Abstract
Nutrient uptake in the yeast Saccharomyces cerevisiae is a highly regulated process. Cells adjust levels of nutrient transporters within the plasma membrane at multiple stages of the secretory and endosomal pathways. In the absence of the ER-membrane-localized chaperone Shr3, amino acid permeases (AAP) inefficiently fold and are largely retained in the ER. Consequently, shr3 null mutants exhibit greatly reduced rates of amino acid uptake due to lower levels of AAPs in their plasma membranes. To further our understanding of mechanisms affecting AAP localization, we identified SSH4 and RCR2 as high-copy suppressors of shr3 null mutations. The overexpression of SSH4, RCR2, or the RCR2 homolog RCR1 increases steady-state AAP levels, whereas the genetic inactivation of these genes reduces steady-state AAP levels. Additionally, the overexpression of any of these suppressor genes exerts a positive effect on phosphate and uracil uptake systems. Ssh4 and Rcr2 primarily localize to structures associated with the vacuole; however, Rcr2 also localizes to endosome-like vesicles. Our findings are consistent with a model in which Ssh4, Rcr2, and presumably Rcr1, function within the endosome–vacuole trafficking pathway, where they affect events that determine whether plasma membrane proteins are degraded or routed to the plasma membrane.
THE capacity of cells to control their repertoire of surface proteins is key to several physiological responses. For example, in response to hormones, cells in ducts and tubules of the kidney maintain water and ion balance by regulating the translocation of water channels (aquaporins) and ion transporters to the cell surface. Neuronal cells are able to alter the number of receptors at synaptic membranes in response to specific stimuli, a process that is important in memory function. Similarly, yeast Saccharomyces cerevisiae cells respond to environmental signals to regulate nutrient uptake by adjusting levels of transport proteins in plasma membrane.
The regulated uptake of amino acids in yeast offers an attractive system for studying the multiple levels of metabolic control that determine the activity of plasma membrane nutrient transporters. Amino acids are transported across the plasma membrane by high- and low-affinity amino acid permeases (AAPs) (Regenberg et al. 1999). The AAPs constitute a protein family comprised of 18 highly homologous members with 12 transmembrane segments (Gilstring and Ljungdahl 2000). A unique member of the AAP family, Ssy1, acts as the receptor component of the Ssy1/Ptr3/Ssy5 (SPS) sensor of extracellular amino acids (reviewed in Forsberg and Ljungdahl 2001). In response to external amino acids, the SPS-sensor induces Stp1/Stp2-dependent transcription of AAP genes (de Boer et al. 2000; Forsberg and Ljungdahl 2001; Nielsen et al. 2001; Andréasson and Ljungdahl 2002). The expression of AAPs is also subject to nitrogen regulation, a process that prevents the utilization of nonpreferred nitrogen sources (e.g., allantoin and proline) when preferred nitrogen sources are present (e.g., ammonia and glutamine) (reviewed by Magasanik and Kaiser 2002).
The downregulation of AAPs and other transporters in the plasma membrane involves their endocytic removal and degradation in the vacuole (reviewed in Dupre et al. 2004). This process requires Rsp5-dependent ubiquitylation, an event often modulated by a phosphorylation (Hein et al. 1995; Hicke et al. 1998; Marchal et al. 1998; Feng and Davis 2000; Kelm et al. 2004). In endosomal compartments, protein cargo destined for vacuolar degradation is sorted into membrane regions that invaginate, leading to the formation of lumenal vesicles (Katzmann et al. 2002). The resulting vesicular compartments, called multi-vesicular bodies (MVBs), fuse with vacuoles. Consequently, the lumenal vesicles, including their cargo and lipids, are degraded. Proteins that reside in the limiting membrane of the MVBs end up in the vacuolar membrane. Recent studies have identified a number of proteins, including those within endosomal sorting complex required for transport (ESCRT) complexes (reviewed by Hurley and Emr 2006), that function in concert and in a highly regulated manner to facilitate the formation of MVBs. Many questions remain regarding the sorting mechanisms that determine whether membrane proteins are directed into MVBs.
Accumulating evidence indicates that many plasma membrane proteins cycle between endosomal compartments and the plasma membrane (Nikko et al. 2003; Bugnicourt et al. 2004; Rubio-Texeira and Kaiser 2006). Gao and Kaiser (2006) have reported that Gse (Gap1 sorting in endosomes) proteins bind and specifically prevent the general AAP (Gap1) from entering late endosomes and thereby restrict Gap1 entrance into the MVB pathway. The finding that assessory proteins selectively influence the inclusion of specific cargo suggests that active sorting mechanisms exist within the endosome–MVB trafficking pathway. This pathway appears to be an important control point that enables cells to adjust levels of nutrient and metabolite transporter by determining whether they are included or excluded from MVBs: inclusion leads to vacuolar degradation; conversely, exclusion enables transporters to cycle back to the plasma membrane.
Newly synthesized AAPs are translocated into the ER membrane and transported via the secretory pathway to the plasma membrane. AAPs depend on the membrane chaperone Shr3 to exit ER (Ljungdahl et al. 1992). In the absence of Shr3, AAPs aggregate in the ER and form large molecular weight complexes that are excluded from COPII transport vesicles (Kuehn et al. 1996; Gilstring et al. 1999; Kota and Ljungdahl 2005). Due to the severely compromised ability of AAPs to exit the ER, shr3 null mutant cells possess low levels of AAP at the plasma membrane and consequently are unable to effectively take up amino acids.
To identify additional proteins that affect the progression of AAPs through the secretory pathway, we selected SSH (high-copy suppressors of shr3Δ) genes. Overexpression of SSH4, SSH5 (also known as RCR2), or SSH6 (also known as RCR1) increases levels of functional AAPs in the plasma membrane and also enhances phosphate and uracil uptake. Conversely, mutants carrying null alleles of these genes exhibit diminished levels of AAPs. SSH4, RCR2, and RCR1 encode membrane proteins; Ssh4 and Rcr2 localize to structures associated with the endosome–vacuole trafficking pathway. Our findings suggest that Ssh4 and Rcr2, and likely Rcr1, affect nutrient transport in yeast by influencing general sorting processes within the endosome–vacuole trafficking pathway that determine whether membrane proteins are targeted to the vacuole or routed to the plasma membrane.
MATERIALS AND METHODS
Media:
Standard media, including YPD, ammonia-based synthetic minimal dextrose (SD) or complete dextrose (SC), was prepared as described previously (Burke et al. 2000). Synthetic minimal and complete media with alternative carbon sources contains 2% galactose (SG, SCG) or 2% raffinose (SR, SCR) in place of dextrose. Synthetic minimal media containing allantoin (SAD) as the sole nitrogen source have been described (Kota and Ljungdahl 2005). Low-phosphate media were prepared according to O'Connell and Baker (1992). Media was made solid with 2% (w/v) Bacto agar (Difco). YPD containing 0.5 mg/ml sulfonylurea (MM) was prepared as described (Jørgensen et al. 1998). Solid YPD medium was supplemented with 200 mg/liter G418 (kan; Invitrogen, San Diego), 100 mg/liter cloNAT (nat; Werner Bioagents), or hygromycin B (hph; Duchefa) for antibiotic selections. Where indicated, 5-fluoroorotic acid (FOA) was added to SD medium to 1 g/liter. For growth assays, equally dense cell suspensions were prepared in water, and 10-fold dilution series were spotted onto desired media and incubated at 30°.
Strains:
Yeast strains used in this work are listed in Table 1. The GAL+ triple-mutant (shr3 leu2 trp1) strain FMAS81:21D was constructed as follows. One allele of SHR3 was deleted by transforming MMDY80 (MATa/α ura3-52/ura3-52 trp1-289/TRP1 leu2-3,112/LEU GAL+) with linearized pPL288 (shr3Δ5∷hisG-URA3-hisG), resulting in strain MMDY81. Correct integration of the deletion construct was verified by Southern blot analysis and tetrad analysis. One Ura+ segregant was propagated on minimal SD containing FOA to attain FMAS81:21D. Strains JKY40, JKY41, and JKY42 were constructed by deleting the entire sequence of SSH4, RCR2, and RCR1 in CAY28 with PCR-amplified hphMX4, natMX4, and kanMX4 cassettes (primers F-ssh4D/R-ssh4D, F-ssh5D/R-ssh5D, and F-ssh6D/R-ssh6D), respectively. JKY43 was obtained similarly by deleting SSH4 in JKY1. Strains JKY47 and JKY48 were constructed in a similar way by deleting the entire sequence of RCR2 or RCR1, respectively, in PLY120. In all cases, the correct integration of SSH4/RCR2/RCR1 deletion cassettes was confirmed by PCR analysis and tetrad analysis to verified that deletion markers segregated 2:2. HFY500 and HFY501 are progeny from the diploid obtained from crossing JKY43 and JKY47. JKY43 was crossed to JKY48; haploid strain HFY502 is derived from this cross. The haploid HFY538 is a segregant from a cross between HFY502 and JKY41.
TABLE 1.
Yeast strains
| Strain | Genotype | Reference |
|---|---|---|
| FMAS81:21D | MATα ura3-52 leu2-3,11trp1-289 2 shr3Δ6 GAL1+ | This work |
| CAY28 | MATα ura3-52 | Andréasson and Ljungdahl (2002) |
| CAY91 | MATaura3-52 ssy1Δ13∷hisG | Andréasson and Ljungdahl (2002) |
| CAY123 | MATaura3-52 stp1Δ51∷AgLEU2 stp2Δ50∷hphMX4 | Andréasson and Ljungdahl (2002) |
| HKY20 | MATaura3-52 lys2Δ201 ssy1Δ13 | Klasson et al. (1999) |
| HFY500 | MATaura3-52 leu2-3,112 shr3Δ6 | This work |
| HFY501 | MATα ura3-52 leu2-3,112 | This work |
| HFY502 | MATaura3-52 shr3Δ6 ssh4Δ∷hphMX rcr1Δ∷kanMX | This work |
| HFY538 | MATaura3-52 ssh4Δ∷hphMX rcr2Δ∷natMX rcr1Δ∷kanMX | This work |
| JKY1 | MATα ura3-52 shr3Δ6 | Kota and Ljungdahl (2005) |
| CEN.PK113-5D/PHO84myc | MATaura3-52 PHO84∷6His-2MYC-loxP-KanMX-loxP MAL2-8c SUC2 | Lagerstedt et al. (2002) |
| JKY6 | MATaura3-52 PHO84∷6His-2MYC pho86Δ∷hphMX4 MAL2-8c SUC2 | Kota and Ljungdahl (2005) |
| JKY40 | MATα ura3-52 ssh4∷hphMX | This work |
| JKY41 | MATα ura3-52 rcr2∷natMX | This work |
| JKY42 | MATα ura3-52 rcr1∷kanMX | This work |
| JKY43 | MATα ura3-52 shr3Δ6 ssh4∷hphMX | This work |
| JKY47 | MATaura3-52 lys2Δ201 leu2-3,112 his3Δ200 rcr2∷natMX | This work |
| JKY48 | MATaura3-52 lys2Δ201 leu2-3,112 his3Δ200 rcr1∷kanMX | This work |
| PLY120 | MATaura3-52 lys2Δ201 leu2-3,112 his3Δ200 | Ljungdahl lab collection |
| PLY126 | MATaura3-52 lys2Δ201 | Kuehn et al. (1996) |
| PLY860 | MATα ura3-52 trp1Δ101∷loxP | Ljungdahl lab collection |
Cloning of SSH4 and SSH5/RCR2:
High-copy suppressors of the shr3 deletion were isolated as follows. Strain FMAS81:21D is unable to grow on SC; shr3 null alleles are conditionally lethal on complex media when present in combination with leu2 or trp1 auxotrophies (Ljungdahl et al. 1992). This strain can grow on SD supplemented with only the required amino acids. Cells of FMAS81:21D cultured in SD (supplemented with uracil, leucine, and tryptophan) were transformed with a GAL1-promoted URA3-marked cDNA library (Liu et al. 1992). The quality of the library was determined prior to transformation by introducing it to Escherichia coli and analyzing insert size and frequency. Inserts ranging in size between 1 and 2.5 kb were found in 22 of 24 clones. Ten independent pools of transformed FMAS81:21D cells were spread onto solid SD, SG, and SR media (containing leucine and tryptophan). Transformed colonies from each pool were carefully scraped from plates, resuspended in individual 15% glycerol stocks, and stored at −80° until needed. The cell density of each stock was determined, and ∼2 × 106 cells were spread onto SCG and SCR media lacking uracil (−ura). From 10 individual pools, four clones growing on SCG (−ura) and four growing on SCR (−ura), respectively (in total, 80 clones), were picked for further evaluation. Plasmids from eight clones that showed increased growth on SCG (−ura) compared to SCR (−ura) and no growth on SC (−ura) were isolated and sequenced using primers annealing to flanking sequences (T7 and POL 95-001). Plasmids pPL501 and pPL507 carrying SSH4 and SSH5 sequences, respectively, were identified.
DNA cloning:
Plasmids used in this study are listed in Table 2. The sequences of oligonucleotide primers are available on request. Plasmids pJK120, pJK121, and pJK122 were created by homologous recombination in yeast; SSH4, RCR2, or RCR1 with their own promoters were PCR amplified from S288C genomic DNA (primers F-SSH4-pRS202/R-SSH4-pRS202, F-SSH5-pRS202/R-SSH5-pRS202, and F-SSH6-pRS202/R-SSH6-pRS202) and cotransformed with BamHI/BglII-restricted pRS202, respectively. BamHI/SalI-restricted pMB50 was cotransformed into yeast with PCR product containing SSH4, RCR2, or RCR1 (primers F-SSH4-pADH1/R-SSH4-pADH1, F-SSH5-pADH1/R-SSH5-pADH1, and F-SSH6-pADH1/R-SSH6-pADH1) amplified from S288C genomic DNA, resulting in plasmids pJK123, pJK124, and pJK125, respectively. Inserting SalI/NotI fragments from pJK120, pJK121, and pJK122 into SalI/NotI-restricted pRS316, respectively, created plasmids pJK126, pJK127, and pJK128. SSH4 was epitope tagged with 6xHA or 3xmyc at the C terminus, immediately prior to the stop codon, creating plasmid pJK129 and pJK131, respectively: the 6xHA and 3xmyc sequences were PCR amplified from plasmids pYM3 and pPL329 (primers F-SSH4-6xHA and R-SSH4-6xHA or F-SSH4-3xMYC and R-SSH4-3xMYC) and cotransformed together with HindIII/ClaI-restricted pJK120 for homologous recombination. RCR2 was epitope tagged at the C terminus in a similar manner with either 6xHA or 3xmyc epitopes: 6xHA and 3xmyc sequences were PCR amplified from plasmids pYM3 and pPL329 (primers F-SSH5-6xHA/R-SSH-6xHA or F-SSH5-3xMYC/R-SSH-3xMYC) and cotransformed together with HindIII/ClaI-restricted pJK121, creating plasmids pJK133 and pJK135, respectively. To express epitope-tagged SSH4 and RCR2 from a CEN plasmid, we constructed pJK130, pJK134, and pJK136 by inserting a NotI/SalI fragment from pJK129 and pJK133 into a NotI/SalI-restricted pRS316 and NotI/XhoI fragment from pJK135 into NotI/XhoI-restricted pRS316, respectively. HA-TAT2 in pJK139 was constructed by homologous recombination: HA-TAT2 with its own promoter was PCR amplified from pAS55 (primers F-HA-TAT2 and R-HA-TAT2) and cotransformed with KpnI/XhoI-restricted pJG4-5. PCR products were digested with DpnI to remove methylated (E. coli derived) DNA prior to transformation into yeast.
TABLE 2.
Plasmids
| Plasmid | Description | Reference |
|---|---|---|
| pCA030 | PAGP1-lacZ in pRS317 | Andréasson and Ljungdahl (2004) |
| pJK120 | SSH4 in pRS202 | This work |
| pJK121 | RCR2 in pRS202 | This work |
| pJK122 | RCR1 in pRS202 | This work |
| pJK123 | PADH1-SSH4 in pMB50 | This work |
| pJK124 | PADH1-RCR2 in pMB50 | This work |
| pJK125 | PADH1-RCR1 in pMB50 | This work |
| pJK126 | SSH4 in pRS316 | This work |
| pJK127 | RCR2 in pRS316 | This work |
| pJK128 | RCR1 in pRS316 | This work |
| pJK129 | SSH4-6xHA in pRS202 | This work |
| pJK130 | SSH4-6xHA in pRS316 | This work |
| pJK131 | SSH4-3xMYC in pRS202 | This work |
| pJK133 | RCR2-6xHA in pRS202 | This work |
| pJK134 | RCR2-6xHA in pRS316 | This work |
| pJK135 | RCR2-3xMYC in pRS202 | This work |
| pJK136 | RCR2-3xMYC in pRS316 | This work |
| pJK139 | HA-TAT2 (2μ TRP1) | This work |
| pAS55 | HA-TAT2 (2μ URA3) | Beck et al. (1999) |
| pPL501 | SSH4 in PGAL-pRS316 | This work |
| pPL507 | SSH5 in PGAL-pRS316 | This work |
| pMB50 | 2μ URA3 PADH1 | Ljungdahl lab collection |
| pJG4-5 | 2μ TRP1 | Gyuris et al. (1993) |
| pYM3 | 6xHA | Knop et al. (1999) |
| pPL329 | 3xMYC | Ljungdahl lab collection |
| pPL288 | shr3Δ5∷hisG-URA3-hisG in pBSII SK(+) | Kuehn et al. (1996) |
| pRS202 | 2μ URA3 | Connelly and Hieter (1996) |
| pRS316 | CEN URA3 | Sikorski and Hieter (1989) |
| pRS317 | CEN LYS2 | Sikorski and Hieter (1989) |
Protein manipulations:
Whole-cell protein extracts were prepared according to Silve et al. (1991). Protein samples from rapamycin-treated cells were obtained as follows. An aliquot of rapamycin stock solution [1 mg/ml in 90% ethanol, 10% Tween-20 (Beck et al. 1999)] was added to cells growing in SC (lacking uracil and tryptophan; OD600 of 2) to a final concentration of 200 ng/ml. Cultures were kept at 30° and 1-ml subsamples were withdrawn at the time points indicated. Protein was prepared and samples were heated for 10 min at 40°, resolved by SDS–PAGE (10% gels), and analyzed by immunoblotting. Blue Native (BN)–PAGE was carried out as described (Kota and Ljungdahl 2005). Protein extracts were prepared from cells grown in SAD at 25°, solubilized with 1.5 μg dodecyl-β-d-malotopyranoside (DM)/μg protein at 4° for 35 min and separated on 4–15% BN gradient gels. High-molecular-weight markers (Amersham Bioscience) were used as standards. Immunoblots were incubated as indicated with primary antibody in blocking buffer diluted as follows: rabbit α-Gap1, 1:15,000; mouse α-Dpm1 monoclonal (Molecular Probes, Eugene, OR), 2 μg/ml; rat α-HA monoclonal (3F10: Roche), 1:1000; mouse α-HA monoclonal (12CA5), 1:5000; mouse α-myc monoclonal (9E10: Roche), 1:2500. Immunoreactive bands were visualized by chemiluminescence emanating from secondary HRP-conjugated antibodies: α-rabbit immunoglobulin (Ig) from donkey, α-mouse Ig from sheep, or α-rat Ig from goat (Amersham Biosciences), using the LAS1000 system (Fuji Photo Film).
β-Galactosidase overlay assay:
Semiquantitative β-galactosidase activity assay was performed with cells grown on solid SD medium (containing 1.3 mm leucine). Low-melting-point agarose (0.5%) was melted in 0.4 m potassium phosphate buffer (pH 7.0), and after slight cooling, 0.2% N-lauroyl sarcosine, 0.05% β-mercaptoethanol, and 0.2 mg/ml X-gal (from 100 mg/ml stock in dimethyl formamide) were added. Approximately 10 ml of the final agarose mixture was poured onto cultured plates and allowed to cool. Plates were incubated at RT until blue precipitate was visible and kept at 4° until photographed.
Fluorescence microscopy:
Cells grown to early log phase in SC (lacking uracil) were processed for indirect immuno-fluorescence analysis (Burke et al. 2000), viewed, and photographed as described in Boban et al. (2006). Antibodies used to visualize epitope-tagged proteins were primary rat α-HA (3F10: Roche) or mouse α-myc (9E10: Roche) diluted 1:400 and 1:300, respectively. Secondary AlexaFluor488-conjugated α-rat or α-mouse antibodies were obtained from Molecular Probes and used in 1:500 dilution.
RESULTS
High-copy suppressors of loss of Shr3 function:
As a result of diminished amino acid uptake capabilities, shr3Δ mutant strains carrying leu2, trp1, or his3 auxotrophies are unable to grow on synthetic complete medium (SC); however, these strains grow well on SD supplemented with only the required amino acids (Ljungdahl et al. 1992). The synthetic lethality on SC is due to the high amino acid content of this medium; the combination of reduced AAP activity and the abundance of competing amino acids, which interfere with the residual uptake mechanisms, effectively inhibits uptake of the required amino acid. Thus, when grown on SC, auxotrophic shr3 mutants cannot synthesize the required amino acid nor can they import them from the external environment at rates sufficient to support growth. This synthetic lethality formed the basis of a high-copy suppression screen for genes that enable auxotrophic shr3 null mutant strains to grow on SC.
A triple deletion strain (shr3Δ leu2Δ trp1Δ) was transformed with a PGAL1-promoted cDNA library (Liu et al. 1992) and transformants able to grow on SC were selected. Plasmids from eight colonies that exhibited consistent and robust growth were rescued and sequenced. Five of the plasmids contained SHR3 and one contained LEU2. The two remaining plasmids contained the open reading frames (ORFs) YKL124w and YDR003w. These genes were designated SSH4 and SSH5, respectively, for suppressor of shr3 null mutation. A database search identified the protein encoded by ORF YBR005w as being 46% identical to Ssh5; SSH4 and SSH5 are ohnologs (Byrne and Wolfe 2005). We conveyed the cloning of the SSH genes to publicly available databases; subsequently it was found that the overexpression of SSH4 suppresses the antiproliferative effect of leflunomide, an immunosuppressant that also inhibits yeast growth (Fujimura 1998b). More recently, the overexpression of YBR005w/SSH6 was reported to confer resistance to the cell-wall-perturbing agent Congo Red, and the gene was redesignated RCR1 (Resistance to Congo Red) and its homolog renamed RCR2 (YDR003w/SSH5) (Imai et al. 2005). Interestingly, the overexpression of RCR2 did not confer resistance to Congo Red, indicating that RCR1 and RCR2 are nonredundant. To avoid confusion, we will refer to SSH5 as RCR2 and SSH6 as RCR1.
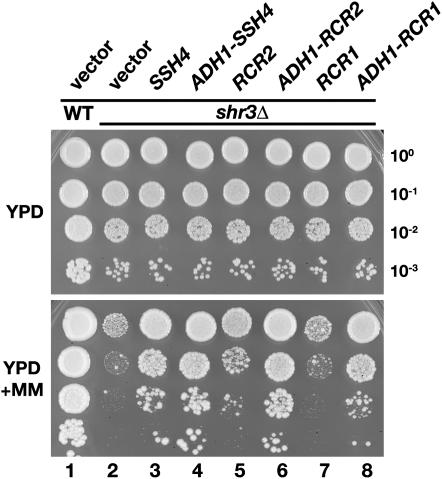
Genomic SSH4, RCR2, and RCR1 sequences with native and PADH1 promoters were inserted into low-copy CEN- and multicopy 2μ-based vectors (see Table 2). These plasmid constructs were individually introduced into an shr3Δ mutant, and the growth characteristics of the resulting strains were assessed on YPD medium in the absence or presence of the herbicide MM (Figure 1). MM inhibits the biosynthetic pathways of leucine, isoleucine, and valine (Falco and Dumas 1985) and thus mimics the synthetic lethal phenotype observed when shr3 and leu2 mutations are combined. As seen in Figure 1, the SHR3 wild-type strain grows well on YPD+MM (dilution series 1), whereas the shr3 null mutant transformed with an empty vector grows poorly (dilution series 2). The shr3 mutants carrying either native or PADH1-promoted SSH4 exhibited robust growth (compare dilution series 2 with 3 and 4). Strains expressing PADH1-promoted RCR2 and RCR1 grew well (dilution series 6 and 8, respectively), whereas strains expressing RCR2 and RCR1 under control of native promoters grew quite poorly (dilution series 5 and 7). These results demonstrate that RCR1, cloned purely on the basis of sequence homology to RCR2, also functions as a high-copy suppressor of shr3Δ. Their ability to suppress shr3 mutations under conditions where amino acid uptake is limiting for growth indicates that the overexpression of SSH4, RCR2, or RCR1 increases amino acid uptake. The fact that we did not isolate a plasmid encoding RCR1 and obtained only one plasmid carrying SSH4 and RCR2, respectively, indicates that the SSH screen was not saturated.
Figure 1.—
SSH4, RCR2, and RCR1 are high-copy suppressors of shr3 deletion. Tenfold dilution series of cell suspensions of CAY28 (wild type) transformed with pRS202 (vector) and JKY1 (shr3Δ) transformed with 2μ plasmids pRS202 (vector), pJK120 (SSH4), pJK123 (ADH1-SSH4), pJK121 (RCR2), pJK124 (PADH1-RCR2), pJK122 (RCR1), or pJK125 (ADH1-RCR1) were spotted onto solid YPD medium and YPD supplemented with MM. Plates were incubated at 30° for 2 days (YPD) or for 3 days (YPD+MM) and photographed.
SSH4, RCR2, and RCR1 were individually disrupted, and strains carrying all combinations of ssh4Δ, rcr2Δ, and rcr1Δ mutations were constructed. The growth of single, double, and triple mutant strains was examined under a variety of conditions. We confirmed that rcr1Δ mutations result in weak Congo Red sensitivity (Imai et al. 2005); however, rcr1Δ strains carrying ssh4Δ and/or rcr2Δ mutations did not exhibit heightened Congo Red sensitivity. Under all other conditions tested, the single, double, and triple mutants grew as well as the wild-type strain. The conditions examined include growth on various nitrogen and carbon sources in the presence of toxic amino acids and analogs at different temperatures (20°–42°) and in the presence of high salt, detergent, or heavy metals.
Overexpression of SSH4, RCR2, and RCR1 increases Gap1 activity:
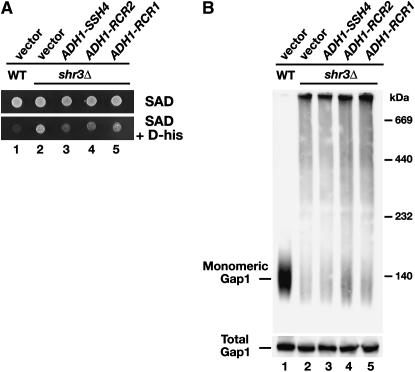
To investigate if the suppressing activity of Ssh4, Rcr2, and Rcr1 is due to their ability to exert an Shr3-like function, we sought to examine the status of Gap1 folding in shr3 mutant cells overexpressing these proteins. To proceed, it was first necessary to determine whether the overexpression of SSH4, RCR2, or RCR1 specifically affect Gap1 transport activity. Gap1 mediates the uptake of toxic d-amino acids (Rytka 1975; Jauniaux and Grenson 1990). Gap1 is expressed at high levels in cells grown on media containing allantoin (SAD) as the sole nitrogen source. Consequently, SHR3 wild-type cells are unable to grow on SAD containing d-histidine (Figure 2A, dilution 1). In contrast, shr3Δ cells exhibit greatly reduced levels of Gap1 transport activity and thus are resistant to the growth-inhibiting affects of d-histidine (Figure 2A, dilution 2). When high-copy plasmids carrying SSH4, RCR2, and RCR1 were introduced into shr3 null mutants, the resulting strains exhibited enhanced d-histidine sensitivity (Figure 2A, dilutions 3–5). The inability to grow efficiently in the presence of d-histidine was not due to a generally slow growth on poor nitrogen medium; all strains grew well and at indistinguishable rates on media without d-histidine (Figure 2A, top). These results indicate that the suppressed strains have increased Gap1-dependent transport activity in their plasma membranes.
Figure 2.—
Suppression of shr3Δ by SSH4, RCR2, and RCR1 is not due to chaperone-like activity in the ER. (A) Suppression of shr3Δ on d-histidine-containing medium. Serial dilutions of cell suspensions of wild-type strain (CAY28) carrying vector control (pRS202) and shr3Δ strain (JKY1) carrying vector control (pRS202) or plasmid-expressing SSH4 (pJK123), RCR2 (pJK124), or RCR1 (pJK125) from the ADH1 promoter were prepared. Equal aliquots of each dilution were applied to SAD or SAD supplemented with d-histidine (0.15%). Plates were incubated at 30° for 2 days and photographed. (B) Analysis of Gap1 aggregation. Protein extracts from strains as in A grown in SAD were prepared and solubilized with DM (1.5 μg DM μg−1 protein) at 4°. Solubilized proteins were separated by BN–PAGE, immunoblotted, and analyzed using antibodies raised against Gap1 (top). Total Gap1 levels were analyzed in protein extracts (20 μg) separated by SDS–PAGE (bottom).
Ssh4, Rcr2, and Rcr1 do not function as ER-membrane-localized chaperones:
In the absence of Shr3, AAPs aggregate in the ER membrane, forming large complexes that can be readily visualized as high-molecular-weight smears on polyacrylamide gels under native conditions (Kota and Ljungdahl 2005). The aggregation state of Gap1 was examined in lysates from wild-type (CAY28) and shr3Δ (JKY1) cells expressing control vector, PADH1-SSH4, PADH1-RCR2, or PADH1-RCR1. Lysates, prepared from cells growing in SAD, were solubilized in the presence of DM, and proteins were separated by Blue Native–PAGE (Figure 2B). Consistent with published results (Kota and Ljungdahl 2005), monomeric Gap1 was readily extracted from membranes prepared from SHR3 wild-type cells (Figure 2B, lane 1), but not from membranes prepared from shr3Δ cells (Figure 2B, lane 2). The overexpression of SSH4, RCR2, or RCR1 did not facilitate monomer solubility (Figure 2B, lanes 3–5). The reduced ability to extract monomeric Gap1 from lysates prepared from shr3 mutants is not a consequence of lower levels of Gap1; similar amounts of Gap1 are present in lysates from all strains (Figure 2B, bottom). On the basis of these results, we conclude that suppression of shr3 by the overexpression of SSH4, RCR2, and RCR1 is not due to increased “chaperone-like” activity in the ER.
Overexpression of SSH4, RCR2, or RCR1 suppress amino acid uptake defects associated with loss of SPS sensor gene expression:
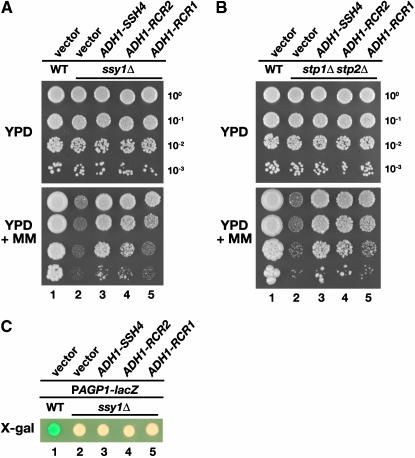
The overexpression of SSY1, encoding the receptor component of the SPS sensor, or the introduction of a multi-copy plasmid with a constitutively active allele of transcription factor STP1 (ASI13-1/STP1Δ131; Andréasson and Ljungdahl 2002), can partly suppress amino acid uptake defects of shr3 mutants (C. Andréasson and P. O. Ljungdahl, unpublished data). This raised the possibility that SSH4, RCR1, and RCR2 suppress shr3 mutations by increasing SPS-sensor-dependent AAP gene expression. This could be accomplished either by augmenting SPS sensor signaling or by promoting nuclear targeting of Stp1 or Stp2; in either case, suppression would be Stp1 and Stp2 dependent. Similar to shr3 mutants, ssy1Δ and stp1Δ stp2Δ cells are unable to grow on YPD media containing MM (Figure 3, A and B, dilution series 2). Plasmids with PADH1-promoted SSH4, RCR2, and RCR1 were introduced into ssy1Δ and stp1Δ stp2Δ mutant strains, and these strains grew markedly well (Figure 3, A and B, lanes 3–5). These findings demonstrate that SSH4, RCR2, and RCR1 suppress the amino acid uptake defects associated with loss of SPS sensor signaling and clearly show that SSY1, STP1, and STP2 are not necessary for the increase in amino acid permease activity resulting from overexpression of SSH4, RCR2, and RCR1.
Figure 3.—
SSH4, RCR2, and RCR1 suppress loss of SPS sensor signaling. (A) SSH4, RCR2, and RCR1 are high-copy suppressors of ssy1Δ. Phenotypic analysis of wild-type (CAY28) and ssy1Δ strains (CAY91) carrying a vector control (pRS202) or expressing SSH4 (pJK123), RCR2 (pJK124), or RCR1 (pJK125) as indicated. Tenfold dilution series of cell suspensions were spotted onto plates containing YPD and YPD supplemented with MM. Plates were incubated at 30° for 2 days (YPD) or for 4 days (YPD+MM) and photographed. (B) SSH4, RCR2, and RCR1 are high-copy suppressors of stp1Δ stp2Δ. Serial dilutions of wild-type (CAY28) and stp1Δ stp2Δ strains (CAY91) carrying a vector control (pRS202) or plasmids expressing SSH4 (pJK123), RCR2 (pJK124), or RCR1 (pJK125) are as indicated. Dilutions of cell suspensions were applied to YPD or YPD with MM as indicated. Plates were incubated as in A and photographed. (C) Expression from the AGP1 promoter was monitored by assessing β-galactosidase activity in cells transformed with a PAGP1-lacZ reporter construct. Wild-type strain PLY126 cotransformed with pCA030 (YCpAGP1-lacZ) and pRS202 (vector), as well as ssy1Δ strain HKY20 carrying pCA030 together with 2μ plasmids expressing SSH4 (pJK123), RCR2 (pJK124), or RCR1 (pJK125) from the ADH1 promoter were applied to solid SD medium supplemented with 1.3 mm leucine. Plates were grown at 30° for 2 days and overlaid with X-gal substrate.
However, the possibility remained that SSH4, RCR2, or RCR1 induce SPS sensor gene expression in a parallel and previously uncharacterized manner that is independent of known SPS-sensing pathway components. To test this, we directly monitored the expression of a plasmid-borne AGP1-promoted β-galactosidase reporter construct (PAGP1-lacZ) in an ssy1Δ background. AGP1 encodes an amino acid permease that is induced by amino acids in an SSY1-dependent manner (Iraqui et al. 1999). In wild-type cells, PAGP1-lacZ is strongly induced on plates containing leucine, and cells turn blue in the presence of the β-galactosidase substrate X-Gal (Figure 3C, spot 1). In ssy1 mutants, the PAGP1-lacZ is not induced, and the cells are white (Figure 3C, spot 2). Cells overexpressing SSH4, RCR2, or RCR1 remain white, indicating that their overexpression does not suppress shr3 mutations by bypassing SPS sensor signaling and inducing AAP gene expression. These results are consistent with genomewide transcription profiling of a strain overexpressing RCR1 (Imai et al. 2005); no alterations in transcription, other than RCR1 itself, were observed.
Ssh4 and Rcr2 localize to the endosome–vacuole trafficking pathway:
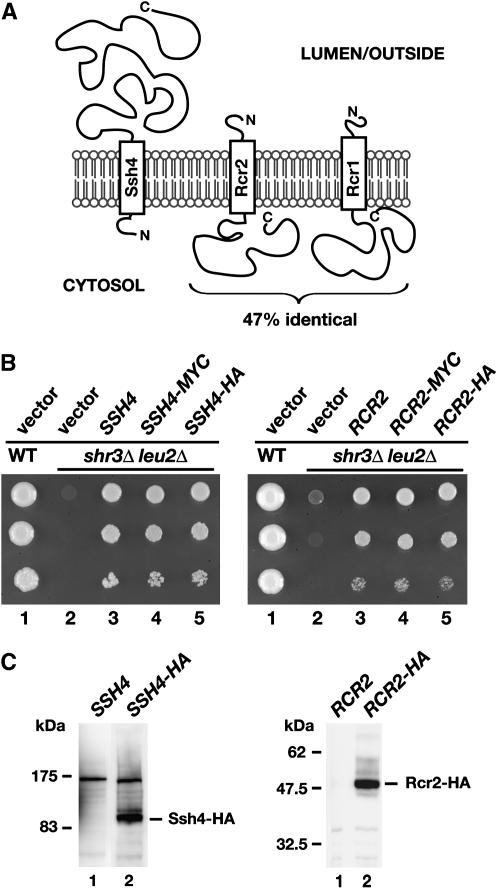
SSH4, RCR2, and RCR1 encode integral membrane proteins with a single membrane-spanning segment (Figure 4A). Ssh4 is comprised of 597 amino acids. Rcr2 and Rcr1 consist of 210 and 213 amino acids, respectively. The membrane-spanning segment in each of these proteins lies close to their N termini. The N terminus of Ssh4 is predicted to face the cytosol, whereas Rcr2 and Rcr1 are type I membrane proteins with their C termini oriented toward the cytoplasm. The topology of Rcr1 has been experimentally verified (Imai et al. 2005). To facilitate localization studies, HA- and myc-epitope tags were individually introduced at the extreme C termini of Ssh4, Rcr2, and Rcr1. The functionality of each of the tagged constructs was determined by examining their ability to suppress the synthetic lethal phenotype exhibited by an shr3 leu2 double mutant on SC. Transformants with multi-copy plasmids with either HA- and myc-tagged alleles of SSH4 and RCR2 grew as well as untagged alleles (Figure 4B; compare dilution series 3 with 4 and 5), indicating that the tagged constructs are fully functional. Ssh4-HA and Rcr2-HA migrate slightly more slowly than proteins of their predicted molecular weights of 72 and 34 kDa, respectively (Figure 4C). We failed to obtain functional tagged versions of Rcr1 (data not shown).
Figure 4.—
SSH4, RCR2, and RCR1 encode membrane proteins. (A) Schematic of secondary structure predictions of Ssh4, Rcr2, and Rcr1. Hydropathy analysis using a window size of 11 amino acid residues indicates that all three proteins harbor a single membrane-spanning segment in close proximity to their N termini (Kyte and Doolittle 1982). The orientation in the membrane is based on the N-best algorithm (TMHMM 1.0 prediction). (B) Functional analysis of epitope-tagged SSH4 and RCR2. Serial dilutions of strain HFY501 (WT: leu2 SHR3) transformed with pRS202 (vector) and strain HFY500 (leu2 shr3Δ) transformed with 2μ plasmids pRS202 (vector), pJK120 (SSH4), pJK131 (SSH4-3xMYC), pJK129 (SSH4-6xHA), pJK121 (RCR2), pJK135 (RCR2-3xMYC), or pJK133 (RCR2-6xHA) were applied to SC (−ura) plates. Plates were incubated at 30° for 3 days and photographed. (C) Immunoblot analysis of whole-cell extracts from strain JKY40 (ssh4Δ) transformed with either pJK120 (SSH4) or pJK130 (SSH4-6xHA) and strain JKY41 (rcr2Δ) transformed with either pJK121 (RCR2) or pJK134 (RCR2-6xHA), grown to log phase in SC (−ura) medium. Antibodies used to visualize Ssh4-HA and Rcr2-HA were α-HA (12CA5, mouse) and α-HA (3E10, rat), respectively.
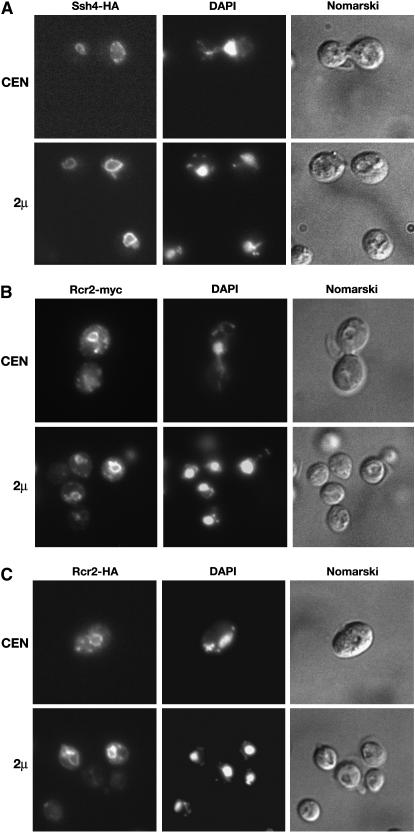
The intracellular location of Ssh4-HA, expressed from either low-copy (CEN) or multi-copy (2μ) plasmids, was determined by indirect immunofluorescence microscopy (Figure 5A). HA-dependent fluorescence was found exclusively around structures corresponding to the vacuole; vacuoles are clearly recognized as major semicircular depressions when cells are observed using Nomarski optics. The pattern of fluorescence was identical in strains carrying CEN (top panels) and 2μ plasmids (bottom panels); however, the fluorescence intensity was notably higher in cells carrying the 2μ plasmid.
Figure 5.—
Localization of Ssh4 and Rcr2. (A) Indirect immunolocalization of Ssh4-HA in strain JKY40 (ssh4Δ) transformed with SSH4-6xHA expressed from either a CEN plasmid (pJK130, top) or a 2μ plasmid (pJK129, bottom). (B) Rcr2-myc was localized in JKY41 (rcr2Δ) transformed with either RCR2-3xMYC expressed from a CEN plasmid (pJK136, top) or from a 2μ plasmid (pJK135, bottom). (C) Indirect immunolocalization of Rcr2-HA was performed with RCR2-6xHA expressed from either a CEN plasmid (pJK134, top) or a 2μ plasmid (pJK133, bottom) in JKY41 (rcr2Δ). Cells expressing Ssh4-HA, Rcr2-myc, or Rcr2-HA were processed for indirect immunofluorescence and viewed by Alexa Fluor 488-dependent fluorescence, DAPI staining, and Nomarski optics as indicated.
The intracellular location of Rcr2 was determined using both myc- (Figure 5B) and HA-epitope-tagged (Figure 5C) constructs. Rcr2-myc- and Rcr2-HA-specific fluorescence was observed primarily in close association with the vacuole. However, both Rcr2-myc- and Rcr2-HA-dependent fluorescence was also observed in small vesicles in the vicinity of the plasma membrane and in large vesicular structures resembling endosomes. The fluorescence associated with expression from the multi-copy 2μ plasmid (bottom panels) showed similar, but more intense fluorescence compared to that observed in cells with the CEN plasmid (top panels). The observation that Rcr2 localizes to endosome-like/vacuolar compartments is consistent with a report that Rcr2 interacts with Ypt7 (Ito et al. 2001). Ypt7 is a small Ras-like GTPase that localizes to the vacuolar membrane; Ypt7 is required for docking and fusion of endosomal vesicles to vacuolar membranes, as well as for homotypic fusion events between vacuolar compartments (Schimmoller and Riezman 1993; Haas et al. 1995).
Steady-state levels of Gap1 and Tat2 increase in strains overexpressing SSH4, RCR2, and RCR1:
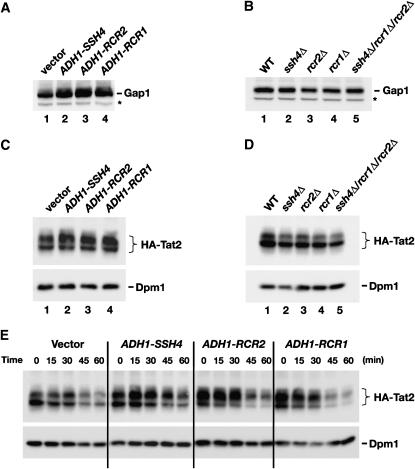
Our localization studies raised the possibility that the suppressors enhance AAP stability by affecting the routing of AAPs within the endosome–vacuole pathway. We examined this possiblity by monitoring the steady-state levels of Gap1 in strains overexpressing SSH4, RCR2, and RCR1. In contrast to our previous results regarding the overexpression of the suppressing genes in shr3Δ mutants, which had no affect on Gap1 levels (Figure 2B), the overexpression of SSH4, RCR2, or RCR1 in an SHR3 wild-type strain resulted in two to threefold higher levels of Gap1 (Figure 6A; compare lane 1 with lanes 2–4). Together, these results are consistent with the notion that the suppressor proteins function post-ER within the endosome–vacuole pathway and that increased suppressor protein levels enhance AAP stability. Gap1 levels were not significantly altered in null mutant strains (Figure 6B).
Figure 6.—
Analysis of Gap1 and Tat2 protein levels. (A) Gap1 levels in whole-cell extracts from a wild-type strain (CAY28) transformed with pRS202 (vector), pJK123 (PADH1-SSH4), pJK124 (PADH1-RCR2), or pJK125 (PADH1-RCR1). Cells were grown to an OD600 of 1.5 in SAD, and proteins in cell lysates were resolved by SDS–PAGE and immunoblotted with α-Gap1 antibodies. The asterisk indicates a nonspecific immunoreactive band unrelated to Gap1 that serves as a loading control. (B) Gap1 levels in whole-cell extracts from wild-type (CAY28), ssh4Δ (JKY40), rcr2Δ (JKY41), rcr1Δ (JKY42), and ssh4Δ rcr2Δ rcr1Δ (HFY538) strains. Cells were grown as in SAD supplemented with uracil, and extracts were prepared and analyzed as in A. (C) Steady-state levels of HA-TAT2 were analyzed in strain PLY860 cotransformed with pJK139 (HA-TAT2) and 2μ plasmid pRS202 (vector), pJK123 (PADH1-SSH4), pJK124 (PADH1-RCR2), or pJK125 (PADH1-RCR1) grown in SC (−ura,−trp) to an OD600 of 2. Cell extracts were resolved by SDS–PAGE and immunoblotted with α-HA (3F10, rat) and α-Dpm1 antibodies. (D) Immunoblotting of extracts from wild-type strain (CAY28), ssh4Δ (JKY40), rcr2Δ (JKY41), rcr1Δ (JKY42), and ssh4Δ rcr2Δ rcr1Δ (HFY538) transformed with plasmid pAS55 (HA-TAT2). Cells were grown SC (−ura) to an OD600 of 2 and proteins were resolved and analyzed as in C. (E) Strain PLY860 cotransformed with plasmids as in C was grown in SC (−ura −trp) to an OD600 of 2. Rapamycin was added and whole-cell extracts were prepared at the time points indicated. Proteins were resolved and analyzed as in C.
To test this notion further, we examined the levels of tryptophan and tyrosine permease Tat2. In contrast to Gap1, Tat2 is expressed and active on medium containing preferred nitrogen sources. We overexpressed SSH4, RCR2, or RCR1 together with a plasmid expressing a functional N-terminal HA-tagged Tat2 under control of its own promoter (Beck et al. 1999). HA-Tat2 migrated as two discrete bands (Figure 6, C–E), a finding consistent with previous observations (Abe and Iida 2003). No background staining occurs on immunoblots developed using the anti-HA 3F10 antibody (see Figure 4C, right, lane 1, for control). Strikingly, the levels of the slower-migrating form of Tat2 (Figure 6C, top band) exhibited clear SSH4, RCR2, and RCR1 dependence. Similar to Gap1 (Figure 6A), cells overexpressing SSH4, RCR2, or RCR1 have more Tat2 than wild-type cells (Figure 6C; compare levels of the slower-migrating form). In contrast, ssh4, rcr2, and rcr1 mutants, as well as the ssh4Δ rcr2Δ rcr1Δ triple-mutant strain, contain 25–50% less Tat2 protein than wild type (Figure 6D). The inverse effects on Tat2 levels—i.e., overexpression vs. deletion of SSH4, RCR2, and RCR1—suggest that the suppressor proteins normally participate in processes that constrain its degradation in the vacuole.
Overexpression of SSH4, RCR2, and RCR1 affects the pattern of rapamycin induced post-translational modifications of Tat2:
Tat2 is downregulated in response to starvation or in the presence of rapamycin (Beck et al. 1999; Loewith et al. 2002; Umebayashi and Nakano 2003). Upon rapamycin treatment, Tat2 is transported from the plasma membrane and from intracellular membranes via the endosomal pathway to the vacuole and degraded, a process that requires ubiquitylation (Beck et al. 1999; Umebayashi and Nakano 2003). We examined rapamycin-induced downregulation of Tat2 in wild-type cells (vector) and in cells overexpressing SSH4, RCR2, or RCR1 (Figure 6E). In all strains, only a fraction of Tat2 remained 60 min after rapamycin addition. However, we noted that rapamycin affected the migration pattern of Tat2 in a manner consistent with rapamycin-induced post-translational modification. At time zero, the bulk of Tat2 in wild-type cells was present in the faster-migrating form (Figure 6E, bottom band), whereas 30 min after the addition of rapamycin, there was a notable shift, and more Tat2 was present in the slower-migrating form (Figure 6E, top band). As expected from the analysis of steady-state levels (Figure 6C), in strains overexpressing SSH4, RCR2, and RCR1 the majority of Tat2 was present in its slower-migrating form (Figure 6C, top band). After the addition of rapamycin, the levels of the faster-migrating form of Tat2 (Figure 6E, bottom band) decreased more rapidily than the slower-migrating form (Figure 6E; compare levels at the 30-min time point). These results suggest that the suppressor proteins either enhance starvation-induced modifications that normally initiate downregulation of Tat2 or affect subsequent events late in the endosomal pathway important for Tat2 degradation.
Overexpression of SSH4, RCR2, or RCR1 enhances the activity of multiple plasma membrane transport systems:
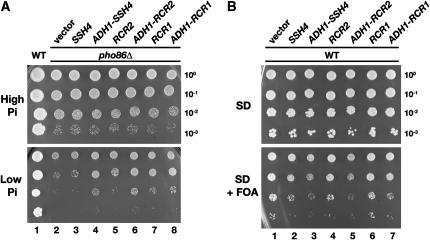
To investigate whether the enhanced stability of AAPs in cells overexpressing SSH4, RCR2, and RCR1 is specific for AAPs, we introduced high-copy versions of SSH4, RCR2, and RCR1 into a pho86 null mutant strain. PHO86 encodes an “Shr3-like” ER-membrane-localized chaperone that facilitates folding of the phosphate transporter Pho84 (Lau et al. 2000; Kota and Ljungdahl 2005). In pho86 mutants, Pho84 aggregates and accumulates in the ER. Consequently, pho86 mutants grow poorly under low-phosphate conditions (Figure 7A, dilution series 2). The overexpression of SSH4, RCR2, or RCR1 enabled pho86 mutants to grow on low-phosphate media (Figure 7A; compare dilution series 2 with 4, 6, and 8).
Figure 7.—
Overexpression of SSH4, RCR2, and RCR1 affects multiple plasma membrane transport systems. (A) Suppression of pho86Δ. Serial dilutions of PHO86 (CEN.PK113-5D/PHO84myc) and pho86Δ strains (JKY6) carrying pRS202 (vector) or plasmids expressing SSH4 (pJK123), RCR2 (pJK124), or RCR1 (pJK125) from the PADH1 promoter, as indicated, were spotted onto high-phosphate (YPD) and low-phosphate (SD 0.2 mm phosphate) media. Plates were incubated at 30° for 3 days and photographed. (B) Growth characteristics on FOA-containing media. Wild-type strain CAY28 was transformed with 2μ plasmids pRS202 (vector), pJK120 (SSH4), pJK123 (ADH1-SSH4), pJK121 (RCR2), pJK124 (ADH1-RCR2), pJK122 (RCR1), or pJK125 (ADH1-RCR1) and 10-fold dilution series were spotted onto SD and SD+FOA (0.1 mg/ml). Plates were incubated at 30° for 3 days and photographed.
Leflunomide is an immunosuppressant that inhibits growth in yeast by interfering with Fur4-dependent uracil uptake (Fujimura 1998a). We examined whether overexpression of SSH4, RCR2, or RCR1 increased uracil uptake by growing strains carrying plasmids with these genes on media supplemented with 5-FOA. FOA is primarily imported into cells by Fur4 (Wittke et al. 1999) and converted intracellularly to the toxic compound 5-fluorouracil. Cells overexpressing SSH4, RCR2, or RCR1 grew notably more poorly on SD-containing FOA compared to cells transformed with the vector control (Figure 7B; compare dilution series 1 with 3, 5, and 6), whereas growth on SD (without FOA; Figure 7B, top) was unaffected. These results are consistent with the idea that overexpression of SSH4 and also of RCR2 and RCR1 leads to increased FOA import, presumably through increased Fur4 activity.
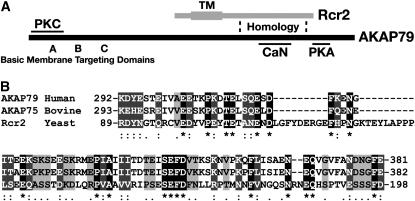
The C terminus of Rcr2 shares sequence similarity with the synaptic scaffold protein AKAP79:
We sought to gain insights into the mechanistic role of Ssh4, Rcr2, and Rcr1 by searching for orthologs in other organisms. A sequence motif within the cytosolic C-terminal portion of Rcr2 (aa 89–198) shares weak but significant homology with the human A-kinase-anchoring protein AKAP79 (Figure 8). The homologous region in AKAP79 contains multiple calcineurin (CaN) inhibitory determinants (aa 315–360; Dell'Acqua et al. 2002). This region of AKAP79 binds and inhibits the phosphatase activity of CaN. The neuronal AKAPs are highly conserved between species. Figure 8B shows a multiple alignment of the homologous regions of Rcr2, human AKAP79 (Carr et al. 1992), and its bovine ortholog AKAP75 (Hirsch et al. 1992). The region of homology is not well conserved in Rcr1, which may explain the nonredundant growth affects (RCR2 vs. RCR1) that we (Figures 3 and 7) and Imai et al. (2005) have observed.
Figure 8.—
The C-terminal region of Rcr2 shows homology to the human scaffold AKAP79. (A) Schematic of the structural features of yeast Rcr2 and human AKAP79. The predicted transmembrane spanning domain (TM) of Rcr2: basic membrane-targeting domains (A–C), PKC, CaN, and cAMP-dependent kinase (PKA) binding regions of AKAP79 are indicated. Sequence homology of Rcr2 and AKAP79 was found in the region indicated between the dashed lines. (B) Multiple alignment of human AKAP79 (aa 292–381), bovine AKAP75 (aa 293–382), and yeast Rcr2 (aa 89–198) sequences (CLUSTAL W, Biology WorkBench 3.2, San Diego Supercomputer Center). Amino acids that are identical in all three proteins (white lettering, solid boxes), as well as amino acids with high similarity (white lettering, darkly shaded boxes) and low similarity (black lettering, lightly shaded boxes) are indicated.
Mammalian AKAPs function as dynamic scaffold molecules that bring phosphatases and kinases close to their targets to control various cellular events, including the stability of plasma membrane proteins (reviewed in Wong and Scott 2004; Dell'Acqua et al. 2006; Smith et al. 2006). A schematic of the structural features of AKAP79 is in Figure 8A. In addition to the CaN inhibitory motif already mentioned, AKAP79 contains basic membrane-targeting domains in the N terminus (Dell'Acqua et al. 1998), a regulatory protein kinase C (PKC)-binding site (aa 31–52; Klauck et al. 1996) and a C-terminally located cAMP-dependent kinase (PKA)-binding domain (aa 391–408; Carr et al. 1992). AKAP79 targets to receptors/channels by binding to associated protein complexes containing MAGUKs, PSD-95, and SAP97 (Colledge et al. 2000).
DISCUSSION
Opposing membrane trafficking pathways determine the residence time and consequently the levels of plasma-membrane-localized nutrient transport systems. Anterograde targeting to the plasma membrane occurs via the secretory pathway, which is responsible for bringing newly synthesized membrane proteins to the plasma membrane. The entry point of the secretory pathway is the endoplasmic reticulum, the compartment where Shr3 is localized. Retrograde transport via the endosomal pathway provides the means to extract and downregulate proteins from the plasma membrane. An important feature of the endosomal pathway is that endocytosed membrane proteins can recycle back to the plasma membrane or, alternatively, enter the MVB pathway that targets them to the vacuole for degradation. Understanding the mechanisms that influence the routing of proteins within the endosomal pathway remains an important and fundamental problem in biology.
In this report we identified SSH4 and RCR2 on the basis of their ability to increase amino acid uptake into an shr3 null mutant strain when overexpressed. The overexpression of the RCR2 homolog RCR1 was also shown to suppress shr3 null mutations. We have experimentally addressed the function of the suppressor gene products, and our evidence supports a model in which Ssh4, Rcr2, and Rcr1 are components of the endosome–vacuole trafficking pathway. Within this pathway the suppressing proteins appear to pleiotropically influence functional expression of several nutrient transport systems. This model is based on several lines of evidence. First, Ssh4 and Rcr2 localize to structures associated with vacuoles, we also observed that Rcr2 localized to larger vesicles around the vacuole, which are probably endosome-like structures. Second, the steady-state levels of two AAPs, Gap1 and Tat2, were higher in cells overexpressing SSH4, RCR2, and RCR1, a finding that accounts for their ability to increase amino acid uptake in shr3 mutant cells. Finally, the suppressing effects were not limited to amino acid permeases; the overexpression of SSH4, RCR2, and RCR1 increased Pho84- and Fur4-dependent phosphate and uracil (FOA) uptake, respectively.
It was critical to address the possibility that these suppressors suppress shr3 mutations by facilitating AAP folding in the ER. However, our results clearly demonstrate that the Ssh4, Rcr2, and Rcr1 exert their effects after AAPs exit the ER. Overexpression of the suppressor genes did not influence shr3Δ-dependent AAP aggregation; thus, these suppressors do not possess membrane-localized chaperone-like activity. Furthermore, in contrast to what was observed in SHR3 wild-type cells, the overexpression of the suppressing genes did not visibly affect the steady-state levels of AAPs in shr3 null mutants (compare Figure 2B, bottom, and Figure 6A). These latter findings indicate that the suppressors exert their effects after AAPs exit the ER.
Our results suggest that similarly to Rcr2, Rcr1 functions post-ER. Recently, Imai et al. (2005) reported that Rcr1 is localized to the ER. These studies relied on observations of cells overexpressing an epitope-tagged construct of Rcr1. Unfortunately, we failed to obtain a functional epitope-tagged allele of RCR1 and thus were unable to confirm this finding. The overexpression of RCR1 results in decreased levels of chitin on the cell surface and thereby confers resistance to the chitin-binding toxin Congo Red (Imai et al. 2005). The relationship between reduced surface content of chitin and increased amounts of active transporters at the plasma membrane is not clear. Alterations in the composition of plasma membrane constituents could be a secondary or compensatory effect (as discussed in the introduction of Imai et al. 2005). Alternatively, RCR1 overexpression could negatively regulate chitin deposition in an inverse manner to the nutrient transporters that we have studied, perhaps by interacting with chitosomes (Bartnicki-Garcia 2006). There are numerous other possibilities to account for the apparent discrepancies between our work and published work regarding Rcr1. Additional experimental work will be required to clarify Rcr1 function.
We have previously isolated spontaneous mutations that suppress the amino-acid-uptake deficiency of cells lacking a functional SPS sensor (Forsberg et al. 2001). Mutations in ASI (Amino acid Sensor Independent) genes were divided into two classes: class I mutations restore transcription of SPS sensor target genes (i.e., ASI1, ASI2, and ASI3/STP1), and class II mutations prevent the downregulation of AAPs via the MVB pathway (i.e., DOA4, BRO1, RSP5, BUL1, VPS20, and VPS36). On the basis of our understanding of the two classes of ASI genes, we examined whether the high-copy suppressors positively affected SPS sensor gene expression or exerted functions indicative of their involvement in the MVB pathway. Our efforts clearly show that SSH4, RCR2, and RCR1 do not affect SPS sensor signaling (Figure 3), and thus these suppressors are not high-copy class I ASI genes. Rather, our results indicate that the high-copy suppressors functionally classify together with class II ASI genes.
The pleiotropic affects observed when the SSH4, RCR2, and RCR1 are overexpressed suggest that they encode components of the endosome–vacuole pathway that participate in common sorting processes that determine the fate of multiple plasma membrane proteins. Thus, the overexpression of Ssh4, Rcr2, and Rcr1 appears to exert effects similar to mutations in ESCRT components that impair the formation of MVBs, i.e., as class II asi mutations. In such mutants, Gap1 is not targeted to the vacuole, but is instead efficiently recycled to the plasma membrane, possibly via trans-Golgi compartments, resulting in increased Gap1 activity (Nikko et al. 2003; Lauwers and André 2006; Rubio-Texeira and Kaiser 2006). Alternatively, Ssh4, Rcr2, and Rcr1 might have a more restricted role, similar to that of the GSE proteins (Gao and Kaiser 2006) that prevent specific types of transporters from entering the MVB pathway.
In the course of examining the effect of overexpressing SSH4, RCR2, and RCR1 on Tat2 levels, we observed that Tat2 migrated as two bands when analyzed by SDS–PAGE. Remarkably, the overexpression of SSH4, RCR2, and RCR1 primarily stabilized levels of the slower-migrating form of Tat2 (Figure 6, C and E, top band). Tat2 is ubiquitylated (Beck et al. 1999; Umebayashi and Nakano 2003) and perhaps also phosphorylated (Schmidt et al. 1998), modifications that are known to affect endocytosis of transporter proteins. Additionally, accumulating evidence indicates that the phosphorylation state of transporters, in addition to Rsp5-dependent ubiquitylation, is critically important for sorting from endosomal compartments to the vacuole (Soetens et al. 2001; Marchal et al. 2002; Blondel et al. 2004; Gadura and Michels 2006). The slower-migrating species of Tat2 might reflect a modified form of the protein that normally would be routed to the vacuole, but fails to be degraded when Ssh4, Rcr2, or Rcr1 are overexpressed.
We found homology between a C-terminal portion of Rcr2 and the neuronal scaffolding protein AKAP79. AKAP79 provides specificity to general enzymes and is thought to regulate the turnover of various surface receptors by combining different sets of enzymes and recruiting them to the correct cellular site (Hoshi et al. 2005; Dell'Acqua et al. 2006). The number of receptors at the synaptic membrane determines how sensitive neurons are to incoming stimuli, and the stability of long-lasting synaptic changes are important for learning. Without the scaffold activity of AKAP79, long-term synaptic changes are not stably maintained, and preventing PKA binding to the AKAP79 ortholog AKAP150 leads to memory dysfunction in rats (Sheng and Lee 2001; Moita et al. 2002).
Scaffolds in protein trafficking have also been described in yeast. Downregulation of the Smf1 manganese transporter, for example, depends on the Bsd2/Tre1/Tre2 scaffolding system. Smf1 is ubiquitylated, internalized, and targeted to the vacuole in response to high intracellular levels of manganese (Liu and Culotta 1999; Eguez et al. 2004). Bsd2, Tre1, and Tre2 possess Rsp5-binding sequences (PPxY), and these scaffolding proteins are thought to bring the Rsp5 ubiquitin ligase and Smf1 together to facilitate sorting of Smf1 to the MVB pathway. Bsd2 also seems to have a more general function in the recognition and vacuolar targeting of misfolded membrane proteins (Liu et al. 1997; Hettema et al. 2004; Stimpson et al. 2006). One possibility is that Rcr2, and perhaps Ssh4 and Rcr1, function in analogy to AKAP79/Bsd2 as scaffolds for assembling components in trafficking of nutrient transporters.
According to our current understanding of scaffold proteins within the endosomal pathway, they appear to ensure the fidelity and selectivity of sorting processes that can proceed even without their assistance. However, in their absence, sorting occurs inefficiently and in an uncontrolled manner. We anticipate that scaffolding proteins are of special importance when cells are challenged by changes in environmental growth conditions. To avoid an overreaction, cells must differentiate between real long-term and transient short-term change. Thus, scaffolding proteins may exert a dampening effect to ensure an accurate response. The identification of binding partners will likely provide important clues to further understanding of how Ssh4, Rcr2, and Rcr1 regulate nutrient transport.
Acknowledgments
Members of the Ljungdahl laboratory and colleagues are acknowledged for constructive comments on the manuscript. We are indebted to Bruno André for his generous gift of α-Gap1 antisera. We thank Michael Hall for plasmid pAS55 expressing HA-Tat2 and Bengt Persson for the strain CEN.PK113-5D/PHO84myc. This work was supported by the Ludwig Institute for Cancer Research and by a grant from the Swedish Research Council (P.O.L.).
References
- Abe, F., and H. Iida, 2003. Pressure-induced differential regulation of the two tryptophan permeases Tat1 and Tat2 by ubiquitin ligase Rsp5 and its binding proteins, Bul1 and Bul2. Mol. Cell. Biol. 23: 7566–7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson, C., and P. O. Ljungdahl, 2002. Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 16: 3158–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andréasson, C., and P. O. Ljungdahl, 2004. The N-terminal regulatory domain of Stp1p is modular and, fused to an artificial transcription factor, confers full Ssy1p-Ptr3p-Ssy5p sensor control. Mol. Cell. Biol. 24: 7503–7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartnicki-Garcia, S., 2006. Chitosomes: past, present and future. FEMS Yeast Res. 6: 957–965. [DOI] [PubMed] [Google Scholar]
- Beck, T., A. Schmidt and M. N. Hall, 1999. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J. Cell Biol. 146: 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel, M. O., J. Morvan, S. Dupre, D. Urban-Grimal, R. Haguenauer-Tsapis et al., 2004. Direct sorting of the yeast uracil permease to the endosomal system is controlled by uracil binding and Rsp5p-dependent ubiquitylation. Mol. Biol. Cell 15: 883–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boban, M., A. Zargari, C. Andréasson, S. Heessen, J. Thyberg et al., 2006. Asi1 is an inner nuclear membrane protein that restricts promoter access of two latent transcription factors. J. Cell Biol. 173: 695–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugnicourt, A., M. Froissard, K. Sereti, H. D. Ulrich, R. Haguenauer-Tsapis et al., 2004. Antagonistic roles of ESCRT and Vps class C/HOPS complexes in the recycling of yeast membrane proteins. Mol. Biol. Cell 15: 4203–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, D., D. Dawson and T. Stearns, 2000. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Sping Harbor, NY.
- Byrne, K. P., and K. H. Wolfe, 2005. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 15: 1456–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr, D. W., R. E. Stofko-Hahn, I. D. Fraser, R. D. Cone and J. D. Scott, 1992. Localization of the cAMP-dependent protein kinase to the postsynaptic densities by A-kinase anchoring proteins. Characterization of AKAP 79. J. Biol. Chem. 267: 16816–16823. [PubMed] [Google Scholar]
- Colledge, M., R. A. Dean, G. K. Scott, L. K. Langeberg, R. L. Huganir et al., 2000. Targeting of PKA to glutamate receptors through a MAGUK-AKAP complex. Neuron 27: 107–119. [DOI] [PubMed] [Google Scholar]
- Connelly, C., and P. Hieter, 1996. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 86: 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer, M., P. S. Nielsen, J. P. Bebelman, H. Heerikhuizen, H. A. Andersen et al., 2000. Stp1p, Stp2p and Abf1p are involved in regulation of expression of the amino acid transporter gene BAP3 of Saccharomyces cerevisiae. Nucleic Acids Res. 28: 974–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Acqua, M. L., M. C. Faux, J. Thorburn, A. Thorburn and J. D. Scott, 1998. Membrane-targeting sequences on AKAP79 bind phosphatidylinositol-4, 5-bisphosphate. EMBO J. 17: 2246–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Acqua, M. L., K. L. Dodge, S. J. Tavalin and J. D. Scott, 2002. Mapping the protein phosphatase-2B anchoring site on AKAP79. Binding and inhibition of phosphatase activity are mediated by residues 315–360. J. Biol. Chem. 277: 48796–48802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Acqua, M. L., K. E. Smith, J. A. Gorski, E. A. Horne, E. S. Gibson et al., 2006. Regulation of neuronal PKA signaling through AKAP targeting dynamics. Eur. J. Cell Biol. 85: 627–633. [DOI] [PubMed] [Google Scholar]
- Dupre, S., D. Urban-Grimal and R. Haguenauer-Tsapis, 2004. Ubiquitin and endocytic internalization in yeast and animal cells. Biochim. Biophys. Acta 1695: 89–111. [DOI] [PubMed] [Google Scholar]
- Eguez, L., Y. S. Chung, A. Kuchibhatla, M. Paidhungat and S. Garrett, 2004. Yeast Mn2+ transporter, Smf1p, is regulated by ubiquitin-dependent vacuolar protein sorting. Genetics 167: 107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falco, S. C., and K. S. Dumas, 1985. Genetic analysis of mutants of Saccharomyces cerevisiae resistant to the herbicide sulfometuron methyl. Genetics 109: 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Y., and N. G. Davis, 2000. Akr1p and the type I casein kinases act prior to the ubiquitination step of yeast endocytosis: Akr1p is required for kinase localization to the plasma membrane. Mol. Cell. Biol. 20: 5350–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg, H., and P. O. Ljungdahl, 2001. Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr. Genet. 40: 91–109. [DOI] [PubMed] [Google Scholar]
- Forsberg, H., M. Hammar, C. Andréasson, A. Moliner and P. O. Ljungdahl, 2001. Suppressors of ssy1 and ptr3 null mutations define novel amino acid sensor-independent genes in Saccharomyces cerevisiae. Genetics 158: 973–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura, H., 1998. a Growth inhibition of Saccharomyces cerevisiae by the immunosuppressant leflunomide is due to the inhibition of uracil uptake via Fur4p. Mol. Gen. Genet. 260: 102–107. [DOI] [PubMed] [Google Scholar]
- Fujimura, H., 1998. b Molecular cloning of Saccharomyces cerevisiae MLF4/SSH4 gene which confers the immunosuppressant leflunomide resistance. Biochem. Biophys. Res. Commun. 246: 378–381. [DOI] [PubMed] [Google Scholar]
- Gadura, N., and C. A. Michels, 2006. Sequences in the N-terminal cytoplasmic domain of Saccharomyces cerevisiae maltose permease are required for vacuolar degradation but not glucose-induced internalization. Curr. Genet. 50: 101–114. [DOI] [PubMed] [Google Scholar]
- Gao, M., and C. A. Kaiser, 2006. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat. Cell Biol. 8: 657–667. [DOI] [PubMed] [Google Scholar]
- Gilstring, C. F., and P. O. Ljungdahl, 2000. A method for determining the in vivo topology of yeast polytopic membrane proteins demonstrates that Gap1p fully integrates into the membrane independently of Shr3p. J. Biol. Chem. 275: 31488–31495. [DOI] [PubMed] [Google Scholar]
- Gilstring, C. F., M. Melin-Larsson and P. O. Ljungdahl, 1999. Shr3p mediates specific COPII coatomer-cargo interactions required for the packaging of amino acid permeases into ER-derived transport vesicles. Mol. Biol. Cell 10: 3549–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyuris, J., E. Golemis, H. Chertkov and R. Brent, 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75: 791–803. [DOI] [PubMed] [Google Scholar]
- Haas, A., D. Scheglmann, T. Lazar, D. Gallwitz and W. Wickner, 1995. The GTPase Ypt7p of Saccharomyces cerevisiae is required on both partner vacuoles for the homotypic fusion step of vacuole inheritance. EMBO J. 14: 5258–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein, C., J. Y. Springael, C. Volland, R. Haguenauer-Tsapis and B. André, 1995. NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol. Microbiol. 18: 77–87. [DOI] [PubMed] [Google Scholar]
- Hettema, E. H., J. Valdez-Taubas and H. R. Pelham, 2004. Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J. 23: 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke, L., B. Zanolari and H. Riezman, 1998. Cytoplasmic tail phosphorylation of the alpha-factor receptor is required for its ubiquitination and internalization. J. Cell Biol. 141: 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch, A. H., S. B. Glantz, Y. Li, Y. You and C. S. Rubin, 1992. Cloning and expression of an intron-less gene for AKAP 75, an anchor protein for the regulatory subunit of cAMP-dependent protein kinase II beta. J. Biol. Chem. 267: 2131–2134. [PubMed] [Google Scholar]
- Hoshi, N., L. K. Langeberg and J. D. Scott, 2005. Distinct enzyme combinations in AKAP signalling complexes permit functional diversity. Nat. Cell Biol. 7: 1066–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley, J. H., and S. D. Emr, 2006. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35: 277–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai, K., Y. Noda, H. Adachi and K. Yoda, 2005. A novel endoplasmic reticulum membrane protein Rcr1 regulates chitin deposition in the cell wall of Saccharomyces cerevisiae. J. Biol. Chem. 280: 8275–8284. [DOI] [PubMed] [Google Scholar]
- Iraqui, I., S. Vissers, F. Bernard, J. O. de Craene, E. Boles et al., 1999. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 19: 989–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., T. Chiba and M. Yoshida, 2001. Exploring the protein interactome using comprehensive two-hybrid projects. Trends Biotechnol. 19: S23–S27. [DOI] [PubMed] [Google Scholar]
- Jauniaux, J. C., and M. Grenson, 1990. GAP1, the general amino acid permease gene of Saccharomyces cerevisiae. Nucleotide sequence, protein similarity with the other bakers yeast amino acid permeases, and nitrogen catabolite repression. Eur. J. Biochem. 190: 39–44. [DOI] [PubMed] [Google Scholar]
- Jørgensen, M. U., M. B. Bruun, T. Didion and M. C. Kielland-Brandt, 1998. Mutations in five loci affecting GAP1-independent uptake of neutral amino acids in yeast. Yeast 14: 103–114. [DOI] [PubMed] [Google Scholar]
- Katzmann, D. J., G. Odorizzi and S. D. Emr, 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3: 893–905. [DOI] [PubMed] [Google Scholar]
- Kelm, K. B., G. Huyer, J. C. Huang and S. Michaelis, 2004. The internalization of yeast Ste6p follows an ordered series of events involving phosphorylation, ubiquitination, recognition and endocytosis. Traffic 5: 165–180. [DOI] [PubMed] [Google Scholar]
- Klasson, H., G. R. Fink and P. O. Ljungdahl, 1999. Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol. Cell. Biol. 19: 5405–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauck, T. M., M. C. Faux, K. Labudda, L. K. Langeberg, S. Jaken et al., 1996. Coordination of three signaling enzymes by AKAP79, a mammalian scaffold protein. Science 271: 1589–1592. [DOI] [PubMed] [Google Scholar]
- Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor et al., 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15: 963–972. [DOI] [PubMed] [Google Scholar]
- Kota, J., and P. O. Ljungdahl, 2005. Specialized membrane-localized chaperones prevent aggregation of polytopic proteins in the ER. J. Cell Biol. 168: 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn, M. J., R. Schekman and P. O. Ljungdahl, 1996. Amino acid permeases require COPII components and the ER resident membrane protein Shr3p for packaging into transport vesicles in vitro. J. Cell Biol. 135: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte, J., and R. F. Doolittle, 1982. A simple method of displaying the hydropathic character of a protein. J. Mol. Biol. 157: 105–132. [DOI] [PubMed] [Google Scholar]
- Lagerstedt, J. O., R. Zvyagilskaya, J. R. Pratt, J. Pattison-Granberg, A. L. Kruckeberg et al., 2002. Mutagenic and functional analysis of the C-terminus of Saccharomyces cerevisiae Pho84 phosphate transporter. FEBS Lett. 526: 31–37. [DOI] [PubMed] [Google Scholar]
- Lau, W. T., R. W. Howson, P. Malkus, R. Schekman and E. K. O'Shea, 2000. Pho86p, an endoplasmic reticulum (ER) resident protein in Saccharomyces cerevisiae, is required for ER exit of the high-affinity phosphate transporter Pho84p. Proc. Natl. Acad. Sci. USA 97: 1107–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauwers, E., and B. André, 2006. Association of yeast transporters with detergent-resistant membranes correlates with their cell-surface location. Traffic 7: 1045–1059. [DOI] [PubMed] [Google Scholar]
- Liu, H., J. Krizek and A. Bretscher, 1992. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics 132: 665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. F., and V. C. Culotta, 1999. Post-translation control of Nramp metal transport in yeast. Role of metal ions and the BSD2 gene. J. Biol. Chem. 274: 4863–4868. [DOI] [PubMed] [Google Scholar]
- Liu, X. F., F. Supek, N. Nelson and V. C. Culotta, 1997. Negative control of heavy metal uptake by the Saccharomyces cerevisiae BSD2 gene. J. Biol. Chem. 272: 11763–11769. [DOI] [PubMed] [Google Scholar]
- Ljungdahl, P. O., C. J. Gimeno, C. A. Styles and G. R. Fink, 1992. SHR3: a novel component of the secretory pathway specifically required for localization of amino acid permeases in yeast. Cell 71: 463–478. [DOI] [PubMed] [Google Scholar]
- Loewith, R., E. Jacinto, S. Wullschleger, A. Lorberg, J. L. Crespo et al., 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10: 457–468. [DOI] [PubMed] [Google Scholar]
- Magasanik, B., and C. A. Kaiser, 2002. Nitrogen regulation in Saccharomyces cerevisiae. Gene 290: 1–18. [DOI] [PubMed] [Google Scholar]
- Marchal, C., R. Haguenauer-Tsapis and D. Urban-Grimal, 1998. A PEST-like sequence mediates phosphorylation and efficient ubiquitination of yeast uracil permease. Mol. Cell. Biol. 18: 314–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal, C., S. Dupre and D. Urban-Grimal, 2002. Casein kinase I controls a late step in the endocytic trafficking of yeast uracil permease. J. Cell Sci. 115: 217–226. [DOI] [PubMed] [Google Scholar]
- Moita, M. A., R. Lamprecht, K. Nader and J. E. LeDoux, 2002. A-kinase anchoring proteins in amygdala are involved in auditory fear memory. Nat. Neurosci. 5: 837–838. [DOI] [PubMed] [Google Scholar]
- Nielsen, P. S., B. van den Hazel, T. Didion, M. de Boer, M. Jørgensen et al., 2001. Transcriptional regulation of the Saccharomyces cerevisiae amino acid permease gene BAP2. Mol. Gen. Genet. 264: 613–622. [DOI] [PubMed] [Google Scholar]
- Nikko, E., A. M. Marini and B. André, 2003. Permease recycling and ubiquitination status reveal a particular role for Bro1 in the multivesicular body pathway. J. Biol. Chem. 278: 50732–50743. [DOI] [PubMed] [Google Scholar]
- O'Connell, K. F., and R. E. Baker, 1992. Possible cross-regulation of phosphate and sulfate metabolism in Saccharomyces cerevisiae. Genetics 132: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regenberg, B., L. During-Olsen, M. C. Kielland-Brandt and S. Holmberg, 1999. Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae. Curr. Genet. 36: 317–328. [DOI] [PubMed] [Google Scholar]
- Rubio-Texeira, M., and C. A. Kaiser, 2006. Amino acids regulate retrieval of the yeast general amino acid permease from the vacuolar targeting pathway. Mol. Biol. Cell 17: 3031–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytka, J., 1975. Positive selection of general amino acid permease mutants in Saccharomyces cerevisiae. J. Bacteriol. 121: 562–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmoller, F., and H. Riezman, 1993. Involvement of Ypt7p, a small GTPase, in traffic from late endosome to the vacuole in yeast. J. Cell Sci. 106(Pt. 3): 823–830. [DOI] [PubMed] [Google Scholar]
- Schmidt, A., T. Beck, A. Koller, J. Kunz and M. N. Hall, 1998. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 17: 6924–6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng, M., and S. H. Lee, 2001. AMPA receptor trafficking and the control of synaptic transmission. Cell 105: 825–828. [DOI] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silve, S., C. Volland, C. Garnier, R. Jund, M. R. Chevallier et al., 1991. Membrane insertion of uracil permease, a polytopic yeast plasma membrane protein. Mol. Cell. Biol. 11: 1114–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, F. D., L. K. Langeberg and J. D. Scott, 2006. The where's and when's of kinase anchoring. Trends Biochem. Sci. 31: 316–323. [DOI] [PubMed] [Google Scholar]
- Soetens, O., J. O. De Craene and B. André, 2001. Ubiquitin is required for sorting to the vacuole of the yeast general amino acid permease, Gap1. J. Biol. Chem. 276: 43949–43957. [DOI] [PubMed] [Google Scholar]
- Stimpson, H. E., M. J. Lewis and H. R. Pelham, 2006. Transferrin receptor-like proteins control the degradation of a yeast metal transporter. EMBO J. 25: 662–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umebayashi, K., and A. Nakano, 2003. Ergosterol is required for targeting of tryptophan permease to the yeast plasma membrane. J. Cell Biol. 161: 1117–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittke, S., N. Lewke, S. Muller and N. Johnsson, 1999. Probing the molecular environment of membrane proteins in vivo. Mol. Biol. Cell 10: 2519–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, W., and J. D. Scott, 2004. AKAP signalling complexes: focal points in space and time. Nat. Rev. Mol. Cell Biol. 5: 959–970. [DOI] [PubMed] [Google Scholar]