Abstract
Prospects for utilizing whole-genome association analysis in autogamous plant populations appear promising due to the reported high levels of linkage disequilibrium (LD). To determine the optimal strategies for implementing association analysis in soybean (Glycine max L. Merr.), we analyzed the structure of LD in three regions of the genome varying in length from 336 to 574 kb. This analysis was conducted in four distinct groups of soybean germplasm: 26 accessions of the wild ancestor of soybean (Glycine soja Seib. et Zucc.); 52 Asian G. max Landraces, the immediate results of domestication from G. soja; 17 Asian Landrace introductions that became the ancestors of North American (N. Am.) cultivars, and 25 Elite Cultivars from N. Am. In G. soja, LD did not extend past 100 kb; however, in the three cultivated G. max groups, LD extended from 90 to 574 kb, likely due to the impacts of domestication and increased self-fertilization. The three genomic regions were highly variable relative to the extent of LD within the three cultivated soybean populations. G. soja appears to be ideal for fine mapping of genes, but due to the highly variable levels of LD in the Landraces and the Elite Cultivars, whole-genome association analysis in soybean may be more difficult than first anticipated.
LINKAGE disequilibrium (LD) is the nonrandom association of alleles at different loci, and is affected by a number of factors. The processes of domestication, population subdivision, founding events, and selection can increase LD throughout the genome or in genomic segments flanking selected loci (Rafalski and Morgante 2004). Recombination decreases LD in a population and can eventually restore equilibrium between loci. LD is the basis of genetic association analysis for the discovery and fine mapping of genes or quantitative trait loci (QTL) in natural populations (Thornsberry et al. 2001; Wilson et al. 2004). Genetic association analysis measures correlations between genetic variants and phenotypic differences on a population basis and thus depends on LD for the detection of significant associations (Flint-Garcia et al. 2003).
LD has been found to have a structure in humans that is best described using a haplotype block model. Haplotype blocks are consecutive sites in high LD flanked by blocks demonstrating historical recombination (Daly et al. 2001; Gabriel et al. 2002; Altshuler et al. 2005). This type of structure can obscure predictable association based purely on physical distance between loci. If the structure of LD is unknown, a significant association of a sequence variant with a trait can place the gene or QTL anywhere within the haplotype block. Phase I of the HapMap project demonstrated that determination of the structure of LD in a subset of populations can identify a subgroup of markers (tag SNPs) that can be useful in sampling most common variations (Altshuler et al. 2005).
Soybean (Glycine max L. Merr.) is a major crop plant grown worldwide on 74 million hectares (Wilcox 2004) and is a species in which there is the potential to apply genetic association analysis for QTL discovery and fine mapping. Soybean was domesticated ∼3000–5000 years ago from the wild species G. soja (Seib. et Zucc.) (Hymowitz 2004). While cultivated soybean is widely known as an autogamous species with outcrossing rates of <1%, the wild progenitor G. soja has been reported to have an outcrossing rate as high as 13% (Fujita et al. 1997). The greater amount of outcrossing in G. soja increases the effective recombination rate, leading to the prediction of an 11-fold lower extent of LD in G. soja as compared to G. max (Flint-Garcia et al. 2003). The largest resource of soybean germplasm is the Asian landraces of G. max that are the most immediate result of domestication. Selections from these landraces became the first introductions grown by North American (N. Am.) farmers and also were the germplasm used for N. Am. cultivar development. This was followed by breeding programs based upon hybridization and selection, resulting in the release of improved cultivars beginning in 1947. Gizlice et al. (1994) analyzed the pedigrees of 258 publicly developed cultivars released between 1947 and 1988 and determined that >86% of the parentage could be traced to only 17 ancestors selected from the introduced landraces. Thus, the current N. Am. soybean germplasm pool, as defined by Gizlice et al. (1994), is the result of several cycles of selection and effective recombination among a relatively small number of selections from the Asian landraces. Our objective was to determine if LD structure and extent varies between different populations of soybean and to determine if the structure and extent of LD is consistent throughout the genome within the individual populations.
MATERIALS AND METHODS
Plant materials:
The plant materials included genotypes from four soybean populations described by Hyten et al. (2006) and listed in supplemental Table S1 at http://www.genetics.org/supplemental/. The first population consisted of 26 G. soja plant introductions from China, Korea, Taiwan, Russia, and Japan. The population of Landraces consisted of 52 Asian plant introductions from China, Korea, and Japan. The G. soja and the Landraces were selected to represent a range of geographic origin and various maturity classes to maximize the diversity sampled. The 17 N. Am. Ancestors were the specific G. max accessions that are estimated to contribute at least 86% of the genes present in the gene pool of N. Am. soybean cultivars (Gizlice et al. 1994). The population of Elite Cultivars consisted of 25 N. Am. cultivars publicly released between 1977 and 1990, which were selected to maximize diversity on the basis of coefficient-of-parentage estimations by Gizlice et al. (1996). Seeds of all genotypes were obtained from the U.S. Department of Agriculture (USDA) Soybean Germplasm Collection (USDA-Agricultural Resource Service, University of Illinois, Urbana, IL). DNA was extracted from bulked leaf tissue of 8–10 G. soja plants or 30–50 G. max plants as described by Keim et al. (1988).
Source of genomic sequences:
Currently, only three regions of contiguous sequence >300 kb in length are publicly available in soybean. Two genomic regions have been deposited in GenBank under accession nos. AX196295, AX196296, AX196297, and AX197417 (http://www.ncbi.nlm.nih.gov). The program bl2seq (http://www.ncbi.nlm.nih.gov) was used for all comparisons of sequences from GenBank. GenBank accession nos. AX196295 and AX196296 completely aligned with a sequence length of 336 kb and were considered one sequence. AX196295 was placed on the genetic map by aligning it to GenBank accession no. BH126500, which is the microsatellite marker BARC-Satt309 mapping to soybean linkage group (LG) G (Song et al. 2004). BARC-Satt309 is tightly linked to the soybean disease resistance gene for soybean cyst nematode (rhg1) (Cregan et al. 1999). The genome region AX196295 will be referred to hereafter as chromosomal region G (CR-G). GenBank accession nos. AX196297 and AX197417 have a 50-kb overlap that forms a complete sequence with a 513-kb total length. AX196297 was placed on the genetic map by aligning it to GenBank accession no. BH126793, which is the microsatellite marker BARC-Satt632 mapping to LG A2 (Song et al. 2004). BARC-Satt632 is tightly linked to the soybean disease resistance gene for soybean cyst nematode (Rhg4) (Cregan et al. 1999). The genome region of AX196297 and AX197417 will be referred to as CR-A2. The third chromosomal region studied was constructed by Graham et al. (2000) and is a BAC contig that has an estimated physical length of 574 kb and is located on LG J. The ends of all BAC clones in the contig have been sequenced and were used for sequence tagged site (STS) development. This BAC contig region will be referred to as CR-J.
SNP discovery and genotyping:
PCR primers were designed throughout the three chromosomal regions with Array Designer 2.0 (Premier Biosoft International, Palo Alto, CA). Primers were used to amplify genomic DNA from the soybean genotypes “Archer,” “Minsoy,” “Noir 1,” “Evans,” “Peking,” and PI 209332. In those instances when a single discrete amplicon was produced, the DNA sequence of each product was determined to verify that it was an STS. PCR and amplification conditions were previously described by Zhu et al. (2003). Forward and reverse sequencing reactions were performed on an ABI 3700 or ABI 3730 using ABI Prism BigDye Terminator version 3.1 cycle sequencing (Applied Biosystems, Foster City, CA). Evenly distributed STSs containing one or more SNPs in the six genotypes were selected throughout the three chromosomal regions for genotyping in each of the 120 individuals composing the four populations. Primer information and positions of the STSs on the three chromosomal regions are listed in supplemental Table S2 at http://www.genetics.org/supplemental/. The genotyping was done via forward and reverse sequencing reactions on the ABI 3700 or ABI 3730 as described above.
Sequence analyses:
Sequence data from each STS were analyzed with SNP-PHAGE (Matukumalli et al. 2006b), which includes the standard DNA analysis software Phred, which estimates the probability of error in base calling, and Phrap that performs sequence alignment and a machine-learning method for SNP discovery (Matukumalli et al. 2006a) and summarizes all data in a MySQL database. The resulting alignments and SNP predictions were visually verified using the Consed viewer (Gordon et al. 1998). SNPs were resequenced if there was any ambiguity as to which allele was present. The pairwise estimates D′ and r2 (Gaut and Long 2003) were calculated using SNPs with a frequency >10% in the individual populations using the software package Haploview v. 3.31 (Barrett et al. 2005). The aggressive tagger mode in Haploview was used to determine tag SNPs, where all common SNPs had a correlation of r2 ≥ 0.8 with one or more of the tag SNPs. SNP data from Hyten et al. (2006) were used to calculate population structure. This data set included the resequencing of 102 randomly chosen genes in the same germplasm used in this study. The SNP information was converted into haplotypes from the 102 loci in each accession and used for the structure analysis. The results were based on a model with correlated allele frequencies among populations in the program Structure 2.0 (Pritchard et al. 2000) with number of populations, K = 2–10, length of burn-in period 50,000, and a run of 500,000 replications of Markov chain Monte Carlo after burn in. Results of Structure were visualized with Distruct software (Rosenberg 2004).
RESULTS
SNP discovery and coverage:
The three regions used for STS development were located in different linkage groups. They consisted of a 336-kb sequenced region (CR-G), a 513-kb sequenced region (CR-A2), and a 574-kb BAC contig with available BAC end sequence (CR-J). A total of 309 PCR primer pairs with amplicons ranging from 500 to 800 bp in length were tested from the three CRs, and 167 (54%) produced a robust STS. Overall, 558 SNPs were discovered in 122 (73%) of the STSs in the diverse group of six G. max genotypes. The sequence analysis of these six genotypes has been demonstrated to discover 93% of the common SNPs (frequency > 0.10) in a diverse G. max germplasm sample (Zhu et al. 2003). The remaining 27% of the STSs were monomorphic in the six genotypes. Sequence diversity was not estimated since the elimination of monomorphic STSs would upwardly bias diversity estimates in the four populations. Seventy-four polymorphic STSs were selected to give maximum coverage across the three chromosomal regions with an average of one STS every 13.5 kb in CR-A2, 12.4 kb in CR-G, and 57.4 kb in CR-J. The lower frequency of STS in CR-J was due to the incomplete sequence data available for this BAC contig.
Population structure:
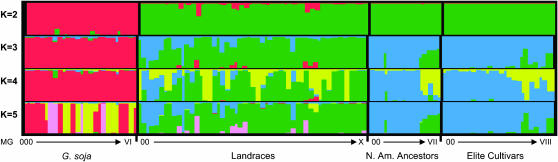
Population structure is commonly evaluated using the software program Structure, which implements a model-based clustering algorithm (Pritchard et al. 2000). Determining the true value of K for population structure from the Hyten et al. (2006) data set was difficult, because there was a lack of a plateau of the ln Pr(X|K) value, which is used to test for significance of the K value. In this study, our sampling strategy presumed four distinct populations, yet the use of any K value from 2 to 5 still resulted in the N. Am. Ancestors and the Elite Cultivars clustering together (Figure 1). Despite the clustering together of the N. Am. Ancestors and the Elite Cultivars, we kept these populations separate for all subsequent analyses since the sampling strategy to obtain the two populations was very different. Of course, the larger K values with multiple clusters occurring within a single population also indicate that population structure may occur in any of the three main populations.
Figure 1.—
Population structure based upon haplotype data of 102 genes of the four soybean populations taken from Hyten et al. (2006). K is the assumption of the number of clusters present (K = 2–5). Each colored vertical line represents an individual that is assigned proportionally to one of the K clusters with the proportions represented by the relative lengths of the K different colors. The individuals within each population are ordered by maturity group (MG) where the earliest-maturing individuals are to the left of each individual population and the latest-maturing individuals are to the right of each individual population.
LD across population samples:
Few summary statistics are available for characterizing LD across large chromosomal regions in large data sets (Gaut and Long 2003). The most common methods are to calculate the pairwise comparison D′ and r2 between all physically linked polymorphic marker combinations and plot these values against distance or in a matrix form (Gaut and Long 2003). D′ is a useful measure for detecting historical recombination, since it will have a value <1 only if all four haplotypes are observed between biallelic loci.
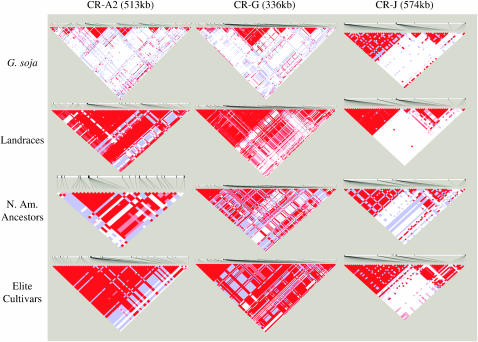
The D′ matrix for the four populations reveals a very different pattern of LD decay between the G. soja and the three cultivated soybean populations (Figure 2). It is apparent that LD has been degraded throughout the three regions in G. soja with only small haplotype blocks still remaining. The haplotype blocks in the G. soja population cover only a small fraction of the three chromosomal regions. Using common methods to define haplotype blocks (Gabriel et al. 2002; Wang et al. 2002; Barrett et al. 2005), G. soja had haplotype blocks with an average block length of 4.8 kb/block that covered 18% of the sequence length (Table 1). The largest haplotype block spanned 25 kb with the majority of blocks spanning <1 kb (data not shown).
Figure 2.—
D′ plots of the three chromosomal regions CR-A2, CR-G, and CR-J in G. soja, Landraces, N. Am. Ancestors, and Elite Cultivars: bright red, D′ = 1 and LOD ≥ 2; shades of pink/red, D′ < 1 and LOD ≥ 2; blue, D′ = 1 and LOD < 2; white, D′ < 1 and LOD < 2.
TABLE 1.
Summary of haplotype blocks in four soybean populations estimated by three methods commonly used to define haplotype blocks
| G. soja | Landraces | N. Am. Ancestors | Elite Cultivars | |
|---|---|---|---|---|
| Blocks estimated using confidence bounds on D′ (Gabriel et al. 2002) | ||||
| Average length per block (kb) | 2.1 | 19.0 | 16.3 | 56.8 |
| Amount of sequence included in blocks (%) | 5 | 36 | 18 | 48 |
| Blocks estimated by four-gamete rule (Wang et al. 2002) | ||||
| Average length per block (kb) | 4.3 | 13.6 | 21.3 | 46.0 |
| Amount of sequence included in blocks (%) | 19 | 35 | 36 | 65 |
| Blocks estimated by Solid Spine of LD (Barrett et al. 2005) | ||||
| Average length per block (kb) | 4.8 | 21.5 | 34.9 | 80.1 |
| Amount of sequence included in blocks (%) | 18 | 42 | 44 | 51 |
The Landrace and N. Am. Ancestor populations had similar-size haplotype blocks, which were on average much larger than those of the G. soja population (Table 1). The largest block in the Landraces spanned a distance of 186 kb, but many blocks were <1 kb in size (data not shown). The largest block in the N. Am. Ancestors spanned 89 kb (data not shown). The average block size and the amount of sequence covered by blocks were the greatest in the Elite Cultivars (Table 1). The average block size in the Elite Cultivars was more than twice that of any of the other populations as estimated by each of the three methods of haplotype block determination (Table 1) and there were only a few blocks that were <1 kb (data not shown).
LD across different genomic regions:
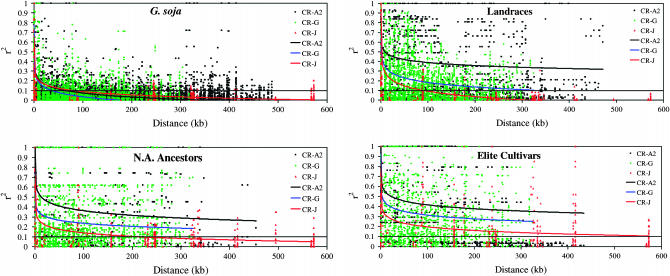
The limited LD in G. soja across the three chromosomal regions is also apparent when r2 is plotted against distance (Figure 3). The average decay of LD in G. soja in the three chromosomal regions analyzed declined to an r2 = 0.1 between 36 and 77 kb (Figure 3). In contrast to G. soja, the other three populations did not demonstrate consistent decay of LD across the three fragments (Figure 3). In CR-A2, LD never decayed below r2 = 0.1 throughout the 513-kb region in the Landraces, N. Am. Ancestors, or the Elite Cultivars. In the CR-G region, LD decay reached r2 = 0.1 at 300 kb in the Landraces but never reached an r2 = 0.1 in the N. Am. Ancestors or in the Elite Cultivars. In both the N. Am. Ancestors and the Elite Cultivars, the decline of r2 in CR-G was greater than in CR-A2. In CR-J, LD decayed to r2 = 0.1 at a distance of ∼90 kb in the Landraces, 212 kb in the N. Am. Ancestors, and 574 kb in the Elite Cultivars (Figure 3).
Figure 3.—
Linkage disequilibrium plots of r2 vs. distance for the three chromosomal regions CR-A2, CR-G, and CR-J in G. soja, Landraces, N. Am. Ancestors, and Elite Cultivars.
Tag SNPs needed for whole-genome association analysis:
Whole-genome association analysis is most efficient in a population with high LD. It is thus apparent that the three G. max populations are more suited for whole-genome association analysis than the G. soja population. However, the small number of the N. Am. Ancestors makes this an impractical population in which to conduct association studies. We therefore determined only the number of tag SNPs needed to detect most of the haplotype block variation in the Landraces and the Elite Cultivars (Table 2).
TABLE 2.
Tag SNPs needed to define allelic variation in Landrace and Elite Cultivars
| Population | Chromosomal region | Size of chromosomal region (kb) | No. of common SNPs (frequency >10%) genotyped | Tag SNPs estimated to capture 100% of allelic variation at r2 > 0.8 | Distribution of tag SNPs (kb/tag SNP) | Estimated tag SNPs required for whole- genome scana |
|---|---|---|---|---|---|---|
| Landraces | CR-A2 | 513 | 96 | 10 | 51 | 13,700 |
| CR-G | 336 | 176 | 36 | 9 | 75,600 | |
| CR-J | 574 | 52 | 16 | 35 | 19,700 | |
| Elite Cultivars | CR-A2 | 513 | 74 | 7 | 73 | 9,600 |
| CR-G | 336 | 149 | 14 | 24 | 29,400 | |
| CR-J | 574 | 51 | 12 | 47 | 14,800 |
Genome euchromatic DNA estimated as 705 Mb (Hymowitz 2004).
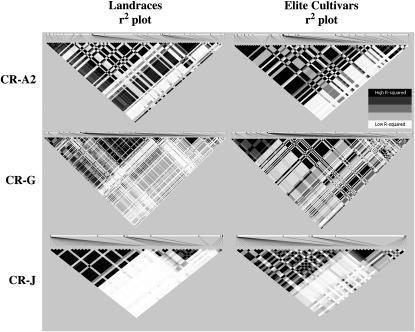
Tag SNPs are defined as a subset of SNPs that capture a large fraction of the allelic variation of all SNP loci (Altshuler et al. 2005). We found that the number of tag SNPs needed to capture 100% of alleles at an r2 > 0.8 in the three regions ranged from a SNP every 9 kb to a SNP every 73 kb, reducing the number of SNPs needed for an effective whole-genome association analysis by 69–91% (Table 2). These data imply that a whole-genome association analysis that will test for most of the haplotype variation of the euchromatic DNA, which was estimated by Hymowitz (2004) to be 64% of the genome or 705 Mb, would require 9600–75,600 SNPs, depending on which of the three fragments is the most representative of the soybean genome (Table 2). Wide variation in the estimated number of SNPs needed to conduct a whole-genome association analysis is due to the differences of LD present among the two populations and the differences in the three genome regions as seen on an r2 plot (Figure 4). With further sampling of genomic LD, a better estimate of the number of SNPs needed for a whole-genome association analysis can be obtained.
Figure 4.—
r2 plots of the three chromosomal regions CR-A2, CR-G, and CR-J in the Landraces and Elite Cultivars, which are likely candidate populations for whole-genome association analysis in soybean: solid, r2 = 1; shadings, 0 < r2 < 1; open, r2 = 0.
DISCUSSION
Variable LD between chromosomal regions and populations:
While effective recombination rate does affect LD decay, many other factors, such as domestication, selection, founding events, population subdivision, and population stratification, can complicate predictions of the extent of LD (Flint-Garcia et al. 2003). The Landraces resulted from domestication, which would be expected to increase LD throughout the entire genome. Furthermore, loci governing traits directly associated with domestication would be associated with extended levels of LD. A possible example is the level of LD on CR-A2, which is more extensive than that in CR-G or CR-J in the Landraces. CR-A2 contains the I locus, which is one of a set of chalcone synthase gene duplications that affect hilum and seed-coat color (Todd and Vodkin 1996). It is likely that the I locus is a gene that was under selection during domestication since nearly all G. soja accessions have completely pigmented seed coats and most landraces have yellow seed coats. Through a sampling of Landraces that contain the different alleles of the I locus, it should be possible to determine if the CR-A2 is a region that was affected by a selective sweep during domestication.
A large effect of domestication on LD has been demonstrated in barley. Wild barley has LD levels that are equal to maize (Morrell et al. 2005), but the LD in the barley landraces is more extensive and most likely due to domestication (Caldwell et al. 2006). Rice, which has also undergone domestication and is a selfing species, has extended amounts of LD ranging up to 115 kb (Garris et al. 2003). However, in soybean, CR-J had LD decay that was almost identical in G. soja and the Landraces. This could suggest that LD in the Landraces may be similar to that of the wild ancestor and that both CR-A2 and CR-G had been targets of selective sweeps with resultant higher levels of LD. Another possibility is that selective forces have acted upon CR-J, which contains several disease-resistant genes, to rapidly decrease the extent of LD via balancing selection.
Before general conclusions about genomewide LD decay in soybean can be reached, more regions need to be characterized, as has been done in Arabidopsis. The first measurement of LD in Arabidopsis was around the FRI locus and it was concluded that LD extended up to 250 kb (Nordborg et al. 2002). A further study of the entire genome indicated that the extent of LD is closer to 50 kb (Nordborg et al. 2005). A similar genome scan of soybean would help determine if the average genome LD is closest to the estimates from the genomic regions CR-A2, CR-G, or CR-J. Such an analysis would greatly assist in the design of future association analysis studies.
The N. Am. Ancestors are the founding population for the Elite Cultivars. An analysis of population structure suggested that there is little structure between these two populations. Compared to the Landraces, these two populations show evidence of increased LD in CR-G and CR-J. A previous study characterized LD in 16 direct introductions to N. Am. (of which 12 accessions were in common with this study) and found LD was extensive and dissipated at 2–3 cM over a 12.5-cM region that encompasses CR-G (Zhu et al. 2003). This result is in agreement with this study, where LD did not dissipate below the r2 = 0.1 threshold in the 17 ancestors in CR-G. Further sequence data in the region surrounding CR-G are needed to confirm LD dissipation in 2–3 cM, which is a distance of 800–1200 kb.
The increased LD in the Elite Cultivars vs. the Landraces and the N. Am. Ancestors may be due to a number of factors. Selection occurring on or near the three chromosomal regions could be responsible for the increased LD. Similarly, an increase in LD due to selection and the bottleneck created by the development of elite inbred lines has also been shown in an elite maize population (Tenaillon et al. 2001) and elite barley populations (Caldwell et al. 2006; Rostoks et al. 2006). Another factor that may have contributed to increased LD in soybean is photoperiod sensitivity (maturity), which resulted in population subdivision in elite soybean cultivars. The structure analysis of 102 genes throughout the genome in the Elite Cultivars revealed a subdivision between early maturing and later-maturing cultivars (Figure 1).
Whole-genome association analysis:
The variable pattern of LD among fragments and populations provides a range of estimates of the number of tag SNPs that will be needed to capture most haplotype variation for a whole-genome association analysis in soybean. Extensive collections of thousands of Asian Landraces and large collections of elite cultivars should provide excellent populations to facilitate whole-genome association analysis. Arabidopsis, which is an autogamous species, has LD extending up to 50 kb and would require one marker within 10 kb of a causative polymorphism to provide reasonable power for a whole-genome scan for QTL discovery (Aranzana et al. 2005). The estimates of LD that we have found in the Landraces and the Elite Cultivars are two to seven times higher than in Arabidopsis, except in the case of CR-J, which was similar to Arabidopsis (Table 2). Still, with the greater extent of LD that we estimated for soybean compared to Arabidopsis, the greater size of the soybean genome implies that 9600–75,600 SNP markers would be needed to successfully identify most common haplotype variation (haplotypes with a minimum allele frequency >10%) in the euchromatic DNA. This result suggests that whole-genome association analysis will require large numbers of markers, even in selfing crop species with high levels of LD. To make such analyses possible, investments in marker discovery as well as in rapid and inexpensive genotyping technologies will be required.
Even with the large number of markers required to assay most of the common variation, there is still likely to be scientific and commercial interest in performing whole-genome scans in soybean. In this regard, an important question is the population in which to attempt such an analysis. While the Elite Cultivars have more extensive LD, thus necessitating fewer markers, we would suggest that the Landraces are the ideal population for whole-genome association studies in soybean. There are an estimated 45,000 unique landraces preserved in soybean germplasm collections around the world, and most have been characterized for many traits (Carter et al. 2004). In contrast, there are only 480 publicly released N. Am. Elite Cultivars that are likely to be readily available for analysis (http://www.ars-grin.gov). Due to the selfing nature of soybean there is likely to be a high degree of population structure, which will increase the likelihood of spurious marker–trait associations. Case-control studies with selection of a proper control population can help control for population structure, hence reducing the rate of false-positive associations (Aranzana et al. 2005). With the larger sampling of Landraces available, several case-control populations can be created for single traits to perform whole-genome scans to discover and then verify positive associations.
There are currently >1100 putative QTL identified in soybean (http://www.soybase.org). All of these putative QTL have been identified via the traditional linkage mapping techniques, but relatively few have been confirmed. The genome location of a QTL mapped with traditional mapping populations of ∼100 individuals generally has a confidence interval of 20–30 cM in size (Stuber et al. 1999), which, in soybean, likely encompasses 8–13 Mb. Given the extent of LD in the Landraces, it should be possible to greatly improve the resolution of QTL position using association analysis. The position of a QTL might be further resolved by fine mapping using a case-control association analysis in G. soja, assuming that genetic variability for the trait in question is available in G. soja. Our results indicate that the resolution of association analysis in G. soja should be <100 kb.
Conclusion:
Our study has determined the level of LD in three chromosomal regions in multiple soybean populations. We have found that LD is highly variable not only among populations but also between different regions of the genome. We have also determined that a large number of SNPs will be required to perform whole-genome association analysis even in a selfing species with relatively extensive LD. A HapMap of soybean will facilitate whole-genome association analysis and will expedite the fine mapping of many currently identified QTL.
Acknowledgments
We thank Tina Sphon, Tad Sonstegard, and the Bovine Functional Genomics Lab–Animal and Natural Resources Institute Beltsville Agricultural Research Center East DNA Sequencing Facility for assistance with the genomic STS sequencing. We thank Charles Fenster, William Kenworthy, and Marla McIntosh for helpful comments on this study. This work was partially supported by United Soybean Board Projects 4212 and 5212. The support of the United Soybean Board is greatly appreciated.
References
- Altshuler, D., L. D. Brooks, A. Chakravarti, F. S. Collins, M. J. Daly et al., 2005. A haplotype map of the human genome. Nature 437: 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranzana, M. J., S. Kim, K. Zhao, E. Bakker, M. Horton et al., 2005. Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet. 1: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett, J. C., B. Fry, J. Maller and M. J. Daly, 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. [DOI] [PubMed] [Google Scholar]
- Caldwell, K. S., J. Russell, P. Langridge and W. Powell, 2006. Extreme population-dependent linkage disequilibrium detected in an inbreeding plant species, Hordeum vulgare. Genetics 172: 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, T. E., R. Nelson, C. H. Sneller and Z. Cui, 2004. Genetic diversity in soybean, pp. 303–416 in Soybeans: Improvement, Production, and Uses, edited by H. R. Boerma and J. E. Specht. American Society of Agronomy, Crop Science Society of America, Soil Science Society of America, Madison, WI.
- Cregan, P. B., J. Mudge, E. W. Fickus, L. F. Marek, D. Danesh et al., 1999. Targeted isolation of simple sequence repeat markers through the use of bacterial artificial chromosomes. Theor. Appl. Genet. 98: 919–928. [Google Scholar]
- Daly, M. J., J. D. Rioux, S. F. Schaffner, T. J. Hudson and E. S. Lander, 2001. High-resolution haplotype structure in the human genome. Nat. Genet. 29: 229–232. [DOI] [PubMed] [Google Scholar]
- Flint-Garcia, S. A., J. M. Thornsberry and E. S. Buckler, IV, 2003. Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 54: 357–374. [DOI] [PubMed] [Google Scholar]
- Fujita, R., M. Ohara, K. Okazaki and Y. Shimamoto, 1997. The extent of natural cross-pollination in wild soybean (Glycine soja). J. Hered. 88: 124–128. [Google Scholar]
- Gabriel, S. B., S. F. Schaffner, H. Nguyen, J. M. Moore, J. Roy et al., 2002. The structure of haplotype blocks in the human genome. Science 296: 2225–2229. [DOI] [PubMed] [Google Scholar]
- Garris, A. J., S. R. McCouch and S. Kresovich, 2003. Population structure and its effect on haplotype diversity and linkage disequilibrium surrounding the xa5 locus of rice (Oryza sativa L.). Genetics 165: 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B. S., and A. D. Long, 2003. The lowdown on linkage disequilibrium. Plant Cell 15: 1502–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizlice, Z., T. E. Carter, Jr. and J. W. Burton, 1994. Genetic base for North American public soybean cultivars released between 1947 and 1988. Crop Sci. 34: 1143–1151. [Google Scholar]
- Gizlice, Z., T. E. Carter, Jr., T. M. Gerig and J. W. Burton, 1996. Genetic diversity patterns in North American public soybean cultivars based on coefficient of parentage. Crop Sci. 36: 753–765. [Google Scholar]
- Gordon, D., C. Abajian and P. Green, 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8: 195–202. [DOI] [PubMed] [Google Scholar]
- Graham, M. A., L. F. Marek, D. Lohnes, P. Cregan and R. C. Shoemaker, 2000. Expression and genome organization of resistance gene analogs in soybean. Genome 43: 86–93. [DOI] [PubMed] [Google Scholar]
- Hymowitz, T., 2004. Speciation and cytogenetics, pp. 97–136 in Soybeans: Improvement, Production, and Uses, edited by H. R. Boerma and J. E. Specht. American Society of Agronomy, Crop Science Society of America, Soil Science Society of America, Madison, WI.
- Hyten, D. L., Q. Song, Y. Zhu, I. Y. Choi, R. L. Nelson et al., 2006. Impacts of genetic bottlenecks on soybean genome diversity. Proc. Natl. Acad. Sci. USA 103: 16666–16671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim, P., T. C. Olson and R. C. Shoemaker, 1988. A rapid protocol for isolating soybean DNA. Soyb. Genet. Newsl. 15: 150–152. [Google Scholar]
- Matukumalli, L., J. Grefenstette, D. Hyten, I.-Y. Choi, P. Cregan et al., 2006. a Application of machine learning in SNP discovery. BMC Bioinformatics 7: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matukumalli, L. K., J. J. Grefenstette, D. L. Hyten, I. Y. Choi, P. B. Cregan et al., 2006. b SNP-PHAGE: High throughput SNP discovery pipeline. BMC Bioinformatics 7: 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell, P. L., D. M. Toleno, K. E. Lundy and M. T. Clegg, 2005. Low levels of linkage disequilibrium in wild barley (Hordeum vulgare ssp. spontaneum) despite high rates of self-fertilization. Proc. Natl. Acad. Sci. USA 102: 2442–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg, M., J. O. Borevitz, J. Bergelson, C. C. Berry, J. Chory et al., 2002. The extent of linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 30: 190–193. [DOI] [PubMed] [Google Scholar]
- Nordborg, M., T. T. Hu, Y. Ishino, J. Jhaveri, C. Toomajian et al., 2005. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K., M. Stephens and P. Donnelly, 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski, A., and M. Morgante, 2004. Corn and humans: recombination and linkage disequilibrium in two genomes of similar size. Trends Genet. 20: 103–111. [DOI] [PubMed] [Google Scholar]
- Rosenberg, N. A., 2004. DISTRUCT: a program for the graphical display of population structure. Mol. Ecol. Notes 4: 137–138. [Google Scholar]
- Rostoks, N., L. Ramsay, K. Mackenzie, L. Cardle, P. R. Bhat et al., 2006. Recent history of artificial outcrossing facilitates whole-genome association mapping in elite inbred crop varieties. Proc. Natl. Acad. Sci. USA 103: 18656–18661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Q. J., L. F. Marek, R. C. Shoemaker, K. G. Lark, V. C. Concibido et al., 2004. A new integrated genetic linkage map of the soybean. Theor. Appl. Genet. 109: 122–128. [DOI] [PubMed] [Google Scholar]
- Stuber, C. W., M. Polacco and M. L. Senior, 1999. Synergy of empirical breeding, marker-assisted selection, and genomics to increase crop yield potential. Crop Sci. 39: 1571–1583. [Google Scholar]
- Tenaillon, M. I., M. C. Sawkins, A. D. Long, R. L. Gaut, J. F. Doebley et al., 2001. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc. Natl. Acad. Sci. USA 98: 9161–9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornsberry, J. M., M. M. Goodman, J. Doebley, S. Kresovich, D. Nielsen et al., 2001. Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28: 286–289. [DOI] [PubMed] [Google Scholar]
- Todd, J. J., and L. O. Vodkin, 1996. Duplications that suppress and deletions that restore expression from a chalcone synthase multigene family. Plant Cell 8: 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N., J. M. Akey, K. Zhang, R. Chakraborty and L. Jin, 2002. Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination, and mutation. Am. J. Hum. Genet. 71: 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox, J. R., 2004. World distribution and trade of soybean, pp. 1–14 in Soybeans: Improvement, Production, and Uses, edited by H. R. Boerma and J. E. Specht. American Society of Agronomy, Crop Science Society of America, Soil Science Society of America, Madison, WI.
- Wilson, L. M., S. R. Whitt, A. M. Ibanez, T. R. Rocheford, M. M. Goodman et al., 2004. Dissection of maize kernel composition and starch production by candidate gene association. Plant Cell 16: 2719–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y. L., Q. J. Song, D. L. Hyten, C. P. Van Tassell, L. K. Matukumalli et al., 2003. Single-nucleotide polymorphisms in soybean. Genetics 163: 1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]