Abstract
The tiger pufferfish (fugu), Takifugu rubripes, is a model fish that has had its genome entirely sequenced. By performing genomewide linkage analyses, we show that the sex of fugu is determined by a single chromosomal region on linkage group 19 in an XX–XY system.
ALTHOUGH the existence of female and male sexes in vertebrates is a conserved phenomenon, there is considerable diversity in the mechanisms that control this choice, either by environmental or genetic factors. Even in the case of animals where sex is determined by the genome, the primary sex-determining gene(s) is not conserved in divergent species (Capel 2000). DMY/dmrt1bY and SRY have been identified as the primary sex-determining genes in the medaka (Oryzias latipes) and in most mammals, respectively (Matsuda et al. 2002). Apart from these cases, the molecular nature of the primary sex-determining gene(s) in the remainder of the vertebrates remains unknown (Schartl 2004).
Teleost fish are an attractive group of organisms with which to study the evolution and mechanisms of sex determination systems. This is because this group of organisms includes different types of sexuality—from hermaphroidism to gonochorism and from environmental to genetic sex determination (Schartl 2004). The tiger pufferfish (fugu), Takifugu rubripes, is a model species of fish that has had its genome entirely sequenced (Aparicio et al. 2002). Since extensive searches in the fugu genome sequence suggested the absence of the previously known sex-determining genes mammalian SRY and medaka DMY (Koopman et al. 2004; Schartl 2004), it follows that the fugu may provide an ideal resource to use in the search for a novel sex-determining gene. However, even the mode of sex determination is poorly understood in this fish and other closely related species. In this article, we characterize the genetic basis of sex determination in fugu.
To investigate whether the choice of developing testicular or ovarian tissues in the fugu is determined by genetic or environmental factors, we analyzed the progeny produced from two independent genetic crosses by taking advantage of a linkage map previously constructed by our laboratory (Kai et al. 2005). To accelerate the search for sex-linked markers, we chose several markers from 22 linkage groups (LGs) and genotyped 15 progeny of each sex from each family. We found that the marker f120 on LG19 cosegregated with phenotypic sex (Figure 1).
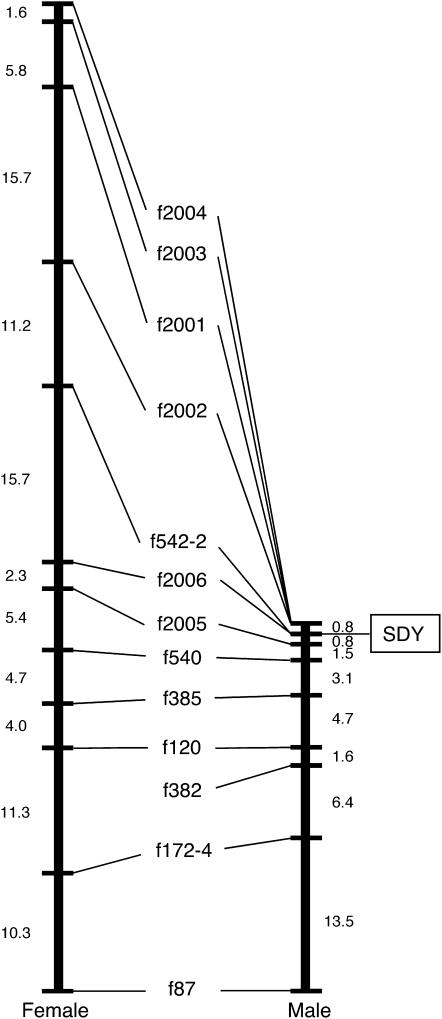
Figure 1.—
Male and female meiotic maps of fugu LG19. The markers f2006, f542-2, and SDY (the locus determining male sex phenotype) showed no recombination. Genetic maps were generated using 75 males and 55 females from family 1. Allelic bridges are indicated by a line connecting female (left) and male (right) linkage maps. Genetic distances in centimorgans between adjacent markers are shown. Genotypes of markers are shown in supplemental data 2 (http://www.genetics.org/supplemental/). To produce progeny from the first family (family 1), we crossed an individual fugu male with an individual fugu female as described previously (Kai et al. 2005). We then created a second family by interspecific crossing. Takifugu niphobles is a species closely related to fugu with a shorter generation time. In our study, an individual fugu male was mated with an individual female T. niphobles and then a single F1 hybrid male was crossed with a second female fugu. Sex was determined by histological examination of the gonads obtained from fish aged 8–10 months. The microsatellite loci were chosen by scanning scaffolds on electronic resources available from the Joint Genome Institute (http://genome.jgi-psf.org/Takru4/Takru4.home.html). Primer sequences for the markers used in this study are shown in supplemental data 1 at http://www.genetics.org/supplemental/ and deposited in DDBJ/EMBL/GenBank with accession nos. AB284949–AB284982. Genotyping with microsatellite markers was performed as described previously (Kai et al. 2005).
The fugu linkage map reported previously contained only five markers anchoring four scaffolds on LG19 (Kai et al. 2005). When analyzing model organisms in which almost the complete genome sequence is available, the candidate region for markers on a particular chromosome can be selected by referring to the genome sequence data. However, this approach currently remains limited for fugu because of the fragmented nature of the draft genome sequence (Aparicio et al. 2002). In contrast, the draft genome sequence of Tetraodon nigroviridis, another model fish, is less fragmented than that of fugu (Ensembl version 36.1d). We therefore assessed whether it might be practical to use the syntenic conservation between the genome of fugu and that of Tetroadon to selectively enrich markers covering the whole range of the putative sex chromosome for fugu. We first compared the whole catalog of genes from these two species and selected fugu genes orthologous to Tetraodon genes on chromosome 11. Of 614 fugu genes identified in this search, 531 genes were contained in 10 major fugu scaffolds (Table 1). We then obtained microsatellite loci from these scaffolds and examined their linkage relationships. These analyses enabled us to anchor 11 fugu scaffolds spanning 10.9 Mb along LG19 and, thus, to establish a genomic tool for analyzing the sex chromosome of fugu (Table 1).
TABLE 1.
Fugu scaffolds anchored to LG19
| Scaffold | Size (bp) | No. of orthologs to Tetraodon genes | Marker |
|---|---|---|---|
| 47 | 1,557,871 | 63 | f2001 |
| 190 | 570,726 | 36 | f2002 |
| 87 | 1,057,823 | 91 | f542-2 |
| 56 | 1,440,700 | 80 | f58 |
| 77 | 1,129,819 | 46 | f67 |
| 33 | 1,758,880 | 66 | f540 |
| 116 | 924,357 | 40 | f385 |
| 318 | 276,150 | Not identified | f120 |
| 169 | 663,033 | 18 | f382 |
| 79 | 1,116,978 | 61 | f172-4 |
| 150 | 723,838 | 30 | f87 |
Scaffolds are arranged along the gene order. The scaffolds mapped to the terminal end of LG19 do not contain telomeric repeats. To identify orthologous genes between fugu and Tetraodon, the whole catalog of predicted genes (FUGU 4.0, Ensembl version 36.4 and TETRAODON 7, Ensembl version 36.1d) were compared using Ensembl Multi MartView (http://www.ensembl.org/) and the best reciprocal matches were considered as being orthologous genes. Detailed data for the genes are shown in supplemental data 3 at http://www.genetics.org/supplemental/.
By using these genetic and physical maps, we were able to add new markers and successfully locate the sex-determining locus at the region where the markers f542-2 and f2006, on scaffolds 87 and 33, respectively, exhibit perfect segregation with phenotypic sex among the 130 progeny from family 1 and the 30 progeny from family 2 (Figure 1; supplemental data 2 at http://www.genetics.org/supplemental/). The sex-linked alleles were inherited from male parents in both families (Figure 1), suggesting that the fugu adopts an XX–XY sex-determining system. Comparison of male and female maps revealed that recombination between markers around the sex-determining locus was reduced during meiosis in males relative to meiosis in females (Figure 1). This could reflect either a general reduction of recombination in male fish (Kai et al. 2005) or region-specific reductions due to the sex-specific differences in DNA sequence close to the sex-determining locus (Peichel et al. 2004).
At present, the draft genome sequences of five species of fish have been fully sequenced or are in the process of being sequenced. Species involved are fugu, medaka, Tetraodon, stickleback (Gasterosteus aculeatus), and zebrafish (Danio rerio). Of these, the sex determination system and sex chromosome has been determined only in medaka and stickleback (Matsuda et al. 2002; Peichel et al. 2004). To compare the sex chromosome between these fish and fugu, we identified medaka and stickleback genes orthologous to the genes surrounding the sex-determining locus in fugu (genes on scaffolds 33, 56, 77, and 87) and ascertained their genomic positions via Ensemble databases. These analyses revealed that none of the genes tested mapped to the sex chromosome of either medaka or stickleback (Table 2). Therefore, it is likely that the sex chromosome in fugu has evolved independently from that of medaka or stickleback.
TABLE 2.
The number and genomic position of medaka and stickleback genes orthologous to genes located around the sex-determining locus of fugu (scaffolds 33, 56, 77, and 87)
| Species | No. of orthologs (genomic position) |
|---|---|
| Medaka | 227 (medaka LG5), 1 (medaka LG15), 46 (not identified) |
| Stickleback | 133 (stickleback LG17), 148 (not identified) |
The sex-determining loci in medaka and stickleback have been placed on medaka LG1 and stickleback LG19, respectively (Matsuda et al. 2002; Peichel et al. 2004). To identify ortholgous genes between fugu and medaka or between fugu and stickleback, the predicted genes (Ensembl version 42.1a for medaka and version 42.1b for stickleback) were compared using Ensembl Multi MartView. Detailed data from this analysis are shown in supplemental data 3 at http://www.genetics.org/supplemental/.
DMRT1 and SOX9 are regarded as conserved key players in the downstream cascade of the sex-determining/differentiation pathway in divergent species including mammals, reptiles, and fish (Capel 2000; Koopman 2005). Existing literature highlights the possibility that these genes may serve as the primary sex-determining genes in some nonmammalian vertebrates (Nanda et al. 1999; Capel 2000; Volff and Schartl 2002). By surveying the fugu genome database, previous studies have identified two sox9 genes (sox9a and sox9b) and one dmrt1 gene in fugu (Koopman et al. 2004; Yamaguchi et al. 2006). To determine whether any of these genes resides in the primary sex-determining locus, we mapped microsatellite loci physically linked to these genes on the meiotic panel. This linkage analysis revealed that none of these genes mapped to the fugu sex chromosome (Table 3). To identify candidates for the sex-determining gene in fugu, we searched the scaffolds around the sex-determining locus (scaffolds 33, 56, 77, and 87) for genes belonging to the sox or dmrt gene families or for genes related to the hormonal pathways involved in sex determination/differentiation. These searches successfully identified amhrII (NEWSINFRUG00000143070) and inhbb (NEWSINFRUG00000130534) on scaffold 33. Since these genes play key roles in the sex differentiation of mammals (Vigier et al. 1989; Brown et al. 2000), it is reasonable to consider that these genes may be strong candidates for the primary sex-determining gene in fugu. Given the freely available nature of the draft fugu genome sequence, a positional cloning strategy should make it possible to accurately identify the sex-determining gene of fugu. Successful identification of the fugu sex-determining gene should provide significant insight into our understanding of the evolution of the XX–XY system.
TABLE 3.
Mapping position of the fugu sox9a, sox9b, and dmrt1 genes
| Gene | Scaffold | LG | Marker |
|---|---|---|---|
| sox9a | 115, 4549 | 5 | f2007, f2008 |
| sox9b | 3 | 1 | f41-2 |
| dmrt1 | 4 | 21 | f378 |
To identify the scaffold that contained fugu sox9a, sox9b, and dmrt1, the coding sequences of these genes (Koopman et al. 2004; Yamaguchi et al. 2006) were BLAST searched against the fugu genome database. The genomic position of these genes was identified using the meiotic panels reported previously (Kai et al. 2005). Genotypes of markers are shown in supplemental data 2 at http://www.genetics.org/supplemental/.
Acknowledgments
We thank Byrappa Venkatesh for critical comments on the manuscript and Akiyuki Ozaki for technical advice. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Aparicio, S., J. Chapman, E. Stupka, N. Putnam, J. M. Chia et al., 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301–1310. [DOI] [PubMed] [Google Scholar]
- Brown, C. W., D. E. Houston-Hawkins, T. K. Woodruff and M. M. Matzuk, 2000. Insertion of Inhbb into the Inhba locus rescues the Inhba-null phenotype and reveals new activin functions. Nat. Genet. 25: 453–457. [DOI] [PubMed] [Google Scholar]
- Capel, B., 2000. The battle of the sexes. Mech. Dev. 92: 89–103. [DOI] [PubMed] [Google Scholar]
- Kai, W., K. Kikuchi, M. Fujita, H. Suetake, A. Fujiwara et al., 2005. A genetic linkage map for the tiger pufferfish, Takifugu rubripes. Genetics 171: 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman, P., 2005. Sex determination: a tale of two Sox genes. Trends Genet. 21: 367–370. [DOI] [PubMed] [Google Scholar]
- Koopman, P., G. Schepers, S. Brenner and B. Venkatesh, 2004. Origin and diversity of the SOX transcription factor gene family: genome-wide analysis in Fugu rubripes. Gene 328: 177–186. [DOI] [PubMed] [Google Scholar]
- Matsuda, M., Y. Nagahama, A. Shinomiya, T. Sato, C. Matsuda et al., 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417: 559–563. [DOI] [PubMed] [Google Scholar]
- Nanda, I., Z. Shan, M. Schartl, D. W. Burt, M. Koehler et al., 1999. 300 million years of conserved synteny between chicken Z and human chromosome 9. Nat. Genet. 21: 258–259. [DOI] [PubMed] [Google Scholar]
- Peichel, C. L., J. A. Ross, C. K. Matson, M. Dickson, J. Grimwood et al., 2004. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr. Biol. 14: 1416–1424. [DOI] [PubMed] [Google Scholar]
- Schartl, M., 2004. A comparative view on sex determination in medaka. Mech. Dev. 121: 639–645. [DOI] [PubMed] [Google Scholar]
- Vigier, B., M. G. Forest, B. Eychenne, J. Bezard, O. Garrigou et al., 1989. Anti-Mullerian hormone produces endocrine sex reversal of fetal ovaries. Proc. Natl. Acad. Sci. USA 86: 3684–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff, J. N., and M. Schartl, 2002. Sex determination and sex chromosome evolution in the medaka, Oryzias latipes, and the platyfish, Xiphophorus maculatus. Cytogenet. Genome Res. 99: 170–177. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, A., K. Y. Lee, H. Fujimoto, K. Kadomura, S. Yasumoto et al., 2006. Expression of the DMRT gene and its roles in early gonadal development of the Japanese pufferfish Takifugu rubripes. Comp. Biochem. Physiol. 1: 59–68. [DOI] [PubMed] [Google Scholar]