Abstract
Traditional microarrays use probes complementary to known genes to quantitate the differential gene expression between two or more conditions. Genomic tiling microarray experiments differ in that probes that span a genomic region at regular intervals are used to detect the presence or absence of transcription. This difference means the same sets of biases and the methods for addressing them are unlikely to be relevant to both types of experiment. We introduce the informatics challenges arising in the analysis of tiling microarray experiments as open problems to the scientific community and present initial approaches for the analysis of this nascent technology.
Introduction
Genomic tiling microarray construction involves the generation of nucleic acid probes that represent a target genomic region and their immobilization on a glass slide (Figure 1a). These probes can either overlap, lay end-to-end, or be spaced at a predefined average distance in genomic space (Figure 1b). A sequence of probes spanning a genomic region is called a ‘tile path’, or a ‘tiling’, and the average distance, in nucleotides, between the centers of neighboring probes is termed the ‘step’ or ‘resolution’ of the tiling. Each probe on a tiling array interrogates the presence of a sequence in a nucleic acid population via hybridization. There are two types of tiling array construction. One type is the oligonucleotide tiling array [1–10]. Such arrays comprise 25–60 bp probes (the choice depending on the manufacturer and/or genome tiling), which are synthesized directly on the slides or prepared in solution and then deposited. Arrays of high density (up to 6.6 million features in <2 cm2) can currently be prepared. The second type of tiling array is constructed using PCR products typically of ~1-kb in length, or bacterial artificial chromosome (BAC) arrays – typically at 1-Mb resolution [11–13].
Figure 1.
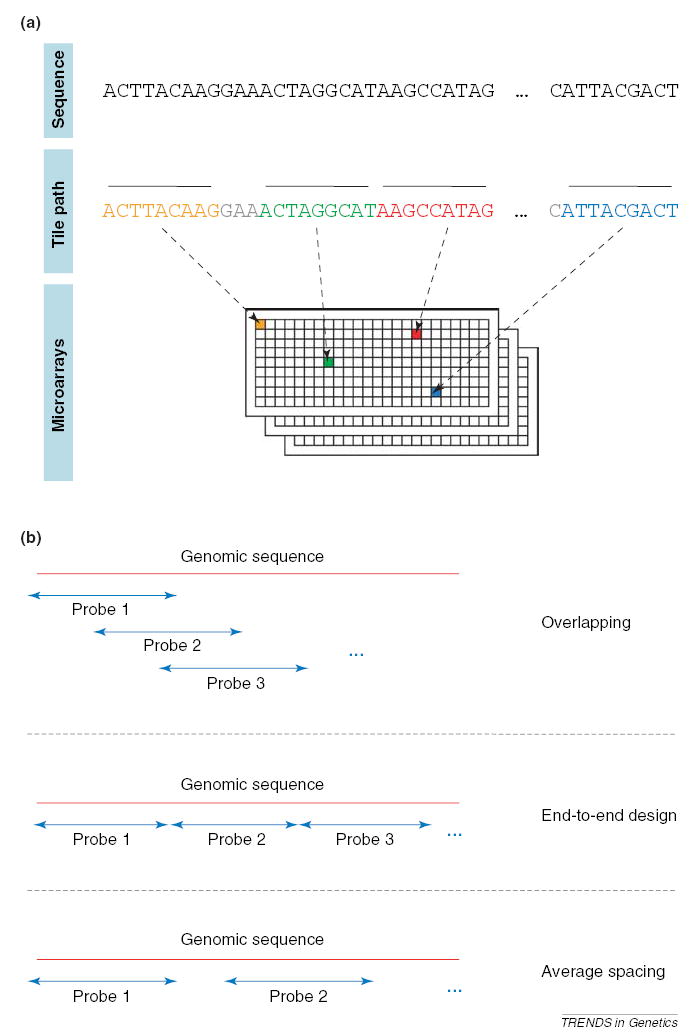
Properties of tiling microarrays. (a) The design of a tiling microarray experiment. Each individual probe in the tiling is indicated by a different color and thick overbar. The probes making up the design constitute a ‘tile path’. Nucleotides not incorporated into probes are grayed. Most array designs randomize the position of the adjacent tiles on the array in an attempt to avoid systematic errors. (b) Tiling designs (tile paths) can be overlapping, end-to-end or spaced.
One caveat of the PCR and BAC tiling arrays is that both the target sequence and its reverse complement sequence are present at each spot, rendering strand specificity impossible without additional experiments. In addition, the ~1-kb PCR fragment microarrays are labor intensive to create and are thus not readily scalable to the study of large genomes at a high resolution. For example, a recent study tiling human chromosome 22 (roughly 1% of the human genome) with PCR products required >20 000 PCR reactions to achieve a 1-kb resolution [13]. A PCR tiling of the entire human genome would require approximately two million PCR reactions at the same resolution and necessitate extensive informatics infrastructure to support the effort. Analytical techniques for such arrays typically follow that of other PCR product-based microarrays and are reviewed elsewhere [14,15]. For these reasons, attention in this manuscript is devoted to discussion of oligonucleotide-based tiling arrays. To focus the discussion further, we will limit our discussion to the application of these arrays to the identification of RNA transcripts. Tiling arrays have several other utilities, including interrogating sequences enriched in chromatin immunoprecipitation DNA (ChIP-chip, reviewed in Refs [16,17]), DNA copy-number alterations (arrayCGH, reviewed in Ref. [18]) and protein-binding motifs (PBMs) [19]. The analyses of these experiments will probably have some common aspects, but their proper study has specialized aspects that cannot be considered here owing to lack of space.
Recent reviews by Johnson et al. [20] and by Mockler and Ecker [21] provide a good general overview of the tiling array technology and its applications. In particular, Johnson et al. raise concerns about low levels of concordance between transcriptional experiments performed in different laboratories. Differences among data sets can arise from several factors, including experimental design, tissues assayed, technological platforms used, and so on. For tiling arrays to reach widespread acceptance, these differences must be identified and resolved. Towards this end, we provide an initial perspective on the tiling microarray experiment from the analytic point of view. In so doing, we provide an introduction to the characteristics of data generated by tiling microarrays, introduce some challenging questions, and give initial views on the analysis of these relatively new types of microarray experiments.
Distribution of signal intensities
For tiling microarrays, a probe representing some genomic sequence is the unit of investigation, and an intensity measurement after hybridization to labeled target is its recorded datum. In theory, this measurement correlates with the number of target nucleic acid molecules that hybridized to that probe during the experiment.
Tiling microarrays built using Affymetrix technology contain a paired ‘mismatch’ probe for each genomic tile probe (http://www.affymetrix.com/). (For convenience, the genomic tile probe that perfectly matches genomic sequence is typically denoted PM and the mismatch probe is similarly denoted MM.) The MM probe is intended to provide a measurement of nonspecific nucleic acid binding to the PM probe and thus the quantity PM – MM typically serves as the intensity measurement for Affymetrix tiling arrays.
In a tiling experiment, as is the case for most microarray experiments, the goal is to identify outliers from the predominant background or noise distribution. It is tempting first to approach this task visually. A priori one might expect that in a transcript-detection experiment, the desired raw intensity distribution might appear bimodal wherein a small peak at higher intensity bins of the histogram contains the transcribed signal distribution and the adjacent, dominant peak comprises the background distribution. Unfortunately, such separation is not realized (Figure 2a) because the majority of transcribed sequences are probably present at levels just above background, in accordance with a transcriptional power-law distribution [22]. This coupled with inherent noise in the background and signal distributions makes a separation between signal and background difficult at the probe level without expensive replicate experiments. Adding to these problems in higher eukaryotes is that the percentage of coding DNA relative to total genomic DNA is very small; this renders distribution fitting procedures, which allow for the separation of mixed distributions, useless without large degrees of separation and/or well-defined functional forms for both background and signal distributions (Figure 2b).
Figure 2.

Intensity distributions. (a) Desired intensity distribution and typical distributions. Intensities are given in arbitrary units. The distribution on the right is from Kapranov et al. [2]. (b) Effect of small signal distributions. In the left plot, the black line marks the background distribution and red shows a hypothetical signal distribution. For the hypothetical signal distribution we have simulated a power-law distribution rather than a Gaussian distribution because it has been suggested that transcript abundances exhibit power-law behavior. Here, the number of measurements comprising the signal distribution is 10% (chosen arbitrarily) of the number of those in the background distribution. On the right is the sum of these two distributions, indicating the difficulty in separating the mixture.
Within-gene variability
Without a clear separation of distributions, identifying transcribed sequences by scanning for stretches of consecutive probes in genomic space exhibiting intensities significantly above those of some background distribution seems reasonable. In fact, this is the approach typically taken in the analysis of tiling array data. Before discussing such methods, an important aspect of tiling array data is worth noting, because it has a major impact on the development of tiling array algorithms. Within a gene present as a single splice variant, we expect that the raw signal intensities measured by its multiple corresponding probes will be equivalent throughout. As exemplified in Figure 3a, in practice, this might not be the case. Intensities measured from probes within different exons of the same gene can vary greatly, and even intensities of genomic nearest neighbor probes (i.e. probes measuring the same annotated exon) can differ by orders of magnitude. Such intergene intensity fluctuations have also been observed with GeneChips® brand arrays, but with these arrays the researcher is typically looking for differences between two or more biological samples so such systematic effects can be avoided by using the ratio of intensity of one sample to intensity of the others. To quantify the intergene fluctuations with tiling arrays on a genomic scale, we looked at the intensity of each probe, p, in a large, human DNA experiment tiling the genome with 36-bp probes with a resolution of 46 bp [1], and compared it with the average intensity for the two neighboring probes of p. We found that for probes lying completely within annotated exons, ~20% of these probes exhibit at least a twofold change in intensity from the average intensity of their two neighboring probes (Figure 3b). Many lesser intensity fluctuations exist. Such fluctuations could potentially be due to complicated populations of splice variants from the same gene, sequence-based probe effects (due to varying binding affinities based on sequence [23]), labeling biases, or from cross-hybridization from sequence-similar transcribed sequences located elsewhere in the genome. As noted earlier, similar problems exist on Affymetrix® GeneChips® brand arrays. The problem is exacerbated, however, on tiling arrays. On GeneChip® brand arrays, exon boundaries of genes are known so outlier detection is straightforward, but on tiling arrays it is difficult to discern outliers because it is unclear which probes to include in outlier detection. For example, should one include a cluster of apparently transcribed probes 20-kb downstream of a transcript? Are they from the same gene, or not? Is a low-intensity probe between other high-intensity probes an outlier, or perhaps an intron? Such questions make tiling-array outlier detection non-straightforward, or worse, impossible.
Figure 3.
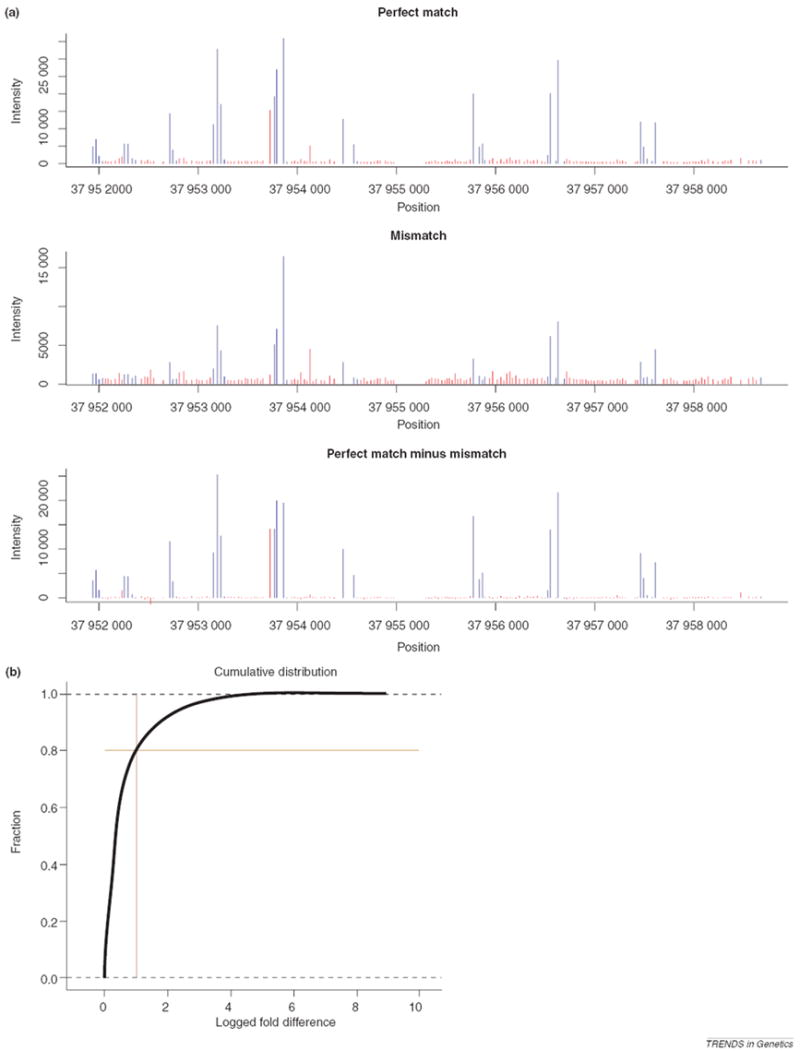
Within-gene intensity fluctuation. (a) Gene exhibiting typical intensity fluctuations. Data shown are for RefSeq gene NM_001001479 from Kapranov et al. [2], using 35-bp probes with a step size of 35 bp. Red bars illustrate probes that lie within introns, whereas blue bars demonstrate probes within exons. Plots for perfect match (PM) intensities, mismatch (MM) intensities and PM – MM differences in intensities are shown. (b) Empirical cumulative distribution function for nearest neighbor intensity fluctuations within RefSeq genes. The x-axis represents the fold difference on a log2 scale between observed intensity and expected intensity, based on the average intensities of neighboring probes. The cross indicates the point at which 20% of all probes show a twofold deviation from their expected intensities. Data are from Bertone et al. [1].
If the intensity fluctuations come from differences in binding energies between probe sequences, there are models for correcting this type of effect in GeneChips® brand arrays, but they are still debated [24] and are as yet untested for tiling microarrays. Another approach for addressing these biases is to deal with them during the probe design procedure. In many designs, the designer is allowed to select probes by shifting their genomic positioning slightly so that the variance of melting temperatures of probes across the whole array is minimized. Probe sets on GeneChips® brand arrays also attempt this. The shifting allowed on tiling arrays is much more constrained because the design must conform to the bounds (or limits) set by the predetermined span of the array. Similar considerations apply in other situations where the probe location is highly constrained, for example, single nucleotide polymorphism (SNP) arrays [25] or arrays with probes spanning splice junctions [7].
Another solution to sequence biases could be to design the arrays with varying probe lengths so as to correct for differences in the melting temperatures of sequences. Such ‘isothermal’ arrays are feasible with the recently developed maskless, photolithography-array synthesis technique [26,27] and are currently under development. Such approaches potentially aid in reducing sequence biases but the likely complicated problem of cross-hybridization remains.
Another design feature is the option to include MM probes on the array. Theoretically, MM probes measure the amount of cross-hybridization to the PM probe from unintended targets. We find that the inclusion of such MM probes reduces within-gene intensity variability somewhat but does not eliminate the effect (Figure 3a). A recent study of GeneChips® brand arrays indicates that ~10% of probes cross-hybridize to multiple genes [28] so it is an interesting question as to how much utility MM probes have for tiling experiments. For a tiling array built for human chromosome 22 [2] on which PM and MM probes are present, we find that PM probe intensities within RefSeq genes significantly correlate with PM probe intensities of the preceding neighboring tile (Spearman ρ = 0.156, P < 10−15) but that PM-MM intensities correlate better (Spearman ρ = 0.175, P < 10−15). It is clear that MM probes help, but there is still much room for improvement.
Tiling microarray normalization
For many applications of tiling arrays, slide-to-slide normalization is not required because there are no technical replicates and we typically are not interested in comparing absolute intensities of genes present on one array with those found on another. However, if technical replicates with the same array design are performed, or if we wish to compare abundances of different genes across the experiment, the measurements of the arrays will have to be scaled to one another before downstream analysis can take place.
For traditional (i.e. nontiling arrays using probes to annotated genes) microarray experiments, it is known that adjustments to the signal distributions can be essential before proper statistical analysis can be undertaken [14,15]. This is partly because there are several experimental parameters that commonly vary from one microarray hybridization to another in the same data set. These variables include, among others, laser strength used to obtain fluorescence measurements, concentration of nucleic acid allowed to hybridize to the array and hybridization times and temperatures. Equally important are so-called intraslide spatial artifacts such as nonuniform hybridization efficiency [29,30] and/or nonuniform background intensities across the microarray surface [31].
A simple adjustment for correcting bias due to differing hybridization concentrations or conditions between microarrays in these conventional gene-based experiments has been to divide each intensity measurement by the median intensity present on the microarray on which it resides [14]. More rigorous approaches that also account for intraslide variability include fitting a loess surface (similar to a sliding window median) to the intensities measured as a function of their physical positioning on the microarray and then using this surface as a normalizing function [32]. These normalization practices assume that at least half of all probes on each microarray produce measured signal due to the presence of hybridized nucleic acid. This is a valid assumption for most conventional microarray studies because typically only annotated genes are represented. However, for most tiling microarray experiments involving large, complex genomes and where poly-A selected RNA is the target, we expect a relatively small number of probes to emit measurable signal due to hybridization. Thus, the median intensity of the microarray is likely to be a background measurement and cannot be used to correct for hybridization-type effects. One might also be tempted to use the uppermost tenth percentile or some other quantile intensity for normalization. Doing so is paramount to ‘guessing’ at how much transcriptional activity there is across the genome, and thus makes the approach less rigorous. This type of median or other quantile correction can, however, still be useful for correcting differing scanning voltages used from array scan to array scan.
Spatial normalizations can be conducted if control probes of identical oligonucleotide sequence are placed uniformly across the surface of the slide because these are all expected to emit the same significant intensity in an ideal experiment. In an Escherichia coli tiling experiment, Selinger et al. [5] printed positive control probes throughout the array and multiplied the intensity of each experimental probe by a correction factor that is a function of the intensities of the four closest control features and their physical distances to the experimental probe. This factor was calculated using Equation 1,
| (Eqn 1) |
where di is the physical distance of control probe i to the experimental probe to be normalized, ci is the intensity of the ith control, and E(c) is the average intensity of all control probes on the array.
An additional point to consider when designing normalization procedures for tiling microarrays is that a single experiment can consist of hundreds of individual microarrays, each containing a distinct set of probes. For instance, one microarray might tile human chromosome 21 and another might tile chromosome 22. An assumption for most standard microarray normalization procedures is that the intensity distributions from array to array are identical. It might be that one chromosome, which is printed on a single slide, is more gene dense than another chromosome, which is printed on a different slide, and the underlying distributions are thus not identical. A clear example of this is seen when contrasting the gene-dense human chromosome 22 with the gene-sparse human chromosome 21. To remedy this issue, all probes can be printed in a random fashion with respect to their chromosomes, strands and locations within their chromosomes. Because the number of probes on each microarray is large, the distributions to be considered should be close to identical. Then, it could be more reasonable to perform slide-to-slide quantile normalization.
Algorithms for tiling microarray analysis
As examples of algorithms for detecting transcription from tiling array data, we review algorithms from three recent human tiling microarray experiments, two of which allow small gaps in genomic space between probes, and one that overlaps the genomic positions. The three examples were taken from studies using three array construction technologies, and therefore represent a good cross-section of the field. The first methods we review were developed for a large-scale experiment using the Affymetrix technology to construct probes spaced, on average, every 35 bp along chromosomes 21 and 22 [2,3]. The second set of methods reviewed were implemented on tiling microarrays built with Nimblegen™ maskless photolithography technology [1] and consisted of 36-bp probes with a resolution of 46 bp (http://www.nimblegen.com/). In both examples, the algorithms presented are independent of data normalizations (i.e. they do not assume any normalizations have been performed) and do not necessarily require replicate hybridizations and are, therefore, general in nature. The third analysis approach we summarize was used in a recent study of human chromosomes 20 and 22 using the Agilent™ technology to deposit 60-bp probes, on average, every 30 bp along those chromosomes [4]. This algorithm requires multiple replicates and is therefore useful when hybridizations are conducted for several tissues [4,6,33].
Analysis of Kampa et al
In an in-depth analysis of human chromosomes 21 and 22 [3], a probe was deemed ‘positive’ using the following procedure. For a given probe, i, all PM-MM pairs are collected within a window, w, of 100 nucleotides, centered at i. For each of these pairs, the difference between PM and MM intensities is calculated. The Hodges–Lehman estimator, or ‘pseudo-median’, is then computed for these PM – MM differences (Figure 4). This estimator is simply the median of pairwise averages amongst the PM minus MM scores within the window and has close ties with rank-sum statistics. Any probe having a Hodges–Lehman estimator above a threshold (defined using bacterial oligonucleotides present on the array but not homologous to any sequences in the human genome) is considered ‘positive’, or transcribed. Using this estimator partly alleviates concerns discussed earlier about within-gene variability because it is notably robust to outliers. Transcribed fragments, or ‘transfrags’, were then constructed from lists of positive probes by merging those that lie in close genomic proximity (within 40 bp – this variable is called ‘maxgap’ in the original publication) to each other and filtering out transfrags <90 bp (this threshold is termed ‘minrun’) in length. This merging of neighboring probes further diminishes the effect of within-gene variability because confidence in measured intensities is increased with increasing amounts of evidence.
Figure 4.

Calculation of the pseudo-median at probe position two. Psuedo-medians are computed in sliding windows (window size three in this example, centered at probe position i). This psuedo-median is simply the median of pairwise averages amongst the PM minus MM scores within the window. See the main text for further details.
Analysis of Bertone et al
Another recent tiling microarray study focusing on the human transcriptome [1] employs an alternative approach for detecting transcription using the binomial theorem. In this work, two procedures are introduced: one algorithm for comparing tiling array data with existing annotation, and another algorithm for identifying transcriptionally active regions (TARs – equivalent but different terminology to the above defined transfrag).
To check the tiling array data against a previously annotated gene, the probes that lie within the exons of that gene were first identified. The number of such probes was denoted as n. For each probe it was recorded whether or not its measured intensity was greater than the median intensity of the slide from which it was measured. By definition, half of the measured intensities of a slide are greater than the median and half are less than the median. To determine if the gene was transcribed, it was determined whether or not the number of intensities within the gene recorded above this median was more than expected by chance alone. The probability, p, of obtaining h probes with above-median intensities out of N probes within the gene is given by Equation 2.
| (Eqn 2) |
Then, it is a matter of choosing the desired false-positive rate and setting the p-value cutoff value accordingly.
A similar approach was taken for finding novel regions of activity within the genome. However, when all stretches of, say, ten adjacent probes are tested for significance the number of hypotheses tested across a mammalian genome reaches into the millions and one would require a low p-value cutoff to weed out the large number of expected false-positives. For example, in the human genome, if one wanted to identify unique stretches of ten probes (each probe a 50 bp oligonucleotide) there would be three million independent tests to perform and if we use a p-value cutoff of 0.01 this would result in an expected 30 000 false-positive regions. To lower the number of false positives obtained, a low p-value cutoff is needed. To achieve such low p-values for novel regions with the above approach, large stretches of adjacent probes or replicate experiments would be required owing to the low statistical power of the test. Mammalian exons are significantly <500 bp on average, and if low p-values are required there will be a significant bias towards the largest transcribed sequences. However, if one is willing to be more stringent when identifying novel regions of interest, then such low p-values can readily be obtained without the use of large stretches or costly replicate experiments. For example, instead of counting the number of probes in a stretch greater than the slide median, the number of probes, h, above the kth percentile (e.g. 80th) of the slide could be counted and this number could be checked for significance with the binomial equation (Equation 3).
| (Eqn 3) |
Regions identified by this method are then merged into a single transcribed region if their genomic coordinates overlap.
The reader should note that the requirement of low P-values is essentially a Bonferroni correction for multiple hypothesis testing. This correction is known to be overly conservative and therefore the above approach probably errs on the conservative side. Another issue with the Bonferroni correction is that it assumes statistical independence of the probes. This is certainly true under the null hypothesis of zero transcription but for probes within the same gene this assumption is probably violated and hence P-values obtained must be considered as merely nominal. To obtain more accurate estimates of false positives, a reasonable approach to take is first to score your data using the above, or any other algorithm, record your ‘hits’, and then randomize your data with respect to genomic location and re-score. The number of hits you obtain from the randomized scoring yields an empirical estimate of false positives.
Another approach towards the multiple testing problem employs the false-discovery rate (FDR) statistic. The FDR technique first requires P-values to be calculated by some statistical method. Then the FDR at a given P-value threshold is simply the threshold multiplied by the number of tests performed, divided by the number of positives obtained at that threshold. This approach can be utilized in traditional microarray experiments as well [34].
Analysis of Schadt et al
In a tiling microarray experiment assessing transcription in six tissues across human chromosomes 20 and 22 [4], probes were first identified as being expressed using robust principal components analysis (PCA). Specifically, the authors used a sliding-window approach with windows of 500 probes (30 bp resolution tiling using 60-bp probes) wherein each window a PCA was performed and the first two dimensions were retained. The window size was selected to ensure that most windows included at least one transcribed sequence, on average. The first principal component was found to agreed closely with average probe intensity across tissues and the second dimension estimated variation across tissues. Next, windows with small values in the second principal component space were discarded. Of the remaining windows, the distance (Mahalanobis distance, MD) from each point in the two-dimensional principal component space (PCS) was computed from the center of the data of the window. The use of the MD is important for controlling for probe-to-probe intensity fluctuations and because it serves as a way of normalizing them all to the same scale. Outliers were identified by comparing these MDs with those obtained by performing the same analysis on a separate set of intron probes (which are thought not to exhibit signal due to the selection of polyA+ RNA by this experiment). MDs significantly larger than those found in the intron distribution (97.5th percentile) were subsequently deemed ‘on’, and the sequences they represent as transcribed. The use of probe sequences not expected to be transcribed as negative controls to aid in identifying transcribed regions is common in tiling experiments. For example, recent reports of tiling the Arabidopsis genome used putative promoter regions to obtain an intensity threshold [8,10], and an E. coli experiment used intensities measured from Bacillus subtilis probes [5] (transcribed sequences were defined as stretches of contiguous probes in genomic space above these thresholds).
The authors of this human tiling go one step further – that is, to group the transcribed segments into putative genes and gene structures. To do this, the authors performed one-dimensional hierarchical clustering of the MDs belonging to probes labeled as ‘on’. Probes that cluster together were grouped to form genes, the simple rationale being that probes from the same gene should exhibit similar intensities.
Comparison of algorithms
As discovered in Johnson et al. [20], tiling experiments probing the same genomic regions often do not produce the same results. There are several possible explanations for these discrepancies, and it is vital for the technology that the causes are identified. One possibility is the employment of different analysis algorithms. For a rigorous comparison, a gold-standard data set where we know exactly which bases are transcribed under some biological condition is required. Unfortunately, no standard exists over a large genomic region. In the absence of a gold standard, we sought to count the number of transcribed regions identified by the algorithms used in Bertone et al. [1] and in Kampa et al. [3] as a function of the respective input parameters (minrun, maxgap, and so on) of the algorithms. Although this does not directly address issues of experimental agreement, it is a step in that direction. In addition to counting the number of regions identified, we also calculated the number of transcribed regions identified in a randomized data set – thus providing an estimate of false positives as a function of algorithm parameterization. These calculations were performed on a single array representing human chromosome 22 from Kapranov et al. [2]. We did not include the analysis in Schadt et al. in our comparison because it requires multiple tissues and is therefore not directly comparable to the other two methods. The two methods we compare use only signal intensities, whereas Schadt et al. used correlations of signals to identify transcription. Clearly, this is an advantage of performing multiple tissue experiments.
Figure 5 plots the number of identified regions versus the number of regions identified in the randomized data for varying parameterizations on a log-log scale. It is clear that both methods demonstrate the ability to find things at a rate significantly favoring the actual data (i.e. many more things are identified than would be expected by chance) and that, for a range of parameterizations, the method of Kampa et al. [3] outperforms that of Bertone et al. [1].
Figure 5.
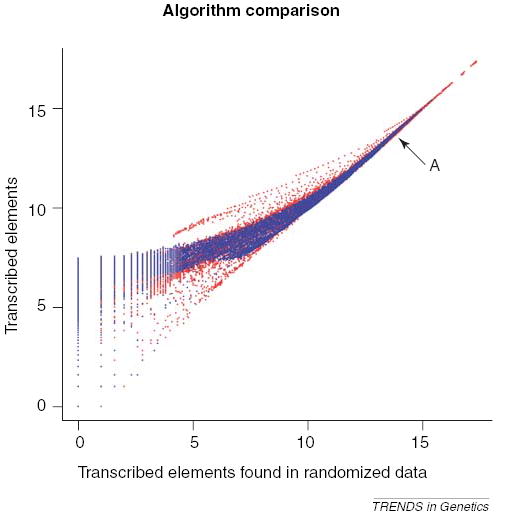
For various algorithm parameterizations, the number of regions identified within human chromosome 22 using the data of Kapranov et al. [2] is plotted against the number of things identified in a randomized data set using the same parameterization (log-log scale). Two algorithms are plotted – the algorithm of Kampa et al. [3] is plotted in red and the method in Bertone et al. [1] is plotted in blue. The arrow labeled ‘A’ points to the parameterization used in Kampa et al. [3].
In the context of Figure 5, the distance a point is above the 45° diagonal correlates with the ability of that parameterization to discriminate real transcription while minimizing the identification of spurious entities. A potentially revealing finding from our analysis is that the parameterization used for these data originally in Kampa et al. yields similar numbers of identified regions in the actual and randomized data. The original work reported a large number of identified regions, but it now seems likely that this number is an overestimate. This is somewhat surprising because the parameterization originally used had sound reasoning behind it based on the average lengths of exons in addition to obtaining an intensity threshold from well-selected bacterial negative controls. The parameterizations yielding significantly more positives than in the randomized set are those that have large windows and require high intensity thresholds.
Concluding remarks
Tiling microarray experiments are an exciting and relatively new application of microarray technology. Because the experiment is inherently different from its gene-centered counterparts, new statistical procedures need to be developed to account for the types of data tiling arrays can generate. We have presented aspects of tiling array data that can make their analysis difficult and have reviewed initial approaches to addressing these issues. The field of tiling array analysis is young and ripe for algorithmic discovery. We hope and expect to see research in this area flourish over the next few years. In particular, we hope to see careful multiplatform and multiprotocol studies where technical reproducibility and sensitivity versus specificity analyses can be carried out. Such studies will require generating a gold-standard genomic region where we understand transcription well – that is, where we know what is truly transcribed and what truly is not. With the advent of the ENCODE (encyclopedia of DNA elements) project [35], which plans to use tiling arrays as a major tool for human genome annotation, there is increasing need for such developments. Such progress will probably expand the robust application of tiling arrays to detailed exon-intron boundary discovery, detection of alternative splicing and single nucleotide polymorphism analysis.
Acknowledgments
We would like to thank Stefan Bekiranov for insightful conversations. This work was supported by NIH grant P50 HG02357.
References
- 1.Bertone P, et al. Identification of novel transcribed sequences in human using high-resolution genomic tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 2.Kapranov P, et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 3.Kampa D, et al. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schadt EE, et al. A comprehensive transcript index of the human genome generated using microarrays and computational approaches. Genome Biol. 2004;5:R73. doi: 10.1186/gb-2004-5-10-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selinger DW, et al. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat Biotechnol. 2000;18:1262–1268. doi: 10.1038/82367. [DOI] [PubMed] [Google Scholar]
- 6.Shoemaker DD, et al. Experimental annotation of the human genome using microarray technology. Nature. 2001;409:922–927. doi: 10.1038/35057141. [DOI] [PubMed] [Google Scholar]
- 7.Stolc V, et al. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- 8.Stolc V, et al. Identification of transcribed sequences in Arabidopsis thaliana by using high-resolution genome tiling arrays. Proc Natl Acad Sci U S A. 2005;102:4453–4458. doi: 10.1073/pnas.0408203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tjaden B, et al. Transcriptome analysis of Escherichia coli using high-density oligonucleotide probe arrays. Nucleic Acids Res. 2002;30:3732–3738. doi: 10.1093/nar/gkf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamada K, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 11.Euskirchen G, et al. CREB binds to multiple loci on human chromosome 22. Mol Cell Biol. 2004;24:3804–3814. doi: 10.1128/MCB.24.9.3804-3814.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martone R, et al. Distribution of NF-kappaB-binding sites across human chromosome 22. Proc Natl Acad Sci U S A. 2003;100:12247–12252. doi: 10.1073/pnas.2135255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rinn JL, et al. The transcriptional activity of human chromosome 22. Genes Dev. 2003;17:529–540. doi: 10.1101/gad.1055203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quackenbush J. Microarray data normalization and transformation. Nat Genet. 2002;32(Suppl):496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
- 15.Leung YF, Cavalieri D. Fundamentals of cDNA microarray data analysis. Trends Genet. 2003;19:649–659. doi: 10.1016/j.tig.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 16.Kirmizis A, Farnham PJ. Genomic approaches that aid in the identification of transcription factor target genes. Exp Biol Med (Maywood) 2004;229:705–721. doi: 10.1177/153537020422900803. [DOI] [PubMed] [Google Scholar]
- 17.Horak CE, Snyder M. ChIP-chip: a genomic approach for identifying transcription factor binding sites. Methods Enzymol. 2002;350:469–483. doi: 10.1016/s0076-6879(02)50979-4. [DOI] [PubMed] [Google Scholar]
- 18.Mantripragada KK, et al. Genomic microarrays in the spotlight. Trends Genet. 2004;20:87–94. doi: 10.1016/j.tig.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 19.Mukherjee S, et al. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson JM, et al. Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 2005;21:93–102. doi: 10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 21.Mockler TC, Ecker JR. Applications of DNA tiling arrays for whole-genome analysis. Genomics. 2005;85:1–15. doi: 10.1016/j.ygeno.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Hoyle DC, et al. Making sense of microarray data distributions. Bioinformatics. 2002;18:576–584. doi: 10.1093/bioinformatics/18.4.576. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L, et al. A model of molecular interactions on short oligonucleotide microarrays. Nat Biotechnol. 2003;21:818–821. doi: 10.1038/nbt836. [DOI] [PubMed] [Google Scholar]
- 24.Wu Z, Irizarry RA. Preprocessing of oligonucleotide array data. Nat Biotechnol. 2004;22:656–658. doi: 10.1038/nbt0604-656b. [DOI] [PubMed] [Google Scholar]
- 25.Cutler DJ, et al. High-throughput variation detection and genotyping using microarrays. Genome Res. 2001;11:1913–1925. doi: 10.1101/gr.197201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albert TJ, et al. Light-directed 5′→3′ synthesis of complex oligonucleotide microarrays. Nucleic Acids Res. 2003;31:e35. doi: 10.1093/nar/gng035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nuwaysir EF, et al. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res. 2002;12:1749–1755. doi: 10.1101/gr.362402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, et al. Detecting false expression signals in high-density oligonucleotide arrays by an in silico approach. Genomics. 2005;85:297–308. doi: 10.1016/j.ygeno.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 29.Kluger Y, et al. Relationship between gene co-expression and probe localization on microarray slides. BMC Genomics. 2003;4:49. doi: 10.1186/1471-2164-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian J, et al. Identification and correction of spurious spatial correlations in microarray data. Biotechniques. 2003;35:42–48. doi: 10.2144/03351bm03. [DOI] [PubMed] [Google Scholar]
- 31.Yin W, et al. Background correction for cDNA microarray images using the TV+L1 model. Bioinformatics. 2005;21:2410–2416. doi: 10.1093/bioinformatics/bti341. [DOI] [PubMed] [Google Scholar]
- 32.Edwards D. Non-linear normalization and background correction in one-channel cDNA microarray studies. Bioinformatics. 2003;19:825–833. doi: 10.1093/bioinformatics/btg083. [DOI] [PubMed] [Google Scholar]
- 33.Ying L, et al. Identification of chromosomal regions containing transcribed sequences using microarrays and computational methods. Proc Am Stat Assn. 2003:4672–4677. [Google Scholar]
- 34.Reiner A, et al. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 35.ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]


