Abstract
Molecular chaperones, such as Hsp40, regulate cellular processes by aiding in the folding, localization, and activation of multi-protein machines. To identify new targets of chaperone action, we performed a multi-copy suppressor screen for genes that improved the slow-growth defect of yeast lacking the YDJ1 chromosomal locus and expressing a defective Hsp40 chimera. Among the genes identified were MID2, which regulates cell-wall integrity, and PKC1, which encodes protein kinase C and is linked to cell-wall biogenesis. We found that ydj1Δ yeast exhibit phenotypes consistent with cell-wall defects and that these phenotypes were improved by Mid2p or Pkc1p overexpression or by overexpression of activated downstream components in the PKC pathway. Yeast containing a thermosensitive allele in the gene encoding Hsp90 also exhibited cell-wall defects, and Mid2p or Pkc1p overexpression improved the growth of these cells at elevated temperatures. To determine the physiological basis for suppression of the ydj1Δ growth defect, wild-type and ydj1Δ yeast were examined by electron microscopy and we found that Mid2p overexpression thickened the mutant's cell wall. Together, these data provide the first direct link between cytoplasmic chaperone function and cell-wall integrity and suggest that chaperones orchestrate the complex biogenesis of this structure.
MOLECULAR chaperones play vital roles in many cellular processes, such as protein folding, degradation, translocation across membranes, and the rearrangement of multi-protein complexes. While molecular chaperones can work independently, they most commonly function as components of large multi-chaperone assemblies. For example, the Hsp70 and Hsp40 chaperones function together to catalyze a variety of essential processes in the cell (Walsh et al. 2004; Hennessy et al. 2005; Mayer and Bukau 2005). One Hsp70–Hsp40 pair, which is the focus of this study, is the Saccharomyces cerevisiae Ssa1 and Ydj1 proteins.
Hsp70's contain an N-terminal ATPase domain and C-terminal substrate-binding domain (Flaherty et al. 1990; Wang et al. 1993; Zhu et al. 1996), and ATP hydrolysis induces a conformational change that allows the substrate-binding domain to bind peptides with high affinity (McCarty et al. 1995; Russell et al. 1999). Repeated cycles of ATP hydrolysis lead to successive rounds of peptide binding and release, which is utilized in all Hsp70-dependent processes such as those listed above. However, Hsp70's are intrinsically weak ATPases and thus require cochaperones such as Hsp40's and nucleotide exchange factors (NEFs) for maximal activity. Hsp40's also bind to peptide substrates and can deliver them to Hsp70 (Wickner et al. 1991; Langer et al. 1992; Szabo et al. 1996; Rudiger et al. 2001; Han and Christen 2003). All Hsp40's contain an ∼70-amino-acid “J domain,” which appears to interact with the ATPase domain of an Hsp70, thus stimulating its ATPase activity (Cheetham and Caplan 1998; Gassler et al. 1998; Suh et al. 1998). The J domain contains four α-helices that form a finger-like projection, and the interaction with Hsp70 is mediated through helix II and an indispensable histidine–proline–aspartic acid (HPD) motif in the loop between helices II and III (Greene et al. 1998). Mutations in the HPD loop and in some residues in helix II abrogate the ability of Hsp40's to stimulate Hsp70 ATP hydrolysis (Wall et al. 1994; Tsai and Douglas 1996; Genevaux et al. 2002; Hennessy et al. 2005). In contrast, NEFs work through a variety of mechanisms to catalyze ADP release from Hsp70 (Shomura et al. 2005), which is required to free bound peptide substrates.
The Hsp70–Hsp40 chaperones can couple with other chaperone machines in the cell. Specifically, Hsp70 and Hsp40 cooperate with the Hsp90 chaperone complex, which is responsible for folding a specific set of client proteins that have neared their native conformation (Terasawa et al. 2005; Zhao and Houry 2005). For example, in vitro reconstitution studies indicate that the progesterone receptor (PR) is first bound by Hsp40, then delivered to Hsp70, and finally presented to the Hsp90 complex. Hsp90, through its interaction with several additional cochaperones, folds PR and remains associated with the receptor until hormone binds (Smith et al. 1992; Hernandez et al. 2002a,b; Pratt and Toft 2003).
Another intricate cellular process, which has been best characterized in the model organism S. cerevisiae, is cell-wall biogenesis (Levin 2005; Lesage and Bussey 2006), but surprisingly few chaperones have been directly implicated in this process (see below and discussion). The yeast cell wall is composed of two distinct layers. The electron-transparent inner layer is composed primarily of β1,3-glucan chains and small amounts of chitin, which are covalently “glued” together by β1,6-glucans (Osumi 1998). In contrast, the electron dense outer layer is composed of glycosylphosphatidylinositol (GPI) and Pir glycoproteins (Osumi 1998), which protect the cell wall and aid in cellular recognition. GPI proteins are attached to the β1,3-glucan chains via β1,6-glucan, whereas Pir glycoproteins are linked directly to β1,3-glucans. Synthesis of β1,3-glucan chains and chitin takes place at the yeast cell wall by known synthases (Douglas et al. 1994; Mazur et al. 1995; Qadota et al. 1996; Santos and Snyder 1997; Valdivia and Schekman 2003), but the location of β1,6-glucan synthesis is unknown, in part because the synthase has not yet been identified (Shahinian and Bussey 2000). β1,6-glucan is entirely localized to the cell wall, suggesting that it is synthesized at this site (Montijn et al. 1999). However, β1,6-glucan synthesis clearly requires the secretory pathway because a mutant allele in the gene encoding an ER luminal Hsp70, BiP, reduces the amount of β1,6-glucan at the cell wall when either of the two ER glucosidases are also disabled (Simons et al. 1998). Mutations in numerous other genes in the secretory pathway also result in reduced β1,6-glucan at the cell wall (Shahinian and Bussey 2000). Finally, overexpression of several Golgi-resident proteins proposed to aid in β-glucan synthesis suppresses the growth defect in strains mutated for the gene encoding Pkc1p, which is involved in maintaining cell-wall integrity (Roemer et al. 1994; Neiman et al. 1997).
A third intricate process in which chaperone machines are essential is exemplified by the tumor-causing virus simian virus 40 (SV40) (Brodsky and Pipas 1998; DeCaprio 1999; Sullivan and Pipas 2002). A single viral-encoded protein, large tumor antigen (TAg), interacts with cellular proteins such as the tumor-suppressors p53 and Rb and co-opts their function to trigger SV40 replication (Ali and DeCaprio 2001). These interactions lead to tumor development in rodents and transformation of rodent cell lines in culture. TAg contains a J domain that interacts with Hsp70 and is indispensable for viral replication and viral-induced cellular transformation (Campbell et al. 1997; Kelley and Georgopoulos 1997; Srinivasan et al. 1997). The J domain of TAg binds to and stimulates Hsp70 ATPase activity, leading to the release of E2F transcription factors from Rb and triggering cell cycle progression (Sullivan et al. 2000). Previous observations indicate that additional unknown factors—perhaps other chaperones—are also probably involved in this process (C. S. Sullivan and J. M. Pipas, unpublished data).
To better define why chaperone activity is required for SV40 function and ultimately to identify these other factors, loss-of-function mutations in the TAg J domain were uncovered in a yeast screen using a chimeric T-Ydj1 protein. This chimera contains the TAg J domain fused to the C terminus of a yeast Hsp40, Ydj1p, and its expression rescues the temperature-sensitive growth defect of ydj1-151 mutant yeast (Fewell et al. 2002). Of the 14 mutant alleles of T-TDJ1 that failed to rescue the temperature sensitivity of ydj1-151, we were intrigued by the K53R mutant because the corresponding lysine maps to the third helix in the J domain, and NMR perturbation experiments suggest that this residue is unaltered by Hsp70 binding (Greene et al. 1998; Landry 2003). However, when this mutation was introduced into TAg, the mutant protein was partially defective for stimulating Hsp70 ATPase activity and for releasing Rb from the E2F complex. A recombinant SV40 engineered to express K53R TAg was replication and transformation deficient (Fewell et al. 2002). Thus a T-YDJ1 construct containing the K53R mutation is a promising genetic tool to explore TAg chaperone function and to identify novel chaperone modulators.
To better define TAg chaperone function, we performed a yeast suppressor screen in ydj1Δ yeast expressing a T-YDJ1 construct with the K53R mutation [T(K53R)-YDJ1]. As reported here, we identified yeast Hsp70 as a suppressor of the K53R allele and uncovered a previously unknown connection between Ydj1p and yeast cell-wall biogenesis. Specifically, we observed cell-wall defects in ydj1Δ and Hsp90 temperature-sensitive mutants and discovered that upregulation of the protein kinase C (PKC) pathway can rescue these defects. These studies are the first to implicate the cytosolic Hsp40 chaperones in maintenance of the yeast cell wall.
MATERIALS AND METHODS
Yeast strains and methods:
S. cerevisiae yeast strains used in this study are listed in Table 1. Unless otherwise indicated, all yeast cultures were grown in yeast extract–peptone–dextrose (YPD) or selective synthetic complete medium (SC) with 2% glucose at room temperature or at 26° (Adams et al. 1997). Cell-wall phenotypes were tested on YP or SC–ura medium with the addition of 0.4 m NaCl, 1 m sorbitol, or 20 μg/ml calcofluor white (CW), and 2% glucose. Yeast transformation was performed by the lithium acetate procedure (Ito et al. 1983). For all serial dilutions, overnight cultures were diluted back to early log phase (0.3–0.4 OD) and allowed to grow 2–5 hr. Cell densities were normalized to the lowest OD and cells were serially diluted 10-fold. Unless specifically indicated, all growth assays were performed at 26°, 30°, 35°, and 37° for 3–7 days.
TABLE 1.
Yeast strains used in this study
| Yeast strain | Genotype | Source |
|---|---|---|
| W303 | MATα ade2-1 leu2-3,112 his3-11,15 trp1-1ura3-1 can1-100 | This lab |
| ACY95b | MATα ade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 ydj1-2∷HIS3 pAV4 | Caplan et al. (1992) |
| JN516 | MATα leu2-3,112 his3-11,15 ura3-52 trp1-Δ1 lys2 SSA1 ssa2∷LEU2 ssa3∷TRP1 ssa4∷LYS2 | Becker et al. (1996) |
| JB67 | MATα leu2-3,112 his3-11,15 ura3-52 trp1-Δ1 lys2 ssa1-45∷URA3 ssa2∷LEU2 ssa3∷TRP1 ssa4∷LYS2 | Becker et al. (1996) |
| p82a | MATaade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 hsc82∷LEU2 hsp82∷LEU2 pTGPD-HSP82 | Nathan and Lindquist (1995) |
| G313N | MATaade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 hsc82∷LEU2 hsp82∷LEU2 pTGPD-HSP82-G313N | Fliss et al. (2000) |
| G170D | MATaade2-1 leu2-3,112 his3-11,15 trp1-1 ura3-1 can1-100 hsc82∷LEU2 hsp82∷LEU2 pTGPD-HSP82-G170D | Nathan and Lindquist (1995) |
| STI1/SSE1 | MATaGAL2 his2-11,15 leu2-3,112 lys1 lys2 trp1Δ1 ura3-52 | Nicolet and Craig (1989) |
| sti1Δsse1Δ | MATaGAL2 his2-11,15 leu2-3,112 lys1 lys2 trp1Δ1 ura3-52 sti1∷HIS3 sse1∷KANR | Liu et al. (1999) |
Molecular techniques:
All plasmids used in this study are listed in Table 2. T-YDJ1 and T(K53R)-YDJ1 constructs were derived from the pOW4 T-YDJ1 plasmid, which is derived from plasmid YCplac33 (Gietz and Sugino 1988) and contains the T-YDJ1 chimeric gene (Fewell et al. 2002) expressed from the alcohol dehydrogenase 1 (ADH) promoter. This chimera consists of amino acids 1–82 of T antigen, which encompasses most of the J domain, and amino acids 71-409 of Ydj1p, which contains the glycine/phenylalanine-rich and zinc-finger-like regions and thus the putative substrate-binding domain (Johnson and Craig 2001; Kim et al. 2001; Fan et al. 2005). pOW4 T(K53R)-YDJ1 was created with the Quikchange site-directed mutagenesis kit (Stratagene, La Jolla, CA) with oligonucleotides 5′-GAGATGAAGAAAAAATGAGGAAAATGAATACTCTG-3′ and 5′-CAGAGTATTCATTTTCCTCATTTTTTCTTCATCTC-3′, using pOW4 T-YDJ1 as a template. The pOW4 T-YDJ1 and pOW4 T(K53R)-YDJ1 vectors were digested with EcoRI and XhoI and the inserts were ligated into pTEF414 (Mumberg et al. 1995) to create TEF414 T-YDJ1 and TEF414 T(K53R)-YDJ1. TEF414 T(H42R)-YDJ1 was created with the Quikchange mutagenesis kit using TEF414 T-YDJ1 as a template and oligonucleotides 5′-AAATGCAAGGAGTTTCGTCCTGATAAAGGAGGAG-3′ and 5′-CTCCTCCTTTATCAGGACGAAACTCCTTGCATTT-3′. The inserts in all constructs were subjected to DNA sequence analysis. An additional mutation, D127G, was detected in all constructs at nucleotide 380 in a nonconserved residue between the glycine/phenylalanine-rich and zinc-finger-like regions of YDJ1 (Caplan and Douglas 1991); however, the presence of this mutation did not affect the growth of yeast-expressing T-Ydj1p.
TABLE 2.
Plasmids used in this study
| Plasmid | Gene | Vector | Reference/source |
|---|---|---|---|
| pCMS39-T-YDJ1 | T-YDJ1 | pTEF414 | This study |
| pCMS41-T(K53R)-YDJ1 | T(K53R)-YDJ1 | pTEF414 | This study |
| pCMS123-T(H42R)-YDJ1 | T(H42R)-YDJ1 | pTEF414 | This study |
| pAV4 | YDJ1 | pRS316 | Caplan et al. (1992) |
| pCMS125-REC102 | REC102 | pRS426 | This study |
| pSR6 | CHS5 | pRS316 | Santos et al. (1997) |
| pSR23 | CHS5 | pRS426 | Santos et al. (1997) |
| p1245 | MID2-HA | YEp352 | Rajavel et al. (1999) |
| p1300 | MID2-GFP | pRS314 | Rajavel et al. (1999) |
| YEp/STM1 | STM1 | YEp213 | Hata et al. (1998) |
| pCMS119-YLR149c | YLR149c | pRS426 | This study |
| SYP1pRS316 | SYP1 | pRS316 | Marcoux et al. (2000) |
| SYP1pRS426 | SYP1 | pRS416 | Marcoux et al. (2000) |
| YEp351-SSA1 | SSA1 | YEp351 | E. Craig |
| pRS426-GPD-(His)6-SSA1 | SSA1 | pGPD426 | McClellan and Brodsky (2000) |
| p1657 | SLG-HA | YEp352 | Rajavel et al. (1999) |
| pSUS1 | SUS1 | pRS316 | Rodriguez-Navarro et al. (2004) |
| pFW46 | CYC8 | pRS316 | Williams and Trumbly (1990) |
| pRT81 | CYC8 | YEp24 | Trumbly (1988) |
| pCMS118-CIS1 | CIS1 | pRS426 | This study |
| pSKN1-IV | SKN1 | YCp50 | Roemeret al. (1993) |
| pThi4ura3 | THI4 | pRS416 | Singleton (1997) |
| pFR22 (YEpU-PKC1) | PKC1 | YEp352 | Roelants et al. (2004) |
| pDLB759 | BCK1 | YEp352 | D. Lew/D. Levin |
| pRS314 BCK1-20 | BCK1-20 | pRS314 | Lee and Levin (1992) |
| pCMS147-BCK1-20 | BCK1-20 | pRS426 | This study |
| pDLB823 | MKK1 | pRS314 | Harrison et al. (2004) |
| pDLB824 | MKK1DD | pRS314 | Harrison et al. (2004) |
| pCMS148-MKK1DD | MKK1DD | pRS426 | This study |
| pDLB758 | MPK1 | YEp352 | D. Lew/D. Levin |
| pNC267 | STE7-myc | 2μ, CYC1 | Zhou et al. (1993) |
| YEp352-Kss1 | KSS1 | YEp352 | Ma et al. (1995) |
| p181HOG1ha3 | HOG1-HA | YEplac181 | Winkler et al. (2002) |
| pCMS155-HOG1-HA | HOG1-HA | pRS426 | This study |
| p111PBS2 | PBS2 | YEplac111 | Winkler et al. (2002) |
| p112PBS2 | PBS2 | YEplac112 | I. Ota |
| pCMS154-Hsp82 | Hsp82 | pRS426 | This study |
The yeast genes REC102, YLR149c, CIS1, and HSP82 were amplified by PCR from genomic wild-type yeast (W303) DNA with primers created against regions 50–200 bp upstream of the TATA box and 15–250 bp downstream of the stop codon. The PCR products were digested with the appropriate restriction enzymes and ligated into pRS426 (Christianson et al. 1992). The sequence of each insert was confirmed by sequence analysis. pRS426 HSP82 had an A493T mutation at a nonconserved residue in the middle region of Hsp82p. This mutation did not alter Hsp82p function since expression of the corresponding protein restored growth in the hsp82 G170D and hsp82 G313N temperature-sensitive strains, and the protein was able to support cell growth as the only copy of Hsp82p in the cell (data not shown). To create a high-copy BCK1-20 expression vector, an ∼6.5-kb fragment containing BCK1-20 under the control of the endogenous promoter was removed from pRS314BCK1-20 (Lee and Levin 1992) using XhoI and NotI and ligated into pRS426. A high-copy version of MKK1DD was created by removing an ∼1.6-kb fragment containing MKK1DD under the control of the endogenous promoter from pDLB824 (Harrison et al. 2004) using EcoRI and SacI and ligating this fragment into pRS426. To obtain the pRS426 HOG1-HA expression plasmid, an ∼2.5-kb fragment containing HOG1-HA under the control of the endogenous promoter was PCR amplified from p181HOG1ha3 (Winkler et al. 2002) and ligated into pRS426. The HOG1 PCR primers encompassed DNA ∼125 bp upstream of the TATA box and introduced two stop codons immediately downstream of the 3′ HA tag. All primer sequences used for this study are available upon request.
High-copy suppressor screen:
To search for high-copy suppressors of the T(K53R)-YDJ1 thermosensitive phenotype in ydj1Δ yeast, a ydj1Δ strain (ACY95b) containing a pRS316-derived CEN-YDJ1 expression vector (pAV4; Caplan et al. 1992), was plated on 5-fluoroorotic acid to select for yeast that had lost pAV4. The surviving ydj1Δ cells were then transformed with TEF414 T(K53R)-YDJ1 and grown on SC–trp medium and expression of T(K53R)-Ydj1p was verified by immunoblot analysis using both the anti-TAg J domain antibody 419 (Harlow et al. 1981) and an antibody against Ydj1p (Caplan and Douglas 1991) (data not shown). Next, a 2μ-URA3 yeast genomic library in the YEp24 vector (Carlson and Botstein 1982) was introduced into these cells and transformants were selected on SC–ura–trp at 35° for 4 days. Approximately 80,000 colonies were screened, and 61 colonies appeared to contain plasmids that suppressed the T(K53R)-YDJ1 growth defect when restreaked onto SC–ura–trp at 35°. The plasmid DNA from these cells was isolated and retransformed into ydj1Δ yeast-expressing T(K53R)-Ydj1p. Upon retransformation, 21 plasmids improved growth to varying extents. Of these, 8 unique plasmids were obtained as assessed by DNA sequence and restriction digest analysis.
Biochemical and immunological methods:
To prepare cellular proteins for immunoblot analysis, 10 ml of yeast were grown to an OD of 0.3–0.9 in the appropriate selective medium at room temperature and the cells were pelleted and resuspended in 0.8 ml of 100 mm Tris, pH 8.0, 20% glycerol, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride (PMSF), 1 μg/ml leupeptin, and 0.5 μg/ml pepstatin A. The cells were lysed by agitation with glass beads and crude protein concentrations were determined by measuring the A280. Protein concentrations in each sample were normalized and total protein was resolved by SDS–PAGE, transferred to nitrocellulose, and immunoblotted with the indicated antiserum. Antiserum against ribosomal protein L3 (a kind gift of J. Warner, Albert Einstein College of Medicine) or Sec61p (Stirling et al. 1992) was used to establish loading controls. Antisera against Sse1p (Goeckeler et al. 2002) and Ydj1p (Caplan and Douglas 1991) were described previously. Antiserum against Hsp82p was provided by A. Caplan, Mount Sinai School of Medicine. Anti-Ssa1p antiserum was prepared in rabbits using a GST-fusion protein that contained amino acids 586–831 of Ssa1p (provided by E. Craig, University of Wisconsin). Antibodies against the HA epitope were obtained from Roche and antibodies against Pkc1p were obtained from Santa Cruz Biotechnology. Bound antibodies were visualized using anti-mouse or anti-rabbit antibodies coupled to horseradish peroxidase and the Supersignal West Pico chemiluminescent substrate kit (Pierce, Rockford, IL). Chemiluminescent signal was detected using a Kodak 440CF Image Station and quantified using Kodak 1D (v. 3.6) software.
Subcellular fractionation of Mid2p was performed by differential centrifugation as previously described (Kabani et al. 2002a) with minor changes. A total of 100 ODs of either wild-type (W303) or ydj1Δ yeast-expressing Mid2p-HA were resuspended in 2 ml PLB (20 mm HEPES, pH 7.4, 100 mm NaCl, 20 mm MgCl2, 1 mm PMSF, 1 μg/ml leupeptin, 0.5 μg/ml pepstatin A) and cells were broken by glass-bead lysis. Unbroken cells were removed by centrifugation at 1400 × g for 5 min and 1 ml of the supernatant (L) was subjected to a medium-speed spin (16,000 × g) for 15 min at 4°. The pellet (P1) was resuspended in 250 μl PLB and half of the supernatant (S1) was saved. The remaining supernatant was subjected to a high-speed spin (150,000 × g) for 15 min at 4°. The resulting pellet (P2) was resuspended in 100 μl PLB and the remaining supernatant (S2) was saved. Total protein concentration in L, S1, P1, S2, and P2 was normalized by Coomassie Brilliant blue staining of polyacrylamide gels. The samples were resolved by SDS–PAGE, and immunoblots were performed as described above.
Indirect immunofluorescence and electron microscopy:
Indirect immunofluorescence microscopy was performed as described previously (Coughlan et al. 2004). Briefly, wild-type (W303) or ydj1Δ yeast strains overexpressing Mid2p-HA were grown to midlog phase, fixed in 3.7% formaldehyde, and treated with 20 μg/ml zymolyase for 45 min at 37°. Cells were incubated with primary antibodies [HA 1:250 and Kar2p (1:250; Brodsky and Schekman 1993)] overnight at 4° and in secondary antibodies (Alexa Fluor 488 goat anti-mouse 1:250 and Alexa Fluor 568 goat anti-rabbit 1:250; Molecular Probes, Eugene, OR) for 2 hr at room temperature. To visualize DNA, the cells were incubated in 2 μg/ml 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) for 5 min at room temperature.
Electron microscopy was performed as previously described (Kaiser and Schekman 1990). In brief, the indicated stains containing either the MID2-HA expression vector or YEp352 (Hill et al. 1986) were fixed in 2.5% glutaraldehyde, processed, sectioned, and affixed to 0.125% formvar-coated grids. Sections counterstained with uranyl acetate and Reynold's lead citrate were examined on JEM-1011 or JEM-1210 (JEOL) transmission electron microscopes. Cell-wall thickness was measured using AMTv542 image capture software (Advanced Microscopy Techniques). A total of 12–16 similarly sized budding cells from each culture were chosen, and the cell-wall thicknesses at 10 points around the mother cell were measured. The average for each cell was calculated and these numbers were averaged for each culture to determine the mean cell-wall thickness.
RESULTS
Identification of suppressors of T(K53R)-YDJ1 and ydj1Δ yeast:
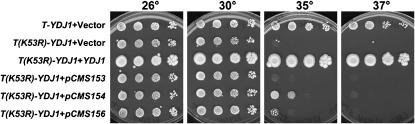
We previously reported on a yeast TAg expression system in which the TAg J domain replaced the homologous domain in a yeast Hsp40, Ydj1p. By PCR mutagenesis of the inserted J domain, new mutations in this domain were isolated, some of which conferred strong phenotypes when inserted into full-length TAg and into SV40 (Fewell et al. 2002). One mutant allele that conferred profound phenotypes was T(K53R)-YDJ1, and to understand better how specific Hsp40 mutations affect yeast cell growth, the T-YDJ1 chimera with the K53R mutation [T(K53R)-YDJ1] was cloned behind a moderate constitutive promoter and introduced into ydj1Δ yeast. We found that expression of wild-type T-YDJ1, but not T(K53R)-YDJ1, restored growth in the ydj1Δ strain at 35° (Figure 1). Next, a high-copy screen was performed to uncover suppressors of the temperature-sensitive phenotype, as described in materials and methods. As noted in the Introduction, one goal of this approach was to better define why the TAg chaperone domain is required for viral replication and function. Upon retransformation, eight unique plasmids were obtained that partially suppressed the T(K53R)-YDJ1 thermosensitive phenotype at 35° (Table 3). Of note, none of the plasmids rescued the temperature sensitive phenotype to the levels seen when T-Ydj1p was expressed (Figure 1). However, plasmid pCMS154 moderately rescued the T(K53R)-YDJ1 phenotype and was the strongest suppressing plasmid uncovered in the screen.
Figure 1.—
pCMS154 in multiple copies moderately rescues the T(K53R)-YDJ1 growth defect at 35°. Ten-fold serial dilutions of ydj1Δ yeast expressing wild-type T-Ydj1p or T(K53R)-Ydj1p and containing a vector control, a Ydj1p expression vector (pAV4), or three plasmids recovered in the screen—pCMS153, pCMS154, and pCMS156—were plated on SC–ura–trp and grown for 5 days at the indicated temperatures. The plasmid pCMS154 contains a yeast genome fragment that includes the genes REC102, CHS5, and MID2 and moderately improves the growth of yeast containing the T(K53R)-YDJ1 expression vector at 35°. The plasmid pCMS156 contained the genes STM1 and YLR149c and led to papillae colony growth at 35°. Plasmid pCMS153 did not improve growth of the ydj1Δ strain containing the T(K53R)-YDJ1 expression vector at 35°.
TABLE 3.
Summary of the results obtained from the screen
| Times recovered | Gene | Plasmid name | Vector | Improves T(K53R)-YDJ1 growth at 35°? | Improves ydj1Δ growth at 30°? |
|---|---|---|---|---|---|
| 6 | REC102 | pCMS125-REC102 | pRS426 | − | − |
| CHS5 | pSR6 | pRS316 | − | − | |
| CHS5 | pSR23 | pRS426 | − | − | |
| MID2-HA | p1245 | YEp352 | +++ | +++ | |
| MID2-GFP | p1300 | pRS314 | ND | +++ | |
| 5 | STM1 | Yep/STM1 | YEp213 | −a | − |
| YLR149c | pCMS119-YLR149c | pRS426 | − | − | |
| 3 | RIM1 | ND | ND | ND | ND |
| SYP1 | SYP1pRS316 | pRS316 | − | − | |
| SYP1 | SYP1pRS426 | pRS426 | + | + | |
| RPS14a | ND | ND | ND | ND | |
| 2 | SSA1 | Yep351-SSA1 | YEp351 pGPD426 ND | ++ | −b |
| SSA1 | pRS426-GPD-(His)6-SSA1 | ND | + | −b | |
| EFB1 | ND | ND | ND | ||
| ERP2 | ND | ND | ND | ||
| 2 | SLG1-HA | p1657 | YEp352 | ++ | +++ |
| 2 | SUS1 | pSUS1 | pRS316 | − | − |
| CYC8 | pFW46 | pRS316 | − | + | |
| CYC8 | pRT81 | YEp24 | ++ | ++ | |
| YBR113w | ND | ND | ND | ND | |
| 1 | YDR020c | ND | ND | ND | ND |
| FAL1 | ND | ND | ND | ND | |
| CIS1 | pCMS118-CIS1 | pRS426 | ++ | − | |
| SES1 | ND | ND | ND | ND | |
| 1 | SKN1 | pSKN1-IV | YCp50 pRS416 | + | − |
| THI4 | pThi4ura3 | ND | − | − | |
| ENP2 | ND | ND | ND |
Eight unique plasmids were isolated in the screen that allowed growth of the ydj1Δ yeast-expressing T(K53R)-Ydj1p at 35° upon retransformation, and the number of times each plasmid was identified is indicated. Select individual genes found on each plasmid isolated in the screen were tested for improved growth of T(K53R)-YDJ1 at 35° and ydj1Δ at 30°. Seven genes improved growth at 35° of ydj1Δ yeast-expressing T(K53R)-Ydj1p and four genes improved the slow-growth phenotype of ydj1Δ yeast at 30°. +++, moderate rescue; ++, weak rescue; +, poor rescue. ND, not determined.
Papillae colonies were detected.
ydj1Δ yeast with high-copy SSA1 expression vectors grew more slowly than empty vector controls.
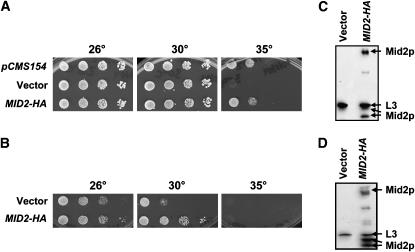
To determine which of the genes in the eight isolated inserts conferred improved growth, 13 individual genes were chosen for further analysis. These included genes encoding chaperones, transcription factors, and proteins of unknown function, and several that had a common link to cell-wall synthesis or integrity (see below). Each gene was amplified by PCR and cloned into a pRS426 vector or was obtained from colleagues (Table 2). Next, the vectors were transformed into the ydj1Δ strain either containing or lacking the T(K53R)-YDJ1 expression vector and tested for their ability to rescue the temperature-sensitive growth defect. It should be noted that ydj1Δ yeast lacking the T(K53R)-Ydj1p expression vector grew poorly at temperatures >26°, and those containing the vector grew poorly >30°, consistent with T(K53R)-Ydj1p exhibiting partial activity (Fewell et al. 2002). However, overexpression of 7 genes suppressed the slow-growth phenotype of T(K53R)-YDJ1 at 35° to varying degrees, and 4 of these (MID2, SLG1/WSC1, CYC8, SYP1) also improved the growth of the ydj1Δ strain lacking the expression vector variably at 30° (Table 3). Notably, ydj1Δ yeast containing pCMS154 (which harbors MID2) or containing a vector engineered specifically for MID2 overexpression conferred the same degree of rescue (Table 3 and Figure 3A). Overexpression of an eighth gene, STM1, did not alter the temperature-sensitive phenotype but led to papillae colony formation [see Figure 1 for data on the abilities of MID2 (pCMS154) and STM1 (pCMS156)-containing plasmids isolated from the screen to suppress the temperature-sensitive growth phenotype].
Figure 3.—
MID2 suppresses the thermosensitive growth defect of ydj1Δ yeast. (A) ydj1Δ yeast expressing T(K53R)-Ydj1p were transformed with the MID2-containing multi-copy plasmid isolated in the screen (pCMS154), an empty vector, or a multi-copy MID2-HA-containing vector, and were serially diluted onto SC–ura–trp. Plates were incubated for 4 days. (B) High-copy MID2-HA also suppresses the growth defect in the ydj1Δ strain lacking the T(K53R)-Ydj1p expression vector, as assessed in A. Mid2p is expressed in the T(K53R)-YDJ1-containing ydj1Δ strain (C) as well as the in the ydj1Δ strain (D) as indicated by immunoblot analysis.
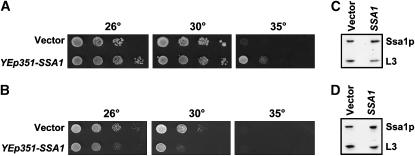
Among the genes tested, only one improved the growth of T(K53R)-Ydj1p-expressing yeast but had a negative effect on ydj1Δ yeast lacking the expression vector. The gene is SSA1, which encodes a yeast Hsp70 that is known to interact with Ydj1p (Figure 2, A and B) (Cyr et al. 1992; Becker et al. 1996; McClellan and Brodsky 2000). SSA1 overexpression in yeast containing YEp351-SSA1 was verified by immunoblot analysis (Figure 2, C and D). One interpretation of this result is that Ssa1p may directly bind the J domain in T(K53R)-Ydj1p and partially repair the lesion conferred by mutating lysine 53 (since only moderate rescue was observed).
Figure 2.—
SSA1 improves the growth of strains exhibiting the T(K53R)-YDJ1 thermosensitive phenotype. Cultures of ydj1Δ yeast expressing T(K53R)-Ydj1p were transformed with a high-copy SSA1-containing vector or an empty vector and were serially diluted onto either SC–ura–trp or SC–ura. Plates were incubated for 4 days. (A and B) High-copy SSA1 allows some growth of ydj1Δ yeast containing the T(K53R)-YDJ1 expression vector at 35°, but slows the growth of ydj1Δ yeast. Immunoblot analysis indicates that Ssa1p is overexpressed (C) 2.1-fold in the ydj1Δ strain expressing T(K53R)-Ydj1p and (D) 2.6-fold in the ydj1Δ yeast strain.
In contrast, four suppressors (MID2, SLG1/WSC1, CYC8, SYP1) were identified that suppressed to varying extents the phenotypes associated with both the deletion of the YDJ1 locus and the phenotype associated with the T(K53R)-YDJ1 expression vector (Table 3; see Figure 3, A and B for an example of studies with the rescue conferred by MID2). To verify the expression of the strongest suppressor, Mid2p, an immunoblot analysis against the HA epitope tag (Figure 3, C and D) was performed and Mid2p resolved as three distinct bands of 40, 47, and 200 kDa. This is consistent with the presence of both immature and highly O-glycosylated forms of Mid2p (Lommel et al. 2004). To determine if the observed improvement of growth was allele specific, MID2-HA was overexpressed in the ydj1Δ strain containing the T(H42R)-YDJ1 expression vector, which contains an H42R mutation in the conserved HPD motif in the TAg J domain (Fewell et al. 2002). We found that MID2-HA overexpression also suppressed the slow-growth phenotype of ydj1Δ yeast containing the T(H42R)-YDJ1 expression vector at 30° (data not shown). Taken together, these results suggest that MID2 suppression is independent of TAg J domain mutant alleles, but instead is the consequence of improved growth of the ydj1Δ strain.
PKC1 and constitutively activated components of the PKC pathway improve the ydj1Δ slow-growth phenotype:
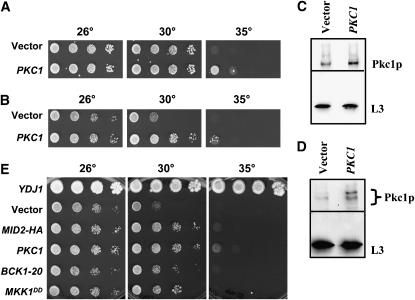
Mid2p and Slg1p/Wsc1p are plasma membrane proteins that sense yeast cell-wall stress and activate the PKC pathway (Philip and Levin 2001), which leads to the transcription of genes involved in cell-wall synthesis (Jung and Levin 1999). Since Mid2p and Slg1p/Wsc1p are components of the PKC pathway and several other of our identified suppressors (Table 3)—Syp1p (Marcoux et al. 2000), Skn1p (Roemer et al. 1994), and Cis1p (Manning et al. 1997)—are genetically linked to PKC1 or MID2, the ability of PKC1 to suppress the ydj1Δ growth phenotype was investigated. A 2μ PKC1 overexpression plasmid was transformed into ydj1Δ cells and ydj1Δ cells expressing T(K53R)-Ydj1p, and moderate rescue of T(K53R)-YDJ1 growth at 35° (Figure 4A) and ydj1Δ growth at 30° (Figure 4B) was observed. The overexpression of Pkc1p in these strains was verified by immunoblot analysis as shown in Figure 4, C and D.
Figure 4.—
Introduction of a high-copy PKC1-containing vector and overexpression of constitutively active components in the PKC pathway improve the slow-growth phenotype of the ydj1Δ strain. A multi-copy PKC1-containing vector and a vector control were transformed into (A) ydj1Δ yeast-expressing T(K53R)-Ydj1p and (B) ydj1Δ yeast, and the transformants were serially diluted 10-fold and incubated for 4 days at the indicated temperatures. An immunoblot analysis indicates (C) 1.8-fold overexpression of Pkc1p in the ydj1Δ strain expressing T(K53R)-Ydj1p and (D) 3.1-fold overexpression of Pkc1p in the ydj1Δ strain. (E) Plasmids containing constitutively active, multi-copy BCK1-20 and MKK1DD alleles transformed into the ydj1Δ strain also improve the ydj1Δ temperature-sensitive defect, although to a lesser extent than MID2 or PKC1.
The PKC-signaling pathway is initiated by the phosphorylation of Pkc1p and terminates with transcription factor activation via the Bck1p, Mkk1p/Mkk2p, and Mpk1 kinases (Levin 2005). To determine if overexpression of downstream members of the PKC pathway also rescued the ydj1Δ slow-growth phenotype, plasmids engineered for the expression of constitutively active BCK1 and MKK1 alleles were introduced into the mutant strain. BCK1-20 has an alanine-to-proline mutation at position 1174, immediately upstream of the kinase domain, which is believed to mimic Bck1p phosphorylated by Pkc1p (Lee and Levin 1992). MKK1DD contains two serine-to-aspartic acid mutations in the kinase domain, which mimics Mkk1p phosphorylation (Harrison et al. 2004). As shown in Figure 4E, overexpression of BCK1-20 or MKK1DD slightly improved the ydj1Δ growth defect. In contrast, overexpression of wild-type Mkk1p or Bck1p failed to confer improved growth (data not shown), consistent with the previously reported necessity of using constitutively active forms of these kinases (Lee and Levin 1992; Harrison et al. 2004). In any event, these results indicate that activation of the PKC pathway can ameliorate the ydj1Δ temperature-sensitive growth phenotype.
In addition to the PKC pathway, yeast have two other cell-wall integrity pathways, the HOG and SVG pathways (Mager and Siderius 2002). Each pathway is activated under different conditions, but cross talk between the HOG and SVG pathways is common. The HOG and SVG pathways even contain some common signaling proteins (Gustin et al. 1998; O'Rourke and Herskowitz 1998; Lee and Elion 1999). To examine if overexpression of HOG and SVG pathway components affects the growth of ydj1Δ yeast, two genes in the HOG pathway (HOG1 and PBS2) and two in the SVG pathway (KSS1 and STE7) were overexpressed in the ydj1Δ strain. Hog1p and Kss1p are MAP kinases that initiate gene expression and Pbs2p and Ste7p are the MAPK kinases that phosphorylate Hog1p and Kss1p, respectively (Brewster et al. 1993; Lee and Elion 1999). Previously, connections between the signaling pathways were examined by overexpressing STE7 in the mkk1Δmkk2Δ strain (Yashar et al. 1995). However, the overexpression of these proteins had no effect on the ydj1Δ slow-growth phenotype (data not shown). These results suggest that rescue of the ydj1Δ slow-growth phenotype is limited to the PKC pathway and cannot be remedied by upregulation of alternative pathways that may sense cell-wall integrity.
MID2 and PKC1 partially suppress the temperature-sensitive phenotype of hsp82 mutant strains:
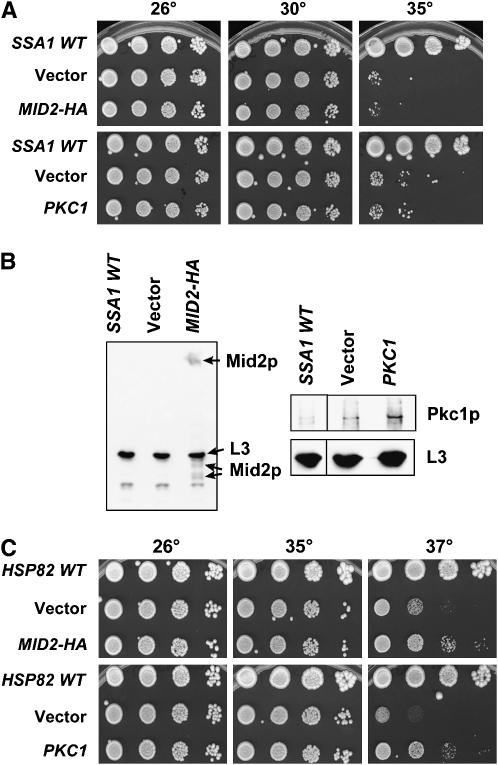
Because Ydj1p functions in multi-protein chaperone complexes with Ssa1p, a cytoplasmic Hsp70, and the yeast Hsp90 homolog, Hsp82p (Cyr et al. 1992; Caplan et al. 1995; Kimura et al. 1995; Becker et al. 1996; McClellan and Brodsky 2000), each of the seven suppressors was also overexpressed in the temperature-sensitive SSA1 strain, ssa1-45, and Mid2p and Pkc1p were overexpressed in the temperature-sensitive hsp82 G313N and hsp82 G170D strains (Bohen and Yamamoto 1993; Nathan and Lindquist 1995; Fliss et al. 2000). None of the suppressors, including MID2 or PKC1 (data not shown; see Figure 5A for data on MID2 and PKC1), rescued the temperature-sensitive phenotype of ssa1-45 yeast, even though immunoblots verified that Mid2p and Pkc1p were overexpressed (Figure 5B). In contrast, overexpression plasmids encoding MID2-HA and PKC1 partially suppressed the growth defect of both hsp82 G313N (Figure 5C) and hsp82 G170D (data not shown). Since the Hsp90 complex contains several other cochaperones, including Sti1p (Hop), Sba1p (p23), and Sse1p (Hsp110) (Chang et al. 1997; Fang et al. 1998; Liu et al. 1999), we also examined whether the introduction of MID2 and PKC1 overexpression plasmids remedied the sse1Δsti1Δ temperature-sensitive phenotype (Liu et al. 1999). No effect on cell growth was observed (data not shown). These data suggest that Mid2p or Pkc1p overexpression affects only the growth of the Hsp40 and Hsp90 mutant strains and that Hsp40 and Hsp90 play a role in yeast cell-wall integrity. These combined data also suggest that the mode of Ssa1p-mediated suppression of the ydj1Δ strain when T(K53R)-Ydj1p is expressed (see above) is distinct.
Figure 5.—
Multi-copy MID2 and PKC1-containing plasmids improve the temperature-sensitive growth defects of select chaperone mutants. ssa1-45 or hsp82 G313N yeast strains were transformed with multi-copy MID2-HA- or PKC1-containing vectors or with an empty vector. These strains and isogenic strains containing a wild-type version of the mutated genes were serially diluted 10-fold on SC–ura for 3–4 days at the indicated temperatures. (A) MID2-HA and PKC1 do not rescue the temperature-sensitive growth phenotype of ssa1-45 yeast. (B) Overexpression of Mid2p-HA (left) and Pkc1p (right) in the ssa1-45 strain were verified by immunoblot analysis. (C) High-copy MID2-HA and PKC1 improve the growth defect of the hsp82 G313N strain.
Because overexpression of Mid2p and Pkc1p improved the growth of hsp82 mutant strains, we investigated whether overexpression of Hsp82p could rescue the ydj1Δ slow-growth phenotype and if, like Ssa1p, Hsp82p also suppressed the T(K53R)-YDJ1 growth defect. To this end, the HSP82 locus was PCR amplified from the yeast genome and inserted in the high-copy pRS426 vector, and the plasmid was transformed into both the ydj1Δ strain and the ydj1Δ strain expressing T(K53R)-Ydj1p. Despite two- to threefold overexpression, HSP82 was unable to rescue either the T(K53R)-YDJ1 or ydj1Δ temperature-sensitive phenotypes (data not shown).
Chaperone levels are tightly regulated (Stone and Craig 1990; Hjorth-Sorensen et al. 2001) and basal chaperone expression levels might dictate whether a given chaperone can be overexpressed. Thus, endogenous concentrations of Ssa1p and Hsp82p were determined in the presence or absence of Mid2p or Pkc1p overexpression in both the wild-type and ydj1Δ strains. Not surprisingly, in ydj1Δ cells containing a vector control, Ssa1p is upregulated 1.6- to 2.7-fold and Hsp82p is upregulated 1.4- to 3.2-fold when compared to the wild-type strain (supplemental Figure S1 at http://www.genetics.org/supplemental/, compare lanes 1 and 4). When Mid2p and Pkc1p are overexpressed, we observed a slight additional upregulation of Hsp82p in the ydj1Δ strain (supplemental Figure S1, lanes 2 and 3). These results suggest that Hsp82p and Ssa1p and likely other gene products are induced to compensate for the lack of Ydj1p. In addition, this may explain why Ssa1p or Hsp82p overexpression alone does not ameliorate the temperature-sensitive phenotype of ydj1Δ yeast (see above).
The ydj1Δ and hsp82 temperature-sensitive strains show phenotypes consistent with cell-wall defects:
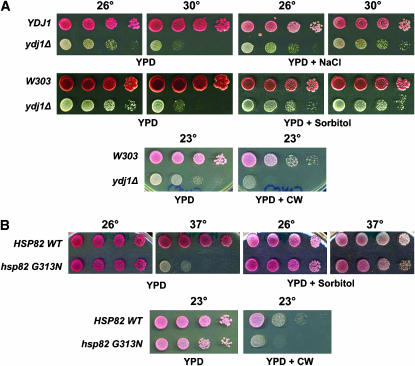
Since the PKC pathway is involved in yeast cell-wall maintenance (Levin 2005), mutation of pathway members leads to cell-wall defects and several detectable growth phenotypes. For example, the poor growth of pkc1Δ yeast is rescued by incubation in osmostabilizing reagents, such as 1 m sorbitol or 0.5 m NaCl (Levin and Bartlett-Heubusch 1992). In addition, pkc1 mutant yeast are sensitive to the cell-wall dye CW (Schmitz et al. 2001), which binds and perturbs the architectural integrity of this structure. Since the introduction of additional copies of MID2 and PKC1 affect the growth of ydj1Δ and hsp82 yeast, it is possible that these mutant strains also have cell-wall defects, as hinted at by previous studies (Lussier et al. 1997; Yang et al. 2006).
To begin to test this hypothesis, ydj1Δ cells with or without a YDJ1 single-copy expression vector (pAV4) were examined on medium supplemented with 0.4 m NaCl. Growth of ydj1Δ yeast was partially restored in the presence of salt (Figure 6A). Next, the growth of wild-type and ydj1Δ yeast was tested in the presence of sorbitol or CW. As expected for strains having cell-wall defects, the growth of ydj1Δ yeast was enhanced on 1 m sorbitol at 30° and was sensitive to CW at 23°. The growth of hsp82 G313N strains was also restored on high sorbitol at 37° and showed sensitivity to CW at 23° (Figure 6B). Notably, the growth of ydj1Δ on sorbitol resembled the growth seen upon overexpression of Mid2p or Pkc1p (Figures 3B and 4B). When Mid2p or Pkc1p was overexpressed on high-sorbitol-containing medium, ydj1Δ cells grew at temperatures as high as 37° (supplemental Figure S2 at http://www.genetics.org/supplemental/). Mid2p and Pkc1p overexpression also rescued the CW sensitivity of the ydj1Δ strain at 30° (data not shown). These results indicate that ydj1Δ and Hsp82 mutant strains have a cell-wall defect and that Mid2p and Pkc1p may repair this defect.
Figure 6.—
The ydj1Δ and hsp82 temperature-sensitive mutant strains have phenotypes consistent with defects in cell-wall synthesis. (A) (Top) Tenfold serial dilutions of ydj1Δ yeast either expressing Ydj1p (“YDJ1”) or lacking the expression vector were plated onto YPD either lacking or containing 0.4 m NaCl, 1 m sorbitol, or 20 μg/ml calcofluor white (CW) at the indicated temperatures for 3 days. (Middle and bottom) Wild-type or ydj1Δ yeast were incubated on control or sorbitol or CW-containing medium as above. (B) HSP82 and hsp82 yeast were grown on either YPD or YPD with either 1 m sorbitol or 20 μg/ml CW at the indicated temperatures for 3 days. All the yeast strains are ade− and have a pink coloration. ydj1Δ yeast also obtained the pink coloration after longer incubations (not shown).
Mid2p is not dramatically mislocalized or aggregated in ydj1Δ yeast:
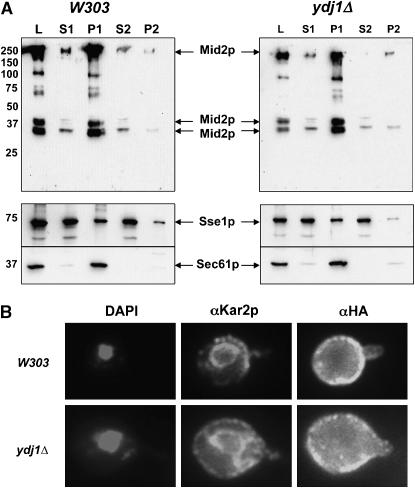
Ydj1p is involved in the translocation of nascent polypeptides into the ER (Caplan et al. 1992) and is able to retain aggregation-prone proteins in solution (Cyr 1995). We therefore hypothesized that Mid2p might be mislocalized or aggregated in ydj1Δ yeast, which would prevent accurate sensing of cell-wall integrity and cause cell-wall defects. But, at least some of the overexpressed Mid2p might escape to the plasma membrane and partially restore cell-wall integrity. To test this hypothesis, the solubility of Mid2p was determined by differential centrifugation in wild-type and ydj1Δ yeast strains overexpressing Mid2p-HA. In both wild-type and ydj1Δ yeast, the majority of the Mid2p-HA localized to the first pellet (P1; 76 and 80%, respectively, of the total protein assayed, i.e., the “load”) (Figure 7A), consistent with Mid2p membrane residence. Sec61p, a component of the ER membrane translocon (Deshaies et al. 1991), fractionated similarly. In wild-type yeast, 14% of the total Mid2p localized to the second pellet (P2), which is nearly identical to the amount of Mid2p found in the P2 from ydj1Δ yeast (15%). Intriguingly, only 1.6% of Mid2p fractionated in the second supernatant (S2), which represents clarified cytosol (the cytosolic Sse1p chaperone was used as a marker) in ydj1Δ cells, compared to 6.3% in wild-type yeast. The small decrease in the levels of soluble cytosolic Mid2p in fractionated lysates prepared from ydj1Δ yeast may be due to an increase in protein aggregation, although only a minor percentage of the total cellular Mid2p is affected.
Figure 7.—
Only a modest increase in Mid2p aggregation is evident in ydj1Δ yeast. (A) Wild-type or ydj1Δ yeast overexpressing Mid2p were lysed and the subcellular fractionation of Mid2p was determined by differential centrifugation. Sec61p, a component of the translocon, is an integral membrane protein, whereas Sse1p, a cytoplasmic chaperone, is primarily in soluble fractions. L, load; S1, first medium-speed supernatant; P1, first medium-speed pellet; S2, second high-speed supernatant; P2, second high-speed pellet. (B) Mid2p localization was determined by indirect immunofluorescence using an antibody against the HA epitope in wild-type or ydj1Δ yeast overexpressing Mid2p. For comparison, the ER was localized using an antibody against the ER-resident chaperone, Kar2p, and the nucleus was visualized with DAPI staining. The distended ER and enlarged cell volume of ydj1Δ yeast is commonly observed.
To investigate whether the subcellular localization of Mid2p was grossly altered when YDJ1 was deleted, indirect immunofluorescence microscopy was performed. Mid2p-HA was overexpressed in wild-type and ydj1Δ yeast, and its location was determined using an antibody against the HA epitope tag. As shown in Figure 7B, Mid2p clearly localized to the plasma membrane in wild-type yeast, which is in agreement with previously published data (Ketela et al. 1999; Rajavel et al. 1999). Some proteins also resided in large intracellular bodies, which may be late secretory vesicles. In ydj1Δ yeast, Mid2p localization was unchanged. Together, these results suggest that Mid2p is not significantly mislocalized or aggregated in the ydj1Δ yeast strain.
In addition to its role in cell-wall integrity, Mid2p has been implicated in actin cytoskeleton rearrangement (Marcoux et al. 2000). Therefore, it was formally possible that Mid2p overexpression rescues actin cytoskeletal defects in the ydj1Δ strain and thus improves growth. To determine if the actin cytoskeleton was perturbed in the ydj1Δ strain, we used fluorescence microscopy to visualize cortical actin patches in wild-type and ydj1Δ yeast. In both strains, phalloidin staining showed several punctate “dots” around the yeast cell periphery (data not shown), characteristic of cortical actin staining (Adams and Pringle 1984). This result suggests that the actin cytoskeleton is not grossly affected in the ydj1Δ strain and that Mid2p overexpression improves the growth of ydj1Δ cells through a different mechanism.
MID2-HA overexpression thickens the cell wall of ydj1Δ yeast:
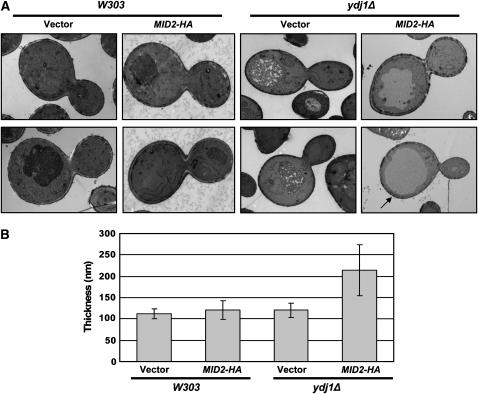
Another reason that Mid2p might improve the growth of ydj1Δ yeast is that Mid2p overexpression may thicken the yeast cell wall (Marcoux et al. 2000), possibly due to an increase in chitin (Ketela et al. 1999). Thus, we hypothesized that extra copies of Mid2p would rescue ydj1Δ growth and the cell-wall defect by strengthening and/or enlarging this structure. To test this hypothesis, wild-type or ydj1Δ cells lacking the T(K53R)-YDJ1 expression vector but containing the MID2-HA overexpression vector or an empty vector control were analyzed by electron microscopy. We first noted that ydj1Δ cells were generally larger than wild-type cells and contained an enlarged vacuole (Figure 8A), which is consistent with previously published data (Caplan and Douglas 1991). Next, the distance between the outside of the cell wall and the plasma membrane was measured in similarly sized budding wild-type or ydj1Δ cells, and the results were quantified as described in materials and methods. Despite the difference in cell size, the cell-wall thickness of the two strains was similar (Figure 8, A and B). This suggests that the cell-wall defect in ydj1Δ yeast arises due to an aberrant cell-wall composition. Strikingly, overexpression of Mid2p in the ydj1Δ cells almost doubled the average thickness of the cell wall from 120 to 214 nm. The increased thickness was seen in the inner electron transparent layer of the cell wall, which consists of β1,3-glucan, β1,6-glucan, and chitin (Osumi 1998). Occasionally, the increased cell-wall thickness was localized to a single region of the yeast cell wall (see the region identified by the arrow in Figure 8A), but in most cells the increased thickness was present throughout the cell periphery. Interestingly, overexpression of Mid2p did not uniformly increase the thickness of the cell wall in the wild-type yeast strain, although yeast with a thicker cell wall in limited regions around the cell were observed in some cases. Overall, we conclude that Mid2p overexpression rescues the ydj1Δ cell-wall defect by increasing its thickness.
Figure 8.—
Overexpression of Mid2p thickens the cell wall of ydj1Δ yeast. Wild-type or ydj1Δ strains containing a MID2-HA expression vector or vector control were analyzed by electron microscopy. (A) Two examples of single budding yeast from each strain are shown. Mid2p overexpression in the ydj1Δ strain thickens the entire cell wall (top) or at select locations (bottom, arrow) compared to the vector control. Wild-type yeast are shown at ×∼25,000 magnification and ydj1Δ yeast are shown at ×∼15,000 magnification. (B) The average cell-wall thickness for each strain was calculated; cell-wall thickness of ydj1Δ cells overexpressing Mid2p vs. those containing the vector control, P < 0.0001.
DISCUSSION
As noted in the Introduction, yeast cell-wall biosynthesis is a highly complex process that is subject to multiple regulatory inputs. Because molecular chaperones have well-described roles in regulating processes as diverse as protein transport, enzyme activation, and the assembly of large multi-protein complexes (Fewell et al. 2001; Young et al. 2003), one might expect many examples in which defects in chaperone function affect cell-wall integrity. Surprisingly, this is not the case, and here we report for the first time that members of the Hsp40 and Hps90 chaperone families are specifically required for cell-wall function. We found that the growth of ydj1Δ and hsp82 temperature-sensitive yeast is improved by conditions that restore the viability of cells with mutations in cell-wall components or regulators of cell-wall biogenesis. We also found that the growth of these strains is hindered by a compound that intercalates into the cell wall. Furthermore, we discovered that the viability of the ydj1Δ and hsp82 temperature-sensitive strains is improved by the overexpression of Mid2p—a cell-wall component that is thought to sense the integrity of this structure and to activate the PKC pathway when cell-wall architecture is compromised—and by the overexpression of Pkc1p and activated components in the PKC pathway, which are known to trigger increased synthesis of cell-wall components. Previous work indicated that the PKC pathway induces the synthesis of cell-wall glycoproteins and proteins involved in cell-wall biosynthesis, including Fks1p and Fks2p (β1,3-glucan synthase) and Chs3p (chitin synthase) (Jung and Levin 1999), and that Mid2p overexpression increases the chitin concentration in the cell wall (Ketela et al. 1999). Indeed, we found that Mid2p overexpression is sufficient to thicken the cell wall in yeast lacking YDJ1, which provides a rationale for why we identified MID2 as the strongest suppressor of the ydj1Δ slow-growth phenotype.
In addition to identifying MID2 and the PKC pathway as suppressors of the slow-growth phenotype of ydj1Δ yeast, our screen uncovered several other genes with links to cell-wall homeostasis and to the PKC pathway (Table 3). For example, Syp1p was discovered as a suppressor of yeast profilin deletion (pfy1Δ) (Marcoux et al. 2000), and Cis1p was uncovered as a suppressor of a cik1Δ yeast strain (Manning et al. 1997). Cik1p cooperates with the Kar3p microtubule motor protein to catalyze karyogamy and chromosome segregation (Page and Snyder 1992). Whereas Syp1p helps polarize actin patches, little else is known about Cis1p. Both screens also uncovered Mid2p and Rom2p—another upstream component of the PKC pathway—as suppressors, which suggests potential links between Syp1p and Cis1p and the PKC pathway. In addition, Cyc8p/Ssn6p, which is found in a corepressor complex with Tup1p, regulates genes under a wide array of stress conditions, including growth in hypertonic media (Proft et al. 2001; Proft and Struhl 2002). Finally, SKN1 is homologous to KRE6, which exhibits synthetic interactions with PKC1, MPK1, and MKK1/MKK2; moreover, Kre6p overexpression can rescue a pkc1Δ lysis defect (Roemer et al. 1994). In contrast, we were surprised to find that the overexpression of CHS5, which is involved in the transport of chitin synthase from the Golgi (Sanchatjate and Schekman 2006), did not rescue the slow-growth phenotype of ydj1Δ yeast (Table 3).
Prior to this study, to our knowledge, there were only two other links between yeast cell-wall integrity and chaperone function. First, it was observed that strains containing mutations in KAR2, which encodes an ER Hsp70, have decreased amounts of β1,6-glucan in the cell wall when ER-resident glucanases are disabled (Simons et al. 1998). Second, some but not all hsp82 mutant alleles exhibit sensitivity to growth on medium containing high sorbitol, which was interpreted as a connection between Hsp90 function and the HOG signaling pathway even though other hsp82 mutants were sensitive to CW (Yang et al. 2006). On the basis of the growing importance of chaperones in nearly every aspect of cell function and maintenance, we anticipate that additional connections between chaperone function and cell-wall architecture will be uncovered.
The Hsp40 and Hsp90 chaperones commonly associate with Hsp70 to engineer distinct cellular processes, and thus we were surprised that overexpression of Mid2p or components in the PKC pathway failed to rescue the ssa1-45 thermosensitive phenotype. One interpretation of this result is that Hsp40 and Hsp90, but not Hsp70, play a role in cell-wall biosynthesis, and we note that in vitro experiments suggested that Ydj1p and Hsp82p may hold PR in a hormone-binding competent state independent of Ssa1p (Hernandez et al. 2002a). However, another interpretation of this result is that allele-specific interactions underlie the ability of Mid2p and Pkc1p to improve the growth of ydj1Δ and hsp82, but not of ssa1-45 yeast. It is important to mention that the G170D and G313N mutations are in the Hsp82 ATPase domain and middle region, respectively (Nathan and Lindquist 1995; Fliss et al. 2000), and have been shown to affect protein stability and Hsp82p client protein activation (Bohen 1995; Lee et al. 2002; Youker et al. 2004). In contrast, the ssa1-45 allele encodes a P417L mutation in the peptide-binding domain of Ssa1p (Becker et al. 1996) and has been shown to compromise protein folding, translocation, and ER-associated degradation (Becker et al. 1996; Kim et al. 1998; Zhang et al. 2001). Moreover, the hsp82 mutations reside in a strain that lacks chromosomal copies of HSP82 and HSC82, whereas the ssa1-45 mutation resides in a strain that lacks the SSA2, SSA3, and SSA4 genes. Unfortunately, no other tight ssa1 temperature-sensitive strains exist in which we can further explore this phenomenon, and it is not clear how the necessary strain backgrounds for these studies affect the effects on cell-wall integrity/signaling that we observed.
Germane to our results, the PKC pathway has been previously linked to Hsp90 function. A yeast two-hybrid screen for Hsp90 clients was performed using yeast expressing an E33A mutant form of the protein that inhibits ATP hydrolysis and thus stabilizes client and cochaperone interactions (Millson et al. 2005). As anticipated, cochaperones such as Sba1p, Sti1p, Sse1p, and Ydj1p were identified, as well as the following kinases: Hog1p (HOG pathway), Kss1p (SVG pathway), Ste11p (several signaling pathways), and Mpk1p/Slt2p (PKC pathway) (Millson et al. 2005). Further, Hsp82 was shown to bind the phosphorylated, stress-activated form of Mpk1p. On the basis of these and other experiments, the authors suggested that Hsp90 folds Mpk1p, which, in turn, is required for proper signaling through the PKC pathway (Millson et al. 2005).
One goal of this study was to better understand why the K53R mutation in SV40 Tag compromises J domain function when inserted into a TAg-Ydj1p chimeric protein (Fewell et al. 2002). As hoped, we identified several genes that rescued to some degree the thermosensitive phenotype of ydj1Δ yeast-expressing T(K53R)-Ydj1p. On the basis of NMR perturbation studies (Greene et al. 1998), it was previously suggested that the solvent-exposed region in helix III that is occupied by the conserved K53 residue would not be in contact with Hsp70. Thus, we were surprised to identify the Hsp70-encoding SSA1 gene in our screen. There are two general explanations for this result. First, it is possible that additional copies of Hsp70 in the cell “fix” the aberrant conformation conferred by the K53R mutation; second, higher levels of Hsp70 might improve the function of a protein or process that lies downstream of the K53R-induced phenotype. At present, we are unable to differentiate between these scenarios.
Another weaker suppressor of the T(K53R)-YDJ1 phenotype that we identified was CIS1. As mentioned above, CIS1 was identified as a suppressor of a cik1Δ phenotype (Manning et al. 1997), and uncovering CIS1 as a weak suppressor of the T(K53R)-YDJ1 phenotype may be indicative of a link between Ydj1p function and microtubule homeostasis. In fact, Ssa1p and Ydj1p were previously suggested to be important for microtubule formation, and, more specifically, strains either containing an ssa1 temperature-sensitive allele (ssa1-134) or that were deleted for YDJ1 showed irregular microtubule assembly at the nonpermissive temperature after nocodazole treatment (Oka et al. 1998). Moreover, both the ssa1-134 and ydj1Δ alleles exhibit synthetic interactions with tub4-1, which is a temperature-sensitive allele in the gene encoding γ-tubulin. Therefore, improved growth of the T(K53R)-YDJ1-containing ydj1Δ strain by CIS1 overexpression may be due to the rescue of a residual microtubule defect.
Because suppressors of the T(K53R)-YDJ1 phenotype could be isolated from the nonbiased screen reported here, we have now begun to examine the effects on T(K53R)-Ydj1p-expressing ydj1Δ yeast when known or suggested TAg interactors are simultaneously produced. For example, we are currently investigating whether BAG1 or HspBP1 expression alter the T(K53R)-YDJ1 thermosensitive phenotype because these mammalian NEFs are known to interact with Hsp70 and to promote ADP dissociation (Raynes and Guerriero 1998; Kabani et al. 2002b; Alberti et al. 2003). Thus, defects in J domain-Hsp70 function evident in T(K53R)-Ydj1p-expressing ydj1Δ yeast might be repaired by producing a factor that enhances the Hsp70 ATPase cycle. We are also testing the effects of expressing Cul7, which, as a member of an SCF ubiquitin ligase complex, helps direct specific proteins for proteasomal degradation (Dias et al. 2002; Ali et al. 2004). Cul7 binds to the TAg J domain in the extended loop between helices III and IV (Kasper et al. 2005), a region that is encoded in the chimeric T-YDJ1 construct used in our studies. In addition to these directed experiments, we also hope in the future to perform a nonbiased suppressor screen using a mammalian, yeast-expression library to uncover new TAg J domain-binding proteins.
Finally, our study suggests a putative connection between TAg J domain function and the PKC pathway. Previous work demonstrated that TAg blocked the apoptotic response upon EGF withdrawal from mouse embryo cells, and chemical inhibition of the PKC pathway also inhibited apoptosis (A. Slinskey and J. M. Pipas, unpublished data). In addition, the amount of an uncharacterized PKC phosphorylated substrate decreased in mouse intestines upon TAg expression (R. Beerman, M. T. Saenz-Robles and J. M. Pipas, unpublished data). While these results suggest a connection between TAg and PKC, much more work is clearly needed to define how the TAg J domain affects the PKC pathway.
Acknowledgments
The authors thank Howard Bussey, Avrom Caplan, Elizabeth Craig, Angel Duran, Loren Field, Vince Guerriero, Jörg Höhfeld, Ed Hurt, David Levin, Daniel Lew, Susan Lindquist, Jürg Nüesch, Irene Ota, Dominick Pallotta, Susana Rodríguez-Navarro, Akira Sakai, Charles Singleton, Dennis Thiele, Jeremy Thorner, Robert Trumbly, and John Warner for reagents. We also thank Sara Hileman for technical assistance with immunofluorescence, Shruthi Vembar and Stacy Hrizo for critical reading of the manuscript, and Craig Scott for assistance with the figures. This work was supported by grant GM75061 from the National Institutes of Health to J.L.B. C.M.W. was supported by a Renal Epithelial Biology training grant (DK61296) and a Mellon Graduate Fellowship.
References
- Adams, A. E., and J. R. Pringle, 1984. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J. Cell Biol. 98: 934–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, A., D. E. Gottschling, C. A. Kaiser and T. Stearns, 1997. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Alberti, S., C. Esser and J. Hohfeld, 2003. BAG-1: a nucleotide exchange factor of Hsc70 with multiple cellular functions. Cell Stress Chaperones 8: 225–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, S. H., and J. A. DeCaprio, 2001. Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin. Cancer Biol. 11: 15–23. [DOI] [PubMed] [Google Scholar]
- Ali, S. H., J. S. Kasper, T. Arai and J. A. DeCaprio, 2004. Cul7/p185/p193 binding to simian virus 40 large T antigen has a role in cellular transformation. J. Virol. 78: 2749–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, J., W. Walter, W. Yan and E. A. Craig, 1996. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell. Biol. 16: 4378–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohen, S. P., 1995. Hsp90 mutants disrupt glucocorticoid receptor ligand binding and destabilize aporeceptor complexes. J. Biol. Chem. 270: 29433–29438. [DOI] [PubMed] [Google Scholar]
- Bohen, S. P., and K. R. Yamamoto, 1993. Isolation of Hsp90 mutants by screening for decreased steroid receptor function. Proc. Natl. Acad. Sci. USA 90: 11424–11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster, J. L., T. de Valoir, N. D. Dwyer, E. Winter and M. C. Gustin, 1993. An osmosensing signal transduction pathway in yeast. Science 259: 1760–1763. [DOI] [PubMed] [Google Scholar]
- Brodsky, J. L., and J. M. Pipas, 1998. Polyomavirus T antigens: molecular chaperones for multiprotein complexes. J. Virol. 72: 5329–5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, J. L., and R. Schekman, 1993. A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J. Cell Biol. 123: 1355–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, K. S., K. P. Mullane, I. A. Aksoy, H. Stubdal, J. Zalvide et al., 1997. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 11: 1098–1110. [DOI] [PubMed] [Google Scholar]
- Caplan, A. J., and M. G. Douglas, 1991. Characterization of YDJ1: a yeast homologue of the bacterial dnaJ protein. J. Cell Biol. 114: 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan, A. J., D. M. Cyr and M. G. Douglas, 1992. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell 71: 1143–1155. [DOI] [PubMed] [Google Scholar]
- Caplan, A. J., E. Langley, E. M. Wilson and J. Vidal, 1995. Hormone-dependent transactivation by the human androgen receptor is regulated by a dnaJ protein. J. Biol. Chem. 270: 5251–5257. [DOI] [PubMed] [Google Scholar]
- Carlson, M., and D. Botstein, 1982. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell 28: 145–154. [DOI] [PubMed] [Google Scholar]
- Chang, H. C., D. F. Nathan and S. Lindquist, 1997. In vivo analysis of the Hsp90 cochaperone Sti1 (p60). Mol. Cell. Biol. 17: 318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham, M. E., and A. J. Caplan, 1998. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones 3: 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero and P. Hieter, 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119–122. [DOI] [PubMed] [Google Scholar]
- Coughlan, C. M., J. L. Walker, J. C. Cochran, K. D. Wittrup and J. L. Brodsky, 2004. Degradation of mutated bovine pancreatic trypsin inhibitor in the yeast vacuole suggests post-endoplasmic reticulum protein quality control. J. Biol. Chem. 279: 15289–15297. [DOI] [PubMed] [Google Scholar]
- Cyr, D. M., 1995. Cooperation of the molecular chaperone Ydj1 with specific Hsp70 homologs to suppress protein aggregation. FEBS Lett. 359: 129–132. [DOI] [PubMed] [Google Scholar]
- Cyr, D. M., X. Lu and M. G. Douglas, 1992. Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J. Biol. Chem. 267: 20927–20931. [PubMed] [Google Scholar]
- DeCaprio, J. A., 1999. The role of the J domain of SV40 large T in cellular transformation. Biologicals 27: 23–28. [DOI] [PubMed] [Google Scholar]
- Deshaies, R. J., S. L. Sanders, D. A. Feldheim and R. Schekman, 1991. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature 349: 806–808. [DOI] [PubMed] [Google Scholar]
- Dias, D. C., G. Dolios, R. Wang and Z. Q. Pan, 2002. CUL7: a DOC domain-containing cullin selectively binds Skp1.Fbx29 to form an SCF-like complex. Proc. Natl. Acad. Sci. USA 99: 16601–16606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas, C. M., F. Foor, J. A. Marrinan, N. Morin, J. B. Nielsen et al., 1994. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-beta-D-glucan synthase. Proc. Natl. Acad. Sci. USA 91: 12907–12911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, C. Y., H. Y. Ren, P. Lee, A. J. Caplan and D. M. Cyr, 2005. The type I Hsp40 zinc finger-like region is required for Hsp70 to capture non-native polypeptides from Ydj1. J. Biol. Chem. 280: 695–702. [DOI] [PubMed] [Google Scholar]
- Fang, Y., A. E. Fliss, J. Rao and A. J. Caplan, 1998. SBA1 encodes a yeast hsp90 cochaperone that is homologous to vertebrate p23 proteins. Mol. Cell. Biol. 18: 3727–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fewell, S. W., K. J. Travers, J. S. Weissman and J. L. Brodsky, 2001. The action of molecular chaperones in the early secretory pathway. Annu. Rev. Genet. 35: 149–191. [DOI] [PubMed] [Google Scholar]
- Fewell, S. W., J. M. Pipas and J. L. Brodsky, 2002. Mutagenesis of a functional chimeric gene in yeast identifies mutations in the simian virus 40 large T antigen J domain. Proc. Natl. Acad. Sci. USA 99: 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty, K. M., C. DeLuca-Flaherty and D. B. McKay, 1990. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature 346: 623–628. [DOI] [PubMed] [Google Scholar]
- Fliss, A. E., S. Benzeno, J. Rao and A. J. Caplan, 2000. Control of estrogen receptor ligand binding by Hsp90. J. Steroid Biochem. Mol. Biol. 72: 223–230. [DOI] [PubMed] [Google Scholar]
- Gassler, C. S., A. Buchberger, T. Laufen, M. P. Mayer, H. Schroder et al., 1998. Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc. Natl. Acad. Sci. USA 95: 15229–15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevaux, P., F. Schwager, C. Georgopoulos and W. L. Kelley, 2002. Scanning mutagenesis identifies amino acid residues essential for the in vivo activity of the Escherichia coli DnaJ (Hsp40) J-domain. Genetics 162: 1045–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R. D., and A. Sugino, 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534. [DOI] [PubMed] [Google Scholar]
- Goeckeler, J. L., A. Stephens, P. Lee, A. J. Caplan and J. L. Brodsky, 2002. Overexpression of yeast Hsp110 homolog Sse1p suppresses ydj1–151 thermosensitivity and restores Hsp90-dependent activity. Mol. Biol. Cell 13: 2760–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, M. K., K. Maskos and S. J. Landry, 1998. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc. Natl. Acad. Sci. USA 95: 6108–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin, M. C., J. Albertyn, M. Alexander and K. Davenport, 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62: 1264–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, W., and P. Christen, 2003. Mechanism of the targeting action of DnaJ in the DnaK molecular chaperone system. J. Biol. Chem. 278: 19038–19043. [DOI] [PubMed] [Google Scholar]
- Harlow, E., L. V. Crawford, D. C. Pim and N. M. Williamson, 1981. Monoclonal antibodies specific for simian virus 40 tumor antigens. J. Virol. 39: 861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, J. C., T. R. Zyla, E. S. Bardes and D. J. Lew, 2004. Stress-specific activation mechanisms for the “cell integrity” MAPK pathway. J. Biol. Chem. 279: 2616–2622. [DOI] [PubMed] [Google Scholar]
- Hata, H., H. Mitsui, H. Liu, Y. Bai, C. L. Denis et al., 1998. Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics 148: 571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy, F., W. S. Nicoll, R. Zimmermann, M. E. Cheetham and G. L. Blatch, 2005. Not all J domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 14: 1697–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, M. P., A. Chadli and D. O. Toft, 2002. a HSP40 binding is the first step in the HSP90 chaperoning pathway for the progesterone receptor. J. Biol. Chem. 277: 11873–11881. [DOI] [PubMed] [Google Scholar]
- Hernandez, M. P., W. P. Sullivan and D. O. Toft, 2002. b The assembly and intermolecular properties of the hsp70-Hop-hsp90 molecular chaperone complex. J. Biol. Chem. 277: 38294–38304. [DOI] [PubMed] [Google Scholar]
- Hill, J. E., A. M. Myers, T. J. Koerner and A. Tzagoloff, 1986. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2: 163–167. [DOI] [PubMed] [Google Scholar]
- Hjorth-Sorensen, B., E. R. Hoffmann, N. M. Lissin, A. K. Sewell and B. K. Jakobsen, 2001. Activation of heat shock transcription factor in yeast is not influenced by the levels of expression of heat shock proteins. Mol. Microbiol. 39: 914–923. [DOI] [PubMed] [Google Scholar]
- Ito, H., Y. Fukuda, K. Murata and A. Kimura, 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, J. L., and E. A. Craig, 2001. An essential role for the substrate-binding region of Hsp40s in Saccharomyces cerevisiae. J. Cell Biol. 152: 851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, U. S., and D. E. Levin, 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 34: 1049–1057. [DOI] [PubMed] [Google Scholar]
- Kabani, M., J. M. Beckerich and J. L. Brodsky, 2002. a Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol. Cell. Biol. 22: 4677–4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani, M., C. McLellan, D. A. Raynes, V. Guerriero and J. L. Brodsky, 2002. b HspBP1, a homologue of the yeast Fes1 and Sls1 proteins, is an Hsc70 nucleotide exchange factor. FEBS Lett. 531: 339–342. [DOI] [PubMed] [Google Scholar]
- Kaiser, C. A., and R. Schekman, 1990. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell 61: 723–733. [DOI] [PubMed] [Google Scholar]
- Kasper, J. S., H. Kuwabara, T. Arai, S. H. Ali and J. A. DeCaprio, 2005. Simian virus 40 large T antigen's association with the CUL7 SCF complex contributes to cellular transformation. J. Virol. 79: 11685–11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, W. L., and C. Georgopoulos, 1997. The T/t common exon of simian virus 40, JC, and BK polyomavirus T antigens can functionally replace the J-domain of the Escherichia coli DnaJ molecular chaperone. Proc. Natl. Acad. Sci USA 94: 3679–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketela, T., R. Green and H. Bussey, 1999. Saccharomyces cerevisiae mid2p is a potential cell wall stress sensor and upstream activator of the PKC1–MPK1 cell integrity pathway. J. Bacteriol. 181: 3330–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. Y., B. Y. Ahn and Y. Cho, 2001. Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen. EMBO J. 20: 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., B. Schilke, E. A. Craig and A. L. Horwich, 1998. Folding in vivo of a newly translated yeast cytosolic enzyme is mediated by the SSA class of cytosolic yeast Hsp70 proteins. Proc. Natl. Acad. Sci. USA 95: 12860–12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, Y., I. Yahara and S. Lindquist, 1995. Role of the protein chaperone YDJ1 in establishing Hsp90-mediated signal transduction pathways. Science 268: 1362–1365. [DOI] [PubMed] [Google Scholar]
- Landry, S. J., 2003. Structure and energetics of an allele-specific genetic interaction between dnaJ and dnaK: correlation of nuclear magnetic resonance chemical shift perturbations in the J-domain of Hsp40/DnaJ with binding affinity for the ATPase domain of Hsp70/DnaK. Biochemistry 42: 4926–4936. [DOI] [PubMed] [Google Scholar]
- Langer, T., C. Lu, H. Echols, J. Flanagan, M. K. Hayer et al., 1992. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature 356: 683–689. [DOI] [PubMed] [Google Scholar]
- Lee, B. N., and E. A. Elion, 1999. The MAPKKK Ste11 regulates vegetative growth through a kinase cascade of shared signaling components. Proc. Natl. Acad. Sci. USA 96: 12679–12684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. C., T. Hon and L. Zhang, 2002. The molecular chaperone Hsp90 mediates heme activation of the yeast transcriptional activator Hap1. J. Biol. Chem. 277: 7430–7437. [DOI] [PubMed] [Google Scholar]
- Lee, K. S., and D. E. Levin, 1992. Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol. Cell. Biol. 12: 172–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage, G., and H. Bussey, 2006. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70: 317–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, D. E., 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69: 262–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, D. E., and E. Bartlett-Heubusch, 1992. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J. Cell Biol. 116: 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. D., K. A. Morano and D. J. Thiele, 1999. The yeast Hsp110 family member, Sse1, is an Hsp90 cochaperone. J. Biol. Chem. 274: 26654–26660. [DOI] [PubMed] [Google Scholar]
- Lommel, M., M. Bagnat and S. Strahl, 2004. Aberrant processing of the WSC family and Mid2p cell surface sensors results in cell death of Saccharomyces cerevisiae O-mannosylation mutants. Mol. Cell. Biol. 24: 46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier, M., A. M. White, J. Sheraton, T. di Paolo, J. Treadwell et al., 1997. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 147: 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, D., J. G. Cook and J. Thorner, 1995. Phosphorylation and localization of Kss1, a MAP kinase of the Saccharomyces cerevisiae pheromone response pathway. Mol. Biol. Cell 6: 889–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager, W. H., and M. Siderius, 2002. Novel insights into the osmotic stress response of yeast. FEMS Yeast Res. 2: 251–257. [DOI] [PubMed] [Google Scholar]
- Manning, B. D., R. Padmanabha and M. Snyder, 1997. The Rho-GEF Rom2p localizes to sites of polarized cell growth and participates in cytoskeletal functions in Saccharomyces cerevisiae. Mol. Biol. Cell 8: 1829–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcoux, N., S. Cloutier, E. Zakrzewska, P. M. Charest, Y. Bourbonnais et al., 2000. Suppression of the profilin-deficient phenotype by the RHO2 signaling pathway in Saccharomyces cerevisiae. Genetics 156: 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, M. P., and B. Bukau, 2005. Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62: 670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur, P., N. Morin, W. Baginsky, M. el-Sherbeini, J. A. Clemas et al., 1995. Differential expression and function of two homologous subunits of yeast 1,3-beta-D-glucan synthase. Mol. Cell. Biol. 15: 5671–5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty, J. S., A. Buchberger, J. Reinstein and B. Bukau, 1995. The role of ATP in the functional cycle of the DnaK chaperone system. J. Mol. Biol. 249: 126–137. [DOI] [PubMed] [Google Scholar]
- McClellan, A. J., and J. L. Brodsky, 2000. Mutation of the ATP-binding pocket of SSA1 indicates that a functional interaction between Ssa1p and Ydj1p is required for post-translational translocation into the yeast endoplasmic reticulum. Genetics 156: 501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millson, S. H., A. W. Truman, V. King, C. Prodromou, L. H. Pearl et al., 2005. A two-hybrid screen of the yeast proteome for Hsp90 interactors uncovers a novel Hsp90 chaperone requirement in the activity of a stress-activated mitogen-activated protein kinase, Slt2p (Mpk1p). Eukaryot. Cell 4: 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montijn, R. C., E. Vink, W. H. Muller, A. J. Verkleij, H. Van Den Ende et al., 1999. Localization of synthesis of beta1,6-glucan in Saccharomyces cerevisiae. J. Bacteriol. 181: 7414–7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg, D., R. Muller and M. Funk, 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122. [DOI] [PubMed] [Google Scholar]
- Nathan, D. F., and S. Lindquist, 1995. Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol. Cell. Biol. 15: 3917–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman, A. M., V. Mhaiskar, V. Manus, F. Galibert and N. Dean, 1997. Saccharomyces cerevisiae HOC1, a suppressor of pkc1, encodes a putative glycosyltransferase. Genetics 145: 637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolet, C. M., and E. A. Craig, 1989. Isolation and characterization of STI1, a stress-inducible gene from Saccharomyces cerevisiae. Mol. Cell. Biol. 9: 3638–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka, M., M. Nakai, T. Endo, C. R. Lim, Y. Kimata et al., 1998. Loss of Hsp70-Hsp40 chaperone activity causes abnormal nuclear distribution and aberrant microtubule formation in M-phase of Saccharomyces cerevisiae. J. Biol. Chem. 273: 29727–29737. [DOI] [PubMed] [Google Scholar]
- O'Rourke, S. M., and I. Herskowitz, 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12: 2874–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi, M., 1998. The ultrastructure of yeast: cell wall structure and formation. Micron 29: 207–233. [DOI] [PubMed] [Google Scholar]
- Page, B. D., and M. Snyder, 1992. CIK1: a developmentally regulated spindle pole body-associated protein important for microtubule functions in Saccharomyces cerevisiae. Genes Dev. 6: 1414–1429. [DOI] [PubMed] [Google Scholar]
- Philip, B., and D. E. Levin, 2001. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol. Cell. Biol. 21: 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt, W. B., and D. O. Toft, 2003. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. (Maywood) 228: 111–133. [DOI] [PubMed] [Google Scholar]
- Proft, M., and K. Struhl, 2002. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 9: 1307–1317. [DOI] [PubMed] [Google Scholar]
- Proft, M., A. Pascual-Ahuir, E. de Nadal, J. Arino, R. Serrano et al., 2001. Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J. 20: 1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota, H., C. P. Python, S. B. Inoue, M. Arisawa, Y. Anraku et al., 1996. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-beta-glucan synthase. Science 272: 279–281. [DOI] [PubMed] [Google Scholar]
- Rajavel, M., B. Philip, B. M. Buehrer, B. Errede and D. E. Levin, 1999. Mid2 is a putative sensor for cell integrity signaling in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 3969–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynes, D. A., and V. Guerriero, Jr., 1998. Inhibition of Hsp70 ATPase activity and protein renaturation by a novel Hsp70-binding protein. J. Biol. Chem. 273: 32883–32888. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro, S., T. Fischer, M. J. Luo, O. Antunez, S. Brettschneider et al., 2004. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116: 75–86. [DOI] [PubMed] [Google Scholar]
- Roelants, F. M., P. D. Torrance and J. Thorner, 2004. Differential roles of PDK1- and PDK2-phosphorylation sites in the yeast AGC kinases Ypk1, Pkc1 and Sch9. Microbiology 150: 3289–3304. [DOI] [PubMed] [Google Scholar]
- Roemer, T., G. Paravicini, M. A. Payton and H. Bussey, 1994. Characterization of the yeast (1→6)-beta-glucan biosynthetic components, Kre6p and Skn1p, and genetic interactions between the PKC1 pathway and extracellular matrix assembly. J. Cell Biol. 127: 567–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudiger, S., J. Schneider-Mergener and B. Bukau, 2001. Its substrate specificity characterizes the DnaJ co-chaperone as a scanning factor for the DnaK chaperone. EMBO J. 20: 1042–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, R., A. Wali Karzai, A. F. Mehl and R. McMacken, 1999. DnaJ dramatically stimulates ATP hydrolysis by DnaK: insight into targeting of Hsp70 proteins to polypeptide substrates. Biochemistry 38: 4165–4176. [DOI] [PubMed] [Google Scholar]
- Sanchatjate, S., and R. Schekman, 2006. Chs5/6 complex: a multi-protein complex that interacts with and conveys chitin synthase III from the trans-Golgi network to the cell surface. Mol. Biol. Cell 17: 4157–4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, B., and M. Snyder, 1997. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J. Cell Biol. 136: 95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, B., A. Duran and M. H. Valdivieso, 1997. CHS5, a gene involved in chitin synthesis and mating in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 2485–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz, H. P., J. Jockel, C. Block and J. J. Heinisch, 2001. Domain shuffling as a tool for investigation of protein function: substitution of the cysteine-rich region of Raf kinase and PKC eta for that of yeast Pkc1p. J. Mol. Biol. 311: 1–7. [DOI] [PubMed] [Google Scholar]
- Shahinian, S., and H. Bussey, 2000. beta-1,6-Glucan synthesis in Saccharomyces cerevisiae. Mol. Microbiol. 35: 477–489. [DOI] [PubMed] [Google Scholar]
- Shomura, Y., Z. Dragovic, H. C. Chang, N. Tzvetkov, J. C. Young et al., 2005. Regulation of Hsp70 function by HspBP1: structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol. Cell 17: 367–379. [DOI] [PubMed] [Google Scholar]
- Simons, J. F., M. Ebersold and A. Helenius, 1998. Cell wall 1,6-beta-glucan synthesis in Saccharomyces cerevisiae depends on ER glucosidases I and II, and the molecular chaperone BiP/Kar2p. EMBO J. 17: 396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton, C. K., 1997. Identification and characterization of the thiamine transporter gene of Saccharomyces cerevisiae. Gene 199: 111–121. [DOI] [PubMed] [Google Scholar]
- Smith, D. F., B. A. Stensgard, W. J. Welch and D. O. Toft, 1992. Assembly of progesterone receptor with heat shock proteins and receptor activation are ATP mediated events. J. Biol. Chem. 267: 1350–1356. [PubMed] [Google Scholar]
- Srinivasan, A., A. J. McClellan, J. Vartikar, I. Marks, P. Cantalupo et al., 1997. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol. Cell. Biol. 17: 4761–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling, C. J., J. Rothblatt, M. Hosobuchi, R. Deshaies and R. Schekman, 1992. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol. Biol. Cell 3: 129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, D. E., and E. A. Craig, 1990. Self-regulation of 70-kilodalton heat shock proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 1622–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh, W. C., W. F. Burkholder, C. Z. Lu, X. Zhao, M. E. Gottesman et al., 1998. Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc. Natl. Acad. Sci. USA 95: 15223–15228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, C. S., and J. M. Pipas, 2002. T antigens of simian virus 40: molecular chaperones for viral replication and tumorigenesis. Microbiol. Mol. Biol. Rev. 66: 179–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, C. S., P. Cantalupo and J. M. Pipas, 2000. The molecular chaperone activity of simian virus 40 large T antigen is required to disrupt Rb-E2F family complexes by an ATP-dependent mechanism. Mol. Cell. Biol. 20: 6233–6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo, A., R. Korszun, F. U. Hartl and J. Flanagan, 1996. A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J. 15: 408–417. [PMC free article] [PubMed] [Google Scholar]
- Terasawa, K., M. Minami and Y. Minami, 2005. Constantly updated knowledge of Hsp90. J. Biochem. 137: 443–447. [DOI] [PubMed] [Google Scholar]
- Trumbly, R. J., 1988. Cloning and characterization of the CYC8 gene mediating glucose repression in yeast. Gene 73: 97–111. [DOI] [PubMed] [Google Scholar]
- Tsai, J., and M. G. Douglas, 1996. A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J. Biol. Chem. 271: 9347–9354. [DOI] [PubMed] [Google Scholar]
- Valdivia, R. H., and R. Schekman, 2003. The yeasts Rho1p and Pkc1p regulate the transport of chitin synthase III (Chs3p) from internal stores to the plasma membrane. Proc. Natl. Acad. Sci. USA 100: 10287–10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall, D., M. Zylicz and C. Georgopoulos, 1994. The NH2-terminal 108 amino acids of the Escherichia coli DnaJ protein stimulate the ATPase activity of DnaK and are sufficient for lambda replication. J. Biol. Chem. 269: 5446–5451. [PubMed] [Google Scholar]
- Walsh, P., D. Bursac, Y. C. Law, D. Cyr and T. Lithgow, 2004. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 5: 567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T. F., J. H. Chang and C. Wang, 1993. Identification of the peptide binding domain of hsc70. 18-Kilodalton fragment located immediately after ATPase domain is sufficient for high affinity binding. J. Biol. Chem. 268: 26049–26051. [PubMed] [Google Scholar]
- Wickner, S., J. Hoskins and K. McKenney, 1991. Function of DnaJ and DnaK as chaperones in origin-specific DNA binding by RepA. Nature 350: 165–167. [DOI] [PubMed] [Google Scholar]
- Williams, F. E., and R. J. Trumbly, 1990. Characterization of TUP1, a mediator of glucose repression in Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 6500–6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, A., C. Arkind, C. P. Mattison, A. Burkholder, K. Knoche et al., 2002. Heat stress activates the yeast high-osmolarity glycerol mitogen-activated protein kinase pathway, and protein tyrosine phosphatases are essential under heat stress. Eukaryot. Cell 1: 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. X., K. C. Maurer, M. Molanus, W. H. Mager, M. Siderius et al., 2006. The molecular chaperone Hsp90 is required for high osmotic stress response in Saccharomyces cerevisiae. FEMS Yeast Res. 6: 195–204. [DOI] [PubMed] [Google Scholar]
- Yashar, B., K. Irie, J. A. Printen, B. J. Stevenson, G. F. Sprague, Jr. et al., 1995. Yeast MEK-dependent signal transduction: response thresholds and parameters affecting fidelity. Mol. Cell. Biol. 15: 6545–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youker, R. T., P. Walsh, T. Beilharz, T. Lithgow and J. L. Brodsky, 2004. Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol. Biol. Cell 15: 4787–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, J. C., J. M. Barral and F. Ulrich Hartl, 2003. More than folding: localized functions of cytosolic chaperones. Trends Biochem. Sci. 28: 541–547. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., G. Nijbroek, M. L. Sullivan, A. A. McCracken, S. C. Watkins et al., 2001. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol. Biol. Cell 12: 1303–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, R., and W. A. Houry, 2005. Hsp90: a chaperone for protein folding and gene regulation. Biochem. Cell Biol. 83: 703–710. [DOI] [PubMed] [Google Scholar]
- Zhou, Z., A. Gartner, R. Cade, G. Ammerer and B. Errede, 1993. Pheromone-induced signal transduction in Saccharomyces cerevisiae requires the sequential function of three protein kinases. Mol. Cell. Biol. 13: 2069–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X., X. Zhao, W. F. Burkholder, A. Gragerov, C. M. Ogata et al., 1996. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272: 1606–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]