Abstract
We have characterized early steps of α2β1 integrin-mediated cell adhesion to a collagen type I matrix by using single-cell force spectroscopy. In agreement with the role of α2β1 as a collagen type I receptor, α2β1-expressing Chinese hamster ovary (CHO)-A2 cells spread rapidly on the matrix, whereas α2β1-negative CHO wild-type cells adhered poorly. Probing CHO-A2 cell detachment forces over a contact time range of 600 s revealed a nonlinear adhesion response. During the first 60 s, cell adhesion increased slowly, and forces associated with the smallest rupture events were consistent with the breakage of individual integrin–collagen bonds. Above 60 s, a fraction of cells rapidly switched into an activated adhesion state marked by up to 10-fold increased detachment forces. Elevated overall cell adhesion coincided with a rise of the smallest rupture forces above the value required to break a single-integrin–collagen bond, suggesting a change from single to cooperative receptor binding. Transition into the activated adhesion mode and the increase of the smallest rupture forces were both blocked by inhibitors of actomyosin contractility. We therefore propose a two-step mechanism for the establishment of α2β1-mediated adhesion as weak initial, single-integrin–mediated binding events are superseded by strong adhesive interactions involving receptor cooperativity and actomyosin contractility.
INTRODUCTION
Integrins are a family of α/β-heterodimeric receptors involved in cell–cell adhesion and cell attachment to the extracellular matrix (Hynes, 1992). In adherent cells, integrins are often arranged into highly organized structures, such as focal complexes, focal adhesions, and fibrillar adhesions (Geiger et al., 2001). In these mature adhesive contacts, integrins are linked to the actin cytoskeleton via their intracellular domains and a multitude of adapter proteins, such as vinculin, talin, and α-actinin (Zamir and Geiger, 2001). Whereas the molecular composition of integrin complexes and signaling pathways controlling their macroscopic assembly or disassembly have been analyzed in detail, less is known about the early molecular events leading to the formation of integrin-based adhesion.
Generally, integrin-mediated adhesion is thought to be initiated on the cell membrane by the engagement of individual integrin dimers with their respective ligand (Lotz et al., 1989; Gallant and Garcia, 2006). The number of receptor–ligand pairs may then grow by increasing the cell–substrate contact area and by receptor diffusion within that zone. Subsequent adhesion strengthening occurs through integrin clustering and linkage to the cytoskeleton. In addition, the ligand affinity of the integrins may increase as a result of intracellular regulatory pathways (Hughes and Pfaff, 1998). At a later stage, the assembly of higher-order integrin–cytoskeletal complexes requires actomyosin-driven contractility under the control of the small guanosine triphosphatase (GTPase) RhoA (Chrzanowska-Wodnicka and Burridge, 1996) and its downstream target Rho-associated kinase (ROCK) (Amano et al., 1997). However, the precise sequence of these events and their relative contribution to the transition from weak initial binding to strong, mature cell–substrate adhesion is still poorly understood, partly because experimental setup to address these questions has not been available.
Single-cell force spectroscopy (SCFS) is well suited for studying the dynamic formation of cellular adhesion because the duration of the cell–substrate interaction can be controlled accurately even for short contact times. In contrast to bulk adhesion assays that measure the mean behavior of the entire cell population, with SCFS the adhesive properties of different cell subpopulations can be distinguished. Furthermore, the effect of inhibitors or augmenters of adhesion can be monitored directly at the single-molecule level. Atomic force microscope (AFM)-based SCFS offers the possibility to measure molecular forces with piconewton accuracy and has been adapted to measure cell–substrate (Zhang et al., 2002; Puech et al., 2005) and cell–cell adhesion (Benoit et al., 2000; Puech et al., 2006). Although other force spectroscopy techniques, such as the biomembrane force probe or optical and magnetic tweezers, offer equal or superior force resolution, the maximum forces that can be detected or applied are frequently below the detachment forces required to remove well-adhering cells. The ability to measure forces with high resolution over a wide range makes AFM a valuable tool to study cellular adhesion forces across dimensions from the single-molecule level to that of the entire cell (Puech et al., 2006).
Here, we have used SCFS to study early events of integrin-mediated cellular adhesion to a structurally well defined collagen type I matrix. Collagens are a protein family containing >20 members, which are divided into fibril-forming and nonfibrillar groups (Hulmes, 2002). Fibrillar collagens, such as collagen type I, are the main component of the ECM where they help resisting tensile forces and provide the major biomechanical scaffold for cell attachment (Kadler et al., 1996). In vertebrate cells four integrins (α1β1, α2β1, α10β1, and α11β1) are the main adhesion-mediating collagen receptors (White et al., 2004). The α-subunits of these receptors contain an I-domain, which is responsible for collagen binding (Tuckwell et al., 1995). Within the I-domain, there is a metal ion-dependent adhesion site (MIDAS), and coordination of the MIDAS with Mg2+ or Mn2+ is necessary for high-affinity collagen binding (Emsley et al., 2000). Based on the spreading behavior of α2β1-expressing cell lines on collagen type I matrices and binding studies using a recombinant α2I-domain, it has been suggested that α2β1 is the main integrin receptor for fibrillar collagen type I (Jokinen et al., 2004). However, α2β1-mediated cell adhesion to collagen type I has not been characterized quantitatively on a single-molecule level, and the dynamics of α2β1-mediated adhesion formation on the cell scale has not been studied. Using the high-force resolution of the AFM, we were able to observe adhesion events mediated by single α2β1 integrins. Examining the formation of cell adhesion over 10 min, we show that cells adhere to the collagen type I in a two-step process: for contact times <60 s, collagen binding is characterized by single-integrin adhesions events. When substrate contact is sustained for >60 s, overall cell adhesion strongly increases as cells switch to an “activated” adhesion state. In activated cells the smallest discrete rupture events rise above the single-integrin level, suggesting the establishment of cooperative integrin receptor binding. Finally, we show that the establishment of strong overall cell adhesion and the increase of the smallest force units are blocked by inhibitors of actomyosin contractility, pointing to an important role for myosin-driven contractility in both processes.
MATERIALS AND METHODS
Cell Culture
Chinese hamster ovary (CHO) cells were maintained in α-minimal essential medium (MEM) containing 10% fetal calf serum, 100 IU/ml penicillin, and 100 μg/ml streptomycin and passaged every 2–3 d or before reaching confluence CHO-A2 cells were stably transfected with human integrin α2 in pAWneo2 as described previously (Nykvist et al., 2000). Before performing SCFS measurements, cells were transferred to serum-free, CO2-independent medium (Dulbecco's modified Eagle's medium [DMEM] containing 20 mM HEPES, pH 7.4) and incubated for 2 h. Cells were then harvested in Hank's buffered salt solution containing 0.05% trypsin, 0.53 mM EDTA at 37°C for 2–3 min. Subsequently, trypsin was inactivated by adding phosphate-buffered saline (PBS) containing soybean trypsin inhibitor (final concentration, 1.3%), and after pelleting at 200 × g for 5 min, cells were resuspended in serum-free, CO2-independent medium. To test the requirement of Ca2+ or Mg2+ on cell adhesion, DMEM was reconstituted from its individual components with the exception of Ca2+ or Mg2+, respectively. To test the effect of specific inhibitors of cell contractility, cells were preincubated with the ROCK inhibitor Y27632 at 10 μM or the myosin light chain kinase (MLCK) inhibitor BDM at 20 mM for 1 h and maintained in the presence of the inhibitor during force measurements. For light microscopy, cells were trypsinized, pelleted, and resuspended as described above, and 25 × 103 cells in a total volume of 3 ml of medium (α-MEM) were added to the collagen matrix. Phase-contrast images were collected after incubating the cells in a humidified atmosphere containing 5% CO2 at 37°C for 90 min.
Surface Decoration with Collagen
Collagen type I matrices were prepared as described previously (Cisneros et al., 2006). Briefly, mica disks (diameter 4 mm) were mounted on glass coverslips (diameter 24 mm) or tissue culture dishes (diameter 35 mm) by using an optical adhesive (OP-29; Dymax, Torrington, CT). Cleaving the top layer revealed an atomically flat mica surface over which buffer containing 50 mM glycine, pH 9.2, 200 mM KCl, and 0.3 μg/ml collagen type I (Cohesion Technologies, Palo Alto, CA) was flushed. After overnight incubation at room temperature, loosely bound collagen was removed by several washes with PBS. The substrates were maintained hydrated to ensure the integrity of the collagen layer. To verify complete coverage of the mica surface, the collagen matrices were imaged by AFM as described previously (Cisneros et al., 2006).
AFM Setup for SCFS
For SCFS, a NanoWizard AFM (JPK Instruments, Berlin, Germany) mounted on top of an Axiovert 200 inverted microscope (Carl Zeiss, Jena, Germany) was used. The AFM head is equipped with a 15-μm Z-range linearized piezoelectric ceramic scanner and an infrared laser. Tipless silicon nitride cantilevers were V-shaped and 200 μm in length, having a nominal spring constant of 0.06 N/m (NP-0; Veeco Instruments, Woodbury, NY). The sensitivity of the optical lever system was calibrated and the cantilever spring constant determined in situ before every experiment by using the thermal noise method (Butt and Jaschke, 1995). Within the reported uncertainty of this method (∼10%), cantilever spring constants were found to be compatible with the manufacturer's specifications. Long-range force spectroscopy was made possible by the use of the CellHesion module (JPK Instruments), extending the vertical range to 100 μm by piezo-driven movements of the sample holder. A closed loop system controlled speed and position of the vertical piezo elements (Puech et al., 2006). Spectroscopy experiments were performed at 37°C by using a temperature-controlled BioCell (JPK Instruments).
Cantilever Decoration, Cell Capture, Adhesion Experiments, and Data Processing for SCFS
Plasma-cleaned cantilevers were functionalized with concanavalin A (Puech et al., 2005) and stored in PBS. Before cells could be attached to the cantilever, they were separated from each other, and the tissue culture dish, which was achieved by a brief trypsination step. Using a biotinylation protocol for cell surface proteins, we verified that even extended trypsin incubation (30 min) did not cleave and remove the α2 or β1 integrin subunits from the cell surface; therefore, it did not interfere with subsequent measurement of α2β1-dependent adhesion (see Supplemental Figure S1). Cells were pipetted into the AFM sample chamber ∼20–30 min after trypsinization. Single cells were captured by pressing the concanavalin A-decorated cantilever onto the cell with a contact force of 500 pN for ∼3 s. The cell was lifted from the surface and allowed to establish firm adhesion on the cantilever for 5 min. To measure the time dependence of cell adhesion, the cell was lowered onto the collagen matrix until reaching a contact force of 1000 pN. During contact (5–600 s), the piezo position was kept constant using the AFM closed loop feedback mode. The cantilever was retracted at constant speed (2.5 μm/s) over pulling ranges ensuring complete separation of cell and surface (10–70 μm). Cell detachment was indicated by the superimposition of trace and retrace baselines. Moderate pulling speeds were chosen to reduce hydrodynamic drag effects on the cantilever (Janovjak et al., 2005). As monitored by light microscopy, the captured cells always detached from the collagen substrate and never from the cantilever during pulling. Usually, two to five force curves were acquired for each cell for contact times ≤120 s and one or two force curves per cell for longer contact times. After each force measurement cycle, the retracted cell was left to recover for 2–3 min before adhering to a different spot on the collagen surface. Parameters, such as detachment force, detachment work, and the number and magnitude of small force steps were extracted from retrace curves using in-house procedures (Puech et al., 2005). Built-in procedures of KaleidaGraph (Synergy Software, Reading, PA) or InStat (GraphPad Software, San Diego, CA) were used for running Student's t test or analysis of variance–Bonferroni tests.
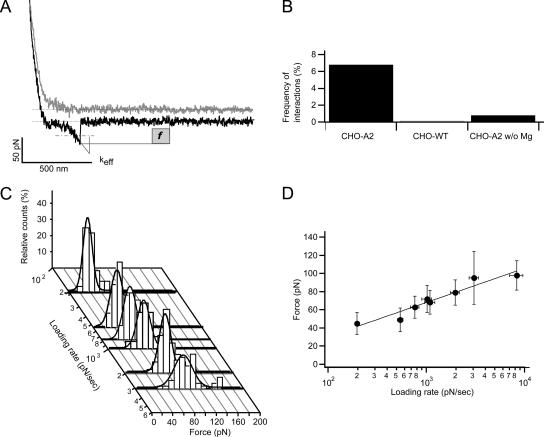
For single-molecule adhesion measurements, 50–80 force curves in total displaying specific binding events were collected at a given pulling speed. Pulling speeds varied between 0.9 and 22 μm/s, corresponding to effective loading rates in the range of ∼200–8000 pN/s. Small rupture events displaying nonlinear loading were detected in <7% of the force curves, ensuring that mainly single-molecule unbinding events were monitored (Benoit et al., 2000; Tees et al., 2001). The measured rupture force was corrected for hydrodynamic drag of the cantilever as described previously (Janovjak et al., 2005). The effective spring constant of the cell/cantilever system, keff, was measured as depicted in Figure 4A. The slope of a straight line fitted through the final third of the nonlinear part of the force curve preceding the point of bond rupture allowed extracting keff. This approximation allowed rendering the influence of local mechanics (molecular unfolding, cell and/or collagen fibril stretching) on the loading of the adhesive bridge. The keff value was then used to calculate the effective loading rate (reff = keff × v) as described previously (Zhang et al., 2002). Mean values of the separation force and of the effective loading rate were used to construct a dynamic force spectrum (DFS), and the data points in the DFS were fitted with a straight line. Applying the following equation to the line fit fm = a × ln(rf) + b with a = kB × T/xu and b = a × ln(1/(a × koff)), where koff is the unstressed dissociation constant and xu is the width of the potential barrier, different energy landscape parameters of the bond (activation barrier width, xu; bond lifetime, 1/koff; and dissociation constant, koff) were extracted according to recent theories of bond rupture under force (Bell, 1978; Evans and Ritchie, 1997).
Figure 4.
Dynamic force spectroscopy of single-integrin unbinding events. (A) Force curve recorded for a cell–substrate contact time of 50 ms and a contact force of ∼200 pN, containing just one single-molecule unbinding event. (f, rupture force; keff, effective spring constant determined for each rupture event from the slope of the line fit shown). Because of hydrodynamic drag, the baselines of trace and retrace curves are slightly shifted. (B) The specificity of the α2β1-mediated interaction was confirmed by analyzing the frequency with which interactions occurred in experiments with CHO-A2 cells and controls (CHO-WT and CHO-A2 cells without Mg2+). Using the same experimental conditions (contact force and time), significantly fewer interactions were detected in control experiments. (C) Distribution of rupture forces measured for different loading rates (n > 50 adhesion events). Force curves were recorded at pulling speeds ranging from 0.9 to 20 μm/s. Values were corrected for hydrodynamic drag as described in Materials and Methods. (D) Dependence of rupture forces (mean ± SD) on the loading rate (mean ± SD). For each loading rate, the mean rupture force was determined from the rupture force distributions shown in C. The mean rupture force values were fitted by a line (r2 = 0.9) and from the obtained fit parameters (slope and y-intercept), bond parameters koff, bond lifetime, and xu were calculated.
RESULTS
α2β1-Mediated Cell Adhesion to Collagen Type I Matrices
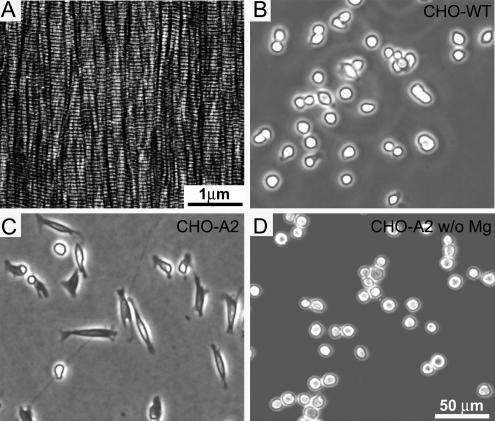
To investigate early steps of integrin-mediated adhesion to collagen, we used structurally well-defined collagen type I matrices (Figure 1A) containing predominantly parallel arrays of collagen fibrils coating the entire support (Jiang et al., 2004). The matrix fibrils displayed the 67 nm D-periodicity frequently observed for collagen type I fibrils in vivo. These ordered collagen matrices are thin (∼3 nm), extremely flat (height variations <1 nm), and promote the adhesion and spreading of different cell types, including fibroblasts and epithelial and endothelial cells (Poole et al., 2005). These matrices are well suited for single-cell adhesion studies because their planarity and structural homogeneity guarantee that differences in adhesion are cell-specific and not caused by variations in the matrix composition or topography.
Figure 1.
α2β1-Mediated cell adhesion and spreading on a collagen type I matrix. (A) AFM topography of an ordered collagen type I matrix. The full range of the gray scale corresponds to a vertical scale of 5 nm. (B) CHO-WT cells lacking endogenous collagen receptors were seeded onto the collagen type I matrix. (C) CHO-A2 cells stably expressing the integrin α2 subunit spread on the collagen matrix. (D) In the absence of Mg2+, CHO-A2 cells do not spread on the collagen matrix. All phase contrast images (B–D) taken 1 h after cell seeding.
Recently, α2β1 has been suggested as a functional cellular receptor for fibrillar collagen type I (Jokinen et al., 2004), but α1β1, α10β1 and α11β1 (Tulla et al., 2001; Zhang et al., 2003) have also been implicated in collagen type I binding. To investigate specifically the role of α2β1 in adhesion to collagen type I, we used a pair of CHO cell lines: CHO wild-type (WT) cells (CHO-WT), which lack endogenous integrin receptors for collagen, and CHO-A2 cells, which stably express the α2 integrin subunit (Nykvist et al., 2000). In these cells, exogenous α2 subunits combine with endogenous β1 subunits to form α2β1 dimers. Thus, CHO-A2 cells express α2β1 as their only collagen-binding integrin.
We seeded CHO-WT and CHO-A2 cells on the collagen type I matrix and compared their spreading behavior. CHO-A2 cells adhered rapidly to the matrix and spread within 5–20 min (Figure 1C). In contrast, CHO-WT cells did not spread and remained rounded even after prolonged contact (>3 h) with the collagen matrix (Figure 1B). In the absence of Mg2+, CHO-A2 cells failed to spread, and they remained rounded (Figure 1D), whereas Ca2+ removal had no influence on cell spreading (data not shown). Fluorescence staining for the α2 subunit in fully spread CHO-A2 cells showed that it colocalized with vinculin and paxillin to classical, elongated focal adhesion contacts (data not shown). Integrins containing αI-domains are the only known Mg2+-dependent collagen receptors, and of this receptor family, CHO-A2 cells exclusively express the α2β1 type. Consequently, we concluded that spreading of CHO-A2 cells on the collagen matrix was mediated by α2β1 integrin.
Adhesion Forces of CHO-A2 and CHO-WT Cells Measured by SCFS
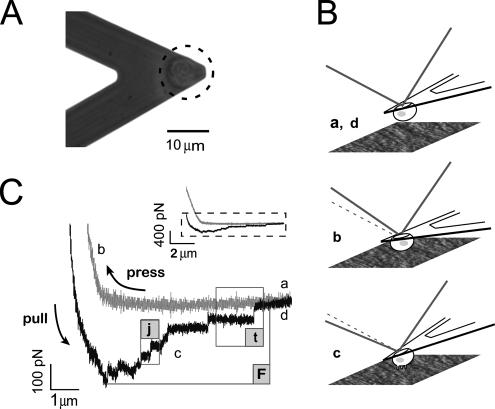
To investigate α2β1-mediated adhesion to collagen in more detail, adhesion of CHO-WT and CHO-A2 cells to collagen type I was compared by SCFS. Single CHO-WT or CHO-A2 cells were attached to concanavalin A-functionalized tipless AFM cantilevers (Figure 2A) (Wojcikiewicz et al., 2004; Puech et al., 2005). Cells were pressed onto the collagen matrix with a predefined contact force for varying times. Subsequently, the cantilever was retracted until the cell was detached from the matrix. A complete force measurement cycle is depicted schematically in Figure 2B. From the corresponding force curve the maximal detachment force (F), corresponding to the maximal cantilever deflection during retraction, was determined (Figure 2C). Force curves usually displayed several small unbinding events, either preceded by a force plateau (“t”) or not (“j”). Sections of the force curve describing j events showed a nearly linear increase preceding the point of rupture and were considered to represent the unbinding of discrete adhesive units under load. In contrast, no force loading (increase in force) occurred before t rupture events. Such t events were interpreted as membrane tether extrusions from a large cell membrane reservoir (Raucher and Sheetz, 1999). Because the force step analysis in this study required constant-rate, nonzero bond loading before rupture, values of t events were excluded. Due to the viscoelastic response of the cell to the cantilever force during substrate contact (Puech et al., 2006), the end of the approach curve and the beginning of the retraction curve were shifted along the force axis (Figure 2C, inset). The viscoelastic deformation of the cell usually occurred within ∼2 s after reaching the predefined contact force and resulted in a decrease of the cell diameter by ∼10–15% and a drop of the effective contact force by several hundred piconewtons (see Supplemental Movie S1). For contact times ≤120 s, usually two to five force cycles per cell were performed, whereas for longer contact times, one or two force curves per cell were recorded. To ensure that the contact history of the cells with the matrix did not influence the force measurements, cells were allowed to rest for 2 to 3 minutes after each force measurement cycle (see Supplemental Figure S2). When testing different spots on the collagen matrix or repeating a force cycle at the same position, similar force curves were obtained, indicating that the matrix integrity was not compromised during the cell pulling experiments (see Supplemental Figure S3).
Figure 2.
Schematic representation of SCFS measurements. (A) Phase-contrast image of a living CHO cell bound to the end of a functionalized cantilever. (B) Force measurement procedure: Cantilever with an attached cell several micrometers above the collagen surface (a and d). The cell is lowered onto the collagen surface until a preset contact force is achieved, and the position of the cantilever is kept at a constant height for a defined contact time (b). Subsequently, the cell is withdrawn from the surface at a set speed (c). Bonds between cell and surface break sequentially until cell and surface are completely separated (d). (C) Representative force curve with steps a, b, c, and d corresponding to those in B for a contact time of 20 s and a pulling speed of 2.5 μm/s. The inset displays the complete approach and retraction curves, whereas the large graph represents a magnified view of the lower part of the same force curve. Multiple unbinding events can be observed (j, jumps in force; t, membrane tethers are pulled out; and F, maximal detachment force).
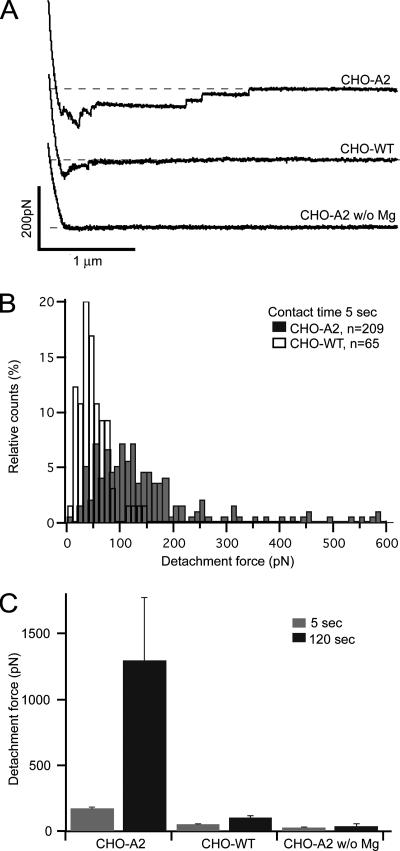
Representative force curves for CHO-A2 (top curve) and CHO-WT (middle curve) cells after a cell–matrix contact time of 5 s are shown in Figure 3A. For CHO-A2 cells, we obtained force curves characterized by maximal detachment forces of up to 600 pN and several single-rupture events. In contrast, CHO-WT force curves displayed drastically reduced detachment forces and usually lacked discrete unbinding steps. In Mg2+-free medium, adhesion of CHO-A2 cells was blocked (Figure 3A, bottom force curve). Conversely, CHO-A2 cell adhesion was fully restored when 0.8 mM Mg2+ was added to the medium (data not shown). Figure 3B displays a histogram of CHO-A2 and CHO-WT cell detachment forces after a contact time of 5 s. CHO-A2 cell detachment forces showed a relatively wide distribution, ranging from ∼40 to 600 pN, whereas CHO-WT cell detachment forces showed a narrower distribution and never exceeded 150 pN. The mean CHO-A2 cell detachment force (189 ± 12 pN; mean ± SD) was almost 4 times higher than for CHO-WT cells (49 ± 7 pN). Between 5 and 120 s of substrate contact, the CHO-A2 mean detachment force increased by almost 1 order of magnitude, whereas the CHO-WT mean detachment force increased only slightly during the same time interval (Figure 3C). In the absence of Mg2+, adhesion of CHO-A2 cells was significantly reduced for both contact times and comparable with CHO-WT cells (Figure 3C). The SCFS measurements demonstrated clearly that for two different contact times CHO-A2 cells adhered more strongly to the collagen matrix than CHO-WT cells and that CHO-A2 cell adhesion was strictly Mg2+ dependent. The requirement of both integrin α2 expression and extracellular Mg2+ showed that adhesion to the collagen matrix was mediated by α2β1. These results were consistent with the different spreading behavior of CHO-A2 and CHO-WT cells on the collagen matrix.
Figure 3.
Measuring α2β1-dependent cell adhesion by SCFS. (A) Representative force distance curves (retrace) for CHO-A2 and CHO-WT and CHO-A2 cells in the absence of Mg2+ after a 5-s collagen matrix contact time. Force curves were recorded at a pulling speed of 2.5 μm/s. For CHO-A2 cells, a series of single-rupture events can be observed. CHO-A2 did not adhere in absence of Mg2+. (B) Distribution of detachment forces for CHO-A2 (50 cells) and CHO-WT (11 cells) after a contact time of 5 s. (C) Comparison of the maximal detachment forces (mean ± SEM) for CHO-WT cells and CHO-A2 cells with and without Mg2+.
Investigating Single α2β1-Mediated Adhesion Events
To further explore α2β1-mediated collagen binding, the SCFS setup was adjusted to perform force measurements with single-molecule sensitivity. To reduce the contact area between cell and matrix, CHO-A2 cells were pressed onto the collagen substrate with low contact force (100–200 pN) and for a short contact time (50–200 msec). Under these conditions, <7% of all force curves displayed unbinding events (Figure 4B), usually in the form of single j event force jumps (Figure 4A). Based on Poisson statistics, at this binding probability (probability that a force curve contains an unbinding event) ∼96% the unbinding events represent the rupture of a single bond (Tees et al., 2001). Thus, the modified SCFS setup allowed us to measure almost exclusively single-molecule adhesion events. Without Mg2+, the binding probability of CHO-A2 cells was decreased to 0.8% (Figure 4B). Because of the low binding probability in the absence of Mg2+, the single-molecule adhesion events could be unequivocally attributed to α2β1. Moreover, under identical contact conditions, the binding probability of CHO-WT cells was only 0.1% (Figure 4B), indicating that the contribution of nonspecific (α2β1-independent) adhesion was minimal. Taken together, these results showed that the small rupture events we observed for CHO-A2 cells corresponded to the unbinding of single α2β1 dimers from the collagen matrix.
Single-Molecule Dynamic Force Spectroscopy
Measuring bond rupture forces over a range of loading rates generates a DFS. This permits the calculation of bond-specific parameters such as koff, 1/koff, and xu (Evans and Ritchie, 1997). To determine these parameters for the α2β1-collagen type I bond, we performed single-molecule adhesion measurements with different loading rates, which were achieved by varying the cantilever retraction speed. Force loading rates were calculated as the product of the pulling speed, v, and the effective spring constant, keff, determined by the composite elastic properties of the cell/cantilever system. The keff was determined for each rupture event as described in Methods and Materials, and a mean keff was calculated for a given pulling speed. Pulling speeds between 0.9 and 22 μm/s yielded effective spring constants between 0.2 and 0.4 mN/m, resulting in loading rates of 180-8800 pN/s. Figure 4C shows the rupture force distributions for different loading rates. In most cases, the rupture forces showed Gaussian distribution (normality test; p > 0.1), indicating that a single class of unbinding events was detected (Evans and Ritchie, 1997; Merkel et al., 1999). With increasing loading rates the mean rupture forces, fm, increased from 38 to 90 pN. Plotting the mean rupture force values versus the logarithm of the respective loading rates yielded a uniformly linear relationship (Figure 4C), which indicated an intermolecular potential with a single activation barrier. Applying Eq. 1 derived by Evans (Evans and Ritchie, 1997) based on the work of Bell (Bell, 1978) to the line fit (see Material and Methods), a dissociation rate koff = 1.3 ± 1.3 s−1, a lifetime of 0.8 ± 0.7 s and a barrier width of 2.3 ± 0.3 Å were determined for the α2β1 integrin/collagen I bond.
Dependence of α2β1-Mediated Adhesion on Contact Time
During the first 120 s of matrix contact, CHO-A2 cell adhesion increased almost 10-fold (Figure 3C). To investigate the time dependence of cell adhesion formation in more detail, we varied the contact time of CHO-A2 cells with the collagen matrix between 5 and 600 s, and we determined the respective detachment forces. For each contact time interval, >10 cells were analyzed, and several force curves were recorded per cell (Supplemental Table S1).
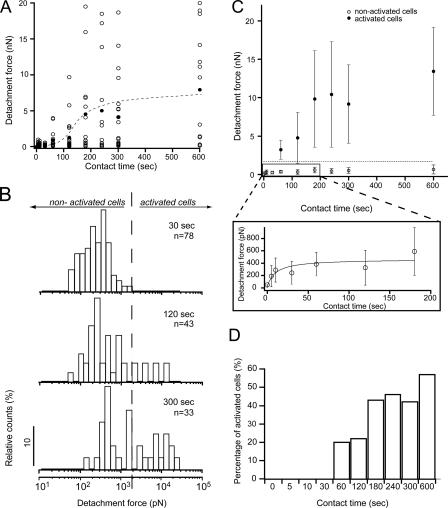
For short contact times (5–10 s), CHO-A2 cells required forces of several hundred piconewtons for matrix detachment (Figure 5A). With increasing contact time the mean detachment force grew slowly until after ∼60 s it increased quickly, reaching ∼5 nN after 180 s. Beyond 180 s, the mean detachment force continued to rise, albeit again more slowly. Thus, recording cell detachment forces over a time course of 10 min revealed a nonlinear buildup of adhesion force across the cell population. Three phases of adhesion formation could be distinguished: a first phase (<60 s) during which forces rose comparatively slowly (“initiation”), a second phase (60–180 s) of rapidly increasing adhesion (“reinforcement”), and a third phase (>180 s) of slow adhesion maturation (“maturation”).
Figure 5.
Dependence of cellular adhesion on substrate contact time. (A) CHO-A2 cell detachment forces were determined for contact times between 5 and 600 s. Open circles in the diagram represent the detachment forces of individual cells for a given contact time, whereas the closed circles denote the corresponding mean detachment forces of all cells tested. For contact times >60 s, the detachment forces show wide variation. The S-shaped hand-drawn trend line (dashed) indicates a nonlinear increase of cellular adhesion with contact time. (B) Detachment force distribution measured for CHO-A2 cells for contact times of 30, 120, and 300 s displayed in a half-logarithmic scale. Two cell populations could be appreciated by eye. The first group contains cells displaying relatively weak adhesion of several hundred piconewtons (nonactivated cells). After a contact time of 30 s, all tested cells fell into the nonactivated group, whereas after longer contact times (120 and 300 s), their relative frequency gradually decreased. Instead, a second cell population displaying detachment forces of up to 20 nN (activated cells) emerged. Dashed line represents cut-off force of 2 nN chosen to separate groups. (C) Detachment forces (mean ± SD) of activated cells (detachment forces >2 nN) and nonactivated cells (detachment forces <2 nN) for different contact times. Dashed line indicates cut-off detachment force of 2 nN. (D) Time-dependent increase in the percentage of activated cells.
Because a single-cell technique was used, detachment forces of individual cells could be compared. Beginning after 60 s of contact time, cell detachment forces showed considerable variation (from ∼0.5 N to 20 nN). This suggested that individual cells were in different adhesive states (Figure 5A). Moreover, for contact times >60 s, cells seemed to be distributed into a low adhesion group (detachment forces below ∼2 nN) and a high adhesion group in which detachment forces ranged from ∼2 to 20 nN. To illustrate the presence of cell populations with different adhesive properties at specific points of the contact time course, detachment force distributions representative for the initiation (30 s), reinforcement (120 s), and maturation (300 s) phases were displayed in half-logarithmic histograms (Figure 5B). During the initiation phase, detachment forces displayed a unimodal distribution and were below 2 nN (Figure 5B, top). During the reinforcement and maturation phases, the detachment force distribution shifted to higher values (Figure 5B, middle and bottom), although some cells still required only low detachment forces. The progressive rearrangement of detachment forces from a unimodal into a wider, multimodal distribution demonstrated the emergence of cell subpopulations displaying increased adhesion during the reinforcement and maturation phases.
To compare cell populations showing high or low adhesion, cells were separated into two groups: low-adhesion, “nonactivated” cells (detachment forces <2 nN), and high-adhesion, “activated” cells (detachment forces between 2 and 20 nN). The cut-off force of 2 nN (Figure 5B, dashed line) was chosen based on the observation that during the initiation phase cell detachment forces never exceeded this value (Figure 5B, top). Thus, the cut-off force separated cells with low detachment forces as a result of weak adhesion established during the initiation phase and cells exceeding these detachment forces due to a rapid increase in cell adhesion during the reinforcement phase. Replotting the mean detachment forces versus contact time separately for both groups emphasizes differences between both groups in terms of time-dependent adhesion formation (Figure 5C). Nonactivated cells were present over the entire time course. The mean adhesion of cells in this group increased during the first 60 s and subsequently reached a plateau of ∼400 pN (Figure 5C, inset). In contrast, activated cells only occurred at the beginning of the reinforcement phase (after 60 s), and their mean adhesion continued to increase all throughout the time interval tested (600 s). Although detachment forces varied widely within the activated population, this group was clearly distinct from the nonactivated population, in which cell detachment forces varied less.
The percentage of cells belonging to the highly adhesive activated group increased with time (Figure 5D). Because the ratio of nonactivated to activated cells changed over time, the differences in adhesion between both groups could not be explained by the presence of two cell subpopulations with intrinsically distinct adhesive properties, for example, due to clonal variations in the expression level of the integrin α2 subunit. Instead, with increasing contact time, cells seemed to switch progressively from a low to a high adhesion state. Monitoring the cell–matrix contact area by transmission light microscopy during the contact period demonstrated that cell spreading fluctuated by ∼10–15% but that it did not increase overall, irrespective of whether cells became activated or not (see Supplemental Figure S4 and Movie S2). Thus, progressive cell spreading did not account for the time-dependent increase in cell adhesion and the switch to the activated state.
The Increase in Overall Cell Adhesion Coincides with an Increase in the Smallest Unbinding Forces
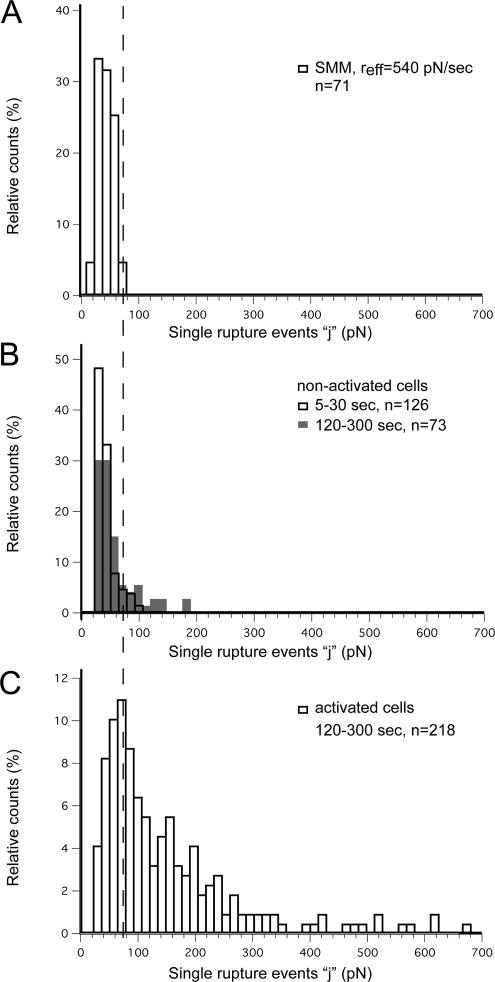
For contact times >5 s, CHO-A2 force curves usually contained several single-rupture (j) events in addition to the main rupture peak denoting the maximal detachment force (Figure 2C). The single-rupture events represented the smallest detectable unbinding force units. When analyzing these events for nonactivated cells (Figure 6B), similar force distributions were obtained during the adhesion initiation (5–30 s) and reinforcement/maturation phases (120–300 s). The force distribution of the single-rupture events in nonactivated cells (46 ± 16 pN; mean ± SD) was also nearly identical to the single-integrin rupture force distribution (47 ± 13 pN; mean ± SD) obtained at a comparable loading rate (∼500 pN/s) during DFS (Figure 6A). Therefore, in nonactivated cells, the smallest force units remained nearly constant with contact time, and they were comparable with the rupture force between a single integrin and the collagen matrix. In contrast, for activated cells, we observed a dramatic shift of the smallest rupture forces to higher values (159 ± 132 pN; mean ± SD) during the adhesion reinforcement/maturation phases (Figure 6C). Consequently, in activated cells the majority of small unbinding events could not be attributed to the rupture of single-integrin–collagen bonds.
Figure 6.
Single-rupture events for activated and nonactivated CHO-A2 cells. (A) Force distribution of single-integrin unbinding events measured in single-molecule experiments (contact time 50–500 ms, contact force 100–200 pN) with a mean loading rate of 540 pN/s. (B) Single separation events (j) measured for nonactivated cells. Unfilled bars represent j event values for short contact times (5–30 s). Gray bars represent j event values for long contact times (180–300 s). (C) Single separation events detected for activated cells (contact times between 180 and 300 s). The cut-off force of 73 pN (dashed line) corresponds to the outer border of the force interval containing 100% of the single-integrin rupture forces shown in the histogram in A.
To compare the smallest rupture events between nonactivated and activated cells, we defined a force interval whose limits of 0 and 73 pN contained 100% of the single-integrin rupture forces measured during DFS. During the initiation phase (5–30 s), >90% of the small rupture events in nonactivated cells fell within this interval, and they were therefore consistent with the unbinding of single integrins from the collagen matrix (Table 1). During the reinforcement/maturation phase (120–300 s), the distribution of rupture forces was shifted slightly toward higher forces. However, the majority of rupture events (75%) still lay within the single-integrin unbinding force interval. We consequently concluded that in nonactivated cells, the majority of the smallest unbinding events represented the rupture of single-integrin–collagen bonds. In activated cells, only 26% of the small rupture events were within the single-integrin unbinding force interval. Thus, the increase in overall cell adhesion of activated cells coincided with the frequent rise of the smallest force units above the single-integrin level. In these cells, the smallest force units showed a wide distribution and reached values as high as 700 pN, or ∼15 times the value determined for the unbinding of a single integrin. The rupture force increase over the single-integrin level suggested that functional adhesive units containing varying numbers of α2β1 receptors had formed and that these receptors displayed cooperative binding.
Table 1.
Percentage of rupture events (“j”) at single-molecule level (<73 pN) detected for different experimental conditions (inhibitors, contact times). The force interval (0,73 pN) encompasses 100% of the single-integrin unbinding events detected at comparable loading rates during single-molecule measurements
| Interval (pN) | SMM (%) | Short (%) | Nonactivated, long (%) | Activated, long (%) | Without inhibitor, long (%) | With ROCK inhibitor, long (%) | With BDM, long (%) |
|---|---|---|---|---|---|---|---|
| <73 | 100 | 91 | 75 | 26 | 29 | 56 | 100 |
| ≥73 | 0 | 9 | 25 | 74 | 71 | 44 | 0 |
Short, contact times from 5 to 30 seconds; Long, contact times from 120 to 300 seconds; SMM, single-molecule measurements.
Actomyosin Contractility Is Required for Establishing Integrin Cooperativity and Overall Cell Adhesion
In cells establishing substrate contact, integrins are frequently clustered into small, dot-like focal complexes that may later mature into larger focal adhesion structures. The maturation of early adhesive sites into focal adhesions involves myosin II-driven contractility regulated by the small GTPase RhoA and its effectors ROCK (Rottner et al., 1999). RhoA activity is also required to bundle actomyosin filaments into large stress fibers that terminate in focal adhesion sites (Chrzanowska-Wodnicka and Burridge, 1996). Therefore, there is a clear link between actomyosin-driven contractility, the formation of mature integrin adhesion complexes, and the establishment of strong overall cell adhesion.
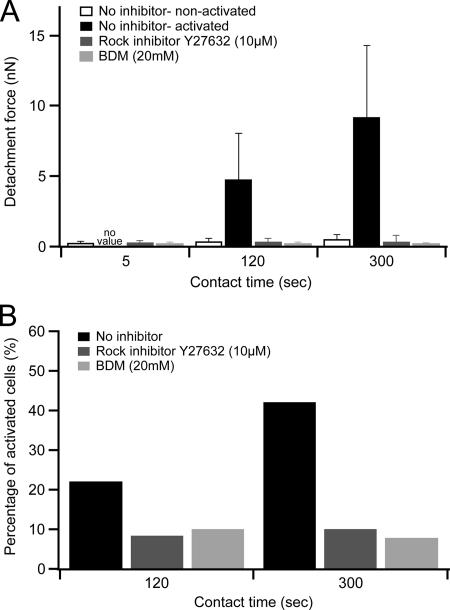
To test whether early CHO-A2 cell adhesion to collagen required actomyosin activity, we performed adhesion assays in the presence of inhibitors blocking actomyosin contractility by different mechanisms. Addition of the myosin inhibitor butandione-2-monoxime (BDM) at 20 mM lead to a reduction of the mean detachment force by >50% at 120 s contact time and by >90% at 300 s (Figure 7A). For the same contact times, the ROCK inhibitor Y27632 also decreased the mean detachment force strongly (120 s, 63% and 300 s, 91%). The detachment force decrease in BDM or ROCK inhibitor-treated cells was mirrored by a reduction in the percentage of activated cells (cells requiring detachment forces >2 nN) from ∼40 to <10% (Figure 7B). Therefore, actomyosin contractility was crucial for cells to establish strong substrate adhesion and to switch into the activated adhesion mode.
Figure 7.
Influence of inhibitors of actomyosin contractility on cell adhesion. (A) CHO-A2 cell detachment forces (mean ± SD) in the presence and absence of the ROCK inhibitor Y27632 at 10 μM or the MLCK inhibitor BDM at 20 mM. (B) Percentage of activated cells in the presence and absence of Y27632 and BDM.
As seen, transition into the activated adhesion mode coincided with a strong increase of the smallest discrete rupture forces. To assess whether actomyosin contractility was required for this force increase, we analyzed the smallest rupture events in inhibitor-treated cells. During the reinforcement/maturation phase (120–300 s), the smallest rupture events in BDM-treated cells fell quantitatively into the single-integrin unbinding force interval, whereas in untreated cells only 29% of unbinding events were consistent with single-molecule unbinding (Table 1). The complete constraint of the small rupture events to the single-integrin level in BDM-treated cells points to an important role for actomyosin contractility in the transition from single to cooperative receptor binding. In ROCK-treated cells, only about half of the small rupture events were found in the single-integrin unbinding class, suggesting that this inhibitor suppressed the establishment of cooperative integrin adhesion less efficiently than BDM.
DISCUSSION
We have used SCFS to characterize the time-dependent formation of α2β1-mediated adhesion to collagen type I. Three consecutive phases of adhesion formation could be distinguished: an initiation phase characterized by comparatively low adhesion, a reinforcement phase of rapidly increasing adhesion, and a maturation phase during which adhesion was consolidated. A similar time-dependent increase of cell adhesion upon plating has been described repeatedly (Lotz et al., 1989; Cohen et al., 2004; Walter et al., 2006) and has been attributed to the concomitant increase of cell–substrate area and new bond formation, subsequent receptor linkage to the cytoskeleton, receptor clustering, and cooperativity (Gallant and Garcia, 2006). However, these processes are interdependent and may temporally overlap, which complicates dissecting their precise contribution to cell adhesion formation.
Operating an AFM in force spectroscopy mode with a living cell attached to the cantilever allowed us to investigate forces from the single-integrin level to overall cell adhesion in the same experimental setup. Importantly, force curves generated during cell detachment contained force information about both overall cell adhesion (maximal detachment force) and individual adhesive units (smallest force steps). Correlating the smallest rupture events with cell-scale adhesion showed that establishment of firm cell adhesion during the reinforcement/maturation phases coincided with the transgression of the smallest rupture forces from the single-integrin level (47 pN) to much higher values (≤700 pN). This rupture force increase indicated a growth of the smallest adhesive units during cell adhesion reinforcement.
Overall adhesive strength (“avidity”) of an adhesion complex results from both the total number of receptor–ligand bonds in that complex and the strength (“affinity”) of each of these bonds (Carman and Springer, 2003). Furthermore, receptor cross-linking and the establishment of receptor cooperativity regulate adhesion strength, because they guarantee that an applied force is distributed more evenly over all bonds. Otherwise, when receptors are uncoupled from each other, receptors experiencing the highest load will become unbound first, and the remaining bonds will then rupture in a sequential, zipper-like manner requiring relatively low forces (Chen and Moy, 2000). At similar loading rates as applied here, protein–ligand rupture forces usually range between ∼20 and 200 pN (Weisel et al., 2003). During adhesion reinforcement, force steps of several hundred piconewtons were measured frequently, which far exceeds the single-molecule rupture force range. Consequently, we attributed the strong single-rupture force increase to the formation of multi-integrin receptor complexes and the establishment of cooperative receptor binding.
During the rupture of integrin clusters several integrin receptors have to unbind simultaneously. Understanding the contribution of individual bonds to force curves detecting multiple bond ruptures is nontrivial. For parallel bond loading, the unbinding forces of single bonds have been proposed to add both linearly (Florin et al., 1994) or nonlinearly (Ratto et al., 2006). In force spectroscopy studies using purified receptor–ligand pairs, quantized peaks representing multiple single-molecule unbinding events have been observed in the rupture force distribution (Florin et al., 1994). However, the force distribution of the smallest rupture events in activated cells showed no clear maxima corresponding to multiples of the single-integrin rupture force of 47 pN. Individual integrin receptors may differ in their linkage to the actin cytoskeleton, resulting in different elastic properties, force loading, and ultimately in slightly different rupture forces of the adhesive bridge. In addition, individual integrin within a cluster may be forced to unbind along different pulling directions. Because it is known that the measured strengths of a bond strongly depends on the pulling direction (Bustamante et al., 2004), this may cause further variations in the unbinding force. Because of the complexity of cooperative integrin binding in the context of a living cell, no conclusion about the exact number of integrin receptors per adhesive unit could be drawn.
Evidence for integrin clustering is often based on the assembly of macroscopic adhesion patches, such as focal complexes and adhesion, which typically occur 10–20 min after cell seeding. However, functional integrin adhesion complexes may first form as small receptor clusters that cannot be resolved by conventional light microscopy (Laukaitis et al., 2001). In agreement, SCFS detected the establishment of cooperative integrin binding as early as 60 s after matrix contact, suggesting that integrins start clustering before this becomes optically detectable.
Establishment of strong α2β1-mediated adhesion could be prevented by inhibiting actomyosin contractility with BDM or Y27632. This was in agreement with studies demonstrating the requirement of myosin II-driven contractility to establish and maintain integrin-containing anchorage sites, such as focal complexes (Rottner et al., 1999) and focal adhesions (Chrzanowska-Wodnicka and Burridge, 1996). Concurrent with the decrease in overall cell adhesion, the smallest rupture force units were reduced to the single-integrin level in inhibitor-treated cells. BDM prevented receptor cooperativity more efficiently than the ROCK inhibitor Y27632, which could be due to the more downstream position of the BDM target myosin II in the RhoA-dependent signaling cascade that controls actomyosin contractility. Because inhibiting actomyosin contractility efficiently prevented the establishment of cooperative binding of integrin receptors, the early events of integrin clustering may be myosin II driven. How could myosin II activity induce integrin clustering? Although unbound integrins are freely diffusive in the membrane plane, ligand binding promotes rapid attachment of integrins to actin filaments (Felsenfeld et al., 1996). Myosin II is an effective F-actin cross-linker (Laevsky and Knecht, 2003), and myosin-driven bundling and alignment of actin filaments carrying ligand-bound integrin complexes could then lead to the clustering of integrin–cytoskeleton complexes (Chrzanowska-Wodnicka and Burridge, 1996). Alternatively, myosin-dependent forces acting on integrin complexes could induce conformational changes in associated mechanosensitive proteins. Internal regions exposed within such mechanosensors could then provide protein–protein interaction interfaces promoting integrin receptor clustering.
Under conditions that minimized cell–substrate contact, cell deadhesion was reduced to the rupture of a single-integrin–collagen bond. At moderate force loading (∼500 pN/s), the α2β1/collagen unbinding force was 47 ± 13 pN, similar to forces measured for other integrin–ligand interactions (for example, 39 ± 8 pN for α5β1-fibronectin; Sun et al., 2005). The strength of α2β1 integrin binding to collagen type I locates at a lower to middle position within the spectrum of known receptor–ligand unbinding forces (Weisel et al., 2003). A comparatively low integrin binding strength would emphasize that strong integrin-mediated cell–matrix adhesion requires high receptor avidity through cooperative receptor binding.
There are several high-affinity bindings sites for α2β1 on fibrillar collagen type I (Knight et al., 2000; Xu et al., 2000). These binding sites share a GER motif, but variations in the flanking sequences may modulate their affinity for α2β1. However, the narrow, unimodal rupture force distribution we obtained during single-molecule measurements suggests that α2β1 bound to all accessible binding sites with similar affinity, consistent with previous observations (Xu et al., 2000).
For the α2β1 integrin–collagen type I bond, we determined a dissociation rate constant koff = 1.3 ± 1.3 s−1. This value is comparable with off rates determined for other cell adhesion molecules (E-cadherin [homotypic]: koff = 1.01 s−1; Panorchan et al., 2006; and E-selectin/carbohydrate koff = 0.72 s−1; Tees et al., 2001) but considerably higher than the off rate determined for integrin α5β1-fibronectin (koff = 0.012–0.13 s−1; Li et al., 2003). Integrin adhesion must be dynamic so that cells can respond quickly to changes in external forces. A high off rate has been suggested to permit the fast remodeling of integrin–matrix interactions (Chrzanowska-Wodnicka and Burridge, 1996). The higher off rate of the α2β1 integrin–collagen type I bond may suggest that this bond is relatively dynamic. Varying off rates may reflect different cellular requirements concerning the turnover rate of specific integrin–ligand pairs.
Several SCFS studies showed that rupturing integrin–ligand bonds requires overcoming at least two activation energy barriers (Zhang et al., 2002; Li et al., 2003). This has been proposed to be a common feature of all receptor–ligand interactions involving a MIDAS motive, where the inner activation barrier seems to represent the rupture between the chelated metal ion and a negatively charged amino acid in the ligand (Li et al., 2003). In contrast, the DFS generated for α2β1–collagen indicated only a single activation barrier. This may reflect differences in the unbinding mechanism between these integrins and their ligands. However, compared with these previous studies using relatively small lymphoid cells, we obtained lower loading rates at comparable pulling speeds. This may be due to the different elastic properties of epithelial CHO cells. Because we were unable to achieve loading rates >10,000 pN/s, possible additional activation barriers may not have been detected.
In conclusion we have characterized α2β1-mediated adhesion from the single-molecule level to overall cell adhesion, and we provide new insight into temporal and mechanistic aspects of early integrin binding events. A current challenge is to perform SCFS measurements while imaging cells by using advanced light microscopic techniques. In this way, the rupture of initial adhesion complexes consisting only of a few receptors could be monitored visually during cell detachment and correlated with the force step information contained in the force curves.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jonne Helenius for helpful discussions, Jyrki Heino (Department of Biochemistry and Food Chemistry, University of Turku, Turku, Finland) for providing reagents, and JPK Instruments for collaborative support. This work was supported by the AO Research Fund, the Deutsche Volkswagenstiftung, and the Free-State of Saxony.
Abbreviations used:
- AFM
atomic force microscopy
- CHO
Chinese hamster ovary
- DFS
dynamic force spectroscopy
- SCFS
single-cell force spectroscopy.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-09-0777) on February 21, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Amano M., Chihara K., Kimura K., Fukata Y., Nakamura N., Matsuura Y., Kaibuchi K. Formation of actin stress fibers and focal adhesions enhanced by Rho-kinase. Science. 1997;275:1308–1311. doi: 10.1126/science.275.5304.1308. [DOI] [PubMed] [Google Scholar]
- Bell G. I. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- Benoit M., Gabriel D., Gerisch G., Gaub H. E. Discrete interactions in cell adhesion measured by single-molecule force spectroscopy. Nat. Cell Biol. 2000;2:313–317. doi: 10.1038/35014000. [DOI] [PubMed] [Google Scholar]
- Bustamante C., Chemla Y. R., Forde N. R., Izhaky D. Mechanical processes in biochemistry. Annu. Rev. Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- Butt H.-J., Jaschke M. Calculation of thermal noise in atomic force microscopy. Nanotechnology. 1995;6:1. [Google Scholar]
- Carman C. V., Springer T. A. Integrin avidity regulation: are changes in affinity and conformation underemphasized? Curr. Opin. Cell Biol. 2003;15:547–556. doi: 10.1016/j.ceb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Chen A., Moy V. T. Cross-linking of cell surface receptors enhances cooperativity of molecular adhesion. Biophys. J. 2000;78:2814–2820. doi: 10.1016/S0006-3495(00)76824-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrzanowska-Wodnicka M., Burridge K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J. Cell Biol. 1996;133:1403–1415. doi: 10.1083/jcb.133.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisneros D. A., Hung C., Franz C. M., Muller D. J. Observing growth steps of collagen self-assembly by time-lapse high-resolution atomic force microscopy. J. Struct. Biol. 2006;154:232–245. doi: 10.1016/j.jsb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Cohen M., Joester D., Geiger B., Addadi L. Spatial and temporal sequence of events in cell adhesion: from molecular recognition to focal adhesion assembly. Chembiochem. 2004;5:1393–1399. doi: 10.1002/cbic.200400162. [DOI] [PubMed] [Google Scholar]
- Emsley J., Knight C. G., Farndale R. W., Barnes M. J., Liddington R. C. Structural basis of collagen recognition by integrin alpha2beta1. Cell. 2000;101:47–56. doi: 10.1016/S0092-8674(00)80622-4. [DOI] [PubMed] [Google Scholar]
- Evans E., Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld D. P., Choquet D., Sheetz M. P. Ligand binding regulates the directed movement of beta1 integrins on fibroblasts. Nature. 1996;383:438–440. doi: 10.1038/383438a0. [DOI] [PubMed] [Google Scholar]
- Florin E. L., Moy V. T., Gaub H. E. Adhesion forces between individual ligand-receptor pairs. Science. 1994;264:415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- Gallant N. D., Garcia A. J. Model of integrin-mediated cell adhesion strengthening. J. Biomech. 2006 doi: 10.1016/j.jbiomech.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Geiger B., Bershadsky A., Pankov R., Yamada K. M. Transmembrane crosstalk between the extracellular matrix–cytoskeleton crosstalk. Nat. Rev. Mol. Cell Biol. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- Hughes P. E., Pfaff M. Integrin affinity modulation. Trends Cell Biol. 1998;8:359–364. doi: 10.1016/s0962-8924(98)01339-7. [DOI] [PubMed] [Google Scholar]
- Hulmes D. J. Building collagen molecules, fibrils, and suprafibrillar structures. J. Struct. Biol. 2002;137:2–10. doi: 10.1006/jsbi.2002.4450. [DOI] [PubMed] [Google Scholar]
- Hynes R. O. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- Janovjak H., Struckmeier J., Muller D. J. Hydrodynamic effects in fast AFM single-molecule force measurements. Eur. Biophys. J. 2005;34:91–96. doi: 10.1007/s00249-004-0430-3. [DOI] [PubMed] [Google Scholar]
- Jiang F., Khairy K., Poole K., Howard J., Muller D. J. Creating nanoscopic collagen matrices using atomic force microscopy. Microsc. Res. Tech. 2004;64:435–440. doi: 10.1002/jemt.20101. [DOI] [PubMed] [Google Scholar]
- Jokinen J., et al. Integrin-mediated cell adhesion to type I collagen fibrils. J. Biol. Chem. 2004;279:31956–31963. doi: 10.1074/jbc.M401409200. [DOI] [PubMed] [Google Scholar]
- Kadler K. E., Holmes D. F., Trotter J. A., Chapman J. A. Collagen fibril formation. Biochem. J. 1996;316:1–11. doi: 10.1042/bj3160001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight C. G., Morton L. F., Peachey A. R., Tuckwell D. S., Farndale R. W., Barnes M. J. The collagen-binding A-domains of integrins α(1)β(1) and α(2)β(1) recognize the same specific amino acid sequence, GFOGER, in native (triple-helical) collagens. J. Biol. Chem. 2000;275:35–40. doi: 10.1074/jbc.275.1.35. [DOI] [PubMed] [Google Scholar]
- Laevsky G., Knecht D. A. Cross-linking of actin filaments by myosin II is a major contributor to cortical integrity and cell motility in restrictive environments. J. Cell Sci. 2003;116:3761–3770. doi: 10.1242/jcs.00684. [DOI] [PubMed] [Google Scholar]
- Laukaitis C. M., Webb D. J., Donais K., Horwitz A. F. Differential dynamics of alpha 5 integrin, paxillin, and alpha-actinin during formation and disassembly of adhesions in migrating cells. J. Cell Biol. 2001;153:1427–1440. doi: 10.1083/jcb.153.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Redick S. D., Erickson H. P., Moy V. T. Force measurements of the alpha5beta1 integrin-fibronectin interaction. Biophys. J. 2003;84:1252–1262. doi: 10.1016/S0006-3495(03)74940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotz M. M., Burdsal C. A., Erickson H. P., McClay D. R. Cell adhesion to fibronectin and tenascin: quantitative measurements of initial binding and subsequent strengthening response. J. Cell Biol. 1989;109:1795–1805. doi: 10.1083/jcb.109.4.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel R., Nassoy P., Leung A., Ritchie K., Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397:50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- Nykvist P., Tu H., Ivaska J., Kapyla J., Pihlajaniemi T., Heino J. Distinct recognition of collagen subtypes by α(1)β(1) and α(2)β(1) integrins. α(1)β(1) mediates cell adhesion to type XIII collagen. J. Biol. Chem. 2000;275:8255–8261. doi: 10.1074/jbc.275.11.8255. [DOI] [PubMed] [Google Scholar]
- Panorchan P., Thompson M. S., Davis K. J., Tseng Y., Konstantopoulos K., Wirtz D. Single-molecule analysis of cadherin-mediated cell-cell adhesion. J. Cell Sci. 2006;119:66–74. doi: 10.1242/jcs.02719. [DOI] [PubMed] [Google Scholar]
- Poole K., Khairy K., Friedrichs J., Franz C., Cisneros D. A., Howard J., Mueller D. Molecular-scale topographic cues induce the orientation and directional movement of fibroblasts on two-dimensional collagen surfaces. J. Mol. Biol. 2005;349:380–386. doi: 10.1016/j.jmb.2005.03.064. [DOI] [PubMed] [Google Scholar]
- Puech P. H., Poole K., Knebel D., Muller D. J. A new technical approach to quantify cell-cell adhesion forces by AFM. Ultramicroscopy. 2006;106:637–644. doi: 10.1016/j.ultramic.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Puech P. H., Taubenberger A., Ulrich F., Krieg M., Muller D. J., Heisenberg C. P. Measuring cell adhesion forces of primary gastrulating cells from zebrafish using atomic force microscopy. J. Cell Sci. 2005;118:4199–4206. doi: 10.1242/jcs.02547. [DOI] [PubMed] [Google Scholar]
- Ratto T. V., Rudd R. E., Langry K. C., Balhorn R. L., McElfresh M. W. Nonlinearly additive forces in multivalent ligand binding to a single protein revealed with force spectroscopy. Langmuir. 2006;22:1749–1757. doi: 10.1021/la052087d. [DOI] [PubMed] [Google Scholar]
- Raucher D., Sheetz M. P. Characteristics of a membrane reservoir buffering membrane tension. Biophys. J. 1999;77:1992–2002. doi: 10.1016/S0006-3495(99)77040-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottner K., Hall A., Small J. V. Interplay between Rac and Rho in the control of substrate contact dynamics. Curr. Biol. 1999;9:640–648. doi: 10.1016/s0960-9822(99)80286-3. [DOI] [PubMed] [Google Scholar]
- Sun Z., Martinez-Lemus L. A., Trache A., Trzeciakowski J. P., Davis G. E., Pohl U., Meininger G. A. Mechanical properties of the interaction between fibronectin and alpha5beta1-integrin on vascular smooth muscle cells studied using atomic force microscopy. Am. J. Physiol. 2005;289:H2526–H2535. doi: 10.1152/ajpheart.00658.2004. [DOI] [PubMed] [Google Scholar]
- Tees D. F., Waugh R. E., Hammer D. A. A microcantilever device to assess the effect of force on the lifetime of selectin-carbohydrate bonds. Biophys. J. 2001;80:668–682. doi: 10.1016/S0006-3495(01)76047-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckwell D., Calderwood D. A., Green L. J., Humphries M. J. Integrin alpha 2 I-domain is a binding site for collagens. J. Cell Sci. 1995;108:1629–1637. doi: 10.1242/jcs.108.4.1629. [DOI] [PubMed] [Google Scholar]
- Tulla M., Pentikainen O. T., Viitasalo T., Kapyla J., Impola U., Nykvist P., Nissinen L., Johnson M. S., Heino J. Selective binding of collagen subtypes by integrin α1I, α2I, and α10I domains. J. Biol. Chem. 2001;276:48206–48212. doi: 10.1074/jbc.M104058200. [DOI] [PubMed] [Google Scholar]
- Walter N., Selhuber C., Kessler H., Spatz J. P. Cellular unbinding forces of initial adhesion processes on nanopatterned surfaces probed with magnetic tweezers. Nano. Lett. 2006;6:398–402. doi: 10.1021/nl052168u. [DOI] [PubMed] [Google Scholar]
- Weisel J. W., Shuman H., Litvinov R. I. Protein-protein unbinding induced by force: single-molecule studies. Curr. Opin. Struct. Biol. 2003;13:227–235. doi: 10.1016/s0959-440x(03)00039-3. [DOI] [PubMed] [Google Scholar]
- White D. J., Puranen S., Johnson M. S., Heino J. The collagen receptor subfamily of the integrins. Int. J. Biochem. Cell Biol. 2004;36:1405–1410. doi: 10.1016/j.biocel.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Wojcikiewicz E. P., Zhang X., Moy V. T. Force and compliance measurements on living cells using atomic force microscopy (AFM) Biol. Proced. Online. 2004;6:1–9. doi: 10.1251/bpo67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Gurusiddappa S., Rich R. L., Owens R. T., Keene D. R., Mayne R., Hook A., Hook M. Multiple binding sites in collagen type I for the integrins α1β1 and α2β1. J. Biol. Chem. 2000;275:38981–38989. doi: 10.1074/jbc.M007668200. [DOI] [PubMed] [Google Scholar]
- Zamir E., Geiger B. Molecular complexity and dynamics of cell-matrix adhesions. J. Cell Sci. 2001;114:3583–3590. doi: 10.1242/jcs.114.20.3583. [DOI] [PubMed] [Google Scholar]
- Zhang W. M., Kapyla J., Puranen J. S., Knight C. G., Tiger C. F., Pentikainen O. T., Johnson M. S., Farndale R. W., Heino J., Gullberg D. α11β1 integrin recognizes the GFOGER sequence in interstitial collagens. J. Biol. Chem. 2003;278:7270–7277. doi: 10.1074/jbc.M210313200. [DOI] [PubMed] [Google Scholar]
- Zhang X., Wojcikiewicz E., Moy V. T. Force spectroscopy of the leukocyte function-associated antigen-1/intercellular adhesion molecule-1 interaction. Biophys. J. 2002;83:2270–2279. doi: 10.1016/S0006-3495(02)73987-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.