger mutant of Glis2 shows a weaker interaction with DNA. Lysates of HEK293 cells expressing FLAG-Glis2 or -Glis2 ZnF mutant were incubated with DNA-cellulose or cellulose beads, and the amount of bound Glis2 was examined by Western blotting by using anti-FLAG antibody. The band intensities were analyzed by densitometry.
To exclude the possibility that the interaction of Glis2 ΔC with DNA is through heterodimer formation with endogenous Glis2, we examined the dimer formation between Glis2 proteins. HA-Glis2 was coexpressed with FLAG-Glis2 or FLAG-Glis2 ΔC in HEK293 cells, and coimmunoprecipitations were examined. However, we detected neither homodimers of Glis2 nor heterodimers of Glis2 and Glis2 ΔC (data not shown). This suggests that dimer formation is not involved in the interaction between Glis2 ΔC and DNA.
In addition, we created a zinc finger mutant of Glis2, by replacing the first cysteine residue of the first Cys2-His2 zinc finger domain with alanine. The Glis2 ZnF mutant exhibits a shift in mobility in SDS-PAGE, probably due to disruption of folding of the first zinc finger domain. This mutation induced a 50% reduction in the interaction with DNA-cellulose (Figure 6C). These data indicate again the specific interaction between Glis2 and DNA and involvement of the zinc finger domain in the interaction.
Glis2 Affects Neuronal Differentiation in Chick Neural Tube
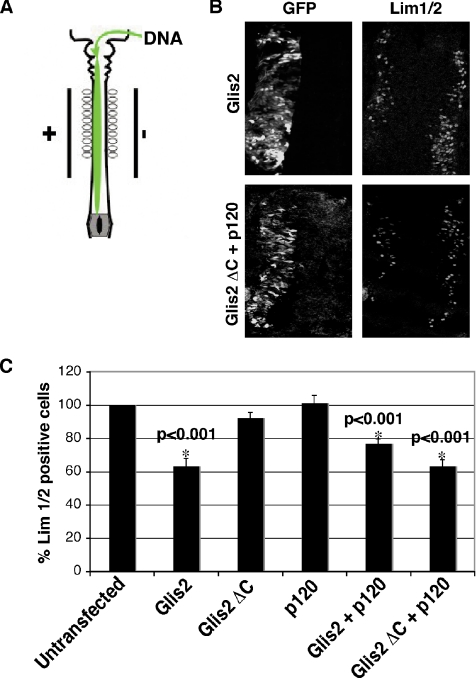
Glis2 is expressed in the neural tube in mouse, chick, and Xenopus embryos (Lamar et al., 2001; Zhang et al., 2002). To investigate the function of Glis2 and Glis2 ΔC, we used in ovo electroporation of the mouse cDNAs in chick neural tube (Figure 7A) (Itasaki et al., 1999). Because only one side of the neural tube is transfected using this technique, the untransfected side can be used as a wild-type control. A previous report showed that expression of Xenopus Glis2 in chick neural tube affected neuronal differentiation (Lamar et al., 2001). We therefore tested the effect of Glis2 and p120 on neuronal differentiation. By using anti-Lim1/2 antibody, a marker of postmitotic neurons, we observed a decrease in the number of Lim1/2-positive cells in the Glis2-expressing side (Figure 7B, top), indicating an inhibition of neuronal differentiation. This effect was also seen when Glis2 ΔC was coexpressed with p120 (Figure 7B, bottom), although expression of Glis2 ΔC alone had no significant effect (Figure 7C). This indicates that the cleaved form of Glis2 is still capable of affecting transcriptional events despite the loss of one zinc finger domain but that it requires p120 for this function. When p120 was coexpressed with full-length Glis2, there was no significant difference in the number of Lim1/2-positive cells compared with neural tubes expressing Glis2 alone (Figure 7C). The decrease in the number of Lim1/2-positive cells in Glis2-, Glis2/p120-, and Glis2 ΔC/p120-expressing neurons was statistically significant compared with the wild-type control (Figure 7C). p120 alone had no effect on the number of Lim1/2-positive cells (Figure 7C). Coexpression with Glis2 ΔC of the N terminus of p120 (amino acids 1–647) that is unable to bind Glis2 did not affect the number of Lim1/2-positive cells (Supplemental Figure S2A).
Figure 7.
Effect of expression of Glis2 and Glis2 ΔC on neuronal differentiation in chick neural tube. (A) Diagram summarizing in ovo electroporation. (B) Staining of Lim1/2, a marker of neuronal differentiation. HH stage 10–12 embryos were electroporated with Glis2, or Glis2 ΔC and p120, and incubated until HH stage 22. Transfected cells were identified by coexpression of GFP. Expression of GFP and Lim1/2 were examined by confocal microscopy using anti-GFP and anti-Lim1/2 antibodies, respectively. (C) Quantification of Lim1/2 staining. Glis2, Glis2 ΔC, p120, Glis2 and p120, or Glis2 ΔC and p120 was transfected in neural tubes. The number of cells positive for Lim1/2 in untransfected and transfected neural tubes was counted from 20 sections of at least six embryos per experiment. Results are expressed as a percentage relative to untransfected cells. Asterisk (*) indicates significantly lower than untransfected cells.
Gli proteins are the downstream targets of Shh signaling. Overexpression of Gli proteins is known to affect dorsoventral patterning of neuronal subtypes in the spinal cord in developing embryos (Briscoe and Ericson, 2001; Persson et al., 2002; Stamataki et al., 2005). Because Glis2 is structurally related to the Gli family of transcription factors and has been shown to interact in vitro with the Gli binding site on DNA (Lamar et al., 2001), we also examined the effect of transfection of Glis2 and Glis2 ΔC on markers of dorsoventral patterning in the neural tube. We examined transfected neural tubes with antibodies to transcription factors Pax6 and Nkx2.2, markers for different neuronal subtypes in the neural tube (Briscoe and Ericson, 2001). However, we did not observe any significant differences in the expression patterns of these markers in neural tubes expressing Glis2 or Glis2 ΔC (Supplemental Figure S2B), whereas expression of Gli1 and Gli3 had a profound effect on the markers' expression pattern (Persson et al., 2002). Therefore, despite the structural similarity to Gli, Glis2 is not able to affect Shh signaling.
DISCUSSION
In this study, we show a novel interaction between p120 and Glis2. Glis2 is the second identified nuclear binding protein of p120, after Kaiso (Daniel and Reynolds, 1999). Both Kaiso and Glis2 are zinc finger-containing transcription factors with repressor activity (Stone et al., 1999; Prokhortchouk et al., 2001). For p120 to interact with either of its nuclear binding partners, it must be free from sequestration by E-cadherin (Figure 3B) (Roczniak-Ferguson and Reynolds, 2003; Kelly et al., 2004). However, there are some clear differences in the modes of interaction of these proteins with p120. First, expression of Glis2 enhances nuclear translocation of p120, which has not been observed for Kaiso. Second, p120 induces cleavage of Glis2 that may regulate Glis2 function. Third, expression of p120 does not affect the interaction between Glis2 and DNA, whereas p120 prevents the binding of Kaiso to DNA (Kelly et al., 2004; Kim et al., 2004; Park et al., 2005).
It has been reported that nuclear localization signal or nuclear export signal sequences of p120 regulate its nucleocytoplasmic shuttling (van Hengel et al., 1999; Roczniak-Ferguson and Reynolds, 2003; Kelly et al., 2004). We found that coexpression of Glis2 and Src also enhances nuclear localization of p120. To assess the importance of Glis2 in this translocation of p120, we tried to knock down expression of Glis2 by using RNAi. Despite several attempts, we were not able to efficiently reduce Glis2 protein levels. Thus, whether Glis2 is essential to recruit p120 into the nucleus remains to be pursued.
Glis2 is homologous to the Gli family of transcription factors. Functional activity of Gli is regulated by C-terminal cleavage. In an analogous manner, Glis2 exhibits C-terminal cleavage that is induced by p120 and Src coexpression. The full-length and cleaved forms of both Glis2 and Gli localize to the nucleus and bind to DNA. In addition, the ubiquitination machinery seems to be involved in cleavage of both Glis2 and Gli family members (Figure 3D) (Jiang and Struhl, 1998). However, the mode of cleavage differs between the two systems. Cleavage occurs between the fourth and fifth zinc finger domains in Glis2, whereas for Gli the cleavage site is outside the zinc finger tandem region. Furthermore, Glis2 cleavage is enhanced by tyrosine kinase Src, whereas serine/threonine kinases, such as glycogen synthase kinase 3β, protein kinase A, and casein kinase 1, are involved in the cleavage of Gli (Ingham and McMahon, 2001; Price and Kalderon, 2002).
According to p120 siRNA data (Figure 2D), p120 is involved in regulation of the expression level of Glis2. It may be due to stabilization of the Glis2 protein via direct interaction with p120. However, it is also possible that the regulation occurs at transcriptional level. Whether p120 induces stabilization or cleavage of Glis2 may depend on the level of other proteins involved in these processes.
We have studied the function of the full-length and cleaved form of Glis2 in two different systems. In the DNA-cellulose binding assay, both full-length and cleaved Glis2 bound to DNA. In chick neural tube, the full-length protein suppressed neuronal differentiation, whereas the cleaved form of Glis2 was only observed to have this effect with coexpression of p120. Thus, despite the loss of one zinc finger domain, the cleaved form is able to bind DNA and affect transcription. We have also shown that the uncoupling of p120 from microtubules and the E-cadherin complex is involved in the cleavage of Glis2. The release of p120 from the E-cadherin complex is also required for binding to Kaiso. The physiological stimuli by which p120 becomes free to interact with these two transcription factors have not yet been clarified. We also examined the fate of the C-terminal cleavage product of Glis2, by making a C-terminally tagged construct. However, preliminary results show the C-terminal product to be highly unstable, suggesting prompt protein degradation after initial cleavage of Glis2 (data not shown). Indeed, the functional significance of the C-terminal fragment after processing of Gli proteins has not been reported.
Because Glis2 has been reported to interact with the Gli binding site (Lamar et al., 2001), we also examined whether Glis2 affects Shh signaling in the chick neural tube. Overexpression of Gli proteins in the neural tube affects dorsal–ventral differentiation patterning (Briscoe and Ericson, 2001; Stamataki et al., 2005). In contrast, no significant effect on patterning was observed when Glis2 was overexpressed (Supplemental Figure S2B). Thus, involvement of Glis2 in Shh signaling has not yet been observed; however, the possibility cannot be ruled out that together with unidentified molecules, Glis2 may play some role in this pathway. Our data indicate that Glis2 negatively regulates neuronal differentiation in the chick neural tube system. Another group has shown that overexpression of Xenopus Glis2 that exhibits 67% homology to mouse Glis2 can promote premature neuronal differentiation in chick neural tube (Lamar et al., 2001). At present, we do not understand this apparent discrepancy between these phenotypes; thus, to fully understand the physiological role of Glis2 in neurogenesis, further genetic examination such as production of a knockout mouse will be required.
Here, we report a novel nuclear partner of p120, and we propose a further functional role of p120 in the nucleus. There are several important questions arising from this study. What are the target DNA sequences of full-length and cleaved Glis2? Are there additional components of the DNA binding complex of full-length and cleaved Glis2? What is the physiological significance of nuclear translocation of p120 by Glis2 and Src? Future studies will lead us to further understand the nuclear function of p120. And finally, how the multiple functions of p120 at cell–cell contacts, in cytoskeletal rearrangements and in the nucleus are regulated and what stimuli coordinate these functions are the long-standing questions to be answered.
Supplementary Material
ACKNOWLEDGMENTS
We thank Julie Pitcher and Dan Cutler (Medical Research Council, Laboratory for Molecular Cell Biology, London) for critical reading of the manuscript. Norberto Serpente (Medical Research Council, Laboratory for Molecular Cell Biology, London) and Anita Mynett (Medical Research Council, National Institute for Medical Research, London) are acknowledged for technical help. We also thank Julie Pitcher and Laura Johnson (Medical Research Council, Laboratory for Molecular Cell Biology, London) for providing technical advice on the DNA-cellulose binding assay, Shun-ichi Nakamura and Taro Okada for pCMV5-FLAG-δ-catenin, and Alan Hall for pRK5-myc-Rho T19N.
Abbreviations used:
- p120
p120 catenin
- Glis2 ΔC
C-terminally cleaved Glis2
- Shh
Sonic Hedgehog
- ZnF
zinc finger.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-10-0941) on March 7, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Alema S., Salvatore A. M. p120 catenin and phosphorylation: Mechanisms and traits of an unresolved issue. Biochim. Biophys. Acta. 2007;1773:47–58. doi: 10.1016/j.bbamcr.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Anastasiadis P. Z., Moon S. Y., Thoreson M. A., Mariner D. J., Crawford H. C., Zheng Y., Reynolds A. B. Inhibition of RhoA by p120 catenin. Nat. Cell Biol. 2000;2:637–644. doi: 10.1038/35023588. [DOI] [PubMed] [Google Scholar]
- Aono S., Nakagawa S., Reynolds A. B., Takeichi M. p120(ctn) acts as an inhibitory regulator of cadherin function in colon carcinoma cells. J. Cell Biol. 1999;145:551–562. doi: 10.1083/jcb.145.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brembeck F. H., Rosario M., Birchmeier W. Balancing cell adhesion and Wnt signaling, the key role of beta-catenin. Curr. Opin. Genet Dev. 2006;16:51–59. doi: 10.1016/j.gde.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Briscoe J., Ericson J. Specification of neuronal fates in the ventral neural tube. Curr. Opin. Neurobiol. 2001;11:43–49. doi: 10.1016/s0959-4388(00)00172-0. [DOI] [PubMed] [Google Scholar]
- Chen X., Kojima S., Borisy G. G., Green K. J. p120 catenin associates with kinesin and facilitates the transport of cadherin-catenin complexes to intercellular junctions. J. Cell Biol. 2003;163:547–557. doi: 10.1083/jcb.200305137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J. M., Reynolds A. B. The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol. Cell. Biol. 1999;19:3614–3623. doi: 10.1128/mcb.19.5.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J. M., Spring C. M., Crawford H. C., Reynolds A. B., Baig A. The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res. 2002;30:2911–2919. doi: 10.1093/nar/gkf398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. A., Ireton R. C., Reynolds A. B. A core function for p120-catenin in cadherin turnover. J. Cell Biol. 2003;163:525–534. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia L. P., Yamamoto M., Zang K., Reichardt L. F. p120 catenin regulates dendritic spine and synapse development through Rho-family GTPases and cadherins. Neuron. 2006;51:43–56. doi: 10.1016/j.neuron.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C. M., Ridley A. J. p120 catenin associates with microtubules: inverse relationship between microtubule binding and Rho GTPase regulation. J. Biol. Chem. 2004;279:6588–6594. doi: 10.1074/jbc.M312812200. [DOI] [PubMed] [Google Scholar]
- Fujita Y., Krause G., Scheffner M., Zechner D., Leddy H. E., Behrens J., Sommer T., Birchmeier W. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat. Cell Biol. 2002;4:222–231. doi: 10.1038/ncb758. [DOI] [PubMed] [Google Scholar]
- Fujita Y., et al. Tomosyn: a syntaxin-1-binding protein that forms a novel complex in the neurotransmitter release process. Neuron. 1998;20:905–915. doi: 10.1016/s0896-6273(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Grosheva I., Shtutman M., Elbaum M., Bershadsky A. D. p120 catenin affects cell motility via modulation of activity of Rho-family GTPases: a link between cell-cell contact formation and regulation of cell locomotion. J. Cell Sci. 2001;114:695–707. doi: 10.1242/jcs.114.4.695. [DOI] [PubMed] [Google Scholar]
- Gumbiner B. M. Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell Biol. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- Hecht A., Vleminckx K., Stemmler M. P., van Roy F., Kemler R. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 2000;19:1839–1850. doi: 10.1093/emboj/19.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan C., Serpente N., Cogram P., Hosking C. R., Bialucha C. U., Feller S. M., Braga V. M., Birchmeier W., Fujita Y. Rap1 regulates the formation of E-cadherin-based cell-cell contacts. Mol. Cell. Biol. 2004;24:6690–6700. doi: 10.1128/MCB.24.15.6690-6700.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D., Anderson K. V. Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development. 2006;133:3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- Ingham P. W. Transducing Hedgehog: the story so far. EMBO J. 1998;17:3505–3511. doi: 10.1093/emboj/17.13.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham P. W., McMahon A. P. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- Ireton R. C., et al. A novel role for p120 catenin in E-cadherin function. J. Cell Biol. 2002;159:465–476. doi: 10.1083/jcb.200205115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itasaki N., Bel-Vialar S., Krumlauf R. ‘Shocking’ developments in chick embryology: electroporation and in ovo gene expression. Nat. Cell Biol. 1999;1:E203–E207. doi: 10.1038/70231. [DOI] [PubMed] [Google Scholar]
- Jamora C., Fuchs E. Intercellular adhesion, signalling and the cytoskeleton. Nat. Cell Biol. 2002;4:E101–E108. doi: 10.1038/ncb0402-e101. [DOI] [PubMed] [Google Scholar]
- Jiang J., Struhl G. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature. 1998;391:493–496. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- Johnson L. R., Scott M. G., Pitcher J. A. G protein-coupled receptor kinase 5 contains a DNA-binding nuclear localization sequence. Mol. Cell. Biol. 2004;24:10169–10179. doi: 10.1128/MCB.24.23.10169-10179.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly K. F., Spring C. M., Otchere A. A., Daniel J. M. NLS-dependent nuclear localization of p120ctn is necessary to relieve Kaiso-mediated transcriptional repression. J. Cell Sci. 2004;117:2675–2686. doi: 10.1242/jcs.01101. [DOI] [PubMed] [Google Scholar]
- Kim S. C., Kim Y. S., Jetten A. M. Kruppel-like zinc finger protein Gli-similar 2 (Glis2) represses transcription through interaction with C-terminal binding protein 1 (CtBP1) Nucleic Acids Res. 2005;33:6805–6815. doi: 10.1093/nar/gki985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. W., Park J. I., Spring C. M., Sater A. K., Ji H., Otchere A. A., Daniel J. M., McCrea P. D. Non-canonical Wnt signals are modulated by the Kaiso transcriptional repressor and p120-catenin. Nat. Cell Biol. 2004;6:1212–1220. doi: 10.1038/ncb1191. [DOI] [PubMed] [Google Scholar]
- Lamar E., Kintner C., Goulding M. Identification of NKL, a novel Gli-Kruppel zinc-finger protein that promotes neuronal differentiation. Development. 2001;128:1335–1346. doi: 10.1242/dev.128.8.1335. [DOI] [PubMed] [Google Scholar]
- McCrea P. D., Turck C. W., Gumbiner B. A homolog of the armadillo protein in Drosophila (plakoglobin) associated with E-cadherin. Science. 1991;254:1359–1361. doi: 10.1126/science.1962194. [DOI] [PubMed] [Google Scholar]
- Noren N. K., Liu B. P., Burridge K., Kreft B. p120 catenin regulates the actin cytoskeleton via Rho family GTPases. J. Cell Biol. 2000;150:567–580. doi: 10.1083/jcb.150.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. I., et al. Kaiso/p120-catenin and TCF/beta-catenin complexes coordinately regulate canonical Wnt gene targets. Dev. Cell. 2005;8:843–854. doi: 10.1016/j.devcel.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Persson M., Stamataki D., te Welscher P., Andersson E., Bose J., Ruther U., Ericson J., Briscoe J. Dorsal-ventral patterning of the spinal cord requires Gli3 transcriptional repressor activity. Genes Dev. 2002;16:2865–2878. doi: 10.1101/gad.243402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. A., Kalderon D. Proteolysis of the Hedgehog signaling effector cubitus interruptus requires phosphorylation by glycogen synthase kinase 3 and casein kinase 1. Cell. 2002;108:823–835. doi: 10.1016/s0092-8674(02)00664-5. [DOI] [PubMed] [Google Scholar]
- Prokhortchouk A., Hendrich B., Jorgensen H., Ruzov A., Wilm M., Georgiev G., Bird A., Prokhortchouk E. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 2001;15:1613–1618. doi: 10.1101/gad.198501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A. B., Daniel J., McCrea P. D., Wheelock M. J., Wu J., Zhang Z. Identification of a new catenin: the tyrosine kinase substrate p120cas associates with E-cadherin complexes. Mol. Cell. Biol. 1994;14:8333–8342. doi: 10.1128/mcb.14.12.8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A. B., Herbert L., Cleveland J. L., Berg S. T., Gaut J. R. p120, a novel substrate of protein tyrosine kinase receptors and of p60v-src, is related to cadherin-binding factors beta-catenin, plakoglobin and armadillo. Oncogene. 1992;7:2439–2445. [PubMed] [Google Scholar]
- Ritt C., Grimm R., Fernandez S., Alonso J. C., Grasser K. D. Basic and acidic regions flanking the HMG domain of maize HMGa modulate the interactions with DNA and the self-association of the protein. Biochemistry. 1998;37:2673–2681. doi: 10.1021/bi972620r. [DOI] [PubMed] [Google Scholar]
- Roczniak-Ferguson A., Reynolds A. B. Regulation of p120-catenin nucleocytoplasmic shuttling activity. J. Cell Sci. 2003;116:4201–4212. doi: 10.1242/jcs.00724. [DOI] [PubMed] [Google Scholar]
- Schaeper U., Gehring N. H., Fuchs K. P., Sachs M., Kempkes B., Birchmeier W. Coupling of Gab1 to c-Met, Grb2, and Shp2 mediates biological responses. J. Cell Biol. 2000;149:1419–1432. doi: 10.1083/jcb.149.7.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamataki D., Ulloa F., Tsoni S. V., Mynett A., Briscoe J. A gradient of Gli activity mediates graded Sonic Hedgehog signaling in the neural tube. Genes Dev. 2005;19:626–641. doi: 10.1101/gad.325905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone D. M., et al. Characterization of the human suppressor of fused, a negative regulator of the zinc-finger transcription factor Gli. J. Cell Sci. 1999;112:4437–4448. doi: 10.1242/jcs.112.23.4437. [DOI] [PubMed] [Google Scholar]
- Surmacz T. A., Bayer E., Rahfeld J. U., Fischer G., Bayer P. The N-terminal basic domain of human parvulin hPar14 is responsible for the entry to the nucleus and high-affinity DNA-binding. J. Mol. Biol. 2002;321:235–247. doi: 10.1016/s0022-2836(02)00615-0. [DOI] [PubMed] [Google Scholar]
- Thoreson M. A., Anastasiadis P. Z., Daniel J. M., Ireton R. C., Wheelock M. J., Johnson K. R., Hummingbird D. K., Reynolds A. B. Selective uncoupling of p120(ctn) from E-cadherin disrupts strong adhesion. J. Cell Biol. 2000;148:189–202. doi: 10.1083/jcb.148.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hengel J., Vanhoenacker P., Staes K., van Roy F. Nuclear localization of the p120(ctn) Armadillo-like catenin is counteracted by a nuclear export signal and by E-cadherin expression. Proc. Natl. Acad. Sci. USA. 1999;96:7980–7985. doi: 10.1073/pnas.96.14.7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelock M. J., Johnson K. R. Cadherin-mediated cellular signaling. Curr. Opin. Cell Biol. 2003;15:509–514. doi: 10.1016/s0955-0674(03)00101-7. [DOI] [PubMed] [Google Scholar]
- Willert K., Jones K. A. Wnt signaling: is the party in the nucleus? Genes Dev. 2006;20:1394–1404. doi: 10.1101/gad.1424006. [DOI] [PubMed] [Google Scholar]
- Xiao K., Allison D. F., Buckley K. M., Kottke M. D., Vincent P. A., Faundez V., Kowalczyk A. P. Cellular levels of p120 catenin function as a set point for cadherin expression levels in microvascular endothelial cells. J. Cell Biol. 2003;163:535–545. doi: 10.1083/jcb.200306001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa M., Kaverina I. N., Wang A., Fujita Y., Reynolds A. B., Anastasiadis P. Z. A novel interaction between kinesin and p120 modulates p120 localization and function. J. Biol. Chem. 2004;279:9512–9521. doi: 10.1074/jbc.M310895200. [DOI] [PubMed] [Google Scholar]
- Yap A. S., Kovacs E. M. Direct cadherin-activated cell signaling: a view from the plasma membrane. J. Cell Biol. 2003;160:11–16. doi: 10.1083/jcb.200208156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Nakanishi G., Kurebayashi S., Yoshino K., Perantoni A., Kim Y. S., Jetten A. M. Characterization of Glis2, a novel gene encoding a Gli-related, Kruppel-like transcription factor with transactivation and repressor functions. Roles in kidney development and neurogenesis. J. Biol. Chem. 2002;277:10139–10149. doi: 10.1074/jbc.M108062200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.