Abstract
The spindle assembly checkpoint (SAC) governs the timing of metaphase-to-anaphase transition and is essential for genome stability. The Caenorhabditis elegans mutant strain gk2 carries a deletion within the mdf-1/MAD1 gene that results in death of the homozygous strain after two or three generations. Here we describe 11 suppressors of the mdf-1(gk2) lethality, 10 identified in an ethyl methanesulfonate (EMS) mutagenesis screen and 1 isolated using the dog-1(gk10) (deletions of guanine-rich DNA) mutator strain. Using time-lapse imaging of early embryonic cells and germline mitotic division, we demonstrate that there are two classes of suppressors. Eight suppressors compensate for the loss of the checkpoint by delaying mitotic progression, which coincides with securin (IFY-1/Pds1) accumulation; three suppressors have normal IFY-1/Pds1 levels and normal anaphase onset. Furthermore, in the class of suppressors with delayed mitotic progression, we have identified four alleles of known suppressors emb-30/APC4 and fzy-1/CDC20, which are components of the anaphase-promoting complex/cyclosome (APC/C). In addition, we have identified another APC/C component capable of bypassing the checkpoint requirement that has not previously been described in C. elegans. The such-1/APC5-like mutation, h1960, significantly delays anaphase onset both in germline and in early embryonic cells.
FAITHFUL transmission of chromosomes to daughter cells is ensured by the spindle assembly checkpoint (SAC) (Hoyt et al. 1991; Li and Murray 1991). This surveillance system generates a “wait” signal, which delays onset of anaphase until all the chromosomes are properly attached to the microtubules at kinetochores. If the wait signal is absent, or if it is ignored, the transmission of exactly one copy of a chromosome to each daughter cell can be compromised. During mitosis, errors in chromosome segregation can result in aneuploidy and chromosomal instability (CIN), which has been recognized as a hallmark of cancers since Boveri (1914; reviewed in Bharadwaj and Yu 2004; Kops et al. 2005). During meiosis, missegregation of chromosomes can result in premature abortion of the fetus or generation of offspring with birth defects (reviewed in Hassold and Hunt 2001).
The interplay between the kinetochores and the spindle is monitored by the SAC components Mad1, Mad2, Mad3, Bub1, Bub3, and Mps1 (reviewed in Cleveland et al. 2003). These components were first identified in Saccharomyces cerevisiae by isolating mutants that failed to arrest in mitosis in the presence of microtubule-depolymerizing drugs (Hoyt et al. 1991; Li and Murray 1991; Weiss and Winey 1996). Although it is still unclear how defective kinetochore–microtubule attachment is detected and how the inhibition of the anaphase-promoting complex/cyclosome (APC/C) by the checkpoint is precisely achieved, it is known that the SAC acts by binding to Cdc20 and inhibiting the activation of the APC/C (Sudakin et al. 2001; reviewed in Karess 2005). Cdc20 is an activator of the APC/C, a large multisubunit E3 ubiquitin ligase that labels an anaphase inhibitor securin protein Pds1 with polyubiquitin chains (reviewed in Page and Hieter 1999). Degradation of the ubiquitinated securin releases the separase protein Esp1. Proteolytic cleavage of the cohesion subunit Scc1 by separase destroys the cohesion between the sister chromatids and triggers the onset of anaphase in mitotic cells (Ciosk et al. 1998).
In Caenorhabditis elegans, the SAC components mdf-1/MAD1, mdf-2/MAD2, san-1/MAD3, bub-1/BUB1, and Y54G9A.6/BUB3 have been identified (Kitagawa and Rose 1999; Oegema et al. 2001; Nystul et al. 2003). MDF-1/Mad1, MDF-2/Mad2, and BUB-1/Bub1 are required in both yeast and C. elegans to promote mitotic delays in the presence of either chemical or mutational disruptions of the microtubule cytoskeleton (Kitagawa and Rose 1999; Encalada et al. 2005). During anoxia-induced suspended animation, embryos lacking functional SAN-1/Mad3 or MDF-2/Mad2 fail to arrest the cell cycle (Nystul et al. 2003). BUB-1/Bub1 was also shown to localize to kinetochores and to have an essential role in kinetochore function as well as a potential role in regulating chromatin cohesion (Oegema et al. 2001; Desai et al. 2003; Monen et al. 2005). In contrast to the yeast MAD and BUB genes, which are essential only in the presence of either chemical or mutational disruptions of the microtubule cytoskeleton (Hoyt et al. 1991; Li and Murray 1991; Wang and Burke 1995), in C. elegans, mdf-1/MAD1 and mdf-2/MAD2 are essential for long-term survival and fertility (Kitagawa and Rose 1999). Loss of function of either of these genes leads to the accumulation of defects including chromosomal abnormalities, X chromosome loss or nondisjunction, defective gonad development, and embryonic lethality (Kitagawa and Rose 1999).
The C. elegans APC/C components emb-27/CDC16, emb-30/APC4, mat-1/CDC27, mat-2/APC1, and mat-3/CDC23 were isolated in forward genetic screens for temperature-sensitive (ts) embryogenesis-defective mutants (abnormal embryogenesis, emb) (Cassada et al. 1981) and ts maternal-effect embryonic lethal (Mel) mutants, which arrest as fertilized one-cell embryos blocked at metaphase of the first meiotic division as metaphase-to-anaphase transition-defective (mat) mutants (Golden et al. 2000; Shakes et al. 2003). The other known C. elegans APC/C subunits, K06H7.6/APC2, M163.4/APC5, F15H10.3/APC-10, and F35G12.9/APC11, were identified through sequence homology (Davis et al. 2002). Furuta et al. (2000) showed that the lethality of the mdf-1 deletion, gk2, can be suppressed by a ts mutation in emb-30(tn377ts)/APC4. Analysis of the vulval cell lineages and mitotic index suggested that mitosis was lengthened in emb-30/APC4 mutants and led to the proposal that delayed anaphase onset can bypass the SAC requirement (Furuta et al. 2000). Our group explored this finding by conducting a screen for suppressors of the mdf-1(gk2) lethal phenotype in search of additional components that function in the metaphase-to-anaphase transition (Kitagawa et al. 2002). Ten suppressors were recovered from the screen and one of them, h1983, was shown to encode the homolog of Cdc20, FZY-1 (Kitagawa et al. 2002). The fzy-1(h1983) hemizygotes had a prolonged metaphase phenotype, supporting the hypothesis that delayed anaphase can rescue the mdf-1(gk2) lethality.
We were interested in determining if all of the mutants that bypass the MDF-1 checkpoint requirement alter the timing of cell cycle progression. We found that the suppressors of mdf-1(gk2) fall into two classes. The major class of suppressors delays mitotic progression in the early C. elegans embryo and germ cells. This class includes two known suppressors and APC/C components, emb-30/APC4 and fzy-1/CDC20, and four new such (suppressors of spindle checkpoint defect) genes. We cloned one of the new such genes that delays mitotic progression and found that it corresponds to an APC5-like gene not previously identified as a component of the APC/C in C. elegans. We also identified a class of suppressors that does not delay mitotic progression, suggesting an alternate mechanism of bypassing the MDF-1 checkpoint requirement.
MATERIALS AND METHODS
C. elegans strains, alleles, and culturing:
The Bristol strain N2 was used as the standard wild-type strain (Brenner 1974). The marker mutations and balancer chromosomes used are listed in the order of chromosomes: LGI, dpy-5(e61), dpy-5(s1300); LGII, dpy-10(e128), unc-4(e120); LGIII, dpy-17(e164), dpy-18(e364), and unc-32(e289); LGIV, dpy-13(e184); and LGV, unc-46(e177), mdf-1(gk2), and nT1(IV;V). The strains used include VC13 dog-1(gk10), KR3627 unc-46(e177) mdf-1(gk2) +/+ + nT1 [let-X], and DG627 emb-30(tn377ts). The CB4856 strain was used for single-nucleotide polymorphism (SNP) mapping as described by Wicks et al. (2001). The AZ212: unc-119(ed3) ruIs32[unc-119(+) pie-1∷GFP∷H2B] III strain was used to visualize the chromosomes in the suppressor mutants. The strains were obtained from the Caenorhabditis Genetics Center. Animals were maintained using standard procedures (Brenner 1974).
Genetic screen for suppressors of the mdf-1(gk2) lethality:
Suppressors of the mdf-1(gk2) lethality were isolated in two separate genetic screens. All the suppressors except for such-4(h2168) were isolated in the ethyl methanesulfonate (EMS) screen as described by Kitagawa et al. (2002). The such-4(h2168) suppressor was isolated in a smaller screen using the dog-1(gk10) mutator strain (Cheung et al. 2002). We constructed a strain of genotype unc-46(e177) mdf-1(gk2) +/+ + nT1[let-X]; dog-1(gk10)/dog-1(gk10). The F1 unc-46 mdf-1 homozygotes (n = 40) were picked and plated individually. After 4 weeks a single plate containing fertile worms was isolated as a suppressor candidate. The suppressor was outcrossed from the dog-1(gk10) background to avoid further accumulation of mutations and the strain of genotype unc-46(e177) mdf-1(gk2) such-4(h2168)/unc-46(e177) mdf-1(gk2) such-4(h2168) was generated. These worms were crossed to N2 males and resegregated several times to eliminate secondary mutations generated in the dog-1(gk10) background.
Genetic analysis:
For analysis of the KR3627 strain (unc-46(e177) mdf-1(gk2) +/+ + nT1 [let-X]), we individually plated five L4 wild-type looking worms at 20° and 25°. The worms were transferred to fresh plates every 12 hr and the plates were scored. The eggs that did not hatch in 24 hr were scored as embryonic arrest. The eggs that hatched but did not reach adult stage were scored as larval arrest. The progeny that developed to adult stage were scored for the ratio of wild-type to unc-46 mdf-1 worms. The observed over expected percentage at 20° was 97% and at 25° was 107%, which indicated that F1 mdf-1(gk2) homozygotes segregated from heterozygous parents developed normally at both temperatures. Similarly, the F2 generation was examined by plating 14 unc-46 mdf-1 worms at the L4 stage and analyzing the progeny. The percentage of fertility was determined by individually plating all progeny that developed to adult stage. The suppressors were characterized in the same manner.
We mapped the such mutations to a chromosome using phenotypic Dpy (Dumpy) markers located in the central regions of the different autosomes and using the CB4856 strain for the SNP mapping procedure as described by Wicks et al. (2001). We sequenced the candidate genes located in the mapped regions. The candidate genes, including 300 bp upstream and downstream, were PCR amplified from genomic DNA isolated from the suppressor strains, purified, and submitted for sequencing (Nucleic Acid and Protein Services; University of British Columbia, UBC). In the case of the mat-2 candidate gene, the full-length cDNA from the h1989 and h1987 suppressors was amplified using reverse transcription–PCR (RT–PCR) and sequenced (Nucleic Acid and Protein Services, UBC).
Immunofluorescence:
For antibody staining we used the protocol available at http://www.genetics.wustl.edu/tslab/protocols.html. Dissected gonads were fixed with 3% formaldehyde, 0.1 m K2HPO4 (pH 7.2), for 1 hr and postfixed with cold 100% methanol for 5 min. After rehydration with 1× PBS containing 0.1% Tween-20 (1× PBS–T) the samples were incubated at 4° overnight with anti-phosphohistone H3 antibody (Upstate Biotechnology, Lake Placid, NY) at 1:200 dilution to visualize mitotic chromosomes. The samples were then incubated with Oregon-green-conjugated anti-rabbit IgG antibody (Molecular Probes, Eugene, OR) at dilution 1:1000. DAPI (1 μg/ml) was added to the secondary antibody solution. The stained gonads were viewed with the Zeiss Axioscope fluorescent microscope with 40× objective. A Retiga 2000R camera (Qimaging) and Openlab 4.0.2 software (Improvision) were used to acquire images.
IFY-1 (APC/C substrate) accumulation in suppressors:
Whole-cell extracts were prepared from synchronized adult gravid worms as described previously (Kitagawa et al. 2002). Fifty micrograms of protein extract per lane were separated by 4–12% SDS–polyacrylamide gel electrophoresis (Invitrogen, San Diego) and transferred to nitrocellulose membrane (Whatman). Membranes were blotted with anti-IFY-1 antibody (Kitagawa et al. 2002) and anti-HCP-3 antibody (Oegema et al. 2001). Antibodies were used at 1:1000 dilution.
DAPI staining:
Animals were shifted at L4 stage from 20° to 25° and adult progeny were prepared for whole-mount DAPI staining. One-day-old synchronized adults were washed with M9 buffer and stained with 150 nm DAPI in ethanol for 90 min at room temperature. Animals were destained overnight in M9 buffer at 4°. Destained animals were mounted on agarose pads and viewed with the Zeiss Axioscope fluorescent microscope with 40× objective. A Retiga 2000R camera (Qimaging) and Openlab 4.0.2 software (Improvision) were used to acquire images.
Identification of the h1988ts, h1959ts, and h1962ts suppressors:
The h1988ts suppressor was positioned using snip-SNPs between C34F11 and K03H9, where fzy-1 is located. The fzy-1 gene, including 300 bp upstream and downstream, was PCR amplified from the h1988ts strain and sequenced (Nucleic Acid and Protein Services, UBC). The h1959ts and h1962ts suppressors were assigned using snip-SNPs to a region where apc-2 and emb-30 genes are located. The complementation analysis was performed by mating emb-30(h1959ts) males to unc-32(e289) emb-30(tn377ts) hermaphrodites at 20° and shifting emb-30(h1959ts)/unc-32(e289) emb-30(tn377ts) progeny at L4 stage to 25° and by mating dpy-17(e164) emb-30(h1962ts) males to unc-32(e289) emb-30(tn377ts) hermaphrodites at 20° and shifting dpy-17(e164) emb-30(h1962ts)/unc-32(e289) emb-30(tn377ts) progeny at L4 stage to 25°. The full-length cDNA of the emb-30/APC4 gene from the h1959ts and h1962ts suppressors was RT–PCR amplified and sequenced (Nucleic Acid and Protein Services, UBC).
Identification of the such-1(h1960) suppressor:
such-1(h1960) was positioned using dpy-17 and dpy-18 markers and snip-SNP mapping procedures between T28D6 and Y41C4A, a small region containing 40 predicted genes. The full-length cDNA of the Y66D12A.17 gene was amplified from both the such-1(h1960) and N2 strains using RT–PCR and sequenced (Nucleic Acid and Protein Services, UBC). The G80A change present in the such-1(h1960) strain was not observed in N2. We also observed that the RT–PCR product is shorter than the predicted cDNA (WormBase WS164) in both N2 and such-1(h1960) strains. Sequencing revealed that 96 bp of the exon III (from 591 to 686) predicted to encode for a protein are spliced out and as a result of this, SUCH-1 is 32 amino acids shorter than predicted in both N2 and such-1(h1960).
Cell division timing of the early embryo:
One-day-old adult gravid hermaphrodites were dissected and embryos were mounted in M9 buffer (Brenner 1974) between a coverslip and a 3% agarose pad. Mineral oil was used to reduce evaporation at the edge of the coverslip. Embryos were observed using a Zeiss Axioscope at 40× magnification. All images were taken with a Retiga 2000R (Qimaging) digital camera and Openlab 4.0.2 software (Improvision). Time points measured were from the nuclear envelope breakdown (NEBD) to the nuclear envelope reformation (NER) as determined by the analysis of differential interference contrast (DIC) images, as described previously (Encalada et al. 2005). To measure the time points from NEBD to anaphase and from anaphase to NER, embryos from hermaphrodites carrying an integrated H2B∷GFP transgene were prepared and visualized using the same procedure.
Online supplemental material:
Supplemental Table 1 (http://www.genetics.org/supplemental/) shows statistical analysis of the data set where NEBD to NER was measured in P0 cells of the suppressors in both wild-type and unc-46(e177) mdf-1(gk2) backgrounds; supplemental Figure 1 (http://www.genetics.org/supplemental/) shows quantification of IFY-1 accumulation in the such mutants. Intensity of protein bands detected with ECL detection kits (GE Healthcare) was quantified using the image analysis software Quantity One (Bio-Rad, Hercules, CA). Relative values of intensity of IFY-1 bands normalized by that of HCP-3 were plotted. The image of the autoradiograph of the ECL reaction is shown in Figure 4. Supplemental Figure 2 (http://www.genetics.org/supplemental/) shows scatter plots of (A) survivability (percentage of fertile progeny; Table 1) and timing of mitotic division in the early embryo (fold increase when compared to N2; Figure 2) and (B) genome stability (defined as percentage of males; Table 1) and timing of mitotic division in the early embryo (fold increase when compared to N2; Figure 2).
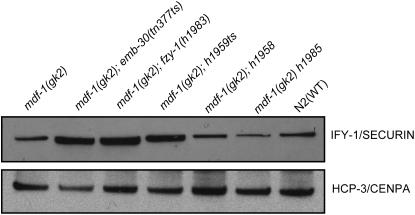
Figure 4.—
IFY-1(APC/C) substrate accumulation in the suppressors. Whole-cell extracts from gravid worms in synchronized culture of the indicated strains were separated by 4–12% SDS–PAGE, transferred to nitrocellulose membrane, and then subjected to Western analysis with anti-IFY-1 antibody and anti-HCP-3 antibody.
TABLE 1.
Suppressors rescue the lethal phenotype of mdf-1(gk2)
| Genotypes | Embryonic arrest (%) | Larval arrest (%) | Adult (%) | Fertile adult (%) | Male (%) |
|---|---|---|---|---|---|
| F2unc-46(e177) mdf-1(gk2) (n = 4133) | 25.2 | 53.4 | 21.4 | 2.5 | 5.1 |
| F2unc-46(e177) mdf-1(gk2) (n = 924) 25° | 97.3 | 2.7 | 0.0 | 0.0. | 0.0. |
| unc-46(e177) mdf-1(gk2); h1959ts (n = 829) | 14.6 | 16.4 | 69.0 | 57.1 | 1.5 |
| unc-46(e177) mdf-1(gk2); emb-30(tn377ts) (n = 1740) 16° | 11.4 | 16.5 | 72.1 | 50.4 | 0.7 |
| unc-46(e177) mdf-1(gk2); h1962ts (n = 455) | 19.4 | 14.5 | 66.1 | 48.1 | 3.1 |
| unc-46(e177) mdf-1(gk2); h1960 (n = 353) | 23.8 | 15.3 | 60.9 | 41.6 | 0.8 |
| unc-46(e177) mdf-1(gk2); h1958 (n = 585) | 24.9 | 22.2 | 52.9 | 40.6 | 1.8 |
| unc-46(e177) mdf-1(gk2); h1988ts (n = 470) | 28.8 | 28.9 | 42.3 | 33.9 | 2.5 |
| unc-46(e177) mdf-1(gk2) h1992 (n = 1135) | 45.3 | 15.0 | 39.7 | 30.7 | 1.8 |
| unc-46(e177) mdf-1(gk2); h1987 (n = 214) | 42.0 | 17.8 | 40.2 | 29.0 | 6.2 |
| unc-46(e177) mdf-1(gk2); fzy-1(h1983) (n = 1437) | 71.2 | 6.9 | 21.9 | 18.4 | 1.9 |
| unc-46(e177) mdf-1(gk2) h1985 (n = 1102) | 51.8 | 19.4 | 28.8 | 16.7 | 4.4 |
| unc-46(e177) mdf-1(gk2); h1989 (n = 989) | 70.1 | 9.3 | 20.6 | 12.6 | 3.9 |
| unc-46(e177) mdf-1(gk2) h2168 (n = 372) | 58.9 | 24.9 | 16.2 | 9.6 | 9.1 |
Shown are the phenotypes of the 11 mutants recovered in the screens for suppressors of the mdf-1(gk2) lethal phenotype. Also included are the two previously known suppressors emb-30(tn377ts) and fzy-1(h1983). The extent of suppression was determined at 20° after at least three backcrosses, unless otherwise indicated.
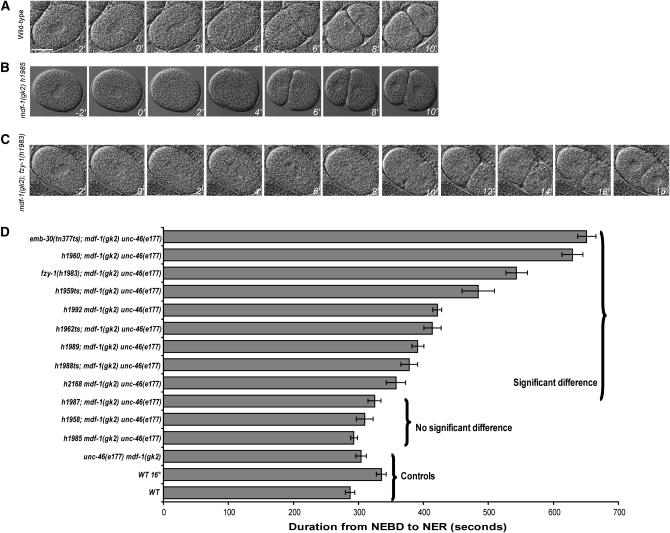
Figure 2.—
Timing of cell division at one-cell stage in suppressors. (A–C) Images of the wild-type, unc-46(e177) mdf-1(gk2) h1985, and unc-46(e177) mdf-1(gk2); fzy-1(h1983) one-cell embryos using time-lapse microscopy. The T = 0 min time point shows cells during NEBD. The embryos were imaged every 2 min from this point until the formation of the two-cell embryo. Pictures were taken every 2 min (bar, 20 μm). (D) Summary of the mitotic timing data from live cell imaging. Bars represent the duration of mitotic division from NEBD to NER in one-cell embryos. Mean durations are plotted in seconds with SEM error bars (n = 10 measurements for each strain). The braces depict the controls, the suppressors that do not cause a significant delay in mitotic progression, and the suppressors that significantly delay the mitotic timing as shown by P-values of Student's t-test statistic (supplemental Table 1 at http://www.genetics.org/supplemental/). All the measurements were performed at 20°, except for emb-30(tn377ts) and wild-type control strains observed at 16°.
RESULTS
Suppressors rescue the lethal phenotype of mdf-1(gk2):
We have previously shown that the mdf-1/MAD1 gene is essential for the long-term survival and fertility of C. elegans (Kitagawa and Rose 1999). mdf-1 homozygotes segregated from the sma-8(n716) +/+ mdf-1(gk2)(V) heterozygous parents had no obvious phenotype at 20°, most likely due to the presence of maternally supplied MDF-1 protein. However, only 21% of F2 mdf-1(gk2) homozygotes grew to adulthood (Kitagawa and Rose 1999). In accordance with these data we observed that unc-46 mdf-1 homozygotes segregated from unc-46(e177) mdf-1(gk2) +/+ + nT1[let-X] heterozygote parents appeared to be normal and grew to adulthood at 20°. Similarly, we observed that only 21% of F2 unc-46 mdf-1 homozygotes developed into adults and the majority of the F3 homozygotes arrested at different developmental stages (Table 1). We also observed that higher temperature intensified the effect; at 25°, 97% of the F2 homozygotes arrested before hatching and the remaining 3% arrested at early larval stages. Thus, the unc-46 mdf-1 homozygotes cannot be maintained beyond the first generation at 25°, while at 20° the strain cannot be maintained beyond the third generation (Table 1).
In our previous EMS mutagenesis screen we isolated 10 suppressors of the mdf-1(gk2) lethal phenotype (Kitagawa et al. 2002). We isolated the h2168 suppressor allele in a small screen using the dog-1(gk10) mutator strain. When dog-1 is mutated or disrupted by RNA interference (RNAi), deletions frequently initiate at dC/dG tracts that are spaced throughout the genome and typically extend a few hundred base pairs upstream of the G tract (Cheung et al. 2002). Of only 80 haploid genomes screened, one plate containing a suppressor mutation was isolated (Table 1). To examine the viability of the suppressor mutations in the absence of the MDF-1 checkpoint, we analyzed all of the suppressor strains for embryonic arrest, larval arrest, and adult progeny (Table 1). We observed that the strongest suppressor allele, h1959ts, decreased the percentage of both embryonic arrest and larval arrest when compared to the F2 unc-46 mdf-1 homozygotes, increasing the percentage of adult progeny (Table 1). On the other hand, the weakest suppressor alleles, h1989 and h2168, resulted in an increase in embryonic arrest and a decrease in larval arrest and yielded the same percentage of adult progeny as the F2 unc-46 mdf-1 homozygotes (Table 1). However, all of the suppressor alleles increased the percentage of fertile progeny from 4- to 23-fold when compared to the F2 unc-46 mdf-1 homozygotes alone (Table 1). We conclude that while none of the suppressors rescues the mdf-1(gk2)-associated phenotypes completely, all of the suppressors allow the strains to be propagated in the absence of the essential SAC component by increasing viability (Table 1).
We tested the fidelity of chromosome segregation in the suppressor strains by scoring the frequency of spontaneous males. C. elegans populations consist largely of self-fertilizing hermaphrodites (5A; XX). During meiosis I, nondisjunction of the X chromosome in the hermaphrodite germline generates gametes with no X chromosome, leading to XO male progeny (5A; XO). Males arise spontaneously at a rate of ∼0.1% at 20° (Hodgkin et al. 1979; Rose and Baillie 1979). We have shown previously that F2 mdf-1 homozygotes have a Him (high incidence of males) phenotype (Kitagawa and Rose 1999). In the F2 unc-46 mdf-1 homozygotes we observed a 50-fold increase in the frequency of males when compared to wild-type animals (Table 1). We also observed that the majority of the suppressor mutations substantially decreased X chromosome missegregation in the absence of the MDF-1 checkpoint. However, none of the suppressors rescued the mdf-1(gk2)-associated Him phenotype completely (Table 1).
The majority of the suppressors delay mitotic progression:
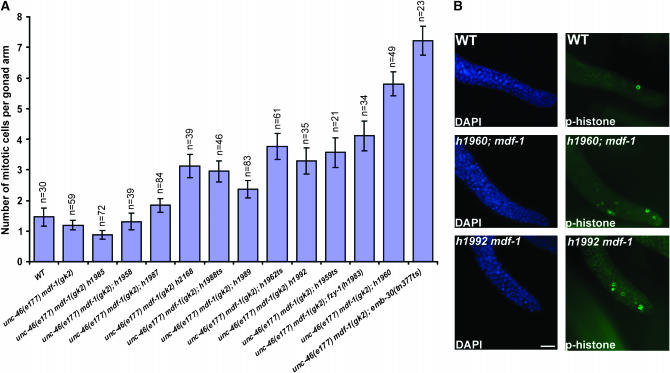
One explanation for the ability of the suppressors to bypass the MDF-1 checkpoint requirement is that they delay anaphase onset. Furuta et al. (2000) showed that in emb-30(tn377ts) gonad arms, there are a significantly increased number of mitotic cells at the permissive temperature (16°), suggesting a mitotic delay in the germ cells. The second identified suppressor, fzy-1(h1983), when analyzed as a hemizygote had a higher mitotic index, suggesting prolonged mitosis in fzy-1(h1983) hemizygotes (Kitagawa et al. 2002). To determine whether all of the suppressors delay mitotic progression, we visualized chromosomes in cells undergoing mitosis in the distal mitotic zone of gravid hermaphrodite gonads by staining with an antibody against phosphorylated histone H3 (Figure 1B). In accordance with the previous data, we observed that unc-46(e177) mdf-1(gk2); emb-30(tn377ts) and unc-46(e177) mdf-1(gk2); fzy-1(h1983) homozygotes had a significantly increased number of cells per single gonad arm when stained with this antibody compared to wild-type or unc-46(e177) mdf-1(gk2) animals (Figure 1A). More importantly, we observed that seven additional suppressors display a variable but significant increase in the number of mitotic cells in the distal arm, suggesting that germ cell mitosis is slowed in these animals (Figure 1A). However, three suppressors display a normal number of stained cells, suggesting normal cell cycle progression. We conclude that germ cell mitosis is slowed in the majority of the suppressors.
Figure 1.—
Mitotic germ cell accumulation in the suppressors. (A) Quantitation of mitotic germ cells in the distal zone of gravid hermaphrodite gonads. Bars represent the mean number of mitotic nuclei per gonad arm with SEM error bars; n, number of gonad arms scored for each suppressor. (B) The representative images of DAPI and anti-phosphohistone H3-stained wild-type, unc-46(e177) mdf-1(gk2); h1960, and unc-46(e177) mdf-1(gk2) h1992 gonads. All the measurements were performed at 20°. Bar, 10 μm.
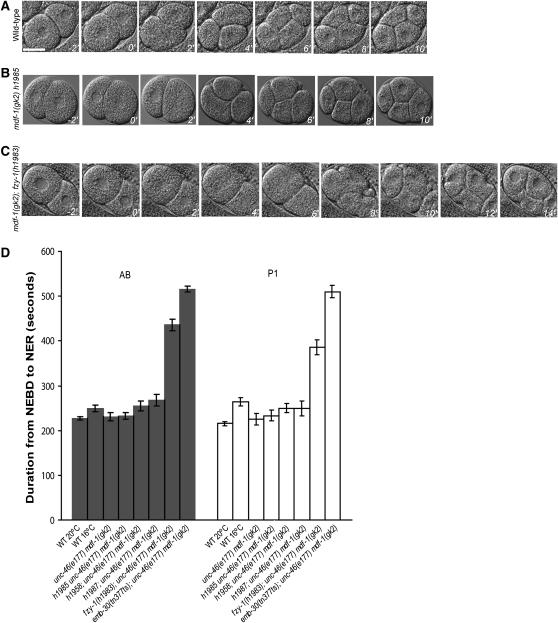
We then examined the timing of mitosis in the early embryo. Recent work from Encalada et al. (2005) has shown that chemical or mutational disruptions of the mitotic spindle activate the spindle checkpoint to delay progression through mitosis in rapidly dividing C. elegans embryonic cells. These results suggested that mdf-1 is required for the cell cycle delay in early embryonic cells in the presence of spindle damage, which prompted us to measure the timing of mitotic division in unc-46(e177) mdf-1(gk2); emb-30(tn377ts) and unc-46(e177) mdf-1(gk2); fzy-1(h1983) early embryos. We analyzed the mitotic progression from NEBD to NER using time-lapse DIC imaging of early embryonic cells as described by Encalada et al. (2005). In the unc-46(e177) mdf-1(gk2); emb-30(tn377ts) embryos the duration from NEBD to NER in P0, AB, and P1 cells was increased approximately twofold when compared to wild-type embryos at 16° (Figures 2 and 3 and supplemental Table 1 at http://www.genetics.org/supplemental/). We also observed that the average time required to progress through mitosis was increased 1.8-fold in the unc-46(e177) mdf-1(gk2); fzy-1(h1983) homozygous one- and two-cell stage embryos when compared to wild-type embryos developing at 20° (Figures 2 and 3 and supplemental Table 1 at http://www.genetics.org/supplemental/). The mitotic delay observed in the early embryonic cells in these two suppressors correlates with an increase in mitotic cells in the germline (Figures 1 and 2). We next measured the NEBD to NER interval duration in all of the suppressors in the unc-46(e177) mdf-1(gk2) background (Figure 2D). The seven suppressors that displayed a significantly elevated number of mitotic cells in the germline also progress through mitosis in a one-cell embryo with a variable but significant delay (Figure 2D). In some mutant embryos, such as h2168, the mean mitotic time was slightly increased to 1.2-fold, while in others, such as h1960, a more dramatic delay of a 2.1-fold increase for the same interval was observed (Figure 2D). Interestingly, we show that the h1987, h1958, and h1985 suppressors have normal timing of mitotic progression in early embryonic cells as well as a normal number of mitotic nuclei in the germline (Figures 1–3 and supplemental Table 1). Our data show that the mitotic timing in the early embryo correlates with the mitotic timing in the germline (Figures 1 and 2).
Figure 3.—
The timing of mitosis in two-cell stage embryos. (A–C) Images of the wild-type, unc-46(e177) mdf-1(gk2) h1985, and unc-46(e177) mdf-1(gk2); fzy-1(h1983) two-cell embryos from Figure 2. The T = 0 min time point shows cells during NEBD in the AB cell. The embryos were imaged every 2 min from this point until the formation of the four-cell embryo (bar, 20 μm). (D) Summary of the mitotic timing data from live-cell imaging. Bars represent the duration of mitotic division from NEBD to NER in two-cell embryos. Mean durations are plotted in seconds with SEM error bars (n = 10 measurements for each strain). The data for h1987, h1958, and h1985 as well as the previously known suppressors emb-30(tn377ts) and fzy-1(h1983) are included. All the measurements were performed at 20°, except for emb-30(tn377ts) and wild-type control strains observed at 16°.
To test the mitotic timing in the suppressor mutants, we isolated all the recessive suppressors not mapping to LGV from the unc-46(e177) mdf-1(gk2) background (Table 2). All of these suppressors are viable at 20° and display a range of different phenotypes (Table 2). As expected, we observed that the mitotic timing in these mutants is independent of the checkpoint (supplemental Table 1 at http://www.genetics.org/supplemental/). Thus, we conclude that the suppressors of the mdf-1(gk2) lethal phenotype fall into two classes: those that delay mitotic division and those that progress through mitosis normally. The suppressors with slowed mitotic cell cycles display mitotic delays in both embryonic and germ cells that may compensate for the absence of the essential checkpoint function. The suppressors with normal mitotic cell cycles in both embryonic and germ cells appear to compensate for the absence of the checkpoint by a different mechanism.
TABLE 2.
Suppressor phenotypes in the absence of mdf-1(gk2)
| Genotypes | Class | Phenotypes |
|---|---|---|
| h1959ts | Recessive | Viable at 20°, some embryonic arrest and mild Him; lethal at 25°, 100% embryonic arrest. |
| h1962ts | Recessive | Viable at 20°, majority of brood consists of unfertilized oocytes and mild Him; sterile at 25°, 100% unfertilized oocytes in F2. |
| h1960 | Recessive | Viable at 20°, no obvious phenotype; viable at 25°, embryonic arrest and Him. |
| h1958 | Recessive | Viable at 20°, no obvious phenotype; viable at 25°, some developmental arrest. |
| h1988ts | Recessive | Viable at 20°, small brood size, embryonic and larval arrest and Him; sterile at 25°, 100% sterile. |
| h1992 | Recessive | Difficulty isolating due to linkage to unc-46 and mdf-1. |
| h1987 | Recessive | Viable at 20°, no obvious phenotype; viable at 25°, developmental arrest. |
| h1985 | Recessive | Difficulty isolating due to linkage to unc-46 and mdf-1. |
| h1989 | Dominant | Difficulty isolating due to phenotype. |
| h2168 | Recessive | Difficulty isolating due to linkage to unc-46 and mdf-1. |
All of the recessive suppressors not located on LGV were isolated from the unc-46(e177) mdf-1(gk20) background and analyzed for phenotypes. The temperature sensitivity was examined by shifting L4 hermaphrodites to 25° and scoring the progeny.
Securin (IFY-1/Pds1) levels correlate with the timing of anaphase onset in the suppressors:
To determine whether the mitotic delays observed in the suppressor mutants are due to the delay in anaphase onset, we constructed suppressor mutant strains that carry ruIs32, an integrated H2B∷GFP transgene (Praitis et al. 2001). In these strains, we analyzed the mitotic progression from NEBD to anaphase onset and from anaphase onset to NER in early embryonic cells as described by Encalada et al. (2005). We successfully constructed seven suppressor strains; however, the remaining suppressors could not be constructed with ruIs32 either because the H2B∷GFP insert is closely linked to the suppressor mutation or because the H2B∷GFP transgene is inviable with these suppressor mutants (Table 3). We determined that the mitotic delay observed in the first class of suppressors is solely due to the delay in anaphase onset. For instance, the 1.6-fold delay in the NEBD to NER interval in the h1988ts suppressor mutant is due to the 2-fold lengthening of the interval from NEBD to anaphase onset (Table 3). In contrast, we did not observe any significant difference in the progression from anaphase onset to NER interval in any of the suppressors when compared to wild-type and mdf-1(gk2) embryos with the H2B∷GFP insert (Table 3). Our data also show that the class of suppressors that display normal mitotic timing in the germline and embryonic cells progresses through anaphase with no delay. We conclude that the mitotic delay observed in the first class of suppressor is due to delayed anaphase onset.
TABLE 3.
Timing of anaphase onset in the suppressor mutants
| NEBD–anaphase | Anaphase–NER | NEBD–NER | |
|---|---|---|---|
| Embryo | s ± SE (n) | s ± SE (n) | s ± SE (n) |
| Wild type | 154.5 ± 4.3 (6) | 143.2 ± 8.2 (6) | 297.7 ± 5.1 (6) |
| F2unc-46(e177) mdf-1(gk2) | 177.2 ± 14.2 (5) | 127.6 ± 6.1 (5) | 304.8 ± 12.9 (6) |
| unc-46(e177) mdf-1(gk2); h1960 | 464.2 ± 13.6 (5)* | 124.2 ± 2.6 (5) | 588.4 ± 14.2 (5)* |
| unc-46(e177) mdf-1(gk2); h1983 | 352.2 ± 5.9 (5)* | 158.2 ± 3.3 (5) | 510.4 ± 5.3 (5)* |
| unc-46(e177) mdf-1(gk2) h1992 | 252.8 ± 11.2 (5)* | 144.6 ± 11.4 (5) | 397.4 ± 7.5 (5)* |
| unc-46(e177) mdf-1(gk2); h1988ts | 344.0 ± 14.1 (5)* | 137.8 ± 8.1 (5) | 481.8 ± 7.2 (5)* |
| unc-46(e177) mdf-1(gk2); h1987 | 178.0 ± 8.1 (5) | 124.6 ± 3.5 (5) | 302.6 ± 8.5 (5) |
| unc-46(e177) mdf-1(gk2); h1958 | 177.5 ± 11.2 (6) | 138.7 ± 4.8 (6) | 316.2 ± 9.5 (6) |
| unc-46(e177) mdf-1(gk2) h1985 | 158.8 ± 8.1 (5) | 149.2 ± 6.6 (5) | 308.0 ± 8.3 (5) |
Suppressor strains that carry ruIs32, an integrated H2B∷GFP transgene (Praitis et al. 2001), were constructed and analyzed for anaphase onset. Asterisks denote a significant difference as shown by P-values of Student's t-test statistic.
We tested the mechanism of anaphase onset timing in the suppressors. Previously, using yeast two-hybrid analysis, we showed that one of the suppressors isolated in the EMS screen, fzy-1(h1983), loses the binding affinity of FZY-1/Cdc20 for securin, IFY-1/Pds1 (Kitagawa et al. 2002). We reasoned that the suppressor mutations with delayed mitosis may be defective in securin IFY-1 destruction, which would delay the activation of separase and hence anaphase onset. To test this, we analyzed IFY-1 levels in the cell extracts prepared from the suppressor mutants (Figure 4). Western blotting showed IFY-1 accumulation in the class of suppressors that delay anaphase onset. On the other hand, the class of suppressors that progress through mitosis normally does not accumulate securin. Furthermore, quantitation of the data revealed that the IFY-1 protein level correlates with the timing of anaphase onset (supplemental Figure 1 at http://www.genetics.org/supplemental/). These data indicate that the first class of suppressors might bypass the checkpoint requirement by reducing the APC/C activity, which results in securin accumulation and delayed anaphase onset. The second class of suppressors most likely rescues the checkpoint lethality by a different mechanism.
Three suppressor mutations correspond to the genes emb-30/APC4 and fzy-1/CDC20:
To genetically map the suppressors, we used phenotypic markers in combination with the snip-SNP mapping procedure as described by Wicks et al. (2001) (see materials and methods). Our mapping data positioned three suppressors to chromosome V, four to chromosome III, and three to the gene cluster of chromosome II (Table 4). Our cell cycle timing analysis showed that the majority of suppressors delay anaphase onset, which prompted us to examine if any of the suppressors represent known downstream targets of the SAC.
TABLE 4.
Positional cloning of such genes
| Alleles of such genes | Mapping and sequencing data |
|---|---|
| LGII | |
| fzy-1(h1988ts) | R378Q in WD40 domain of FZY-1. |
| such-3(h1989) | Located in gene cluster on II. No sequence changes in the scc-1, ify-1, fzy-1, and emb-27 genes or in the mat-2 cDNA. |
| such-5(h1987) | Located in gene cluster on II. No sequence changes in the scc-1, ify-1, fzy-1, and emb-27 genes or in the mat-2 cDNA. |
| LGIII | |
| emb-30 (h1959ts) | M480I in nonconserved region of EMB-30. |
| emb-30 (h1962ts) | T265I in WD40 domain of EMB-30. |
| such-1 (h1960) | G27D in semiconserved region of Y66D12A.17. |
| such-6 (h1958) | Closely linked to dpy-17. No sequence changes in the apc-11 coding region. |
| LGV | |
| such-2(h1992) | Located in gene cluster on V. Not duplicated mdf-1 as determined by PCR and RT–PCR. No sequence changes in the apc-10 coding region. |
| such-4(h2168) | Maps to the right of unc-46. Not duplicated mdf-1 as determined by PCR and RT–PCR. |
| such-7(h1985) | Maps to the right of unc-46. Not duplicated mdf-1 as determined by PCR and RT–PCR. |
The h1988ts suppressor was mapped to the gene cluster of chromosome II, where the known SAC effector and mdf-1 suppressor, fzy-1, is located. Sequencing of fzy-1 from the h1988ts suppressor strain identified a single-base-pair change from guanine to adenine at position 1133 of the cDNA sequence in exon IX. This change caused an R378Q alteration in the amino acid sequence in the nonconserved region within the WD40 domain (Table 4). The fzy-1(h1988ts) allele is more severe than the previously identified fzy-1(h1983) allele (Kitagawa et al. 2002). fzy-1(h1988ts) homozygotes can be maintained at 20° but display severely reduced brood size (22 progeny) of which 49% arrest as embryos, 20% arrest as larvae, and 31% develop into adults. The strain also has a Him phenotype (5% of the adult progeny are males). At 25° the fzy-1(h1988ts) homozygotes are 100% sterile (Table 2).
The h1959ts and h1962ts suppressors were positioned in the gene cluster on chromosome III, where the known APC/C components apc-2 and emb-30 are located. These suppressor alleles are temperature sensitive and cannot be maintained at 25° (Table 2). The map position, delayed mitosis, and temperature-sensitive phenotype of the h1959ts and h1962ts suppressors suggested that these might be new alleles of the emb-30/APC4 gene. Both of the suppressors failed to complement emb-30(tn377ts). Sequencing of emb-30 from the h1959ts suppressor strain identified a single-base-pair change from guanine to adenine at position 1440 of the cDNA sequence in exon VII. This change caused an M480I alteration in the amino acid sequence within the nonconserved region. Similarly, sequencing of the h1962ts suppressor identified a single-base-pair change from cytosine to thymine at position 796 of the cDNA sequence in exon V. This change caused a T265I alteration in the amino acid sequence in the nonconserved region within the WD6 domain. The emb-30(h1959ts) allele is a new conditional allele that belongs to the same class as the previously identified suppressor allele emb-30(tn377ts) (Furuta et al. 2000). The emb-30(h1959ts) homozygotes are easily maintained at 20°; the brood size is normal (237 progeny) with 18% embryonic arrest, 2% larval arrest, and 80% progeny that develop into adults, of which 0.8% are males (Table 2). At 25° the emb-30(h1959ts) homozygotes are 100% embryonic lethal (Table 2). Interestingly, the second emb-30(h1962ts) allele does not belong to any of the previously described classes (Furuta et al. 2000) (Table 2).
The sterility of the emb-30(h1962ts) allele is specific to sperm:
We observed that dpy-17(e164) emb-30(h1962ts) worms had a characteristic phenotype at restrictive temperature as the majority of the brood at 25° consisted of unfertilized oocytes (Table 5). We further analyzed the emb-30(h1962ts) phenotype in the absence of the dpy-17(e164) background and observed that emb-30(h1962ts) worms laid mainly unfertilized oocytes at both permissive and restrictive temperatures (Table 5). The emb-30(h1962ts) homozygotes can be maintained at 20°, as all of the adult progeny (23%) segregated from the emb-30(h1962ts) mothers cultured at 20° produced ∼30% fertilized oocytes (Table 5). However, at 25° all of the adult progeny (18%) segregated from the emb-30(h1962ts) mothers laid 100% unfertilized oocytes (Table 5). In the emb-30(h1962ts) broods at both permissive and restrictive temperatures we also observed a higher incidence of males and some embryonic and larval arrest (Table 5). We conclude that the emb-30(h1962ts) allele is a new conditional allele that results in sterility at 25° due to unfertilized oocytes.
TABLE 5.
The emb-30(h1962ts) mutant phenotype
| Parental genotype | Brood sizea | Unfertilized oocytes (%) | Embryonic arrest (%) | Larval arrest (%) | Adult (%) | Males (%) |
|---|---|---|---|---|---|---|
| Temperature 25° | ||||||
| dpy-17(e164) emb-30(h1962ts) | 49 | 74.5 | 7.1 | 1.2 | 17.2 | 2.2 |
| dpy-17(e164) emb-30(h1962ts) × N2 ♂ | 56 | 0 | 0 | 0 | 100 | 50.8 |
| dpy-17(e164) control | 139 | 37.2 | 0 | 0 | 62.8 | 0.3 |
| emb-30(h1962ts) | 74 | 73.5 | 6.7 | 2.2 | 17.6 | 3.3 |
| emb-30(h1962ts) × N2 ♂ | 111 | 1.8 | 1.2 | 1.4 | 95.6 | 49.1 |
| Temperature 20° | ||||||
| dpy-17(e164) emb-30(h1962ts) | 117 | 35.2 | 2.4 | 4.1 | 58.3 | 1.0 |
| dpy-17(e164) emb-30(h1962ts) × N2 ♂ | 170 | 0.6 | 2.8 | 1.2 | 95.4 | 50.0 |
| dpy-17(e164) control | 244 | 26.7 | 0 | 0 | 73.3 | 0.1 |
| emb-30(h1962ts) | 285 | 70.3 | 4.3 | 1.9 | 23.5 | 1.3 |
| emb-30(h1962ts) × N2 ♂ | 243 | 17.0 | 1.5 | 0.9 | 80.6 | 45.7 |
Brood size is defined as the total number of shelled eggs and unfertilized oocytes laid by a single hermaphrodite.
Many mutations that eliminate spermatogenesis or cause production of defective sperm in hermaphrodites are recognized by unfertilized, round, brown oocytes that lack an eggshell, resulting in self-sterile hermaphrodites (reviewed in L'Hernault 2006). To analyze the nature of the sterility in the emb-30(h1962ts) hermaphrodites we mated several L4 emb-30(h1962ts) hermaphrodites to wild-type males at both restrictive and permissive temperatures. If these hermaphrodites have defective spermatogenesis, their oocytes would be fertilized by wild-type male sperm. We observed that wild-type sperm were capable of fertilizing emb-30(h1962ts) oocytes at both temperatures and in all strains tested (Table 5). We also observed that the majority of the broods consisted of fertilized oocytes that developed into fertile adults (Table 5). In contrast, the wild-type sperm did not rescue the tn377ts and h1959ts alleles of the emb-30 gene, which result in an embryonic arrest phenotype at restrictive temperature. We conclude that the sterility observed in the emb-30(h1962ts) mutant is a consequence of defective spermatogenesis.
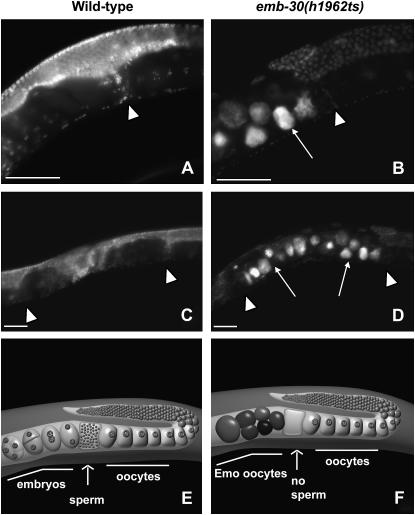
To analyze whether emb-30(h1962ts) hermaphrodites produce defective sperm incapable of fertilization or a limited amount or no sperm at restrictive temperature, 1-day-old adult progeny from emb-30(h1962ts) mothers at 25° were fixed and stained with DAPI (Figure 5). If the emb-30(h1962ts) hermaphrodites produced sperm incapable of fertilization, we would expect to see many sperm accumulated in the spermatheca. On the contrary, we observed that all of the examined emb-30(h1962ts) hermaphrodites segregated from mothers cultured at 25° (20/20) contained no detectable sperm (Figure 5, B and D). We also observed that emb-30(h1962ts) oocytes pass through spermatheca, resume meiosis, continue cycling, and become highly polyploid, endomitotic oocytes (Emo) (Figure 5). The Emo phenotype is common to ovulated but unfertilized oocytes in C. elegans (McCarter et al. 1999). These results provide further evidence that emb-30(h1962ts) hermaphrodites produce normal oocytes and defective sperm.
Figure 5.—
The emb-30(h1962ts) allele has an Emo phenotype at 25°. Animals were shifted in L4 stage from 20° to 25° and their adult progeny were DAPI stained as young adults. Images show partial gonad and partial uterus (A and B) or the entire uterus (C and D). In wild-type hermaphrodites (A and C) the highly condensed haploid sperm nuclei can be seen in the spermatheca (arrowhead). Left of the spermatheca, embryos are located within the uterus (A and C). In emb-30(h1962ts) animals (B and D), there is no sperm DNA detected in the spermatheca (arrowhead) and the uterus is filled with Emo oocytes only (arrows). (E) Schematic of a single wild-type adult hermaphrodite gonad arm including half of the uterus. A maturing oocyte is pushed into the spermatheca, successfully fertilized by sperm, and pushed into the uterus where it develops into an embryo. (F) Schematic of a single emb-30(h1962ts) adult hermaphrodite gonad arm including half of the uterus. A maturing oocyte is pushed into the spermatheca. The lack of sperm results in a failure to fertilize. The unfertilized oocytes pass through spermatheca into the uterus, continue cycling, and become Emo, highly polyploid, round, brown oocytes that lack an eggshell. Bars, ∼50 μm.
The such genes identify new mdf-1/MAD1 interactors:
Seven suppressor mutations do not appear to be alleles of known C. elegans metaphase-to-anaphase transition pathway components. We mapped the h1987 and h1989 suppressors to the gene cluster on chromosome II, where many known downstream targets of the SAC are located. However, sequencing of these suppressor strains revealed no nucleotide changes in the scc-1, ify-1, fzy-1, emb-27, and mat-2 candidates (Table 4). Similarly, the h1958 suppressor strain contained no sequence changes in the apc-11 gene located in the region where the suppressor was positioned (Table 4). The h1960 suppressor was assigned to a region on the right arm of chromosome III, where no known components are located (Table 4). Furthermore, while the h1992 suppressor, located in the gene cluster of chromosome V and closely linked to mdf-1(gk2), revealed no sequence alteration in the apc-10 candidate, the h1985 and h2168 suppressors were assigned to the regions where no known components are located (Table 4). In addition, neither of the analyzed suppressors shows any obvious phenotypes in wild-type background at 20°; they are also viable at 25° (Table 2). Our sequencing and mapping data suggest that these seven suppressors identify novel mdf-1/MAD1 interactors and we have named them such (for suppressor of spindle checkpoint defect): such-1(h1960), such-2(h1992), such-3(h1989), such-4(h2168), such-5(h1987), such-6(h1958), and such-7(h1985) (Table 4). We further focused on finding the molecular basis of the such-1(h1960) suppressor, which results in a significant delay of anaphase onset and maps to the right arm on LGIII where no known APC/C components are located (Table 4).
The such-1(h1960) corresponds to Y66D12A.17, an APC5-like gene:
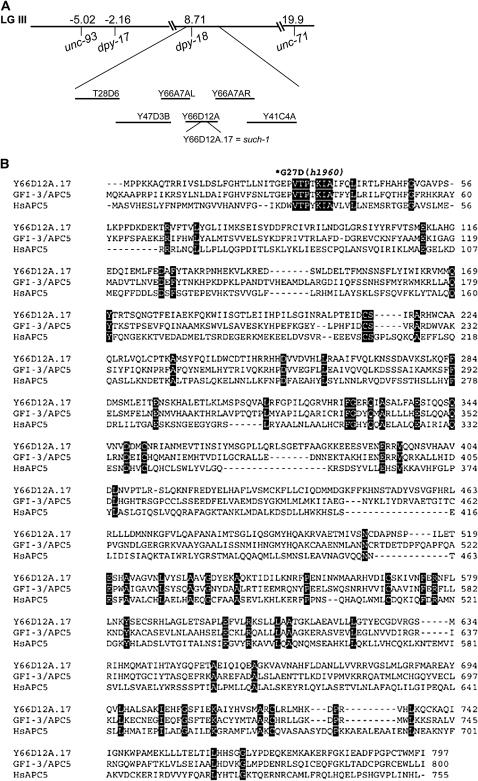
We positioned the such-1 suppressor using snip-SNP mapping between T28D6 and Y41C4A, a small region containing 40 predicted genes (Figure 6A). We observed that one of the genes within this region is Y66D12A.17, a previously unknown gene that appears to be weakly similar to the human anaphase-promoting complex subunit APC5 (WormBase WS164). The such-1(h1960) suppressor displays a delayed mitosis phenotype in both germline and early embryonic cells as a result of delayed anaphase onset (Figures 1–3 and Table 3). We would expect to see this phenotype as a consequence of reduced APC/C activity. Sequence analysis of the coding region of Y66D12A.17 from the such-1(h1960) strain revealed a single-base-pair change from guanine to adenine at position 80 of the cDNA sequence in exon I. This change caused a G27D alteration in the amino acid sequence in the semiconserved region (Table 4 and Figure 6B). We therefore refer to Y66D12A.17 as such-1. The such-1(h1960) allele has no obvious phenotypes in the wild-type background at 20°; the brood size is normal (260 progeny) with 7% embryonic arrest, 93% progeny that develop into adults, and 0.2% males (Table 2). The such-1(h1960) homozygotes are also viable at 25°, but higher temperature intensified the phenotype as the brood size decreased (172 progeny), embryonic arrest increased to 46%, 15% of progeny arrested as larvae, and only 39% of progeny developed into adults (Table 2). We observed that at 25° such-1(h1960) also results in a Him phenotype (3.6%).
Figure 6.—
Molecular cloning and DNA sequence analysis of the such-1/Y66D12A.17 gene. (A) Mapping and cloning of the such-1(h1960) suppressor. such-1 was positioned using snip-SNP mapping procedures between T28D6 and Y41C4A. (B) The Y66D12A.17 gene product GFI-3/Apc5 and H. sapiens Apc5 proteins were aligned using ClustalW. Common residues have a black background and the asterisk denotes the amino acid residue substituted in such-1(h1960).
There are nine APC/C subunits identified to date in C. elegans; some were isolated in forward genetic screens for ts emb mutants (Cassada et al. 1981) and ts mat mutants (Golden et al. 2000; Shakes et al. 2003) and some were identified through sequence homologies (Davis et al. 2002). gfi-3/M163.4 was reported to encode APC5 (Zachariae et al. 1998) and to share 11% identity and 20% similarity with S. cerevisiae Apc5p (Davis et al. 2002). Our RT–PCR analysis of the such-1 gene from both the wild-type (N2) and the h1960 strains revealed that 96 bp of the exon III predicted to encode for a protein (WormBase WS164) are spliced out. As a result of this, SUCH-1 is 32 amino acids shorter than predicted (Figure 6B). When we aligned the expressed SUCH-1 with S. cerevisiae Apc5p, we also observed 11% identity and 20% similarity. This would suggest that C. elegans has two APC5-like genes. We also observed that GFI-3 and SUCH-1 are 37% identical and 57% similar. We next aligned GFI-3 and SUCH-1 to the Homo sapiens homolog of APC5. We observed that, while GFI-3 is 19% identical and 39% similar, SUCH-1 is 19% identical and 40% similar to the H. sapiens APC5 (Figure 6B). These data further indicate that SUCH-1 is a paralog of GFI-3 and that both genes are Apc5p homologs in C. elegans. The recovery of the such-1(h1960) allele in our suppressor screen underscores the usefulness of this screen for identifying new components of the metaphase-to-anaphase transition pathway.
DISCUSSION
The spindle assembly checkpoint prevents aneuploidy by delaying the metaphase-to-anaphase transition until all chromosomes are successfully attached to microtubules at kinetochores. In this study we have analyzed mutations that bypass the MDF-1/Mad1 checkpoint requirement for survival and demonstrated that the majority of these mutants delay mitotic divisions in germline and early embryonic cells. This class includes suppressor alleles of the known SAC effectors, emb-30/APC4 and fzy-1/CDC20, a newly identified such-1/APC5-like gene, and potentially three new SAC effectors. We also have identified a second class of suppressors that works through an unknown mechanism, which neither delays anaphase onset nor accumulates securin. Our mapping and sequencing data suggest that components of the APC/C are unlikely to be responsible for this class of suppressors.
The mdf-1 checkpoint is essential in normally growing C. elegans. In the absence of MDF-1, chromosome missegregation accumulates, leading to highly penetrant maternal-effect lethality/sterility (Kitagawa and Rose 1999). Previous work has shown that the reduction of activity in SAC effectors, emb-30(tn377ts) and fzy-1(h1983), can suppress the lethality and sterility associated with the absence of MDF-1 (Furuta et al. 2000; Kitagawa et al. 2002). Here we describe three new alleles of fzy-1 and emb-30. emb-30 has been well studied in C. elegans. There are 17 alleles of emb-30 grouped to five different classes on the basis of their mutant phenotype (Furuta et al. 2000). The emb-30(h1959ts) allele is a new conditional allele, which belongs to the same class as the previously identified suppressor allele emb-30(tn377ts) (Furuta et al. 2000). However, the emb-30(h1962ts) allele identified in the suppressor screen does not belong to any of the previously described classes. It displays a spermatogenesis defect. Previous work by Furuta et al. (2000) described oocyte-specific alleles in the emb-30 gene. However, oocytes in emb-30(h1962ts) animals appear normal and can be fertilized by wild-type sperm and develop into adult progeny. The h1962ts mutation responsible for the sperm defect is the first mutation to be reported in the highly divergent WD6 domain of EMB-30 (Furuta et al. 2000). We show that a mutation in this domain bypasses the MDF-1 checkpoint requirement and results in defective spermatogenesis.
Normally, activation of the SAC in response to spindle defects delays mitotic division in the early embryo and germline (Kitagawa and Rose 1999; Encalada et al. 2005). The first demonstration that mitotic delay could compensate for the lack of checkpoint was that mutations in the emb-30/APC4 and fzy-1/CDC20 genes have the ability to suppress the mdf-1(gk2) lethal phenotype (Furuta et al. 2000; Kitagawa et al. 2002). In this article, we report on these and additional mutants that cause delayed mitosis, which correlates with securin accumulation. It is likely that the anaphase onset delays observed in these mutants allow more time for proper kinetochore attachment, thus preventing defects in chromosome segregation.
We expected those suppressors with longer mitotic delays to improve chromosome segregation fidelity and result in better viability. In general this was true as we observed that an increase in mitotic delay correlated with a decrease in X chromosome missegregation and, to a lesser extent, with an increase in viability. For instance, such-1 delays mitosis from 304 (mdf-1) to 629 sec, decreases X chromosome missegregation from 5 to 0.8%, and improves viability from 2 to 42% (Table 1 and Figure 2). However, there are exceptions; for example, for the alleles of emb-30, the mitotic delay correlates better with the reduction in X chromosome missegregation than it does with the increase in viability (Table 1 and Figure 2). When suppressor mutations were removed from the mdf-1 background, we observed a wide range of viability reduction associated with the suppressor mutations (Table 2). Thus, it is reasonable to speculate that the viability of the suppressor strains is affected by other consequences of a suppressor mutation in addition to mitotic delay.
The cell cycle of early embryonic cells in C. elegans is rapid, consists entirely of S phase and mitosis, and lacks gap phases. The cell divisions in the early C. elegans embryo are asymmetric and asynchronous (Figures 2A and 3A), which is crucial for normal development and cell fate specification. It is unclear whether or not C. elegans embryos have functional S- and M-phase checkpoints. However, several lines of evidence suggest that the S-phase checkpoint (atl-1 and chk-1) is functional and developmentally controlled by preferential activation in the P1 blastomere to generate the 2-min asynchrony (Brauchle et al. 2003). Attenuated asynchrony in chk-1(RNAi) and div-1 mutants (DNA polymerase-α) results in sterility and embryonic lethality, respectively (Encalada et al. 2000; Brauchle et al. 2003). Recently, Holway et al. (2006) showed that the S-phase checkpoint is actively silenced during the DNA damage response in the early C. elegans embryo to ensure normal timing of cell division, even in the presence of heavily damaged chromosomes. In contrast, the duration of mitosis is consistent in early C. elegans embryonic cells (Encalada et al. 2000, 2005 and this report) and the activated mitotic checkpoint will delay progression through mitosis in response to either chemical or mutational disruption of the embryonic microtubule cytoskeleton (Encalada et al. 2005). The average duration of these delays does not exceed 2.5-fold even after treatment with the microtubule-destabilizing drug nocodazole (Encalada et al. 2005). In accordance with these data, we did not observe >2.1-fold mitotic delays in the suppressors. We also observed that the first two embryonic cell divisions in the suppressor mutants remained asynchronous (Figures 2 and 3). Our observations suggest that an M-phase delay is tolerated by a C. elegans embryo better than an S-phase delay. For instance, the such-1(h1960) suppressor delays progression through mitosis >2-fold, yet it is viable (Table 2). The defects observed in such-1(h1960) embryos, as well as the rest of the suppressor mutations (Table 2), are mild compared to the embryonic lethality associated with S-phase delays in the presence of replication problems (Encalada et al. 2000; Brauchle et al. 2003; Holway et al. 2006). These data suggest that the SAC might not be involved in developmental regulation contributing to the asynchrony of the early embryonic cells. Thus, the suppressors that delay mitotic progression might shed light on the relationship between S- and M-phase checkpoints, the embryonic sensitivity to delayed interphase, and the embryonic tolerance to 2-fold mitotic delays.
Three suppressors, such-5, -6, and -7, rescue the mdf-1 lethality without delaying anaphase onset. The normal mitotic timing observed in these mutants suggests that these suppressors rescue the lethality by an alternate mechanism. Indeed, we showed that these suppressors do not accumulate securin, further suggesting normal APC/C activity. It is possible that mutations in the components downstream of APC/C would rescue mdf-1 lethality without securin accumulation; however, we would expect mutants in the downstream components to delay anaphase onset. In mouse cells, it was shown that lethality associated with deletion of the Mad2 gene can be suppressed by the deletion of p53 to yield viable Mad2−/− p53−/− cells, which exhibit extraordinarily high level of CIN (Burds et al. 2005). The authors showed that the deletion of p53 did not alter mitotic timing in Mad2−/− cells (Burds et al. 2005). In support of these findings, Wang et al. (2004) showed that haploid loss of the p53 gene rescued the embryonic lethality in mice caused by targeted deletion of Brca1, exon 11 (Brca1Δ11/Δ11), and animals developed into adults exhibiting increased tumorigenesis and chromosomal abnormalities. They further showed that Brca1 controls the spindle checkpoint by regulating the expression of MAD2. Both studies speculated that p53 might have a role in sensing ploidy. It is possible that mutations that result in defective apoptosis could rescue mdf-1 lethality and progress through mitosis normaly; however, ced-3 and cep-1 mutants are not capable of rescuing the mdf-1(gk2) lethality in C. elegans (our unpublished data). Alternatively, mutations in genes involved in checkpoint activity could partialy compensate for MDF-1 loss without delaying mitotic timing.
In the case of the such genes that delay mitotic progression, mapping and sequencing suggest that they will identify genes not previously known to function in the metaphase-to-anaphase transition in C. elegans. This is supported by the finding that such-1(h1960) is a new gene not previously described to have a role in the metaphase-to-anaphase transition. It is also likely that mutational inactivation of processes other than the metaphase-to-anaphase transition—for example, transcriptional regulation—could result in delayed mitosis. In human cancer cells lesions in the retinoblastoma pathway lead to Mad2 overexpression through the E2F family of transcription factors. The aberrantly expressed Mad2 displayed a significant, twofold longer mitosis, as a result of delayed degradation of securin and cyclin B (Hernando et al. 2004).
Our analysis has identified a new APC/C component that rescues mdf-1(gk2) lethality. such-1 encodes an APC5-like product not previously described. Many of the APC/C subunits have been identified in C. elegans (reviewed in Yeong 2004). All of the subunits when inactivated by dsRNA resulted in embryonic lethality at the meiotic one-cell stage, with the exception of apc-5/M163.4 and apc-10/F15H10.3, which arrested later in development (Davis et al. 2002). The authors proposed that the C. elegans apc-5 and apc-10 components do not function as a part of the meiotic APC/C. We observed that the C. elegans genome has two functional copies of APC5: gfi-3/M163.4, which was previously described (Zachariae et al. 1998; Davis et al. 2002), and such-1/Y66D12A.17 described here. The latter suppresses mdf-1(gk2) lethality and delays anaphase onset, which clearly demonstrates its role in the metaphase-to-anaphase transition. Furthermore, we found that the apc-10/F15H10.3 gene has a paralog as well, the Y48G1C.12 gene located on chromosome I. Our analyses revealed that apc-5 and apc-10 are the only known APC/C components in C. elegans that have paralogs. Thus, it is possible that the functional redundancy of the paralogs allows embryos to progress through meiosis.
We describe here two classes of suppressors that can rescue phenotypic defects resulting from a loss of the essential metaphase-to-anaphase checkpoint. When separated from mdf-1(gk2) the suppressors are viable, but exhibit chromosomal abnormalities and variable reductions in viability. Despite rescuing the viability of animals in the absence of the MDF-1 checkpoint, all of the suppressors still display high levels of chromosomal instability. Thus, we believe that in addition to increasing our understanding of the regulation of chromosome segregation and early embryonic development this collection of mutants could also provide us with new valuable insights into cancer development.
Acknowledgments
We are grateful to Donald Riddle and David Baillie for generously sharing experimental equipment and to Nigel O'Neil for all his support. We thank the C. elegans Gene Knockout Consortium for providing deletion mutants (http://ko.cigenomics.bc.ca/) and the Caenorhabditis Genetics Center for providing the strains. We are grateful to A. Desai for sharing the anti-HCP-3 antibody. We are indebted to Sanja Tarailo, Berdjis Bahrami, and Shir Hazir for their technical assistance in the study and Marko Graovac for Figure 5, E and F. We also thank the Rose Lab members and our reviewers for helpful discussion and comments on the manuscript. This work was supported by a University Graduate Fellowship to M.T., by the Cancer Center Support grant CAO21765 from the National Cancer Institute and the American Lebanese Syrian Associated Charities to R.K., and by a grant from the Canadian Institutes for Health Research to A.M.R.
References
- Bharadwaj, R., and H. Yu, 2004. The spindle checkpoint, aneuploidy, and cancer. Oncogene 23: 2016–2027. [DOI] [PubMed] [Google Scholar]
- Boveri, T., 1914. Zur Frage der Entstehung maligner Tumoren. Gustav Fischer, Jena, Germany.
- Brauchle, M., K. Baumer and P. Gonczy, 2003. Differential activation of the DNA replication checkpoint contributes to asynchrony of cell division in C. elegans embryos. Curr. Biol. 13: 819–827. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burds, A. A., A. S. Lutum and P. K. Sorger, 2005. Generating chromosome instability through the simultaneous deletion of Mad2 and p53. Proc. Natl. Acad. Sci. USA 102: 11296–11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassada, R., E. Isnenghi, M. Culotti and G. von Ehrenstein, 1981. Genetic analysis of temperature-sensitive embryogenesis mutants in Caenorhabditis elegans. Dev. Biol. 84: 193–205. [DOI] [PubMed] [Google Scholar]
- Cheung, I., M. Schertzer, A. M. Rose and P. M. Lansdorp, 2002. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat. Genet. 31: 405–409. [DOI] [PubMed] [Google Scholar]
- Ciosk, R., W. Zachariae, C. Michaelis, A. Schevchenko, M. Mann et al., 1998. An ESP/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 93: 1067–1076. [DOI] [PubMed] [Google Scholar]
- Cleveland, D. W., Y. Mao and K. F. Sullivan, 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signalling. Cell 112: 407–421. [DOI] [PubMed] [Google Scholar]
- Davis, E. S., L. Wille, A. B. Chestnut, P. L. Sadler, D. C. Shakes et al., 2002. Multiple subunits of the Caenorhabditis elegans anaphase-promoting complex are required for chromosome segregation during meiosis I. Genetics 160: 805–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., S. Rybina, T. Muller-Reichert, A. Shevchenko, A. Shevchenko et al., 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17: 2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encalada, S. E., P. R. Martin, J. B. Phillips, R. Lyczak, D. R. Hamill et al., 2000. DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Dev. Biol. 228: 225–238. [DOI] [PubMed] [Google Scholar]
- Encalada, S. E., J. Willis, R. Lyczak and B. Bowerman, 2005. A spindle checkpoint functions during mitosis in the early Caenorhabditis elegans embryo. Mol. Biol. Cell 16: 1056–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta, T., S. Tuck, J. Kirchner, B. Koch, R. Auty et al., 2000. EMB-30: an APC-4 homologue required for metaphase to anaphase transition during meiosis and mitosis in Caenorhabditis elegans. Mol. Biol. Cell 11: 1401–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden, A., P. L. Sadler, M. R. Wallenfang, J. M. Schumacher, D. R. Hamill et al., 2000. Metaphase to anaphase (mat) transition-defective mutants in Caenorhabditis elegans. J. Cell Biol. 151: 1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold, T., and P. Hunt, 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2: 280–291. [DOI] [PubMed] [Google Scholar]
- Hernando, E., Z. Nahle, G. Juan, E. Diaz-Rodriguez, M. Alaminos et al., 2004. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature 430: 797–802. [DOI] [PubMed] [Google Scholar]
- Hodgkin, J., H. R. Horvitz and S. Brenner, 1979. Nondisjunction mutants of the nematode C. elegans. Genetics 91: 67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holway, A. H., S. H. Kim, A. LaVolpe and W. M. Michael, 2006. Checkpoint silencing during the DNA damage response in Caenorhabditis elegans embryos. J. Cell Biol. 172: 999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt, M. A., L. Totis and B. T. Roberts, 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell 66: 507–517. [DOI] [PubMed] [Google Scholar]
- Karess, R., 2005. Rod-Zw10-Zwilch: a key player in the spindle checkpoint. Trends Cell Biol. 15: 386–392. [DOI] [PubMed] [Google Scholar]
- Kitagawa, R., and A. M. Rose, 1999. Components of the spindle-assembly checkpoint are essential in Caenorhabditis elegans. Nat. Cell Biol. 1: 514–521. [DOI] [PubMed] [Google Scholar]
- Kitagawa, R., E. Law, L. Tang and A. M. Rose, 2002. The Cdc20 homolog, FZY-1, and its interacting protein, IFY-1, are required for proper chromosome segregation in Caenorhabditis elegans. Curr. Biol. 12: 2118–2123. [DOI] [PubMed] [Google Scholar]
- Kops, G. J., B. A. Weaver and D. W. Cleveland, 2005. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer 5: 773–785. [DOI] [PubMed] [Google Scholar]
- L'Hernault, S. W., 2006. Spermatogenesis, in WormBook, edited by The C. elegans Research Community (doi/10.1895/wormbook.1.7.1; http://www.wormbook.org).
- Li, R., and A. W. Murray, 1991. Feedback control of mitosis in budding yeast. Cell 66: 519–531. [DOI] [PubMed] [Google Scholar]
- McCarter, J., B. Bartlett, T. Dang and T. Schedl, 1999. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 205: 111–128. [DOI] [PubMed] [Google Scholar]
- Monen, J., P. S. Maddox, F. Hyndman, K. Oegema and A. Desai, 2005. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat. Cell Biol. 12: 1148–1155. [DOI] [PubMed] [Google Scholar]
- Nystul, T. G., J. P. Goldmark, P. A. Padilla and M. B. Roth, 2003. Suspended animation in C. elegans requires the spindle checkpoint. Science 302: 1038–1041. [DOI] [PubMed] [Google Scholar]
- Oegema, K., A. Desai, S. Rybina, M. Kirkham and A. A. Hyman, 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153: 1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, A. M., and P. Hieter, 1999. The anaphase promoting complex: new subunits and regulators. Annu. Rev. Biochem. 68: 583–609. [DOI] [PubMed] [Google Scholar]
- Praitis, V., E. Casey, D. Collar and J. Austin, 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, A. M., and D. L. Baillie, 1979. Effect of temperature and parental age on recombination and nondisjunction in Caenorhabditis elegans. Genetics 92: 409–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakes, D. C., P. L. Sadler, J. M. Schumacher, M. Abdolrasulnia and A. Golden, 2003. Developmental defects observed in hypomorphic anaphase-promoting complex mutants are linked to cell cycle abnormalities. Development 130: 1605–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin, V., G. K. T. Chan and T. J. Yen, 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, MAD2. J. Cell Biol. 154: 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., and J. D. Burke, 1995. Checkpoint genes required to delay cell division in response to nocodazole respond to impaired kinetochore function in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 6838–6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. H., H. Yu and C. X. Deng, 2004. A requirement for breast-cancer-associated gene 1 (BRCA1) in the spindle checkpoint. Proc. Natl. Acad. Sci. USA 101: 17108–17113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, E., and M. Winey, 1996. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol. 132: 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks, S. R., R. T. Yeh, W. R. Gish, R. H. Waterston and R. H. A. Plasterk, 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28: 160–164. [DOI] [PubMed] [Google Scholar]
- Yeong, F. M, 2004. Anaphase-promoting complex in Caenorhabditis elegans. Mol. Cell. Biol. 24: 2215–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae, W., A. Shevchenko, P. D. Andrews, R. Ciosk, M. Galova et al., 1998. Mass spectrometric analysis of the anaphase-promoting complex from yeast: identification of a subunit related to cullins. Science 279: 1216–1219. [DOI] [PubMed] [Google Scholar]