Abstract
The yeast Oxa1 protein is involved in the biogenesis of the mitochondrial oxidative phosphorylation (OXPHOS) machinery. The involvement of Oxa1 in the assembly of the cytochrome oxidase (COX) complex, where it facilitates the cotranslational membrane insertion of mitochondrially encoded COX subunits, is well documented. In this study we have addressed the role of Oxa1, and its sequence-related protein Cox18/Oxa2, in the biogenesis of the F1Fo-ATP synthase complex. We demonstrate that Oxa1, but not Cox18/Oxa2, directly supports the assembly of the membrane embedded Fo-sector of the ATP synthase. Oxa1 was found to physically interact with newly synthesized mitochondrially encoded Atp9 protein in a posttranslational manner and in a manner that is not dependent on the C-terminal, matrix-localized region of Oxa1. The stable manner of the Atp9-Oxa1 interaction is in contrast to the cotranslational and transient interaction previously observed for the mitochondrially encoded COX subunits with Oxa1. In the absence of Oxa1, Atp9 was observed to assemble into an oligomeric complex containing F1-subunits, but its further assembly with subunit 6 (Atp6) of the Fo-sector was perturbed. We propose that by directly interacting with newly synthesized Atp9 in a posttranslational manner, Oxa1 is required to maintain the assembly competence of the Atp9-F1-subcomplex for its association with Atp6.
INTRODUCTION
Oxa1 is a protein of the mitochondrial inner membrane that is involved in the biogenesis of complexes of the oxidative phosphorylation machinery (OXPHOS). Oxa1 is best characterized for its role in the biogenesis of the cytochrome oxidase complex (COX), as it was initially identified in the yeast Saccharomyces cerevisiae as an assembly factor of this complex (Oxidase assembly 1; Bauer et al., 1994; Bonnefoy et al., 1994). COX is the terminal electron acceptor complex and thus a key component of the mitochondrial OXPHOS machinery. Analysis of oxa1 null mutant (Δoxa1) mitochondria indicated a complete loss of detectable COX complexes, and hence oxa1 mutants display a respiratory deficient phenotype (Bauer et al., 1994; Bonnefoy et al., 1994). Further analysis of a temperature-sensitive oxa1 mutant, the pet ts1402 strain, indicated that the COX complex assembly defect observed in the absence of Oxa1 function is attributed to a failure of the mitochondrially encoded COX subunits, subunits 1 and 2 (Cox1 and Cox2, respectively), key subunits of the COX complex, to become inserted into the membrane in the absence of the Oxa1 protein (Hell et al., 1997, 1998, 2001). A direct role for Oxa1 in facilitating the membrane insertion process of these proteins was further indicated by the demonstration that Oxa1 physically interacts in a cotranslational manner with the mitochondrially encoded COX subunits, i.e., as they were being synthesized on ribosomes within the mitochondrial matrix and undergoing membrane insertion (Hell et al., 1998, 2001). In addition evidence for a direct interaction of Oxa1 with the mitochondrial ribosomes, which is supported through the matrix-localized C-terminal region of Oxa1, has also been presented (Jia et al., 2003, Szyrach et al., 2003).
Closer examination of oxa1 null mutant mitochondria has demonstrated that the function of Oxa1 is not limited to the assembly of the COX complex, but that it is also involved in the biogenesis of other OXPHOS complexes. Published reports indicate the enzyme activities of the cytochrome bc1 and F1Fo-ATP synthase complexes are significantly reduced in the Δoxa1 mitochondria (Altamura et al., 1996; Szyrach et al., 2003). However, in contrast to the COX complex, which is completely absent in Δoxa1 mitochondria, the cytochrome bc1 complex and F1Fo-ATP synthase complex activities, although strongly reduced, are clearly still detectable in Δoxa1 mitochondria (Altamura et al., 1996). Consistently, the insertion of mitochondrially encoded subunits of these complexes, cytochrome b and Atp6, respectively, was found to be only partially impaired in mitochondria isolated from the pet ts1402 oxa1 strain, in contrast to Cox1 and Cox2 subunits, where the insertion process was almost completely blocked (Hell et al., 2001). Thus the presence of measurable levels of cytochrome bc1 and ATP synthase enzymes in Δoxa1 mitochondria suggests that, in contrast to the COX complex, Oxa1 does not play an essential role in the assembly of these OXPHOS complexes. It is presently unknown whether the translocase function of Oxa1 is required for the biogenesis of the ATP synthase complex or whether Oxa1 may function in some other capacity to assist in the assembly of this complex.
Oxa1 is a member of the conserved Oxa1/YidC/Alb3 protein family, which is found throughout prokaryotes and eukaryotes (Yen et al., 2001; Stuart, 2002; Kuhn et al., 2003). The bacterial members (YidC) of this family have been shown to mediate the insertion of a number of membrane proteins including subunits of the cytochrome oxidase (CyoA) and subunit c (equivalent to mitochondrial Atp9) of the F1Fo-ATP synthase complexes (van Bloois et al., 2004, 2006; van der Laan et al., 2004, 2005; Celebi et al., 2006; Kol et al., 2006; du Plessis et al., 2006). The Alb3 protein members are localized in the thylakoid membrane system of chloroplasts and the role of Alb3 in the insertion of a number of photosynthetic membrane proteins has been documented (Sundberg et al., 1997; Moore et al., 2000; Gerdes et al., 2006). Thus the Oxa1/YidC/Alb3 family represents an evolutionarily conserved group of proteins involved in facilitating critical steps to ensure the integration of proteins into diverse biological membrane systems (Kuhn et al., 2003). Recently the Oxa2/Cox18 protein was proposed to form a new subfamily of the Oxa1/YidC/Alb3 proteins. Members of this group, represented by Cox18 in the yeast S. cerevisiae, and Oxa2 in Neurospora crassa, have been shown to play a critical role in the biogenesis of the COX complex (Souza et al., 2000; Saracco and Fox, 2002; Funes et al., 2004; Preuss et al., 2005). The Cox18 protein is required for the assembly of the COX complex, as it has been reported to be involved in facilitating the export of the C-terminal region of the Cox2 protein from the matrix across the inner membrane to the intermembrane space (Saracco and Fox, 2002). Thus the role of Cox18/Oxa2 proteins in protein translocation has been proposed to overlap with that of its sequence-related Oxa1. Oxa1 however has been proposed to play a pivotal role in the cotranslational insertion of the mitochondrially encoded COX subunits, whereas the Cox18/Oxa2 protein may be functionally different, possibly by being limited to performing posttranslational insertion events (Preuss et al., 2005). In this respect, it is noteworthy that the matrix-localized C-terminal region of Oxa1 which supports the ribosome association is not conserved among the Cox18/Oxa2 protein family members.
The precise function of Oxa1 and its sequence-related Cox18/Oxa2 protein, in the assembly of the ATP synthase is currently unknown and is the central theme of this study. The F1Fo-ATP synthase complex can be divided into two functionally distinct parts, the hydrophilic F1-part, which performs the ATP synthesis and hydrolysis events, and the membrane-embedded Fo-sector, which facilitates the passage of protons across the inner membrane. The F1-sector is composed entirely of nuclear-encoded proteins, whereas the Fo-complex is assembled from both nuclear and mitochondrially encoded proteins. The mitochondrially encoded Atp6, Atp8, and Atp9 are all subunits of the Fo-sector. The Atp9 subunit spans the inner membrane twice in an Nout-Cout orientation and assembles to form a homo-oligomeric ring structure composed of 9–12 subunits. The Atp9 oligomer directly interacts and cooperates with Atp6 to mediate the transport of protons across the membrane, the driving force for ATP synthesis by the coupled F1-sector (Stock et al., 1999; Devenish et al., 2000; Velours and Arselin, 2000; Capaldi and Aggeler, 2002). During the assembly of the ATP synthase complex, the newly synthesized Atp9 has been shown to oligomerize and form an F1-containing subcomplex before associating with the Atp6 protein (see Ackerman and Tzagoloff, 2005 for recent review). The assembly of Atp6 with the F1-Atp9 subcomplex thus represents a late step in the assembly pathway of the F1Fo-complex. The inner membrane protein Atp10 has been shown to act as an Atp6-specific chaperone and by directly interacting with the newly synthesized Atp6 protein Atp10 coordinates the assembly of Atp6 with the F1-Atp9 subcomplex (Herrmann et al., 1994; Tzagoloff et al., 2004).
The involvement and possible overlapping functions of Oxa1 and Cox18/Oxa2 in the assembly of the yeast S. cerevisiae ATP synthase therefore were directly analyzed in this study. We present data here demonstrate that Oxa1 plays an important, but nonessential, role in the assembly of the Fo-sector of the ATP synthase complex. Our data indicate that Cox18/Oxa2, unlike Oxa1, is not required for ATP synthase assembly, however. In addition, our results shed light on how Oxa1 may function to mediate the assembly of the Fo-sector. We demonstrate that Oxa1 can directly interact with Atp9 after its translation in mitochondria and in a manner that does not require the matrix-exposed C-terminal region of Oxa1. We propose that the interaction of Atp9 with Oxa1 is necessary to ensure the efficient and correct assembly of Atp9 oligomers and in such a manner that they are competent to subsequently assemble together with Atp6 in the membrane. In the absence of Oxa1, Atp9 fails to correctly assemble with Atp6 efficiently, and this has severe consequences for the levels of enzymatically active F1Fo-ATP synthase complexes in oxa1-deficient mutants. Finally, we show here that Oxa1 can be purified with ATP synthase complex, highlighting the affinity that Oxa1 has for this complex.
MATERIALS AND METHODS
Yeast Strains
Yeast strains used in this study were wild-type W303-1A (Mata, leu2, trp1, ura3, his3, ade2), rho0 W303-1A (Mata, leu2, trp1, ura3, his3, ade2), and the oxa1-null mutant, Δoxa1 (W303-1A, leu2, trp1, ura3, ade2, OXA1::KANr), whereby the complete OXA1 open reading frame (ORF) was replaced by the kanamycin resistance gene (KANr) using homologous recombination (Wach et al., 1994). Construction of the strains Δcox18 and Δoxa1, Δcox18 was performed by the introduction of the HIS3 gene into the COX18 locus of wild-type and Δoxa1::KANr cells, resulting in the replacement of the entire COX18 ORF. Correct homologous recombination of the HIS3 or KANr gene at their respective gene locus was verified by PCR analysis (results not shown). Construction of the Δimp1 strain was performed by introduction of the LEU2 auxotrophic gene into the wild-type cells, resulting in the replacement of the ORF of the IMP1 ORF (Cruciat et al., 2000). Strains expressing full-length Oxa1 and Oxa1His from the galactose-inducible GAL10 promoter have been previously described (Jia et al., 2003). Construction of the strain expressing histidine-tagged C-terminally truncated Oxa1 (Oxa1Δ83His) was achieved as follows. A DNA fragment encoding for a C-terminally truncated Oxa1 (codons 1-318) followed by 12 histidine-encoding codons and a translational stop codon was generated by PCR. This fragment was then cloned into a yeast-integrating vector Yip351-GAL10 (LEU2) and downstream of the galactose-inducible promoter GAL10. The resulting plasmid Yip351-GAL10-OXA1Δ83His was cloned into the yeast genome of the oxa1-null mutant at the LEU2 locus after linearization with BstEII.
H2O2 Treatment and Dispersal of Atp9 Oligomer to Monomers
Mitochondria (200 μg) in 20 mM Tris-HCl, pH 8.0, buffer were mixed with H2O2, final concentration 4% (vol/vol). The samples were shaken at 25°C for 10 min. SDS sample buffer (Laemmli, 1970) was added, and samples were heated at 95°C for 5 min before loading the gel. This treatment with H2O2 resulted in an efficient dispersal of the SDS-resistant Atp9 oligomers into monomeric subunits (results not shown).
Digitonin Solubilization of Mitochondria and Clear Native Gel Electrophoresis
Clear native-PAGE (CN-PAGE) analysis of F1Fo-ATP synthase complexes after solubilization with digitonin was performed essentially as described previously (Wittig and Schägger, 2005). Mitochondria (200 μg of protein) were lysed with 36 μl of digitonin buffer (34 mM potassium acetate, 34 mM HEPES-KOH, pH 7.4, 11.4% glycerol, 1 mM phenylmethylsulfonyl fluoride) and digitonin (at concentrations indicated) for 30 min on ice and subjected to a clarifying spin (JA-25.50 rotor, Beckman Avanti J-25 system (Fullerton, CA); 30 min, 30,000 × g). The supernatants were collected, and 2 μl of 50% glycerol was added to the samples before electrophoresis. Samples were analyzed by CN-PAGE using a 3.5–10% gradient gel.
In Gel Oligomycin-sensitive ATPase Activity Assay
F1Fo-ATPase activity assay was performed after CN-PAGE, as previously described (Wittig and Schägger, 2005). Native gels were incubated for 3 h in a buffer containing 270 mM glycine, 35 mM Tris-HCl, pH 8.4. In a parallel assay, additional oligomycin (5 μg/ml) was supplemented in the buffer. The ATP hydrolysis assay was carried out in 270 mM glycine, 35 mM Tris-HCl, 8 mM ATP, 14 mM MgSO4, 0.2% Pb(NO3)2, pH 8.4, and stopped after 5–10 min by a 30 min incubation in 50% methanol, 50% water, followed by a 30 min incubation in water.
In Organello Labeling of Mitochondrial Translation Products
In organello labeling of mitochondrial translation products was performed in isolated mitochondria for 30 min at 25°C, essentially as described previously (Hell et al., 2001). After translation and chase in the presence of cold methionine and puromycin for 30 min, mitochondria were lysed in SDS-sample buffer (Laemmli, 1970). Translation products were analyzed by SDS-PAGE and autoradiography.
Ni-NTA Purification of Oxa1His Complex
Mitochondria harboring the GAL10 expressed Oxa1His derivative (200 μg) were solubilized in 200 μl of TNT buffer (1% Triton X-100, 300 mM NaCl, 60 mM Tris-HCl, pH 7.4) for 5 min on ice. After a clarifying spin (20,860 × g, 15 min at 4°C), the supernatants were incubated for 1 h at 4°C with the Ni-NTA beads (equilibrated in the TNT buffer containing 30 mM imidazole). The beads were washed with TNT-imidazole buffer, and purified proteins were eluted with SDS sample buffer containing 5% (vol/vol) β-mercaptoethanol and 0.5 M imidazole followed by SDS-PAGE analysis. When indicated, mitochondria were initially solubilized with 0.1% SDS, and a buffer containing 0.1% SDS instead of Triton X-100 was used.
Gel Filtration Analysis
Mitochondria (200 μg protein) were solubilized in 200 μl of buffer containing 1% Triton X-100, 300 mM NaCl, and 60 mM Tris-HCl, pH 7.4, for 5 min on ice. After a clarifying spin (100,000 × g, 15 min at 4°C), the supernatant was loaded to the Superose 12 column. Samples were run with buffer containing 0.1% Triton X-100, 300 mM NaCl, and 60 mM Tris-HCl, pH 7.4, at the speed of 0.1 ml/min, and 0.5-ml/fraction eluent was collected. Proteins were trichloroacetic acid (TCA) precipitated from each fraction and analyzed by SDS-PAGE. Immunoblotting was performed to analyze the presence of Atp9, Atp6, and Atpβ.
Miscellaneous
Mitochondria were isolated from cultures grown at 30°C in YP-0.5% lactate, 2% galactose medium. Isolation of mitochondria was performed as described previously (Jia et al., 2003).
RESULTS
The Assembly of the F1Fo-ATP Synthase Is Perturbed in the Absence of Oxa1, But Not Cox18
To investigate the requirement for Oxa1 and Cox18 in the biogenesis of the ATP synthase enzyme, we analyzed both the enzymatic activity and assembly state of ATP synthase complexes in mitochondria isolated from S. cerevisiae yeast mutants deficient in Oxa1 and/or Cox18 (Figure 1A). Mitochondria isolated from the single deletion strains Δoxa1 and Δcox18 and also from the double gene knockout strain Δoxa1, Δcox18 were solubilized with the detergent digitonin, and the resulting extracts were subjected to clear native PAGE (CN-PAGE) analysis. The oligomycin-sensitive enzymatic activity of the ATP synthase complexes was assayed after CN-PAGE and using an in-gel ATP hydrolysis assay, in the presence or absence of the F1Fo-ATP synthase specific inhibitor oligomycin (Figure 1A). In parallel, mitochondria isolated from the isogenic wild-type strain and an independent cytochrome oxidase–deficient strain, the Δimp1 strain, were also analyzed, as controls.
Figure 1.
Characterization of assembly defects of ATP synthase in Δoxa1 and Δcox18 mutants. (A) Mitochondria from wild-type (WT), Δoxa1, Δcox18, Δoxa1, Δcox18, and Δimp1 strains were solubilized with buffer containing 2% digitonin and subjected to CN-PAGE followed by in-gel ATPase activity assay in the absence or presence of oligomycin (5 μg/ml, + or − oligom, as indicated). The position of the dimeric (Vdim) and monomeric (Vmon) forms of the F1Fo-ATP synthase are indicated. (B) Mitochondria from WT and Δoxa1 strains were solubilized with either 1 or 2% digitonin as indicated subjected to CN-PAGE, and Western blotted. ATP synthase complexes were detected with an anti-F1 antiserum. The presence of F1-containing subcomplexes in the Δoxa1 mitochondria are indicated by an asterisk (*). (C) Mitochondria from rho0, WT, Δoxa1, Δcox18, Δimp1, and Δcox18, Δoxa1 strains were subjected to SDS-PAGE and Western blotted using antisera against F1 subunit β (Atpβ), Fo subunits Atp4, Atp6, Atp9, and the inner membrane protein Tim23. For the analysis of the Atp9 levels, mitochondria were treated with H2O2 (see Materials and Methods) to disperse SDS-resistant Atp9 oligomers to monomeric proteins, before the SDS-PAGE analysis.
Under the digitonin lysis conditions used, the active and oligomycin-sensitive F1Fo-ATP synthase enzyme complex from wild-type mitochondria was solubilized and resolved on CN-PAGE as a combination of dimeric and monomeric complexes, consistent with previously published observations (Arnold et al., 1998; Saddar and Stuart, 2005). The observed monomeric F1Fo-ATP synthase complexes are thought to be a result of a dissociation of some ATP synthase dimers by the increased detergent concentrations (Arnold et al., 1998). Enzymatically active F1Fo-ATPase complexes were also detected in the Δoxa1 mitochondrial extract. However, the levels of these complexes were strongly reduced when compared with that in wild-type mitochondria (∼20% of wild-type levels; Figure 1A). Observation of both dimeric and monomeric forms of the complex in the Δoxa1 mitochondria indicated that the ATP synthase complexes could fully assemble, albeit at reduced levels, in the absence of Oxa1. Importantly, the F1Fo-ATP synthase complex in Δoxa1 mitochondria could also be inhibited through the addition of oligomycin (Figure 1A). This inhibition is an indicator of the correct and functional assembly of the coupled F1Fo-complexes in the Δoxa1 mitochondria, because oligomycin acts specifically on the Fo-sector, where it is thought to bind to the region of Atp9 that interfaces with Atp6 (John and Nagley, 1986; Nagley et al., 1986). Uncoupled F1-complexes are not inhibited through the addition of oligomycin. In the Δoxa1 mitochondria, a low level of smaller molecular weight complexes displaying oligomycin-insensitive ATPase activity were also detected, indicating the possible presence of partially assembled F1-intermediates in the absence of Oxa1 (results not shown).
Enzymatically active F1Fo-ATP synthase complexes, which were oligomycin sensitive, were clearly detected in the Δcox18 mutant at the levels that were only slightly reduced when compared with the wild-type control mitochondria. Indeed the levels of active ATP synthase complexes in the Δcox18 mitochondria were similar to those of an independent COX assembly mutant, the Δimp1 strain (Figure 1A). Imp1 is a protease involved in the maturation of the Cox2 protein, and Δimp1 mutants are deficient in COX complexes (Nunnari et al., 1993). Subunits of the ATP synthase complex do not undergo Imp1-specific proteolytic events; hence, the assembly of this complex is not directly affected in the Δimp1 mutants. The slight reduction of ATP synthase levels in the Δcox18 and Δimp1 mutants may therefore be due to an indirect effect in response to the absence of the COX complex activity (Figure 1A).
The activity levels of the ATP synthase complex, and its sensitivity to oligomycin, in the Δcox18, Δoxa1 mitochondrial were comparable to those isolated from the Δoxa1 strain (Figure 1A). Thus, despite sharing sequence homology, the Cox18 and Oxa1 proteins do not appear to bear overlapping functions with regards to the assembly of the ATP synthase complex. Deletion of the COX18 gene did not abolish the ability of a significant, albeit reduced, level of F1Fo-ATP synthase complex to assemble in the absence of Oxa1.
To demonstrate that the strongly reduced levels of enzymatically active ATP synthase complexes in the Δoxa1 mitochondria reflected an overall reduction in the protein levels of the complex, rather than a lack or inhibition of its activity, the protein levels of the assembled ATP synthase in Δoxa1 mitochondria was further analyzed after CN-PAGE analysis by Western blotting and immunedecoration with antisera against the purified F1-complex (Figure 1B). Consistent with the enzyme activity measurements, the level of the assembled ATP synthase dimers and monomers were significantly reduced, but remained detectable in the Δoxa1 mitochondria (Figure 1B). Furthermore in the Δoxa1 mitochondria, lower molecular weight F1-complexes, corresponding to partially assembled F1-containing complexes were also observed, indicating a defect in the assembly of the ATP synthase complexes in the absence of Oxa1. This level of partially assembled F1-complexes was not observed in the wild-type control mitochondria. A similar accumulation of partially assembled F1-complexes was also observed in the Δoxa1, Δcox18 double mutant, but not, however, in the Δcox18 single null mutant mitochondria (results not shown).
Taken together, the CN-PAGE results indicate a strong reduction in both the protein and enzyme activity levels of the assembled F1Fo-ATP synthase complex in the absence of Oxa1, but not in the absence of Cox18. We conclude therefore that Oxa1, unlike Cox18, plays an important role in the biogenesis of the enzymatically active F1Fo-ATP synthase complex.
Fo-Sector Subunits Atp4 and Atp6, But Not Atp9, Are Strongly Reduced in the Absence of Oxa1
Given that Oxa1 has been described to be involved in facilitating the insertion of proteins into the inner membrane, we analyzed directly the steady state levels of ATP synthase subunits of the membrane embedded Fo-sector in Δoxa1 mitochondria. Mitochondria isolated from the single null mutant strains, Δoxa1 and Δcox18 and also from the double knockout strain Δcox18, Δoxa1 were analyzed by SDS-PAGE and Western blotting. Wild-type, COX-deficient Δimp1 and rho0 (contain no mtDNA) mitochondria were also analyzed in parallel as controls. Immunedecoration was subsequently performed to assess the steady state levels of the Fo-sector subunits Atp4, Atp6, and Atp9 and also the F1-sector subunit, Atpβ (Figure 1C). The level of Tim23, an inner membrane protein and component of the TIM23 translocase, was analyzed as a loading control for the mitochondria.
Consistent with the CN-PAGE results, the levels of Atp4, Atp6, and Atp9 were not adversely affected in the Δcox18 mitochondria, because they were similar to those observed in the wild-type and Δimp1 mitochondria. In the Δoxa1 mitochondria, however, the steady state levels of Atp4 and Atp6 were significantly reduced. Although reduced relative to wild type, the levels of Atp9 in the mitochondria isolated from the Δoxa1 strain were not so dramatically affected as those of Atp6 or Atp4 proteins. The levels of Atp4, Atp6, and Atp9 in the Δcox18, Δoxa1 double mutant were similar to those of the Δoxa1 single mutant, again supporting the conclusion that Cox18 does not contribute to the assembly or stability of the ATP synthase in the absence of Oxa1. Note we were unable to assess the steady state levels of the other mitochondrially encoded Fo-ATP synthase subunit, Atp8, because an antibody specific for the Atp8 protein was not available. In contrast to the Fo-subunits, the F1-sector subunit Atpβ was not so significantly reduced in the Δoxa1 mitochondria. Thus it would appear that the reduction in assembled F1Fo-ATP synthase complexes in the absence of Oxa1 reflects the limiting levels of Fo-sector, rather than F1-sector subunits.
Oxa1 Associates Specifically with Newly Synthesized Atp9
We investigated next whether Oxa1 directly plays a role in the assembly of the ATP synthase complex by physically interacting with mitochondrially encoded subunits of the Fo-sector of the ATP synthase complex in a posttranslational manner and before their assembly into the F1Fo-ATP synthase complex. Three of the Fo-sector subunits are encoded by the mitochondrial DNA: Atp6, Atp8, and Atp9. To address a possible association between Oxa1 and these subunits of the Fo-sector, we performed an in organello translation in the presence of [35S]methionine using mitochondria isolated from the wild-type strain or from strains expressing authentic Oxa1 or the Oxa1His-tagged derivatives (Figure 2A). The pattern of protein translation products was similar between the different mitochondrial types with the exception of a number of higher molecular mass radiolabeled bands (range 55–75 kDa), which correspond to SDS-resistant forms of the mitochondrially synthesized proteins. Two prominent bands (70 and 72 kDa) were observed in the mitochondria harboring the overexpressed Oxa1 and Oxa1His proteins, respectively. A weaker 70-kDa band was observed in the wild-type mitochondria. The shift in the electrophoretic mobility of this band in the mitochondria harboring the His-tagged derivative of Oxa1 suggested that these additional radiolabeled bands represent an SDS-resistant complex between a mitochondrially encoded protein and Oxa1. The mobility difference between the two forms of this band (between the Oxa1 and Oxa1His mitochondria) are accounted for by the presence of the additional 12 histidine residues (1.7 kDa in mass) of the Oxa1His derivative.
Figure 2.
Oxa1 associates specifically with newly synthesized Atp9. (A) In organello translation was performed in the presence of [35S]methionine for 30 min at 25°C in mitochondria isolated from the wild-type (WT) yeast strain, a strain overexpressing Oxa1 (GAL-Oxa1) and from a strain expressing histidine-tagged Oxa1 (GAL-Oxa1His). Incorporation of [35S]methionine was stopped by addition of excess unlabeled methionine, and the mitochondria were solubilized in SDS buffer and subjected to SDS-PAGE. The bands marked with an asterisk (*) indicated the complex formed by Oxa1 and a translation product. The positions of the molecular mass standards and mitochondria-encoded proteins, Var1, Cox1, Cox2, cytochrome b (Cytb), Cox3, Atp6, Atp8, and Atp9, are indicated. (B) In organello translation was performed in mitochondrial harboring overexpressed Oxa1 or Oxa1His as described in A. The complex formed between Oxa1His and the radiolabeled translation product(s) were purified by Ni-NTA chromatography and either loaded directly on the gel (−TCA) or after TCA precipitation (+TCA). The mobility of the SDS-resistant Oxa1His-Atp9 complex and the monomeric Atp9 protein after release by TCA precipitation are indicated.
To further verify that this radiolabeled band represents an Oxa1-containing complex and to ascertain the identity of the radiolabeled associating protein, we took advantage of its SDS-stability and the formation of the complex in mitochondria harboring the His-tagged version of Oxa1. In organello translation was performed in mitochondria bearing the Oxa1His protein, and after translation mitochondria were lysed in an SDS-containing buffer, and the solubilized proteins were subjected to Ni-NTA chromatography. Mitochondria containing overexpressed wild-type Oxa1, i.e., nontagged Oxa1, were analyzed in parallel as a control. The Ni-NTA purified proteins were analyzed either directly by SDS-PAGE or after a TCA-precipitation step. The TCA precipitation step has been previously described to disperse SDS-resistant complexes of mitochondrial translation products (Herrmann et al., 1994; Tzagoloff et al., 2004). The SDS-resistant radiolabeled complex was recovered on the Ni-NTA beads, when translation was performed in the mitochondria bearing the His-tagged Oxa1 derivative, but not when translation was performed in the control mitochondria bearing the expressed nontagged version of Oxa1 (Figure 2B). This result confirms that the radiolabeled complex was indeed an Oxa1His-containing complex. TCA precipitation of this purified material resulted in the release of one radiolabeled species, which from its electrophoretic behavior was determined to be Atp9, using a gel system that resolves the Atp9 protein from the closely migrating Atp8 protein. In addition, using this gel system, the affinity-purified radiolabeled Atp9 protein was shown to comigrate with endogenous Atp9 protein, which was recognized by Atp9-specific antibodies (results not shown, and also see Figure 6). No evidence to suggest that this complex contained Atp6 (or the other mitochondrially encoded proteins), in addition to Atp9, was obtained. Thus we conclude that after translation in mitochondria, newly synthesized Atp9, remains in association with Oxa1, where it can form an SDS-resistant complex.
Figure 6.
Oxa1 interacts with Atp9 before its assembly into an F1Fo-ATP synthase complex. In organello translation in the presence of [35S]methionine was performed in mitochondria bearing overexpressed Oxa1 or Oxa1His (Oxa1 or Oxa1His) or in mitochondria harboring a histidine-tagged Atpβ subunit (AtpβHis), as indicated. After a chase reaction as described in Figure 2, the mitochondria were reisolated and lysed in Triton X-100 containing buffer and subjected to Ni-NTA chromatography, as described in Figure 5D. The Ni-NTA bound material (indicated by B) and a sample corresponding to 10% of the total material applied to the Ni-NTA beads were analyzed by SDS-PAGE, Western blotting, and autoradiography (right panels, indicated by 35S), followed by immunedecoration with Atp9 and Atp6 specific antisera (α-Atp9 and α-Atp6, respectively; left panel). The electrophoretic positions of Atp6, Atp8, and monomeric Atp9 are indicated. The position of the 72-kDa Oxa1-Atp9 SDS-resistant oligomer is indicated by an asterisk (*).
Assembly of Atp9 Oligomer Is Affected in Absence of Oxa1
Given that Oxa1 associates with newly synthesized Atp9 in a posttranslational manner, we next addressed if Oxa1 is involved in the assembly of Atp9 into the F1Fo-ATP synthase complex. During its assembly pathway, Atp9 forms a homo-oligomeric complex, which then associates with F1-sector subunits (Herrmann et al., 1994; Tzagoloff et al., 2004). We term this F1-containing Atp9-assembly intermediate here the Atp9-F1 subcomplex. Formation of the Atp9-F1 subcomplex has been shown to precede the Atp10-dependent step of coassembly of Atp9 with Atp6 (Herrmann et al., 1994; Tzagoloff et al., 2004). As reported earlier, the steady state levels of Atp9, in contrast to other Fo-subunits, appear not to be drastically reduced in the absence of Oxa1. We hypothesized therefore that Atp9 can assemble to an intermediate stage and be at least partially stabilized in the absence of Oxa1.
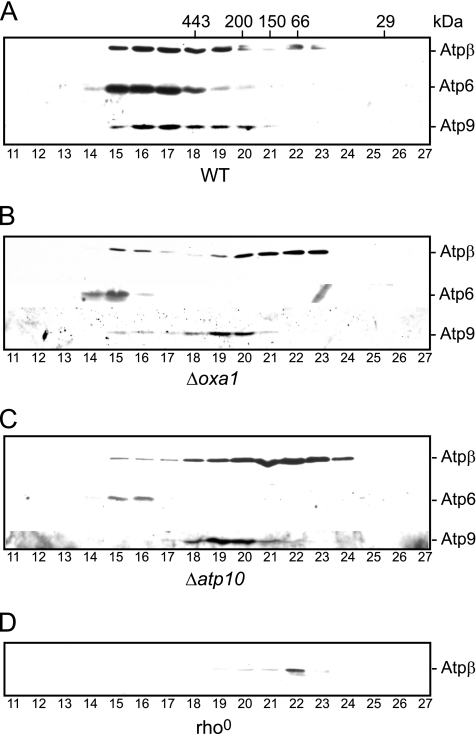
We therefore addressed the assembly state of the endogenous Atp9 in the Δoxa1 mitochondria and compared it to that of Atp9 accumulated in the Δatp10 mitochondria (Figure 3). Wild-type and rho0 mitochondria were also analyzed in parallel as controls. Mitochondria were lysed using Triton X-100, a detergent that maintains the stability of the monomeric form (but not the dimeric form) of the F1Fo-ATP synthase, and the assembly state of the Atp9 subunit was analyzed by size exclusion chromatography (Figure 3). In wild-type mitochondria, the majority of Atp9 was present in the monomeric ATP synthase complex, because it coeluted from the column together with Atp6 and Atpβ, in fractions 15–18, corresponding to a complex of ∼550 kDa. Some Atp9 eluted in later fractions together with a low level of Atpβ (fractions 19 and 20), with a molecular mass of ∼300 kDa, and most likely corresponds to the previously described Atp9-F1 subcomplex. Atp6 was found exclusively with the fully assembled ATP synthase complex, however (Figure 3A).
Figure 3.
Atp9 assembles into an F1-containing subcomplex in the absence of Oxa1. Mitochondria from wild-type (WT; A), Δoxa1 (B), Δatp10 (C), and rho0 (D) strains were solubilized with 1% Triton X-100 containing buffer and subjected to size exclusion chromatography on Superose 12 column. Fractions (11–27) eluted from the column were TCA precipitated and subjected to SDS-PAGE followed by Western blotting with antisera against F1 subunit Atpβ and Fo subunits Atp6 and Atp9. The elution profile of the molecular mass standards 443, 200, 150, 66, and 29 kDa are indicated in the wild-type panel. Note the exposure time for Atp6 in the Δoxa1 and Δatp10 mitochondria was significantly longer than that for the wild-type control, because of the strongly decreased levels of Atp6 in these mutant mitochondria.
In the Δoxa1 mitochondria, the majority of Atp9 eluted from the column with some of the Atpβ protein, in fractions 19 and 20, which correspond to the ∼300-kDa Atp9-F1 subcomplex. Only a minor fraction of Atp9 eluted from the column in the fractions (fractions 15–18) corresponding to fully assembled F1Fo-ATP synthase, as indicated by the coelution of Atp6 and Atpβ (Figure 3B). The observed low level of fully assembled F1Fo-ATP synthase complexes in the Δoxa1 strain is consistent with the previous CN-PAGE results (Figure 1). Hence only a small fraction of the total Atpβ subunit was present in the fractions corresponding to the fully assembled F1Fo-ATP synthase complex; the majority of Atpβ eluted from the column either with Atp9-F1 subcomplex (fractions 19 and 20) or later in F1-complex of ∼150 kDa (fractions 21–23). The latter F1-complex was similar in mass as an Atpβ-containing assembly intermediate that could be observed in the rho0 mitochondria, which lack all of the mtDNA-encoded Fo-sector subunits, Atp9, Atp8, and Atp6 (Figure 3D). We conclude from these results that in the Δoxa1 mitochondria, the Atp9 protein, which does not assemble into the complete F1Fo-ATP synthase complex, accumulates in an Atp9-F1 subcomplex. The behavior of the Atp9 in the Δoxa1 mitochondria was similar to that in the Δatp10 mutant mitochondria, where the Atp9 protein also accumulated in a 300-kDa complex (Figure 3C). As mentioned earlier, assembly of Atp9 into its homo-oligomer, followed by its association with F1-subunits, occurs in an Atp10-independent manner; but further assembly of this oligomer with Atp6 does not occur in the absence of Atp10, thus preventing the complete assembly of the F1Fo-ATP synthase complex. The similarity of the behavior of Atp9 in Δoxa1 and the Δatp10 mitochondria, i.e., competence to assemble in a stable manner in an Atp9-F1 subcomplex, would suggest that further assembly of the ATP synthase in the absence of Oxa1 is limited by the step of coassembly of the Atp9 oligomer with the Atp6 protein.
Newly Synthesized Atp9 Can Oligomerize in the Absence of Oxa1, But Assembly with Atp6 Is Perturbed
In a second independent approach we tested the ability of newly synthesized Atp9 to assemble in the absence of Oxa1, by analyzing its ability to form SDS-resistant oligomers in Δoxa1 mitochondria after its synthesis (Figure 4). When [35S]methionine-labeling of mitochondrial translation products is performed in isolated wild-type mitochondria, in addition to the eight mitochondrially encoded proteins, two additional slower migrating radiolabeled bands, with electrophoretic mobilities corresponding to the masses of 48- and 54-kDa products, are observed (Figure 4A), as has been previously reported (Herrmann et al., 1994; Tzagoloff et al., 2004). These 48- and 54-kDa products correspond to oligomers of Fo-subunits that are released from fully or partially assembled F1Fo-ATP synthase complexes, but fail to be completely depolymerized by the SDS, due to the very hydrophobic nature of the Atp9-Atp9 interactions in the Fo-oligomer. These SDS-resistant Atp9 complexes can be dispersed into individual monomeric polypeptides through TCA precipitation (Herrmann et al., 1994; Tzagoloff et al., 2004), where it has been established that the 48-kDa product is composed of Atp9 proteins, whereas the 54-kDa product is generated from a later stage assembly intermediate as it contains Atp6, in addition to Atp9 (Tzagoloff et al., 2004). In wild-type mitochondria the level of the 54-kDa oligomer was observed to be greater than that of the 48-kDa complex, indicating efficient assembly of Atp9 with the Atp6 protein in the presence of Oxa1 (Figure 4B). The level of the 54-kDa complex observed, but of not the 48-kDa oligomer, was partially affected in the presence of oligomycin, indicating the binding of the ATP synthase–specific inhibitor may adversely affect the formation of the Atp9-Atp6 complex (Figure 4B). Assembly of the 48-kDa Atp9-containing complex was observed in the Δoxa1 mitochondria, indicating the ability of Atp9 to oligomerize in the absence of Oxa1, a result consistent with the previously described gel filtration analysis. In contrast however, to the wild-type control mitochondria, formation of the 54-kDa complex was perturbed in the absence of Oxa1, because a significant decrease in the amount of the 54-kDa product relative to the 48-kDa complex was observed (Figure 4B).
Figure 4.
Assembly of the ATP synthase complex is impaired in Δoxa1 mitochondria. Mitochondrial from wild type (A and B) and Δoxa1 (B) strains were subjected to translation and radiolabeled for 30 min at 25°C followed by a chase period in the presence of excess cold methionine, an ATP-regenerating system and in the absence or presence of oligomycin, as indicated. Mitochondria were reisolated, lysed and subjected to SDS-PAGE, Western blotting, and autoradiography. The positions of the 54-kDa SDS-resistant complex between Atp9 and Atp6, and the 48-kDa Atp9-oligomer, are indicated, by 54k and 48k, respectively. (B) The levels of both the 54-kDa (dark gray bars) and 48-kDa (light gray bars) complexes were quantified by Phosphorimaging and are each expressed as a percentage of the total, i.e., the sum of the 54- and 48-kDa signals.
In summary these data support the conclusion that the oligomerization of newly synthesized Atp9 protein may occur in the absence of Oxa1, but its further assembly into a Fo-sector, a step that involves its interaction with the Atp6 protein, is perturbed in the Δoxa1 mitochondria.
Oxa1 Copurifies with Components of the F1Fo-ATP Synthase
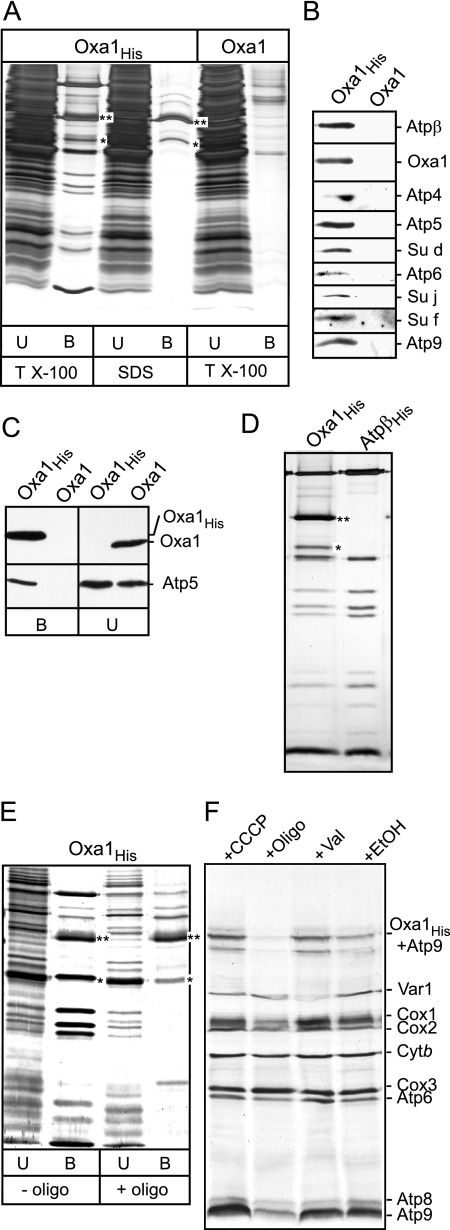
Given that Oxa1 associates with newly synthesized Atp9 in a posttranslational manner, we investigated whether the Oxa1 complex was also associated with the pre-existing assembled ATP synthase complex in mitochondria (Figure 5). To do so, we used the derivative of Oxa1 bearing a C-terminal histidine tag (Oxa1His) expressed in the Δoxa1 strain and performed affinity purification of the Oxa1His and its associating proteins, using metal chelating chromatography. Mitochondria harboring the Oxa1His derivative were lysed with Triton X-100, and the Oxa1His protein was purified on Ni-NTA agarose beads. Oxa1His and copurifying proteins were analyzed by SDS-PAGE followed by silver staining (Figure 5A). In addition to Oxa1His (38 kDa), a number of other polypeptides were observed to copurify with Oxa1His. We verified the identity of these Oxa1-associating proteins as subunits of the ATP synthase complex by either MALDI-TOF analysis (Atpα and Atpβ subunits) or through Western blot analysis using subunit specific antisera (Atpα, Atpβ, Atp4, Atp5, Atp6, and Atp9 and subunits f, j, and d; Figure 5B, Su f, Su j and Su d). The recovery of these ATP synthase subunits on the Ni-NTA agarose beads was specific for the presence of the His-tagged version of the protein, because they were not recovered when mitochondria bearing the nontagged version of Oxa1 were used as a control (Figure 5B). Furthermore, the recovery of the ATP synthase subunits on the beads appeared to be due to their specific association with Oxa1His, because when mitochondrial lysis and purification was performed using the more stringent detergent SDS, only Oxa1His was recovered on the beads (Figure 5A).
Figure 5.
The ATP synthase complex can be copurified together with Oxa1. (A) Mitochondria isolated from strains expressing Oxa1His or Oxa1 were solubilized with buffer containing either 1% Triton X-100 (TX-100) or 0.1% SDS as indicated and incubated with Ni-NTA beads as described in Materials and Methods. Twenty percent of the unbound material (U) and the Ni-NTA beads purified material (indicated by B) were analyzed by SDS-PAGE and silver-staining. (B) Mitochondria isolated from strains expressing Oxa1His or Oxa1 (GAL-Oxa1 and GAL-Oxa1His, respectively) were solubilized with buffer containing 1% Triton X-100, as indicated, and were incubated with Ni-NTA beads. The Ni-NTA–purified material was analyzed by SDS-PAGE and Western blotting with antisera against ATP synthase subunits Atpβ, Atp4, Atp5, Atp6, Su d, Su j, Su f, Atp9, and Oxa1 as indicated. (C) Mitochondria harboring expressed Oxa1 or Oxa1His proteins were lysed in Triton X-100 containing buffer and were subjected to Ni-NTA purification. The purified material (indicated by B) and 10% of the unbound material (U) were subjected to SDS-PAGE, Western blotting, and immunedecoration with Oxa1 and Atp5 antisera as indicated. (D) Mitochondria harboring expressed Oxa1His (200 μg protein) or histidine-tagged Atpβ (AtpβHis; 50 μg protein) were solubilized with buffer containing 1% Triton X-100 as indicated and incubated with Ni-NTA beads. The Ni-NTA–purified material was analyzed by SDS-PAGE and silver staining. (E) Mitochondria harboring Oxa1His were solubilized with buffer containing 1% Triton X-100 in the absence or presence of oligomycin (10 μg/ml) as indicated. Samples were further analyzed as described in A. (F) In organello translation was performed in Oxa1His containing mitochondria in the presence of CCCP (100 μM), oligomycin (10 μg/ml +oligo), valinomycin (0.5 μM, +Val), or ethanol (+EtOH) as a control, for 30 min at 25°C. After the addition of cold methionine, mitochondria were reisolated and subjected to SDS-PAGE and autoradiography. The position of the radiolabeled Atp9-Oxa1His SDS-resistant complex is indicated. The asterisks (** and *) in A, C, and D indicate the positions of Oxa1His and a C-terminal degradation product of Oxa1His, respectively.
Western blotting of both the Ni-NTA purified and unbound material indicated that the recovery of Oxa1His on the beads was complete (Figure 5C). Approximately 5% of the Atp5 protein was recovered with Oxa1His on the Ni-NTA beads, thus giving an indication of the level of ATP synthase complexes that can be found in association with Oxa1 under these purification conditions (Figure 5C). Our findings here suggest that the ATP synthase complex purified together with Oxa1His may represent an assembled F1Fo-complex. Analysis of the electrophoretic profile of the subunits, which copurified with Oxa1His, was similar to that of the ATP synthase complex purified in parallel under the same Triton X-100 lysis conditions by using the His-tagged version of the β-subunit of the F1-sector (Mueller et al., 2004; Figure 5D), suggesting that an intact or very late assembly intermediate of the ATP synthase complex was associated with Oxa1.
The recovery of the ATP synthase components with the Oxa1His was affected by the presence of the F1Fo-ATP synthase specific inhibitor, oligomycin. When mitochondria harboring the Oxa1His derivative were incubated with oligomycin before lysis and Ni-NTA agarose chromatography, the ATP synthase subunits failed to copurify with the Oxa1His protein (Figure 5E). The recovery of the Oxa1His protein itself on the Ni-NTA beads was not affected by the oligomycin (Figure 5E). As mentioned earlier, oligomycin binds to a hydrophobic region of Atp9 that interfaces with Atp6 and by doing so blocks the transfer of H+ ions between these proteins. Thus the binding of oligomycin may interfere with the ability of Atp9 to bind in a stable manner with the Oxa1 protein. As shown previously (Figure 4B), oligomycin also appears to affect the ability of newly synthesized Atp9 and Atp6 proteins to assemble together in a stable manner. Pretreatment of mitochondria with carbonyl cyanide m-chlorophenylhydrazone (CCCP) before detergent lysis and Ni-NTA purification, did not adversely affect the ability of the Oxa1 protein to remain in association with the ATP synthase complex (results not shown). CCCP is a potent dissipater of the proton motive force and hence indirectly inhibits the F1Fo-ATP synthase, because of the lack of an available pH gradient to drive the forward ATP synthesis reaction. We therefore consider it likely that the ATP synthase–specific inhibitor oligomycin interferes with the Fo-sector–Oxa1 interface directly, rather than indirectly through an inhibition of the ATP synthesis activity of the F1Fo-complex. In support of this conclusion, formation of the previously described SDS-resistant complex between Oxa1 and newly synthesized radiolabeled Atp9 was also adversely affected when the in organello translation reaction was performed in the presence of oligomycin (Figure 5F). Formation of the Atp9-Oxa1 complex was not affected when translation was performed in the presence of CCCP or valinomycin (dissipates the Δψ aspect of proton motive force; Figure 5F). These results support the conclusion that oligomycin acts directly on the Atp9-Oxa1 interface to disrupt this interaction and is not acting indirectly through modulation of the electrochemical gradient and hence the activity of the ATP synthase enzyme.
In summary, we demonstrate that Oxa1 can physically interact with the assembled ATP synthase complex in a specific manner. Furthermore, association of Oxa1 with the ATP synthase appears to involve binding at the Atp9 interface because, like the newly synthesized Atp9-Oxa1 interaction, it is perturbed by the presence of oligomycin. It is important to note that there are multiple copies of Atp9 subunit and only one Atp6 subunit per Fo-sector. It is therefore conceivable that in a given Fo-sector Atp9-Oxa1 and Atp9-Atp6 interactions may occur, and this may explain why Atp6 is copurified with the other ATP synthase subunits bound to the Oxa1His protein
Association of Oxa1 with Newly Synthesized Atp9 Can Occur before It Assembles into an F1Fo-ATP Synthase Complex
Given that Oxa1 can associate with the assembled ATP synthase complex, we next addressed whether the observed association between the newly synthesized Atp9 and the Oxa1 protein was possibly due to its prior assembly into an assembled F1Fo-ATP synthase complex or whether Atp9's association with Oxa1 occurs before the complete assembly of Atp9. We therefore compared the amount of newly synthesized Atp9, which can be found in association with Oxa1His with that associated with the F1-ATP synthase complex, using mitochondria harboring the AtpβHis derivative (Figure 6). Association of radiolabeled Atp9 with the AtpβHis protein would be observed if the in organello synthesized Atp9 protein can assemble with the F1-sector after synthesis in isolated mitochondria.
In organello translation in the presence of [35S]methionine was performed in mitochondria bearing the Oxa1His protein or AtpβHis protein and after translation, mitochondria were lysed in buffer containing Triton X-100 (detergent conditions that maintain the assembled F1Fo-ATP synthase complex intact) and the solubilized proteins were subjected to Ni-NTA chromatography. When translation was performed in the mitochondria bearing the Oxa1His derivative, both the SDS-resistant Oxa1His-Atp9 72-kDa complex and monomeric Atp9 were recovered on the Ni-NTA beads, as previously described (Figure 6, right panels). The level of newly synthesized Atp9 found in association with the AtpβHis protein, however, was significantly lower than that observed together with the Oxa1His protein (Figure 6, right panels), indicating that the majority of the radiolabeled Atp9 purified with Oxa1His had not been assembled into an F1Fo-ATP synthase complex. On the other hand, immunedecoration with Atp9-specific antiserum showed that significantly more of the endogenous (i.e., pre-existing/assembled) Atp9 was recovered with the purified AtpβHis protein, than with the Oxa1His protein (Figure 6, left panel). A similar observation was made also for the preexisting Atp6 protein (Figure 6, left panel). The recovery of the Atp9 and Atp6 proteins with the AtpβHis protein further confirms the ability of this protein to affinity-purify the assembled ATP synthase complex, as was previously indicated in Figure 5D. It is important also to note here that the radiolabeled Atp9 protein affinity-purified with AtpβHis and Oxa1His comigrated with the immunedecorated endogenous Atp9 protein using a gel system where Atp9 can be resolved from Atp8, thus providing further verification for the identity of the radiolabeled Atp9 protein associated with Oxa1. Finally, the recovery of both newly synthesized and pre-existing Atp9 on the Ni-NTA beads was specific for the presence of the histidine-tagged Oxa1 or Atpβ derivatives, because no Atp9 was recovered on the beads when control mitochondria bearing nontagged versions of these proteins was analyzed in parallel (Figure 6).
Thus comparing the efficiencies of recovery of the radiolabeled versus endogenous Atp9 proteins with the Oxa1His and AtpβHis proteins, we conclude that the majority of the radiolabeled Atp9 protein associated with Oxa1His is not assembled in to a F1Fo-ATP synthase complex. Rather, a significant fraction of the purified radiolabeled Atp9 protein represents an intermediate on the assembly pathway, which accumulates with Oxa1 after synthesis in isolated mitochondria.
Newly Synthesized Atp9 Can Associate with C-Terminally Truncated Oxa1
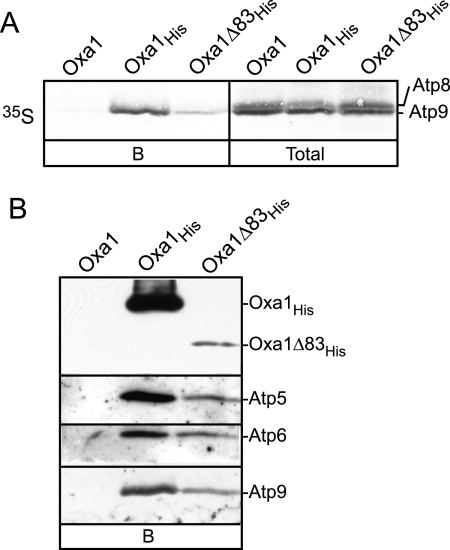
Finally, we investigated whether the C-terminal matrix located region of Oxa1 is required for the interaction of Oxa1 with newly synthesized Atp9 or with the endogenous ATP synthase subunits. In organello translation was performed in mitochondria carrying expressed C-terminally His-tagged 83 amino acids truncated Oxa1 (Oxa1Δ83His). After translation, the mitochondria were solubilized by Triton X-100 containing buffer and subjected to Ni-NTA affinity purification (Figure 7). Both newly synthesized and endogenous Atp9 could be purified on the Ni-NTA beads together with Oxa1Δ83His (Figure 7, A and B). In addition, immunedecoration with Atp5 and Atp6 antisera indicated the ability of other ATP synthase subunits to copurify with the Oxa1Δ83His protein. Thus, as was observed with the full-length Oxa1His derivative, the ATP synthase can be purified together with the C-terminally truncated Oxa1Δ83His derivative. The efficiency of purification of ATP synthase subunits appeared to be lower with the Oxa1Δ83His derivative than with the full-length Oxa1His protein. However, immunedecoration of the Ni-NTA purified material with an N-terminal specific Oxa1 antiserum indicated that the levels of Oxa1Δ83His recovered on the beads was drastically lower than that of the full-length Oxa1His protein (Figure 7B). Immunedecoration of total mitochondrial protein with the N-terminal Oxa1 antiserum indicated that the overall steady state levels of Oxa1Δ83His were strongly reduced relative to Oxa1His and this accounts for the reduced level of Oxa1Δ83His, which was recovered quantitatively, on the Ni-NTA beads (results not shown). Thus we conclude the reduced level of radiolabeled and assembled Atp9 recovered with the Oxa1Δ83His derivative reflects the lowered levels of Oxa1Δ83His present in the mitochondria, rather than an impaired affinity of the C-terminally truncated Oxa1 for the Atp9 protein. It would appear therefore that the addition of the C-terminal histidine tag affected the stability of the Oxa1Δ83His derivative, because previous studies have shown that the levels of the Oxa1Δ83 protein, i.e., nontagged, can be comparable to the authentic Oxa1 protein (Jia et al., 2003; Szyrach et al., 2003).
Figure 7.
The C-terminal region of Oxa1 is not required for the Atp9-Oxa1 interaction. (A and B) Mitochondria were isolated from yeast strains expressing Oxa1, Oxa1His, or Oxa1Δ83His derivatives under control of the GAL10 promoter. (A) In organello translation in the presence of [35S]methionine was performed in the mitochondria, and samples were further analyzed as described in Figure 6. (B) Immunoblotting of Atp5 was performed, in addition to Atp6 and Atp9, and the presence of Oxa1His and Oxa1Δ83His on the Ni-NTA beads were detected using an Oxa1 antibody specific for the N-terminal region of Oxa1.
Taken together, we conclude that the C-terminal region of Oxa1 is not required for the observed association between Oxa1 and the newly synthesized Atp9 or the endogenous ATP synthase components.
DISCUSSION
We report here that Oxa1 associates with and supports the assembly of the F1Fo-ATP synthase complex in yeast mitochondria. In the absence of Oxa1, a strong decrease in the levels of F1Fo-ATPase enzyme activity was observed, and this decrease corresponds to a reduction in the overall protein levels of the assembled F1Fo-ATP synthase complex. The severely reduced levels of F1Fo-ATP synthase complex observed in the Δoxa1 mitochondria appear to be directly due to the absence of Oxa1 and are not due to an indirect consequence of the absence of the assembled COX complex in these mitochondria.
It is important to note, however, that although strongly reduced in levels, assembled and enzymatically functional ATP synthase complexes can still be detected in the Δoxa1 mitochondria, suggesting that a low level of Oxa1-independent ATP synthase assembly can occur. This is in contrast to the COX complex where an essential role for Oxa1 in facilitating the insertion of mitochondrially encoded subunits, Cox1 and Cox2, is well documented. Our findings here, however, do not support a role for the Oxa1-related protein, Cox18, in the assembly of the ATP synthase complex, even in the absence of Oxa1. We conclude therefore that although the presence of Oxa1 is important for the biogenesis of the F1Fo-ATP synthase complex, it is clearly not essential and that level of Oxa1-, Cox18-independent assembly of this complex can occur. Cox18 has been proposed to play a role in the posttranslational insertion of the C-terminal region of Cox2, which becomes inserted into the membrane with two transmembrane segments (TMs) to attain a Nout-Cout orientation. Translocation of the C-terminal domain of Cox2 (∼150 residues) to the intermembrane space ensues in a posttranslational manner. Atp9 also contains two TM segments and attains a Nout-Cout membrane topology similar to that of Cox2 (but does not contain an extended hydrophilic intermembrane space exposed C-terminal tail) but clearly does not depend on Cox18 for its insertion of assembly events. We conclude therefore that Cox18 does not represent a common posttranslational translocase for all mitochondrially encoded proteins.
We demonstrate here that Oxa1 can directly associate with the newly synthesized Atp9 protein before its assembly into an intact F1Fo-ATP synthase complex. Furthermore, the observed interaction of Atp9 with Oxa1 does not require the matrix-localized C-terminal, basic tail of Oxa1, a region of the protein that is limited to the Oxa1-family members, i.e., not conserved among the Cox18/Oxa2 proteins. Thus the presence of the C-terminal tail region of Oxa1 does not account for the reason that Oxa1, but not Cox18, is required for the assembly of Atp9 into the ATP synthase complex, but rather may reflect some sequence specific differences in their transmembrane segments or N-terminal regions. It is important to note that the observed physical association of Oxa1 with newly synthesized Atp9 in a posttranslational manner is in marked contrast to that previously observed for Oxa1 and its COX subunit substrates, the Cox1, Cox2, and Cox3 subunits, which transiently interact with Oxa1 and only in a cotranslational manner. After completion of synthesis, the full-length, membrane-inserted COX subunits do not remain in association with Oxa1. It has been previously suggested that proteins that coordinate events downstream of membrane insertion, i.e., the subsequent assembly of membrane-inserted polypeptides, are more likely to bind to these proteins in a posttranslational and stable manner (Tzagoloff et al., 2004). Binding of assembly proteins or chaperones to substrate proteins in this manner is argued to maintain the competence of their substrates for subsequent assembly events. We propose therefore that the observed posttranslational and stable association between Atp9 and Oxa1 indicates a postmembrane insertion role for Oxa1 in the assembly of Atp9 into the Fo-sector.
Consistently, the results presented here demonstrate the ability of Atp9 to assemble into its oligomeric form and to associate with F1-sector subunits in the absence of Oxa1. Indeed the size exclusion chromatography experiments demonstrate that the Atp9-F1-subcomplex present in the Δoxa1 mitochondria is similar to those in the Δatp10 strain, a mutant reported to be defective in the step of integration of the Atp6 protein into late Atp9-F1-assembly intermediates (Tzagoloff et al., 2004). Taken together these data support a model where the activity of Oxa1 is required downstream of the translation and membrane insertion events for the mitochondrially encoded Atp9 subunit. The involvement of Oxa1 in the assembly of the mitochondrial F1Fo-ATP synthase complex may differ therefore from that reported for the YidC (the prokaryotic homolog of Oxa1) and the bacterial ATP synthase complex. Current evidence supports a role of YidC in the membrane insertion step of ATP synthase subunit c (the prokaryotic equivalent of Atp9; van Bloois et al., 2004; van der Laan et al., 2005; Kol et al., 2006). On the basis of the demonstrated posttranslational interaction between newly synthesized Atp9 and Oxa1 and the ability of Atp9 to assemble into the Atp9-F1 subcomplex in the absence of Oxa1, we propose that the mitochondrial Oxa1 acts in a postmembrane insertion step for Atp9. Furthermore, from the data presented it appears that the assembly of Atp6 into the accumulated Atp9-F1-subcomplex represents the step that is adversely affected in the Δoxa1 mitochondria.
By interacting with the newly synthesized Atp9 oligomer in a posttranslational manner, Oxa1 may serve to maintain the Atp9 oligomer in a “competent state” to coassemble with the Atp10-chaperoned Atp6 protein. Thus in the absence of either Oxa1 or Atp10, the efficient assembly of the Atp6-Atp9 containing F1Fo-sector, would be hindered, due to a lack of Atp9- or Atp6-specific chaperones, respectively. Alternatively, it is possible that in the absence of Oxa1, the problem may lie at the level of the newly synthesized Atp6 protein, for example, that it fails to become efficiently inserted into the inner membrane and hence does not associated with, an otherwise competent Atp9 oligomer. Available data, however, would favor that insertion of Atp6 is not severely blocked when the translocase activity of Oxa1 is compromised. The insertion of newly synthesized Atp6 was only partially affected (decreased by ∼35%, relative to wild type) in the pet ts1402 oxa1 mutant mitochondria, conditions where the translocase activity of Oxa1 was severely affected, as judged by inhibition of Cox2 insertion (which was ∼95% inhibited; Hell et al., 2001). For these reason we favor a role for Oxa1 in the assembly of the Fo-sector of the ATP synthase is related to its ability to associate with Atp9 in a posttranslational manner. We propose that features of Oxa1 distinct from those required for its translocation activity of the COX subunits, are necessary to chaperone the newly synthesized Atp9 protein and by doing so support its coassembly with Atp6.
A dichotomy in Oxa1's functions therefore appears to exist, whereby the translocase activity of Oxa1 required for the COX subunits, is distinct from another function of Oxa1 that is required in the assembly of Atp9 into the F1Fo-ATP synthase complex. In support of this proposal, an oxa1 mutant, bearing a mutation in the first TM region of Oxa1 has been reported to significantly affect cytochrome oxidase formation, whereas the assembly of the ATP synthase complex was relatively unaffected (Lemaire et al., 2004). Indeed other oxa1 mutants, which bear mutations within the intermembrane space exposed loop region between the second and third transmembrane segments of the Oxa1 protein, also display a strong reduction in COX levels and yet have normal assembly levels of the F1Fo-ATP synthase complex (Bushman and Stuart, unpublished results). Taking this together with the data reported here, we argue that the translocase activity of Oxa1 is not directly required for the Fo-sector subunits of the mitochondrial ATP synthase complex.
We report here also that a subpopulation of the ATP synthase complex can be copurified with a C-terminal His-tagged Oxa1 protein. The subunit profile of the ATP synthase complex which was found in association with Oxa1His was very similar to that affinity purified directly using a His-tagged Atpβ subunit. These findings suggest that an almost- or completely assembled ATP synthase complex can exist in physical association with the Oxa1 protein. The binding of Oxa1 to the ATP synthase complex most likely involves the Fo-sector and more specifically occurs at the Atp9 interface, because the copurification of these complexes was sensitive to pretreatment of the mitochondria with oligomycin before the detergent lysis step. The observed coassociation of ATP synthase with the Oxa1His, however, was not artificially caused by the elevated levels of the Oxa1 derivative in these mitochondria (expressed under the control of GAL10 promoter and hence levels of Oxa1 were ∼50 times greater than endogenous levels; results not shown). A similar copurification of F1Fo-subunits was also observed when the Oxa1His derivative was expressed at endogenous levels under control to Oxa1's own promoter region. Under these conditions, significantly lower levels of ATP synthase subunits were, however, recovered with Oxa1His on the Ni-NTA beads, but these levels paralleled the lower content of the Oxa1His protein in the sample (result not shown).
Why does Oxa1 associate with the ATP synthase complex and does this interaction have any bearing on the role of Oxa1 in the biogenesis of the ATP synthase complex? The ability of Oxa1 to bind to the assembled ATP synthase complex, most likely reflects the innate ability of Oxa1 to interact with the Atp9 protein. Common features of the newly synthesized Atp9 protein and the oligomeric Atp9 assembled in the F1Fo-ATP synthase, appears to support the observed Oxa1 associations, because both interactions are sensitive to the pretreatment with oligomycin. The ability of the ATP synthase to physically interact with another inner membrane protein complex is not unique to Oxa1. It has been previously described that the F1Fo-ATP synthase complex can be found with the Mdl1 protein, an ATP-binding cassette transporter (ABC) protein, involved in peptide export across the inner membrane (Galluhn and Langer, 2004). We did not observe association of Mdl1 with the Oxa1His protein under the lysis conditions used in this study (results not shown).
It is presently unclear from our studies whether the observed association between these complexes is for the benefit of Oxa1 or of the ATP synthase complex (or possibly even for their mutual benefit). The Oxa1-ATP synthase association may be to support the activity of Oxa1, either directly in a bioenergetic manner or indirectly to regulate the activity of Oxa1 as a translocase for the COX subunits, possibly to maintain a balance between the biogenesis levels of COX and ATP synthase complexes. It is equally plausible that the Oxa1-ATP synthase association serves to benefit the ATP synthase complex during its final steps of assembly. The association of Oxa1 with the Atp9 may interfere with the H+ pumping activity of the complex, because the oligomycin sensitivity of binding may indicate that Oxa1 binds at the interface of the Atp6/Atp9 proteins. The stable association of Oxa1 with the Fo-sector in this manner may ensure that the ATP synthase complex remains inactive (i.e., not leaky to H+ transfer, which may dissipate of the membrane potential) until the complete assembly of a tightly coupled F1Fo-ATP synthase complex is achieved. At any given time therefore, a small percentage of mitochondrial ATP synthase complexes, completing their assembly may be found in association with Oxa1, their assembly chaperone.
In summary, we demonstrate here an important role for Oxa1, but not Cox18, in the assembly of the F1Fo-ATP synthase complex. Oxa1 directly interacts with Atp9 after its translation in mitochondria, and we propose that this interaction with Oxa1 is necessary to chaperone Atp9 and ensure the efficient and correct assembly into an active and coupled F1Fo-ATP synthase complex. Further experiments to investigate the molecular basis for the physiological relevance of the observed Oxa1-ATP synthase association are currently underway in our laboratory.
ACKNOWLEDGMENTS
We thank Dr. Jean Velours (Bordeaux) for the generous gift of the Atp4 and Atp9 antibodies and Dr. Rod Devenish (Monash) for the Atp5 and Su d subunit antibodies. We are grateful to Dr. David Mueller (The Chicago Medical School, Chicago, IL) for giving us the yeast strain expressing the histidine-tagged Atpβ protein and Dr. Sharon Ackerman (Wayne State School of Medicine, Detroit, MI) for the kind gift of the Δatp10 strain. This research was supported by funding from the National Science Foundation Grant M.C.B. 0347025 and U.S. Public Health Service Grant R01GM61573 to R.A.S.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-10-0925) on March 7, 2007.
REFERENCES
- Ackerman S. H., Tzagoloff A. Function, structure, and biogenesis of mitochondrial ATP synthase. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:95–133. doi: 10.1016/S0079-6603(05)80003-0. [DOI] [PubMed] [Google Scholar]
- Altamura N., Capitanio N., Bonnefoy N., Papa S., Dujardin G. The Saccharomyces cerevisiae OXA1 gene is required for the correct assembly of cytochrome c oxidase and oligomycin-sensitive ATP synthase. FEBS Lett. 1996;382:111–115. doi: 10.1016/0014-5793(96)00165-2. [DOI] [PubMed] [Google Scholar]
- Arnold I., Pfeiffer K., Neupert W., Stuart R. A., Schägger H. Yeast mitochondrial F1Fo-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J. 1998;17:7170–7178. doi: 10.1093/emboj/17.24.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer M., Behrens M., Esser K., Michaelis G., Pratje E. PET1402, a nuclear gene required for proteolytic processing of cytochrome oxidase subunit 2 in yeast. Mol. Gen. Genet. 1994;245:272–278. doi: 10.1007/BF00290106. [DOI] [PubMed] [Google Scholar]
- Bonnefoy N., Chalvet F., Hamel P., Slonimski P. P., Dujardin G. OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes controls cytochrome oxidase biogenesis. J. Mol. Biol. 1994;239:201–212. doi: 10.1006/jmbi.1994.1363. [DOI] [PubMed] [Google Scholar]
- Capaldi R. A., Aggeler R. Mechanism of the F1Fo-type ATP synthase, a biological rotary motor. Trends Biochem. Sci. 2002;27:154–160. doi: 10.1016/s0968-0004(01)02051-5. [DOI] [PubMed] [Google Scholar]
- Celebi N., Yi L., Facey S. J., Kuhn A., Dalbey R. E. Membrane biogenesis of subunit II of cytochrome bo oxidase: contrasting requirements for insertion of N–terminal and C-terminal domains. J. Mol. Biol. 2006;357:1428–1436. doi: 10.1016/j.jmb.2006.01.030. [DOI] [PubMed] [Google Scholar]
- Cruciat C., Brunner S., Baumann F., Neupert W., Stuart R. A. The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J. Biol. Chem. 2000;275:18093–18098. doi: 10.1074/jbc.M001901200. [DOI] [PubMed] [Google Scholar]
- Devenish R. J., Prescott M., Roucou X., Nagley P. Insights into ATP synthase assembly and function through the molecular genetic manipulation of subunits of the yeast mitochondrial enzyme complex. Biochim. Biophys. Acta. 2000;1458:428–442. doi: 10.1016/s0005-2728(00)00092-x. [DOI] [PubMed] [Google Scholar]
- du Plessis D. J., Nouwen N., Driessen A. J. Subunit a of cytochrome o oxidase requires both YidC and SecYEG for membrane insertion. J. Biol. Chem, 2006;281:12248–12252. doi: 10.1074/jbc.M600048200. [DOI] [PubMed] [Google Scholar]
- Funes S., Nargang F. E., Neupert W., Herrmann J. M. The Oxa2 protein of Neurospora crassa plays a critical role in the biogenesis of cytochrome oxidase and defines a ubiquitous subbranch of the Oxa1/YidC/Alb3 protein family. Mol. Biol. Cell. 2004;15:1853–1861. doi: 10.1091/mbc.E03-11-0789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluhn D., Langer T. Reversible assembly of the ATP-binding cassette transporter Mdl1 with the F1Fo-ATP synthase in mitochondria. J. Biol. Chem. 2004;279:38338–38345. doi: 10.1074/jbc.M405871200. [DOI] [PubMed] [Google Scholar]
- Gerdes L., Bals T., Klostermann E., Karl M., Philippar K., Hunken M., Soll J., Schunemann D. A second thylakoid membrane-localized Alb3/OxaI/YidC homologue is involved in proper chloroplast biogenesis in Arabidopsis thaliana. J. Biol. Chem. 2006;281:16632–16642. doi: 10.1074/jbc.M513623200. [DOI] [PubMed] [Google Scholar]
- Hell K., Herrmann J., Pratje E., Neupert W., Stuart R. A. Oxa1p mediates the export of the N- and C-termini of pCoxII from the mitochondrial matrix to the intermembrane space. FEBS Lett. 1997;418:367–370. doi: 10.1016/s0014-5793(97)01412-9. [DOI] [PubMed] [Google Scholar]
- Hell K., Herrmann J. M., Pratje E., Neupert W., Stuart R. A. Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc. Natl. Acad. Sci. USA. 1998;95:2250–2255. doi: 10.1073/pnas.95.5.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K., Neupert W., Stuart R. A. Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J. 2001;20:1281–1288. doi: 10.1093/emboj/20.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann J. M., Stuart R. A., Craig E. A., Neupert W. Mitochondrial heat shock protein 70, a molecular chaperone for proteins encoded by mitochondrial DNA. J. Cell Biol. 1994;127:893–902. doi: 10.1083/jcb.127.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L., Dienhart M., Schramp M., McCauley M., Hell K., Stuart R. A. Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal region of Oxa1. EMBO J. 2003;22:6438–6447. doi: 10.1093/emboj/cdg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John U. P., Nagley P. Amino acid substitutions in mitochondrial ATPase subunit 6 of Saccharomyces cerevisiae leading to oligomycin resistance. FEBS Lett. 1986;207:79–83. doi: 10.1016/0014-5793(86)80016-3. [DOI] [PubMed] [Google Scholar]
- Kol S., Turrell B. R., de Keyzer J., van der Laan M., Nouwen N., Driessen A. J. YidC-mediated membrane insertion of assembly mutants of subunit c of the F1Fo ATPase. J. Biol. Chem. 2006;281:29762–29768. doi: 10.1074/jbc.M605317200. [DOI] [PubMed] [Google Scholar]
- Kuhn A., Stuart R., Henry R., Dalbey R. E. The Alb3/Oxa1/YidC protein family: membrane-localized chaperones facilitating membrane protein insertion? Trends Cell Biol. 2003;13:510–516. doi: 10.1016/j.tcb.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lemaire C., Guibet-Grandmougin F., Angles D., Dujardin G., Bonnefoy N. A yeast mitochondrial membrane methyltransferase-like protein can compensate for oxa1 mutations. J. Biol. Chem. 2004;279:47464–47472. doi: 10.1074/jbc.M404861200. [DOI] [PubMed] [Google Scholar]
- Moore M., Harrison S., Peterson E. C., Henry R. Chloroplast Oxa1p homolog albino3 is required for post-translational integration of the light harvesting chlorophyll-binding protein into thylakoid membranes. J. Biol. Chem. 2000;275:1529–1532. doi: 10.1074/jbc.275.3.1529. [DOI] [PubMed] [Google Scholar]
- Mueller D. M., Puri N., Kabaleeswaran V., Terry C., Leslie A. G., Walker J. E. Ni-chelate-affinity purification and crystallization of the yeast mitochondrial F1-ATPase. Protein Expr. Purif. 2004;37:479–485. doi: 10.1016/j.pep.2004.06.035. [DOI] [PubMed] [Google Scholar]
- Nagley P., Hall R. M., Ooi B. G. Amino acid substitutions in mitochondrial ATPase subunit 9 of Saccharomyces cerevisiae leading to oligomycin or venturicidin resistance. FEBS Lett. 1986;195:159–163. doi: 10.1016/0014-5793(86)80152-1. [DOI] [PubMed] [Google Scholar]
- Nunnari J., Fox T. D., Walter P. A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science. 1993;262:1997–2004. doi: 10.1126/science.8266095. [DOI] [PubMed] [Google Scholar]
- Preuss M., Ott M., Funes S., Luirink J., Herrmann J. M. Evolution of mitochondrial Oxa proteins from bacterial YidC. Inherited and acquired functions of a conserved protein insertion machinery. J. Biol. Chem. 2005;280:13004–13011. doi: 10.1074/jbc.M414093200. [DOI] [PubMed] [Google Scholar]
- Saddar S., Stuart R. A. The yeast F1Fo-ATP synthase: analysis of the molecular organization of subunit g and the importance of a conserved GXXXG motif. J. Biol. Chem. 2005;280:24435–24442. doi: 10.1074/jbc.M502804200. [DOI] [PubMed] [Google Scholar]
- Saracco S. A., Fox T. D. Cox18p is required for export of the mitochondrially encoded Saccharomyces cerevisiae Cox2p C-tail and interacts with Pnt1p and Mss2p in the inner membrane. Mol. Biol. Cell. 2002;13:1122–1131. doi: 10.1091/mbc.01-12-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza R. L., Green-Willms N. S., Fox T. D., Tzagoloff A., Nobrega F. G. Cloning and characterization of COX18, a Saccharomyces cerevisiae PET gene required for the assembly of cytochrome oxidase. J. Biol. Chem. 2000;275:14898–14902. doi: 10.1074/jbc.275.20.14898. [DOI] [PubMed] [Google Scholar]
- Stock D., Leslie A.G.W., Walker J. E. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- Stuart R. A. Insertion of proteins into the inner membrane of mitochondria: the role of the Oxa1 complex. Biochem. Biophys. Acta. 2002;1592:79–87. doi: 10.1016/s0167-4889(02)00266-5. [DOI] [PubMed] [Google Scholar]
- Sundberg E., Slagter J. G., Fridborg I., Cleary S. P., Robinson C., Coupland G. ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell. 1997;9:717–730. doi: 10.1105/tpc.9.5.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyrach G., Ott M., Bonnefoy N., Neupert W., Herrmann J. M. Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J. 2003;22:6448–6457. doi: 10.1093/emboj/cdg623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Barrientos A., Neupert W., Herrmann J. M. Atp10p assists assembly of Atp6p into the F0 unit of the yeast mitochondrial ATPase. J. Biol. Chem, 2004;279:19775–19780. doi: 10.1074/jbc.M401506200. [DOI] [PubMed] [Google Scholar]
- van Bloois E., Haan G. J., de Gier J. W., Oudega B., Luirink J. F1Fo ATP synthase subunit c is targeted by the SRP to YidC in the E. coli inner membrane. FEBS Lett. 2004;576:97–100. doi: 10.1016/j.febslet.2004.08.069. [DOI] [PubMed] [Google Scholar]
- van Bloois E., Haan G. J., de Gier J. W., Oudega B., Luirink J. Distinct requirements for translocation of the N-tail and C-tail of the Escherichia coli inner membrane protein CyoA. J. Biol. Chem. 2006;281:10002–10009. doi: 10.1074/jbc.M511357200. [DOI] [PubMed] [Google Scholar]
- van der Laan M., Bechtluft P., Kol S., Nouwen N., Driessen A. J. F1Fo ATP synthase subunit c is a substrate of the novel YidC pathway for membrane protein biogenesis. J. Cell Biol. 2004;165:213–222. doi: 10.1083/jcb.200402100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan M., Nouwen N. P., Driessen A. J. YidC—an evolutionary conserved device for the assembly of energy-transducing membrane protein complexes. Curr. Opin. Microbiol. 2005;8:182–187. doi: 10.1016/j.mib.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Velours J., Arselin G. The Saccharomyces cerevisiae ATP synthase. J. Bioenerg. Biomembr. 2000;32:383–390. doi: 10.1023/a:1005580020547. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pohlmann R., Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wittig I., Schägger H. Advantages and limitations of clear-native PAGE. Proteomics. 2005;5:4338–4346. doi: 10.1002/pmic.200500081. [DOI] [PubMed] [Google Scholar]
- Yen M. R., Harley K. T., Tseng Y. H., Saier M. H., Jr Phylogenetic and structural analyses of the oxa1 family of protein translocases. FEMS Microbiol. Lett. 2001;204:223–231. doi: 10.1111/j.1574-6968.2001.tb10889.x. [DOI] [PubMed] [Google Scholar]