Abstract
In most eukaryotes, recombination of homologous chromosomes during meiosis is necessary for proper chromosome pairing and subsequent segregation. The molecular mechanisms of meiosis are still relatively unknown, but numerous genes are known to be involved, among which are many mismatch repair genes. One of them, mlh1, colocalizes with presumptive sites of crossing over, but its exact action remains unclear. We studied meiotic processes in a knockout line for mlh1 in zebrafish. Male mlh1 mutants are sterile and display an arrest in spermatogenesis at metaphase I, resulting in increased testis weight due to accumulation of prophase I spermatocytes. In contrast, females are fully fertile, but their progeny shows high rates of dysmorphology and mortality within the first days of development. SNP-based chromosome analysis shows that this is caused by aneuploidy, resulting from meiosis I chromosomal missegregation. Surprisingly, the small percentage of progeny that develops normally has a complete triploid genome, consisting of both sets of maternal and one set of paternal chromosomes. As adults, these triploid fish are infertile males with wild-type appearance. The frequency of triploid progeny of mlh1 mutant females is much higher than could be expected for random chromosome segregation. Together, these results show that multiple solutions exist for meiotic crossover/segregation problems.
MEIOSIS is the cell division that produces haploid cells from diploid stem cells. This process consists of two division steps, from which the first, meiosis I, is the reduction division. The two homologs of each chromosome separate in this step. In most eukaryotic organisms, during the prophase of meiosis I recombination occurs between homologs, which is essential for the formation of stable bivalents. This in turn is necessary for proper alignment and spindle attachment in metaphase I and subsequent cell division.
In prophase I five different stages are recognized: leptotene, zygotene, pachytene, diplotene, and diakinesis (Hunt and Hassold 2002; Marcon and Moens 2005). In leptotene, a large number of double-strand breaks are generated in each chromosome (Marcon and Moens 2005). The homologs are still apart, but begin to search for homology. Synapsis, close association of homologs by binding of synaptonemal complex (SC) proteins, starts in zygotene and is completed in pachytene. During synapsis the double-strand breaks are being repaired. Only a fraction, on average one or two breaks per pair, is repaired via homologous recombination with a nonsister chromatid of the homolog. These crossing-over sites become visible in diplotene, when desynapsis takes place. Finally, in diakinesis, crossing overs are stabilized as chiasmata, which are then the only sites that keep the chromosome pair together.
The involvement of mismatch repair (MMR) genes in meiosis first became noted in the mouse, knockouts for several of them displaying fertility problems (Wei et al. 2002). The MMR proteins function in similar types of mutS and mutL heterodimeric complexes as they do in repair of replication errors in mitotic cells (Kolas et al. 2005). However, the mutS complex that acts in recombination, mutSγ, consists of msh4 and msh5, genes that do not appear to play any role in mismatch repair (Wei et al. 2002). MutSγ binds in foci together with replication protein A (RPA) to the DNA during synapsis in zygotene and pachytene (de Vries et al. 2005; Marcon and Moens 2005). The frequency of these foci in mammals is 5- to 10-fold higher than the eventual number of crossing-over sites (Kneitz et al. 2000). MSH4 and MSH5 promote synapsis, as the mouse mutants display synapsis defects (de Vries et al. 1999; Edelmann et al. 1999; Kneitz et al. 2000). As a result of that, both sexes do not have gametes and are sterile. This is much alike the phenotype of the knockout for one of the synaptonemal complex proteins, SYCP1 (de Vries et al. 2005). In contrast, Caenorhabditis elegans msh4 and msh5 do not act before pachytene stage and are essential for crossing over, a function more like mlh1 in mammals (Zalevsky et al. 1999; Kelly et al. 2000).
The mutL complex involved in meiosis is also different from the MMR mutL complex and consists of MLH1 and MLH3, although a minor role for MLH3 in MMR has been reported (Harfe et al. 2000; Cannavo et al. 2005). The MLH1/MLH3 complex is present in distinct foci on the synapsed elements during pachytene, at a later stage than the MSH4/MSH5 complex. After MLH3 binding MLH1 is recruited (Lipkin et al. 2002; Kolas et al. 2005). The MLH1/MLH3 foci coincide in timing, number, and position with the presumptive sites of crossing over (Marcon and Moens 2003; Moens 2006). The idea is therefore that the complex stabilizes a limited number of recombination sites for repair by crossing over, while the other sites will be repaired via different mechanisms. Mlh1 and mlh3 knockout mice have very similar phenotypes in respect to meiosis (Baker et al. 1996; Edelmann et al. 1996; Woods et al. 1999; Eaker et al. 2002; Lipkin et al. 2002). Both sexes are sterile, but where males lack spermatozoa completely, females do have oocytes, which, however, hardly ever finish meiosis (Edelmann et al. 1996; Woods et al. 1999; Lipkin et al. 2002). The meiosis defect in these animals also occurs at a later time point than in msh4 and msh5 knockout mice. Synapsis is normal, but the frequency of crossing overs is dramatically reduced (Baker et al. 1996; Guillon et al. 2005), and chromosomes are mostly present as univalents instead of bivalents in metaphase I (Baker et al. 1996; Woods et al. 1999).
Zebrafish as a vertebrate model organism has been very important for the study of early embryonic development, due to its frequently mentioned advantages of external fertilization, high numbers of progeny, and easily traceable development of transparent embryos. These advantages apply similarly to meiosis research, but this direction has hardly been followed so far in zebrafish. We used our recently developed efficient reverse genetic procedure to generate a knockout for mlh1 in the fish (Wienholds et al. 2003; Wienholds and Plasterk 2004). We show here that the necessity of mlh1 for recombination during meiosis in zebrafish is similar to that in mammals, but that its absence in female meiosis results in a different bypass of meiotic problems.
MATERIALS AND METHODS
Generation of mlh1 mutant fish:
Two amplicons, covering exons 2–4 and 8–10 of the zebrafish mlh1 gene, respectively, were used for target-selected mutagenesis (Wienholds et al. 2003; Wienholds and Plasterk 2004). The obtained mutant fish in TL background was outcrossed with an AB fish, and heterozygous offspring was subsequently incrossed. Genotyping was done by amplification and resequencing, using exon 10-specific forward (5′-AGTGAAGGGCTTCATCTCC-3′) and reverse (5′-AAGTAGTGCATCTATTGAAAATG-3′) primers.
Western blot was performed using standard procedures with a monoclonal anti-human MLH1 antibody (BD Biosciences Pharmingen, Franklin Lake, NJ) and an anti-α-actin antibody (Abcam, Cambridge, UK). Human Jurkat control lysate was provided with the anti-MLH1 antibody.
Histology and morphometry:
Testes for histological and morphometric evaluation were dissected, fixed in 4% glutaraldehyde, and subsequently weighed. They were embedded in 2-hydroxyethyl methacrylate, and 4-μm sections were stained with toluidine blue.
For morphometric analysis, the weights of various testicular tissue components were determined by light microscopy using a 441-intersection grid placed in the ocular of the light microscope. Fifteen fields chosen randomly (6615 points) were scored for each animal at 400× magnification. Staining or cutting artifacts were rarely seen and were not considered in the total number of points utilized to obtain weights. Points were classified as one of the following: spermatogonia, primary spermatocytes, secondary spermatocytes, spermatids, spermatozoa, empty spaces, abnormal meiotic figures, apoptosis, and others (somatic cells, blood and lymphatic vessels, and connective tissue). The weight (milligrams) of each testis component was determined as the product of the volume density (%) and the testis weight. To obtain a more precise measure of testis weight the testis capsule, the efferent ductules, and connective tissue associated with testes were excluded from the testis weight using image analysis. For that, six longitudinal testis sections were made and evaluated (∼80 μm apart from each other) for each animal, considered to represent the entire testes. All data are presented as the mean ± SEM and analyzed via ANOVA (Newman–Keuls test). The significance level in comparisons was considered to be P < 0.05.
TUNEL labeling was performed on tissue fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned at 5 μm thickness. A commercial in situ cell-death detection kit (In Situ Cell Death detection kit, POD; Roche Diagnostics, Mannheim, Germany) was used. After labeling, the slides were counterstained with Mayer hematoxylin and mounted.
Spermatocyte spreads and immunocytology:
Spermatocyte spreads were performed as described previously (Moens 2006). Testes were dissected and suspended in 50 μl phosphate-buffered saline (PBS). One microliter of cell suspension was added to 30 μl of 1:2 PBS:mQ water on each well of a multiwell slide. After settling for 20 min, the cells were fixed for 3 min in 2% paraformaldehyde (PFA) + 0.03% SDS, fixed for another 3 min in 2% PFA at pH 7.5–8, and washed in 0.4% Photo-Flo 200 (Kodak, Rochester, NY) or Agepon (AGFA, Mortsel, Belgium) and dried.
For immunocytology, the slides were blocked in 10% antibody dilution buffer and incubated in primary antibodies in antibody dilution buffer (10% goat serum, 3% bovine serum albumin, 0.05% Triton X-100 in PBS) for 2–3 hr or overnight. After washing, the cells were treated with secondary antibodies for 1 hr at 37°, washed and dried, and mounted in ProLong Antifade mounting agent (Molecular Probes, Carlsbad, CA) or DAPI vectashield (Vector Labs, Burlingame, CA). Primary antibodies for SYCP1, SYCP3, and SMC3 for visualization of SCs, for MLH1, and CREST serum to detect centromeres were described previously (Moens 2006).
Synapsis was visualized by silver staining, by putting a few drops of 40% silver nitrate on the slide and covering it with a piece of fine nylon mesh. Slides were stained in a 60° oven for 30–60 min until they turned brown.
SNP sequencing:
SNPs for detection of aneuploidy were selected from a large set of amplicons previously used for SNP verification (Guryev et al. 2006). A nearly genomewide set of 96 SNPs was compiled on two criteria: (1) being polymorphic between a homozygous mutant female and a wild-type male and (2) showing a quantitative relationship to artificial mixtures of parental DNA in 1:1 and 1:2 proportions. This set covered 22 of 25 zebrafish chromosomes in the Zv4 genome assembly of Ensembl. SNP database numbers are provided in supplemental Table 2 at http://www.genetics.org/supplemental/. Embryonic DNA was amplified and sequenced for these SNPs. Peak area proportions were determined visually. They were partly confirmed by calculating sequence trace peak areas electronically (V. Guryev, personal communication; h094.niob.knaw.nl/peakarea).
Chromosome spreads:
Twenty-four-hour postfertilization (hpf) embryos were dechorionated and incubated for 90 min in colchicine to arrest cells in metaphase. After hypotonic treatment in 1.1% sodium citrate and fixation in 3:1 methanol:acetic acid, cells were suspended in 50% HAc and spread onto glass slides. Slides were mounted and stained with DAPI vectashield (Vector Labs).
RESULTS
Generation of mlh1 knockout zebrafish:
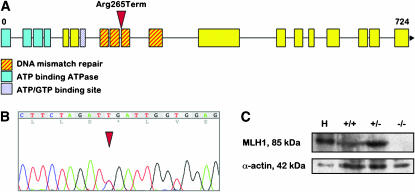
The mlh1 gene in zebrafish was annotated on the basis of homology with other species and consists of 19 coding exons. Using an ENU-driven target-selected mutagenesis screen, we isolated an individual that has a C-to-T transition in exon 10, thereby introducing a stop codon at position 265 (Figure 1, A and B). This individual was crossed out, and the heterozygous offspring subsequently crossed in to generate homozygous mutant zebrafish. Homozygous mutants were obtained with the expected frequency of 25%, and they had normal appearance. The sex ratio in the homozygote population was not significantly different from the other genotypes. The knockout phenotype was confirmed at the protein level by Western blotting, where no full-length MLH1 protein was detectable in testes of a homozygous mutant male (Figure 1C). As expected, homozygous mutant fish develop tumors, which become visible at ∼6 months of age (H. Feitsma and E. Cuppen, unpublished results).
Figure 1.—
The generation of mlh1 knockout zebrafish. (A) Genomic organization of the zebrafish mlh1 gene. The position of the ENU-induced point mutation introducing a premature stop codon is marked by an arrowhead. (B) Sequence trace showing the C-to-T transition in the heterozygous founder fish. (C) Western blot of testes proteins and human Jurkat control lysate (H) stained with antibodies for MLH1 (85 kDa) and α-actin (42 kDa). MLH1 protein is completely absent in mlh1−/− testes.
Male mutant phenotype:
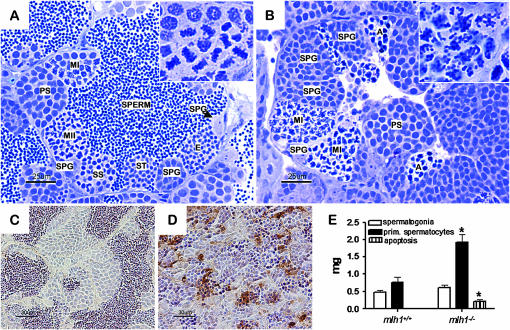
Male mlh1 mutant zebrafish showed normal mating behavior and could induce egg lays from females, but none of the eggs became fertilized. Histological analysis of the testes revealed that postmeiosis I stages of spermatogenesis, that is, secondary spermatocytes, spermatids, and spermatozoa, as visible in the wild type (Figure 2A), were completely absent in the mutant (Figure 2B). In metaphase of meiosis I, wild-type primary spermatocytes showed strongly condensed chromosomes perfectly aligned on the metaphase plate (Figure 2A, inset). In contrast, in mutant primary spermatocytes, chromosomes were dispersed throughout the nucleus (Figure 2B, inset). Also, in mutant testis groups of cells with strongly condensed nuclei could be observed, which are cells in apoptosis (Figure 2B). The high incidence of apoptotic spermatocytes in mutants was confirmed by TUNEL staining (Figure 2D), while wild-type testis showed very low levels of apoptosis (Figure 2C).
Figure 2.—
Testis histology of mlh1−/− zebrafish. (A and B) Cross section of seminiferous tubule in mlh1+/+ (A) and mlh1−/− (B) zebrafish, showing spermatogenic cysts with different types of germ cells, spermatogonia at different mitotic divisions (SPG), primary spermatocytes (PS), first meiotic division (MI), secondary spermatocytes (SS), second meiotic division (MII), spermatids (ST), spermatozoa (SPERM), and apoptotic spermatocytes (A). Note the absence of postmeiosis I stages and the presence of apoptotic spermatocytes with strongly condensed nuclei in mlh1−/− testis. Inset shows a magnified view of the first meiotic division. In the wild type, chromosomes are aligned at the cell equator just prior to division. In the mutant, chromosomes are randomly distributed throughout the nucleus. (C and D) TUNEL staining of mlh1+/+ (C) and mlh1−/− fish (D). High numbers of apoptotic spermatocytes can be seen in mutant testis. Wild-type testis shows a very low incidence of apoptosis. (E) Morphometric analysis of testes sections. The mutant shows a large increase in amounts of spermatocytes and apoptotic cells, but not of spermatogonia.
Quantitative morphometric analysis of testis tissue components (six males for each group) showed that apoptotic germ cells were indeed more prominent in the mutant (Figure 2E; supplemental Table 1 at http://www.genetics.org/supplemental/), and a high incidence of abnormal meiotic figures was observed. More strikingly, we observed a strong accumulation (∼150%) of primary spermatocytes in mutant testis, whereas the number of spermatogonia did not increase significantly (Figure 2E; supplemental Table 1). As a result, absolute and relative (expressed as percentage of body weight) testis weight was higher in mutants. For all parameters, no difference between wild-type and heterozygous animals was observed (data not shown).
Synapsis and crossovers:
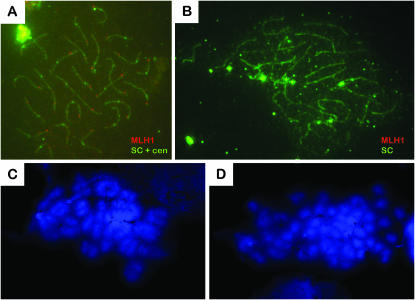
To analyze further the progress of prophase I in mlh1 mutant males, we performed immunocytology experiments on meiotic chromosome spreads of spermatocytes to follow synapsis of homologous chromosomes. The formation of the SC, which starts from the chromosome ends in zygotene and is completed in early pachytene (Figure 3A), was normal in mutants (Figure 3B). MLH1 foci, which are at the positions of crossing over, can clearly be seen in wild-type spermatocytes (Figure 3A). Their number and distribution per spermatocyte were similar to previous findings (Moens 2006), where 1 or occasionally 2 foci per synapsed pair were seen, with in total 147 foci on 140 SCs analyzed. As expected, foci were completely absent in the mutant (Figure 3B).
Figure 3.—
Chiasmata formation but not synapsis is defective in mlh1−/− spermatocytes. (A and B) Twenty-five synapsed pairs of chromosomes are visible in pachytene spermatocytes of both wild-type (A) and mutant males (B). SCs and centromeres (cen) are stained with green fluorescence, labeled with FITC. MLH1 is fluorescent red with rhodamine. One or two distinct MLH1 foci per synapsed pair can be seen in wild-type cells, but are absent in mutants. (C and D) DAPI-stained metaphase spermatocytes have bivalents in wild types (C) and mostly univalents in mlh1−/− mutants (D).
In diplotene, synaptonemal complex proteins have dissociated from the chromosomes. In the wild-type situation, bivalents that are held together by one or two chiasmata per chromosome pair could be seen (Figure 3C), but in the mutant most chromosomes were present as univalents (Figure 3D). This indicates a failure in the stabilization of crossovers.
Female mutant phenotype:
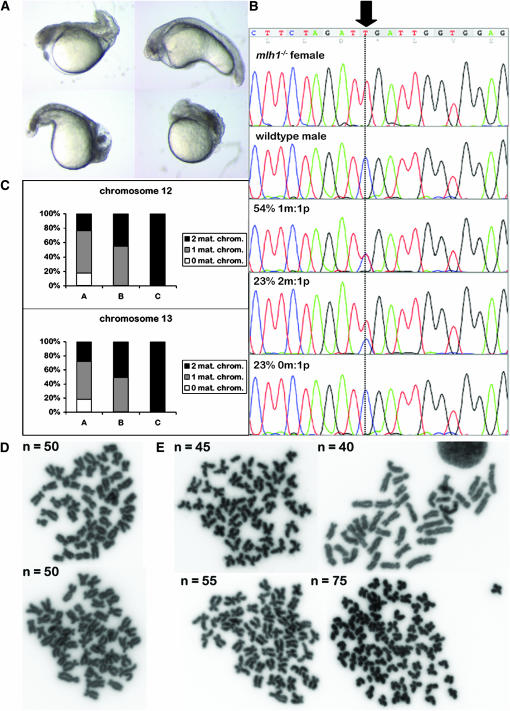
Mlh1−/− female zebrafish also showed normal mating behavior. They had average-size clutches that were normally fertilized. Ovary histology revealed no differences from wild type, including no increased apoptosis (data not shown). Fertilized eggs started to develop normally, but embryos showed high rates of dysmorphology and lethality within the first days postfertilization (dpf) (Figure 4A). Only 1–20% per cross were still alive and healthy at 7 dpf.
Figure 4.—
Progeny from mlh1−/− females is severely aneuploid. (A) At 24 hpf, embryos from an mlh1−/− female are strongly malformed and show necrosis and apoptosis. (B) Quantitative genotyping of a single-nucleotide polymorphism that has different alleles in a mutant female (top section) and in a wild-type male (second section) and its progeny. Only 54% of progeny is heterozygous at this position (third section). Forty-six percent has either no or two maternal chromosomes (bottom two sections). m, maternal; p, paternal. (C) Quantitative SNP typing of two chromosomes in embryos from four crosses that were assigned to different categories. Category A (n = 407) contained all embryos that died before day 4 or were unable to hatch. Category B (n = 63) embryos had hatched at 4 dpf but were clearly malformed and/or did not develop a swim bladder. Category C (n = 65) contained only healthy embryos at 4 dpf. The frequency of 0, 1, or 2 maternal chromosomes is plotted for each category. (D and E) Chromosome spreads of cells of 24-hpf embryos. Embryos from wild-type females (D) have the normal diploid number of 50 chromosomes per cell. Embryos from mutant females (E) have abnormal chromosome numbers, up to 75, which is equal to triploidy.
We crossed a mutant female with a wild-type male and genotyped 44 of their progeny at 24 hpf for the point mutation in the mlh1 gene. Heterozygosity at this position for all embryos was expected, but in contrast to that we observed that a large fraction have two or no copies of the maternal allele, as quantified by the peak height in the sequence trace (Figure 4B), indicating chromosomal missegregation. To extend this observation to other chromosomes, we selected a set of SNPs that were distributed throughout the genome (Guryev et al. 2006) and homozygously different between the parental mlh1−/− female and the wild-type male. This set covered 22 of 25 zebrafish chromosomes. Quantitative SNP analysis on 16 4-day-old embryos of a cross of these two animals revealed that all 16 deviated severely from the normal diploid chromosome number (Table 1), showing missegregation of on average 14 of 22 chromosomes. This effect could always be assigned to the maternal contribution of chromosomes, as in all cases only a single paternal copy was present for each chromosome. As a comparison, only 3 cases of 36 progeny from heterozygous females were aneuploid and no aneuploids were found in 30 progeny from wild-type females (Table 1). A second category of SNPs, being heterozygous in the mutant female, was used to be able to distinguish the two maternal homologs of a chromosome. We found that in trisomies always both maternal alleles were present, indicating a meiosis I defect and not meiosis II (data not shown).
TABLE 1.
Aneuploidy and triploidy in progeny of mlh1−/− females
| Maternal genotype (no. of embryos)a | No. of polymorphic markersb | No. of chromosomes coveredb | % euploidc | % aneuploidc | % triploidc |
|---|---|---|---|---|---|
| −/− (16) | 48 | 22 | 0 | 75 | 25 |
| +/− (36) | 31/51/40d | 19/21/17d | 92 | 8 | 0 |
| +/+ (30) | 34 | 16 | 100 | 0 | 0 |
Progeny of three outcrosses with females of the indicated genotype and wild-type males was genotyped for 96 SNP markers. The number of embryos assayed per genotype is given in parentheses.
The number of polymorphic loci and hence the number of chromosomes covered differs between crosses because different founders were used and zebrafish are highly outbred.
Progeny was classified as euploid when all markers gave the expected 1:1 heterozygote ratio for all SNPs and as aneuploid when at least one marker revealed only the paternal allele or when the ratio between male vs. female alleles was 1:2. Triploidy was assigned to those embryos for which all polymorphic markers had a 1:2 ratio between male and female alleles.
Three different crosses were used.
Surprisingly, 4 of the 16 mutant progeny had two maternal copies of each chromosome tested, together with one set of paternal chromosomes, which strongly suggests a complete triploid genome (Table 1). To determine the effect of triploidy on embryonic survival, we assigned categories of embryos with increasing survival rates and typed embryos from five pair crosses for two SNPs on two different chromosomes (Figure 4C). Category A (n = 407) contained all embryos that died before day 4 or were unable to hatch. Category B (n = 63) embryos had hatched at 4 dpf but were clearly malformed and/or did not develop a swim bladder. Finally, category C (n = 65) contained only healthy embryos at 4 dpf. Embryos from category A had no or two maternal copies of the analyzed chromosomes in ∼50% of the cases. For a single embryo this would mean that it has no or two maternal copies of around half of its chromosomes. Embryos that survived longer in general had more chromosomes. Category B embryos had one or two maternal chromosomes and embryos from C had two maternal copies of both chromosomes in 100% of the cases. The latter will thus most likely have two maternal copies of all chromosomes and will therefore be the triploid embryos. Indeed, the fraction of embryos in category C is similar to the fraction of triploids found in the more extensively genotyped set.
To confirm the abnormal chromosome numbers, we performed chromosome counts in cells of 24-hpf embryos of similar crosses. Offspring from wild-type females had the normal diploid number of 50 chromosomes per cell (Figure 4D). In mutant female progeny different cells of one embryo had the same chromosome number, but this number varied greatly between embryos (Figure 4E). We also observed cases of 75 chromosomes per cell, confirming the frequent existence of fully triploid embryos.
Triploid progeny:
From a new cross of an mlh1−/− female with a wild-type male, we followed the embryos of the triploid category over a longer period. From 20 healthy embryos at 7 dpf, 17 survived until adulthood with normal appearance, and all were males. Partial SNP typing confirmed triploidy in 16 of these animals. These 16 males had greatly reduced fertility. Some were completely infertile, others were able to fertilize at maximum 3% of eggs, and all eggs that were fertilized developed dysmorphologically and died before 7 dpf.
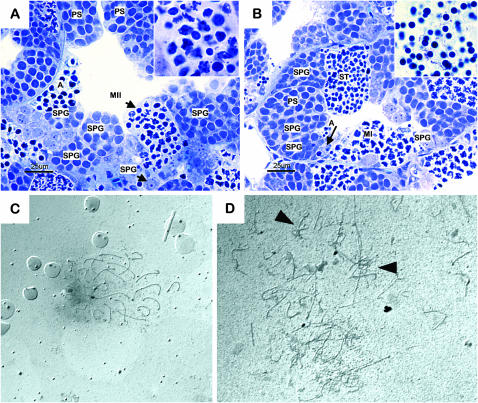
Morphological analysis of testes showed that mitotic proliferation of germ cells appeared normal in the triploid males. However, although some germ cells were able to go through both meiotic divisions and in some animals a few mature sperm were found, overall only very low counts of spermatids and spermatozoa were observed (Figure 5, A and B). Furthermore, abundant apoptosis of primary spermatocytes was seen (Figure 5, A and B), as well as an unusual cellular composition of cysts during the second meiotic division, in which cells had lost synchrony and were in different developmental stages (Figure 5A, inset). Silver-stained spermatocyte spreads revealed that synapsis was strongly disorganized in triploid animals (Figure 5D) as compared to wild type (Figure 5C). Obviously, >25 chromosomal structures are present in the triploid, and irregular synapsis figures can be seen.
Figure 5.—
Triploid males have low fertility due to synapsis problems. (A and B) Cross sections of seminiferous tubules in infertile (A) and fertile (B) triploid zebrafish, with spermatogonia (SPG), primary spermatocytes (PS), second meiotic division (MII), and apoptotic germ cells (A). Inset in A: magnification of second meiotic division; note the concomitant presence of secondary spermatocytes, cells in second meiotic division, and spermatids. Inset in B: magnification of spermatozoa in the efferent ductules. (C and D) Synapsis in triploid spermatocytes (D) is strongly disorganized as compared to that in wild type (C). Irregular synapsis figures are indicated by arrowheads.
DISCUSSION
To obtain better insight into the role of MMR proteins in meiotic processes, we have generated zebrafish that are deficient in MLH1 protein. We found that male mlh1 mutants are sterile because spermatogenesis is arrested at the primary spermatocyte stage. A defect in chiasmata formation causes chromosomes to be unpaired at metaphase I, and therefore no proper chromosome alignment, spindle attachment, and subsequent cell division are possible. This indicates that the function of MLH1 in zebrafish is similar to that in mammals and is associated with stabilization of recombination sites that are repaired via crossing over. The time point of arrest in mutant males is also similar to what was reported in mice (Eaker et al. 2002). The defect in formation of crossing overs occurs already in late pachytene, but it is recognized as a problem by the cell only in metaphase, when the high number of univalents does not allow cell division to progress. Apparently, after synapsis, spindle assembly is the next checkpoint for meiotic division.
We observed a large accumulation of primary spermatocytes in mutant testes. This was also reported for mouse, albeit not in a quantitative manner (Eaker et al. 2002). The accumulation reflects a prolonged period during which mutant cells are arrested, eventually leading to apoptotis. We did not observe a significant difference in the amount of spermatogonia, indicating that the mitotic stages of spermatogenesis proceed normally. Even though the fate of spermatocytes in zebrafish resembles that in mouse mlh1 loss-of-function mutants, a difference was observed with respect to the mutants' testis weight, which increased in zebrafish but decreased in mice (Edelmann et al. 1996; Lipkin et al. 2002). A possible explanation for this difference may be found in a different relative capacity of Sertoli cells to remove dead germ cells, which could be lower in zebrafish. On the other hand, there was very little cellular debris in the tubular lumen of zebrafish mutant testes, indicating that the Sertoli cells' capacity was not oversaturated.
In females, the situation in zebrafish is markedly different from that in mammals, as female mlh1 mutant zebrafish show normal fertility. This seems, however, rather a result of zebrafish biology than of the existence of different mechanisms in zebrafish. Mouse female mlh1 mutants do have a normal ovary and fully developed oocytes. A small part of these oocytes is able to ovulate and extrude the first polar body, which means in fact that they can finish meiosis I, despite the lack of chiasmata (Edelmann et al. 1996; Woods et al. 1999). The result is an aneuploid oocyte, which, upon fertilization, is hardly able to finish the second meiotic division and subsequent mitotic divisions (Woods et al. 1999). Oocytes from zebrafish mlh1 mutants show the same error-prone finishing of meiosis I resulting in aneuploid eggs. The difference in this model is that after fertilization the embryo makes little use of its own genome initially, but instead develops on maternally supplied mRNAs and proteins from the yolk. Transcription from the zygotic genome does not commence before ∼3 hpf (Duffy et al. 2005) and maternal supplies are active until much later (Wagner et al. 2004). In this respect, severe aneuploidy would be tolerable during the first hours of development. Only after the embryonic genome “starts up” will problems caused by abnormal chromosomal content become manifest. This has also been seen in other cases, such as for the futile cycle mutant, in which oocytes fail to undergo nuclear division but continue cell division, resulting in many anucleate cells (Dekens et al. 2003).
The difference between sexes in this case is more striking, but this has been reported for many meiotic mutants (reviewed in Hunt and Hassold 2002). Male meiosis is an ongoing, regulated process that can be highly selective on gametes as they are produced in large numbers in successive generations during the male's reproductive life. Female meiosis is much more programmed, producing only one generation of oocytes during development, which is stored in dictyotene until ovulation. To reproduce, the female needs oocytes of this pool to finish meiosis, but that comes with a cost of higher error rates. Recent studies on zebrafish female meiosis suggest that also in zebrafish, oogenesis takes place predominantly in young females (N. Kochakpour, personal communication). The differences in female fertility phenotypes in the different MMR mutants can also partly be explained as a difference in timing of the defects. In mlh1 mutants, for both sexes the problem occurs in pachytene, but the defect becomes noted only in metaphase. In oogenesis, however, dictyotene arrest is in diplotene, so just prior to the problem. The first meiotic division is finished only at ovulation. This explains why female mutants still have adult germ cells and males do not. It contrasts with msh4 and msh5 mouse mutants, where the defect already occurs in zygotene and early pachytene, and well before diplotene, and where female mutants do not have oocytes. We predict that female msh4 mutants in zebrafish would also be fully sterile.
We found that progeny from mlh1−/− females had high levels of aneuploidy. The overall frequency of missegregation of chromosomes was close to 50%, which suggests random chromosome segregation. That is consistent with the observed unpairing of homologs in male zebrafish and male and female mice, due to the absence of chiasmata. As a result, chromosomes are dispersed throughout the nucleus in metaphase of MLH1-deficient spermatocytes. In the subsequent division each chromosome might randomly go to either side, which would result in an average missegregation frequency of 50%. However, the chance then for all chromosomes to go to one side, which would result in triploid embryos, would be (0.5)50 = <10−15. We find triploids at a much higher frequency of up to 25%. Several processes could be proposed to explain the high frequency of triploids. Possibly, mechanisms exist to circumvent meiotic problems and one of them could be complete skipping of the first meiotic division when it is strongly delayed due to, for example, spindle defects. In this case, however, we do also see many cases where embryos are close to triploidy but miss one or more chromosomes. Another explanation could be that chromosomes have a tendency to go to one spindle pole. An important characteristic of female meiosis is that both divisions have the spatially unequal result of an oocyte of original size and a small polar body. If chromosomes are not aligned at the metaphase plate but are randomly spaced, they might not be pulled properly to the polar body pole. Then it would be spatially more likely that most chromosomes stay within the oocyte. In line with this, human embryos resulting from maternal meiosis I missegregation commonly had extra chromosomes rather than missing chromosomes (Kuliev and Verlinsky 2004). In addition to this, monopolar spindles are frequent in female gametes (Woods et al. 1999), but absent in males. More generally, one could also speculate that it could be an advantage to keep most chromosomes in the oocyte and less in the polar body, since the latter will be degraded anyway. In our study we did, however, also see embryos with <25 chromosomes from the mother, which indicates that this does not hold true for all cases.
Interestingly, triploid but not aneuploid zebrafish embryos were found to develop normally. The fact that they were all males is striking, but the numbers are too small to draw conclusions, as sex ratios in zebrafish are known to vary highly between crosses. Breeding of more triploid zebrafish is necessary to verify if this is a fully penetrant phenotype. Polyploid individuals of many lower animal species and plants are viable (Meneely 1994; Marian 1997; Henry et al. 2005), but in mammals they generally are not. However, in humans, where triploidy is often due to double sperm fertilizations, triploid embryos reach further developmental stages than most aneuploid ones, and rare cases of shortly surviving triploid humans have been reported (Hasegawa et al. 1999).
Triploidy in fish can be induced by applying early pressure or heat shock of oocytes to block the second meiotic division (Marian 1997; Mizgireuv et al. 2004). In zebrafish, gametogenesis has not been studied before in triploids, but in rainbow trout (Carrasco et al. 1998) and sea bass (Felip et al. 2001) it was shown to result in compromised fertility. As expected, we now also see in zebrafish that spermatogenesis is impaired due to disorganized synapsis, which is a logical consequence of the odd chromosome number. Apparently, spermatogenesis is not completely blocked, but the few resulting spermatozoa are aneuploid, resulting in nonviable embryos upon fertilization.
Taken together, we illustrate the versatility of zebrafish as a model for studying meiotic processes and defects. Our results on meiotic segregation problems in zebrafish show interesting similarities to mechanisms underlying human miscarriages, especially with respect to the female origin of aneuploidies and the substantial fraction of triploids within those.
Acknowledgments
The authors thank E. de Bruijn for help obtaining mutant fish, V. Guryev for providing SNP primers and SNP quantification tools, M. Verheul for technical assistance, N. Kochakpour for kindly sharing her findings on zebrafish female meiosis, and M. Tijsterman for critically reading the manuscript. This work was supported by funds from the Cancer Genomics Center (Nationaal Regie Orgaan Genomics), the European Union-funded FP6 Integrated Project ZF-MODELS, and the Brazilian Coordenação de Aperfeiçoamento de Pessoal de Nivel Superior foundation.
References
- Baker, S. M., A. W. Plug, T. A. Prolla, C. E. Bronner, A. C. Harris et al., 1996. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 13: 336–342. [DOI] [PubMed] [Google Scholar]
- Cannavo, E., G. Marra, J. Sabates-Bellver, M. Menigatti, S. M. Lipkin et al., 2005. Expression of the MutL homologue hMLH3 in human cells and its role in DNA mismatch repair. Cancer Res. 65: 10759–10766. [DOI] [PubMed] [Google Scholar]
- Carrasco, L. A., S. Doroshov, D. J. Penman and N. Bromage, 1998. Long-term, quantitative analysis of gametogenesis in autotriploid rainbow trout, Oncorhynchus mykiss. J. Reprod. Fertil. 113: 197–210. [DOI] [PubMed] [Google Scholar]
- Dekens, M. P., F. J. Pelegri, H. M. Maischein and C. Nusslein-Volhard, 2003. The maternal-effect gene futile cycle is essential for pronuclear congression and mitotic spindle assembly in the zebrafish zygote. Development 130: 3907–3916. [DOI] [PubMed] [Google Scholar]
- de Vries, S. S., E. B. Baart, M. Dekker, A. Siezen, D. G. de Rooij et al., 1999. Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev. 13: 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries, F. A., E. de Boer, M. van den Bosch, W. M. Baarends, M. Ooms et al., 2005. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 19: 1376–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy, K. T., M. F. McAleer, W. R. Davidson, L. Kari, C. Kari et al., 2005. Coordinate control of cell cycle regulatory genes in zebrafish development tested by cyclin D1 knockdown with morpholino phosphorodiamidates and hydroxyprolyl-phosphono peptide nucleic acids. Nucleic Acids Res. 33: 4914–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaker, S., J. Cobb, A. Pyle and M. A. Handel, 2002. Meiotic prophase abnormalities and metaphase cell death in MLH1-deficient mouse spermatocytes: insights into regulation of spermatogenic progress. Dev. Biol. 249: 85–95. [DOI] [PubMed] [Google Scholar]
- Edelmann, W., P. E. Cohen, M. Kane, K. Lau, B. Morrow et al., 1996. Meiotic pachytene arrest in MLH1-deficient mice. Cell 85: 1125–1134. [DOI] [PubMed] [Google Scholar]
- Edelmann, W., P. E. Cohen, B. Kneitz, N. Winand, M. Lia et al., 1999. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat. Genet. 21: 123–127. [DOI] [PubMed] [Google Scholar]
- Felip, A., F. Piferrer, M. Carrillo and S. Zanuy, 2001. Comparison of the gonadal development and plasma levels of sex steroid hormones in diploid and triploid sea bass, Dicentrarchus labrax L. J. Exp. Zool. 290: 384–395. [DOI] [PubMed] [Google Scholar]
- Guillon, H., F. Baudat, C. Grey, R. M. Liskay and B. de Massy, 2005. Crossover and noncrossover pathways in mouse meiosis. Mol. Cell 20: 563–573. [DOI] [PubMed] [Google Scholar]
- Guryev, V., M. J. Koudijs, E. Berezikov, S. L. Johnson, R. H. Plasterk et al., 2006. Genetic variation in the zebrafish. Genome Res. 16: 491–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe, B. D., B. K. Minesinger and S. Jinks-Robertson, 2000. Discrete in vivo roles for the MutL homologs Mlh2p and Mlh3p in the removal of frameshift intermediates in budding yeast. Curr. Biol. 10: 145–148. [DOI] [PubMed] [Google Scholar]
- Hasegawa, T., N. Harada, K. Ikeda, T. Ishii, I. Hokuto et al., 1999. Digynic triploid infant surviving for 46 days. Am. J. Med. Genet. 87: 306–310. [DOI] [PubMed] [Google Scholar]
- Henry, I. M., B. P. Dilkes, K. Young, B. Watson, H. Wu et al., 2005. Aneuploidy and genetic variation in the Arabidopsis thaliana triploid response. Genetics 170: 1979–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, P. A., and T. J. Hassold, 2002. Sex matters in meiosis. Science 296: 2181–2183. [DOI] [PubMed] [Google Scholar]
- Kelly, K. O., A. F. Dernburg, G. M. Stanfield and A. M. Villeneuve, 2000. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics 156: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneitz, B., P. E. Cohen, E. Avdievich, L. Zhu, M. F. Kane et al., 2000. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 14: 1085–1097. [PMC free article] [PubMed] [Google Scholar]
- Kolas, N. K., A. Svetlanov, M. L. Lenzi, F. P. Macaluso, S. M. Lipkin et al., 2005. Localization of MMR proteins on meiotic chromosomes in mice indicates distinct functions during prophase I. J. Cell Biol. 171: 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuliev, A., and Y. Verlinsky, 2004. Meiotic and mitotic nondisjunction: lessons from preimplantation genetic diagnosis. Hum. Reprod. Update 10: 401–407. [DOI] [PubMed] [Google Scholar]
- Lipkin, S. M., P. B. Moens, V. Wang, M. Lenzi, D. Shanmugarajah et al., 2002. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat. Genet. 31: 385–390. [DOI] [PubMed] [Google Scholar]
- Marcon, E., and P. Moens, 2003. MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics 165: 2283–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcon, E., and P. B. Moens, 2005. The evolution of meiosis: recruitment and modification of somatic DNA-repair proteins. BioEssays 27: 795–808. [DOI] [PubMed] [Google Scholar]
- Marian, L. A., 1997. Production of triploid transgenic zebrafish, Brachydanio rerio (Hamilton). Indian J. Exp. Biol. 35: 1237–1242. [PubMed] [Google Scholar]
- Meneely, P. M., 1994. Sex determination in polyploids of Caenorhabditis elegans. Genetics 137: 467–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizgireuv, I. V., I. G. Majorova, V. M. Gorodinskaya, V. V. Khudoley and S. Y. Revskoy, 2004. Carcinogenic effect of N-nitrosodimethylamine on diploid and triploid zebrafish (Danio rerio). Toxicol. Pathol. 32: 514–518. [DOI] [PubMed] [Google Scholar]
- Moens, P. B., 2006. Zebrafish: chiasmata and interference. Genome 49: 205–208. [DOI] [PubMed] [Google Scholar]
- Wagner, D. S., R. Dosch, K. A. Mintzer, A. P. Wiemelt and M. C. Mullins, 2004. Maternal control of development at the midblastula transition and beyond: mutants from the zebrafish II. Dev. Cell 6: 781–790. [DOI] [PubMed] [Google Scholar]
- Wei, K., R. Kucherlapati and W. Edelmann, 2002. Mouse models for human DNA mismatch-repair gene defects. Trends Mol. Med. 8: 346–353. [DOI] [PubMed] [Google Scholar]
- Wienholds, E., and R. H. Plasterk, 2004. Target-selected gene inactivation in zebrafish. Methods Cell Biol. 77: 69–90. [DOI] [PubMed] [Google Scholar]
- Wienholds, E., F. van Eeden, M. Kosters, J. Mudde, R. H. Plasterk et al., 2003. Efficient target-selected mutagenesis in zebrafish. Genome Res. 13: 2700–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods, L. M., C. A. Hodges, E. Baart, S. M. Baker, M. Liskay et al., 1999. Chromosomal influence on meiotic spindle assembly: abnormal meiosis I in female Mlh1 mutant mice. J. Cell Biol. 145: 1395–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky, J., A. J. MacQueen, J. B. Duffy, K. J. Kemphues and A. M. Villeneuve, 1999. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics 153: 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]