Abstract
Plant species may remain morphologically distinct despite gene exchange with congeners, yet little is known about the genomewide pattern of introgression among species. Here we analyze the effects of persistent gene flow on genomic differentiation between the sympatric sunflower species Helianthus annuus and H. petiolaris. While the species are strongly isolated in testcrosses, genetic distances at 108 microsatellite loci and 14 sequenced genes are highly variable and much lower (on average) than for more closely related but historically allopatric congeners. Our analyses failed to detect a positive association between levels of genetic differentiation and chromosomal rearrangements (as reported in a prior publication) or proximity to QTL for morphological differences or hybrid sterility. However, a significant increase in differentiation was observed for markers within 5 cM of chromosomal breakpoints. Together, these results suggest that islands of differentiation between these two species are small, except in areas of low recombination. Furthermore, only microsatellites associated with ESTs were identified as outlier loci in tests for selection, which might indicate that the ESTs themselves are the targets of selection rather than linked genes (or that coding regions are not randomly distributed). In general, these results indicate that even strong and genetically complex reproductive barriers cannot prevent widespread introgression.
NATURAL hybridization is frequent in many groups of plants and animals (Arnold 1997), yet in most instances the hybridizing species remain morphologically distinct because of selection against hybrids (Coyne and Orr 2004). Less is known about the maintenance or pattern of molecular genetic differences between hybridizing species, although theory indicates that neutral or advantageous alleles may move across species boundaries unless they are tightly linked to loci that contribute in some way to reproductive isolation (Barton 1979; Harrison 1990). Thus, if reproductive barriers have a complex genetic basis, it might be that much of the genome will be protected from gene flow. Indeed, Coyne and Orr (2004, p. 41) write that “Our guess is that morphologically distinct taxa showing rampant gene exchange at many loci will be rare.” They go on to argue (p. 42) that studies of hybrid zones have shown that “much of the genome cannot move between species because it is linked to divergently selected alleles.” Alternatively, if few loci contribute to isolation or hybrids are numerous, then more extensive genetic exchange is predicted (Rieseberg and Wendel 1993; Wu 2001).
Empirical data on interspecific gene flow are often difficult to interpret because shared polymorphisms between species may be interpreted to result from introgression and/or from the joint retention of ancestral alleles (Gottlieb 1972; Heiser 1973; Rieseberg and Wendel 1993; Hey et al. 2004; Muir and Schlötterer 2005; Lexer et al. 2006; Patterson et al. 2006). The introgression hypothesis is favored if (1) the species hybridize frequently (both now and in the past); (2) associations are found between linked markers (Doebley 1989; Hey et al. 2004; Scotti-Saintagne et al. 2004a); (3) genetic distances vary across the genome (Stump et al. 2005; Turner et al. 2005) and are highest near loci that contribute to species differences or reproductive isolation; and (4) populations or species with sympatric or parapatric distributions are less divergent on average than those with a history of allopatry. In contrast, the retention of ancestral polymorphism is typically viewed as more parsimonious for pairs of taxa that (1) lack a history of hybridization, (2) are strongly isolated reproductively, and (3) show equivalent divergence across loci and between sympatric and allopatric populations (Muir and Schlötterer 2005). Of course, in species that are exchanging genes, these hypotheses need not be mutually exclusive.
When introgression does occur, an important question concerns the lengths of chromosomal segments that are protected from interspecific gene flow. Again, theory is informative: gene flow near an isolation locus should be inversely proportional to the selection:recombination ratio (Barton 1979). Empirical data generally accord with this prediction in that rates of introgression tend to be reduced near quantitative trait loci (QTL) for hybrid sterility (Rieseberg et al. 1999) or ecological differences (Scotti-Saintagne et al. 2004b; Rogers and Bernatchez 2005), as well as in areas of low recombination such as chromosomal inversions and centromeres (Rieseberg et al. 1999; Noor et al. 2001; Stump et al. 2005; Turner et al. 2005). However, few studies have measured the lengths of protected chromosomal segments (although see Scotti-Saintagne et al. 2004a and Turner et al. 2005).
The annual sunflower species Helianthus annuus and H. petiolaris are well suited for comparisons of genomic patterns of introgression and differentiation. The two species form numerous hybrid zones across the Great Plains of central North America. Although many of the zones have originated recently as a consequence of human disturbance, phylogenetic data imply that the species have long engaged in hybridization because they have given rise to three stabilized hybrid species that arose between 60,000 and 210,000 generations before present (Schwarzbach and Rieseberg 2002; Welch and Rieseberg 2002; Gross et al. 2003).
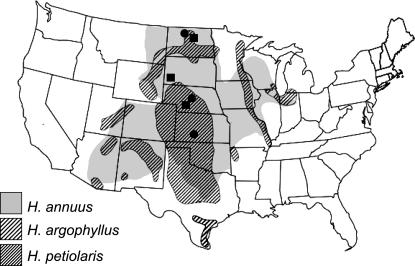
Despite the long history of hybridization, the two species remain morphologically (Rosenthal et al. 2002, 2005), karyotypically (Burke et al. 2004; Lai et al. 2005b), and ecologically distinct (Gross et al. 2004). H. annuus is the larger and more robust of the two species and predominates in the more mesic soils of the eastern Great Plains, while H. petiolaris is more frequent in the drier, sandier soils of the western Great Plains and intermountain regions (Figure 1). Comparative mapping studies indicate that the species differ by a minimum of eight translocations and three inversions (Rieseberg et al. 1995; Burke et al. 2004) and that first-generation hybrids average <1% seed set (Ungerer et al. 1998). Pollen-sterility QTL often map to chromosomal breakpoints and are underdominant, observations that imply that the rearrangements have direct effects on hybrid sterility (Rieseberg et al. 1999; Lai et al. 2005b). However, QTL in these regions are highly epistatic (Gardner et al. 2000; Lai et al. 2005b), indicating that gene incompatibilities may have accumulated near the chromosomal breakpoints as well, as has been predicted by theory (Noor et al. 2001; Navarro and Barton 2003). Regardless of mechanism, analyses of patterns of molecular marker introgression across four narrow hybrid zones between the two species (Rieseberg et al. 1999; Buerkle and Rieseberg 2001) indicate that rates of introgression across the rearranged chromosomes are approximately half that across collinear chromosomes.
Figure 1.—
The likely prehistoric distribution of three Helianthus species. Note that recently colonized areas are readily diagnosed because sunflowers are restricted to human-disturbed sites. Collections of H. annuus are indicated by squares and H. petiolaris by circles. Collections of H. argophyllus are not shown because the symbols would obscure its entire range.
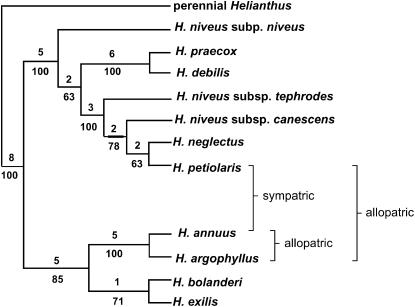
In this article, we analyze patterns of genetic differentiation at 108 microsatellite loci and DNA sequence variation at 14 nuclear genes across six populations of H. annuus and H. petiolaris. To better distinguish between segregating ancestral polymorphisms and the products of introgression, we also compare levels of differentiation at many of these loci with a third, historically allopatric sunflower species, H. argophyllus. Phylogenetically sister to H. annuus (Figure 2; Schilling and Heiser 1981; Chandler et al. 1986; Schilling et al. 1987; Rieseberg et al. 1991; Rieseberg 1991), H. argophyllus is a narrow endemic, found only on the coastal plain of southern Texas (Figure 1). The species appears to have been completely isolated geographically until the mid-20th century, when it came into contact with a weed form of H. annuus; hybrid swarms between the two species were first reported in the early 1950s (Heiser 1951). However, no contact or hybridization has been reported between H. argophyllus and H. petiolaris.
Figure 2.—
Phylogenetic tree for Helianthus section Helianthus based on restriction site analyses of chloroplast DNA and nuclear ribosomal genes (redrawn from Rieseberg 1991). Three homoploid hybrid sunflower species are excluded from this tree so that information on geographical relationships of the focal taxa can be provided.
Specific questions that we address with these data include:
Is there evidence of widespread gene flow between the sympatric sunflower species, H. annuus and H. petiolaris, despite the very strong sterility barrier?
Are sympatric species less differentiated genetically than historically allopatric congeners?
Do the locations of chromosomal rearrangements and/or positions of QTL for pollen sterility predict patterns of genomic differentiation?
Do microsatellites associated with genes show greater differentiation than those from anonymous DNA regions?
Do any of the loci exhibit the footprint of positive selection?
MATERIALS AND METHODS
Collections and DNA isolations:
Although the geographic distributions of H. annuus and H. petiolaris overlap broadly across the central and western United States, populations of the two species often are locally allopatric because of differences in habitat preferences. For this study, we collected achenes from three locally allopatric populations of each species (H. petiolaris: NDM, Minot, ND, 40 km east of Minot near Highway 2; NEO, O'Neil, NE, 16 km south of O'Neil on Highway 281; KSG, Great Bend, KS, junction of southeast 100 avenue and southeast 70 road; H. annuus: NDC, Cross Ranch, ND, 1.6 km north of entrance to Cross Ranch State Park; SDD, Hermosa, SD, 1.6 km south of Hermosa on Highway 79; NEG, Gothenburg, NE, 4.8 km south of Gothenburg on Highway 701; Figure 1). All of these populations are distant enough from the alternative species (>2 km) so that no direct contact is likely (no early generation hybrids were detected), but near enough to a contact zone (<35 km) such that isolation by distance is unlikely. Populations chosen for sampling were large (>1000 plants) and achenes were collected from 60 plants that were evenly spaced across the populations.
For H. argophyllus, four individuals were sampled from each of 12 populations: ARG1, Guadaloupe County, Texas, south of Luling on Highway 10 just west of intersection with Highway 80; ARG2, Gonzales County, Texas, between Nixon and Leesville, on Country Road 131 just north of intersection between Highway 80 and Highway 97; ARG4, Victoria County, Texas, between Victoria and Coleto Creek Reservoir under bridge of Highway 59 and railroad tracks; ARG6, Aransas County, Texas, between Lamar and Fulton on Highway 35 just south of bridge over Copano Bay; ARG7, Aransas County, Texas, on Highway 35 just south of Estes; ARG8, Nueces County, Texas, on Mustang Island near port Aransas; ARG9, Nueces County, Texas, on beach access Road 6 near Padre Island Balli Park; ARG11, Kleberg County, Texas, on Highway 77 at bridge over Los Olmos Creek; ARG12, Brooks County, Texas, south of Falfurrias on Highway 297; ARG13, Refugio County, Texas, on Highway 77 north of bridge over Aransas river; ARG14, Goliad County, Texas, intersection of Highway 183 and Mañahuilla Creek; ARG15, Gonzales County, Texas, between Westhoff and Smiley on Highway 87.
Achenes collected from H. annuus and H. petiolaris populations (above) were germinated in the Indiana University greenhouses, and DNA was isolated from a total of 268 juvenile plants (∼55 plants/population). For H. argophyllus, field-collected leaf tissue was employed for DNA isolations: 12 populations × 4 individuals/population = 48 plants total. All extractions were performed using a QIAGEN (Valencia, CA). Dneasy 96 plant kit.
Microsatellite analyses:
Primer sequences for microsatellites from anonymous genomic regions and from the flanking regions of expressed sequence tags (ESTs) were obtained from Tang et al. (2002) and the Compositae Project (CGP) database (http://compgenomics.ucdavis.edu/), respectively. In all, 352 anonymous microsatellites and 186 EST-associated microsatellites were screened for clean amplification in both H. annuus and H. petiolaris. From these initial screens, 44 anonymous and 64 EST-associated microsatellites were chosen for the populational surveys. All 108 markers have been placed on genetic linkage maps of H. annuus (Gedil et al. 2001; Burke et al. 2002; Tang et al. 2002; Yu et al. 2003; S. K. Knapp, unpublished results) and/or H. petiolaris (Burke et al. 2004; Lai et al. 2005b), so inferences may be made about patterns of genomic differentiation. For comparative purposes, 55 of the EST-associated microsatellites were also analyzed in H. argophyllus. Primers were synthesized by MWG Biotech (High Point, NC), Invitrogen (Grand Island, NY), or PE Biosystems (Foster City, CA). The 5′-end of each forward primer was labeled with one of three fluorescent dyes (6FAM, HEX, or NED) or “5′ tailed” with an M13 universal primer (Schuelke 2000) that was labeled with one of four dyes (6FAM, VIC, NED, or PET).
Microsatellites were amplified using a touchdown PCR protocol designed to reduce nonspecific primer associations and subsequent arbitrary fragment amplification (Don et al. 1991). An initial denaturing cycle of 3 min at 95° was followed by 10 touchdown cycles (the annealing temperature drops 1° each cycle) of 30 sec at 94°, 30 sec at the annealing temperature, and 45 sec at 72°. The touchdown cycles were followed by an additional 29 cycles of 30 sec at 94°, 30 sec at the final annealing temperature, and 45 sec at 72°, which in turn were followed by a 20-min final elongation period at 72°. The annealing temperature in the first touchdown cycle was 10° above the final annealing temperature. The final annealing temperature ranged from 48° to 60° for different microsatellite loci, depending on the best average amplification.
Microsatellite variation was assayed via electrophoresis on an ABI 3730 capillary sequencer (Applied Biosystems, Foster City, CA). Amplification products of nonoverlapping size and color were pooled and diluted 1:60 with ddH2O. One microliter of the diluted PCR product was added to a 9-μl mixture of 0.09 μl of the GenSize (St. James, NY). R500 ROX size standard. Samples were denatured for 5 min at 95° and cooled on ice before loading onto the 3730. Chromatographs of genotypic data were generated and fragment lengths were scored using GENEMAPPER 3.0 (Applied Biosystems).
Sequence analyses:
Eight genes from rearranged chromosomes and six from collinear chromosomes (Lai et al. 2005a) were chosen for sequencing to assess whether the low differentiation between H. annuus and H. petiolaris was the result of interspecific gene flow rather than allele size convergence due to stepwise mutation at microsatellite loci. Sequence data were obtained from six individuals of each species, with two plants sampled per population.
When feasible, PCR products treated with ExoSAP-IT (USB, Cleveland) were employed as templates for sequencing. Sequencing reactions were carried out in a total volume of 10 μl containing 2 μl of water, 3 μl of 5 mm MgCl2, 2 μl of 2 μm primer, 2 μl of PCR product (10–20 ng of DNA), and 1 μl of ABI Big Dye version 3.1. Sequencing reactions were performed with the following program: 1 min at 96° followed by 24 cycles of 10 sec at 96°, 5 sec at 50°, and 4 min at 60°. Reactions were then cooled to 4°, purified with the magnetic bead CleanSeq kit from Agencourt, and loaded on an ABI 3730 capillary sequencer (Applied Biosystems).
For individuals that were heterozygous for length mutations, fragments were cloned using the Topo-TA cloning kit with DH5α-T1R one-shot chemically competent cells (Invitrogen, Carlsbad, CA). Sequencing reactions were performed on purified plasmids (QIAGEN QIAprep Miniprep) using the same protocol as for PCR products.
Genetic differentiation and diversity measures:
For the microsatellite data, global FST across all six populations of H. annuus and H. petiolaris, and interspecific FST from pooled samples of conspecific populations, were calculated for each locus according to Weir and Cockerham (1984). To assess the proportion of variance in FST attributable to genetic differences between species, among conspecific populations, and within populations, hierarchical analyses of molecular variance (AMOVA) (Excoffier et al. 1992) were carried out for each locus using ARLEQUIN 2.0. The proportion of variance in each hierarchical class was tested by permuting individual genotypes.
Interspecific FST's and expected heterozygosities were also calculated for anonymous and EST-associated microsatellites independently, and differences in means between the two kinds of microsatellites were tested by t-tests and Mann–Whitney tests, as implemented by SPSS version 13.0 (SPSS, Chicago). Because the magnitude of FST (or GST) may depend on the variability of a locus, we also calculated a standardized measure of differentiation, G′ST, which was developed by Hedrick (2005) to facilitate comparisons among loci that differ in variability. Confidence intervals for interspecific FST's and G′ST's were calculated by resampling (with replacement) over all loci.
For the sequence data, the average number of nucleotide substitutions per site (Dxy; Nei 1987), interspecific FST (Hudson et al. 1992), and Gamma′ST (Hedrick 2005) were calculated for each locus using the DnaSP software package (version: 4.10.8; Rozas et al. 2003). Gamma′ST is a standardized measure of GammaST (Nei 1982), equivalent to G′ST for nucleotide data. Also, hierarchical AMOVA (Excoffier et al. 1992) was carried out for the sequence data using ARLEQUIN 2.0.
Analyses of factors that might predict patterns of genomic differentiation:
We analyzed five factors that might influence patterns of genomic differentiation in this hybridizing species pair. These included (1) the class of microsatellites (anonymous vs. EST-associated microsatellites); (2) the kind of linkage group (rearranged vs. collinear); (3) tight linkage to pollen-sterility QTL (Lai et al. 2005b); (4) tight linkage to chromosomal breakpoints (Burke et al. 2004; Lai et al. 2005b); and (5) tight linkage to QTL for interspecific phenotypic differences (Rieseberg et al. 2003; Lexer et al. 2005). We defined tight linkage as being within 5 cM of the most likely QTL positions or chromosomal breakpoints. Eight microsatellites were excluded from these and subsequent analyses because of unstable map positions across different H. annuus maps. Likewise, the 14 sequenced loci were excluded because of small sample sizes.
To estimate the effects of these factors on genomic differentiation, we first analyzed each factor separately in one-way ANOVAs with mean interspecific G′ST (after arcsine-square-root transformation) as the dependent variable (supplemental Table S1 at http://www.genetics.org/supplemental/). The minimum-effect sizes detectable with the sample sizes employed were tested using the computer software PASS (http://www.ncss.com/index.htm) with power at the standard value of ≥0.8 (corresponding to β < 0.2) and type I (α) error at 0.05.
Because the comparisons are not strictly independent, we also performed a multi-way ANOVA with each of the factors listed above included as main effects. All analyses were performed using SPSS version 13.0 (SPSS).
Autocorrelation of genetic differentiation along linkage groups:
To test for correlations between mean interspecific G′ST's and genetic map distances, we employed Moran's index (Sokal and Oden 1978) or spatial autocorrelation, following Scotti-Saintagne et al. (2004a). Observed Moran's indexes were compared to the null distribution of G′ST values constructed from 1000 permutations. Map distances employed for this analysis are from the RHA801 × RHA280 population described in Tang et al. (2002), which is the densest map currently available for H. annuus (supplemental Table S1 at http://www.genetics.org/supplemental/).
Detection of loci under selection:
We employed a coalescent simulation developed by Beaumont and Nichols (1996), which analyzes the distribution of FST values and detects outlier loci—loci that behave differently from most other loci in a given sample, presumably due to selection. Briefly, the program FDIST2 (http://www.rubic.reading.ac.uk/-mab/software/fdist2.zip) was used to simulate a null distribution of FST values (conditional on heterozygosity) under an infinite-alleles model and a symmetrical two-island model of population structure. Simulations employed the mean observed FST and the same sample sizes used in the empirical study. For comparisons with the simulated distribution, data from conspecific populations were pooled to avoid violating the assumption of symmetric migration among islands. Outlier loci were detected by comparing the empirical distribution of FST's with a simulated distribution derived from 20,000 paired values of FST and heterozygosity. Because we were searching for candidate loci that would be subject to additional testing in future studies, we did not apply the extremely conservative Bonferroni correction for multiple comparisons. Instead, we report all loci with an uncorrected P-value of <0.01, with the expectation of at least one spurious test result. The possible homology of outlier loci was determined by Blast searches against GenBank using Blast on the NCBI website (http://www.ncbi.nlm.nih.gov).
RESULTS
Microsatellite variation within and among populations of H.annuus and H. petiolaris:
Genetic diversity is high within populations of both species, with mean expected heterozygosities over all microsatellite loci ranging from 0.59 to 0.71 (Table 1). Diversity is significantly greater for anonymous than for EST-associated microsatellites, as has been reported in previous studies of Helianthus (Pashley et al. 2006) and other organisms (Cho et al. 2000; Cherdsak et al. 2004; Woodhead et al. 2005). Levels of genetic diversity are significantly affected by the number of microsatellite repeats (anonymous loci, mean number of repeats = 11.91; EST-associated loci, mean = 5.83; P < 0.001, one-tailed t-test), which are known to correlate strongly with mutation rate (Petit et al. 2005), but not by the length of repeat motifs. However, EST-associated microsatellites are seemingly more likely to have experienced significant selection in the recent past (see below), which might also have contributed to the reduced diversity. Furthermore, selection against frameshift mutations in microsatellite motifs located in protein-encoding regions may reduce diversity of some EST-associated microsatellites (Metzgar et al. 2000).
TABLE 1.
Comparison of gene diversity and interspecific genetic differentiation between anonymous and EST-associated microsatellites
| Anonymous microsatellites | EST-associated microsatellites |
P-value
|
||
|---|---|---|---|---|
| Mann–Whitney | t-test | |||
| Interspecific genetic differentiation | ||||
| FST | 0.050 (±0.046) | 0.092 (±0.081) | 0.003 | 0.001 |
| Hedrick's G′ST | 0.470 (±0.338) | 0.489 (±0.323) | 0.769 | 0.819 |
| Genetic diversity | ||||
| No. of alleles | 18.7 (±8.35) | 10.5 (±4.32) | 0.000 | 0.000 |
| Expected heterozygosity | ||||
| H. annuus | ||||
| NDC | 0.776 (±0.156) | 0.636 (±0.214) | 0.000 | 0.000 |
| NEG | 0.767 (±0.142) | 0.595 (±0.223) | 0.000 | 0.000 |
| SDD | 0.782 (±0.153) | 0.668 (±0.184) | 0.000 | 0.001 |
| H. petiolaris | ||||
| KSG | 0.767 (±0.171) | 0.658 (±0.177) | 0.000 | 0.002 |
| NDM | 0.658 (±0.240) | 0.544 (±0.217) | 0.002 | 0.012 |
| NEO | 0.772 (±0.135) | 0.682 (±0.172) | 0.001 | 0.004 |
The numbers in parentheses are standard deviations.
AMOVA (Table 2) indicates that only a small fraction of gene diversity is attributable to differences among populations within species (anonymous loci: 2.7% and EST-associated loci: 3.6%) or among species (anonymous loci: 4.3% and EST-associated loci: 8.6%). Nonetheless, AMOVA of individual loci indicates that 98 and 91% of loci were significantly differentiated among all populations and among conspecific populations, respectively (P < 0.05; 89 and 66% of loci remained significant after Bonferroni correction, respectively). Likewise, 38% of loci exhibited significant differentiation among species, all of which remained significant after Bonferroni correction for multiple comparisons.
TABLE 2.
AMOVA for anonymous vs. EST-associated microsatellite loci
| Source of variation | d.f. | Sum of squares | Variance components | % of variation | Fixation indexes | P |
|---|---|---|---|---|---|---|
| Anonymous microsatellites | ||||||
| Between species | 1 | 262.7 | 0.76 | 4.3 | FCT = 0.043 | 0.000 |
| Among populations within species | 4 | 235.3 | 0.48 | 2.7 | FSC = 0.029 | 0.000 |
| Within populations | 530 | 8,645.6 | 16.31 | 93.0 | FST = 0.071 | 0.000 |
| Total | 535 | 9,143.6 | 17.55 | 100.0 | ||
| EST-associated microsatellites | ||||||
| Between species | 0 | 577.3 | 1.82 | 8.6 | FCT = 0.086 | 0.000 |
| Among populations within species | 4 | 347.5 | 0.77 | 3.6 | FSC = 0.040 | 0.000 |
| Within populations | 530 | 9,881.0 | 18.64 | 87.8 | FST = 0.122 | 0.000 |
| Total | 535 | 10,805.8 | 21.24 | 100.0 | ||
Overall, FST between H. annuus and H. petiolaris averaged 0.08 ± 0.01 (supplemental Table S1 at http://www.genetics.org/supplemental/), with significantly greater differentiation observed for EST-associated microsatellites than for anonymous loci. This difference is mostly due to the lower average allelic diversity for EST-associated microsatellites, since no difference in interspecific genetic differentiation between these two classes of loci was observed for Hedrick's G′ST, which is robust to differences in allele numbers among loci. It is noteworthy that the average G′ST across all loci (0.47 ± 0.03) is approximately six times that of FST (supplemental Table S1 at http://www.genetics.org/supplemental/), indicating that estimates of genetic differentiation based on the latter should be viewed cautiously when highly polymorphic loci are studied (Hedrick 2005).
Sequence variation in H. annuus and H. petiolaris:
A much larger fraction of sequence variation is attributable to differences among species (39%) than is true for the microsatellite data (see above). However, this might be due to small sample sizes rather than to a real difference in how diversity is partitioned. Indeed, while average interspecific FST (0.39 ± 0.05) is significantly higher for sequence than microsatellite data (supplemental Table S2 at http://www.genetics.org/supplemental/), mean Gamma′ST (0.30 ± 0.04) shows the opposite pattern (supplemental Table S2 at http://www.genetics.org/supplemental/).
Comparisons with the historically allopatric species, H.argophyllus:
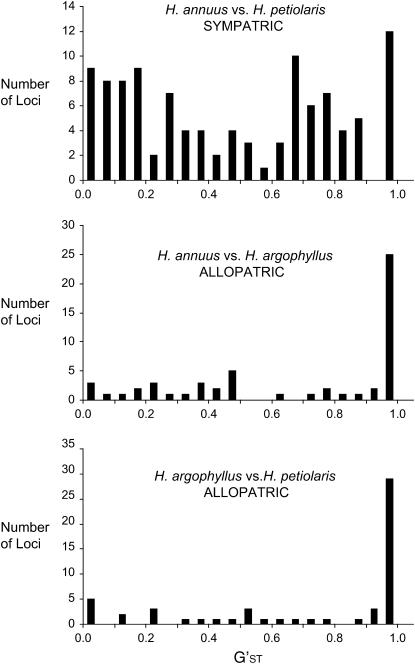
Genetic distances (both FST and G′ST) among the historically allopatric species (H. annuus vs. H. argophyllus and H. petiolaris vs. H. argophyllus) were significantly greater than for the sympatric species pair, H. annuus vs. H. petiolaris (Table 3; Figure 3). This is despite the closer phylogenetic relationship and weaker reproductive barrier between H. annuus and H. argophyllus (F1 fertility ranges from 5 to 50%; Heiser 1951).
TABLE 3.
Interspecific genetic differentiation and 95% confidence intervals over 55 microsatellite loci analyzed in all three Helianthus species
| Comparison | Mean FST (95% C.I.) | Mean G′ST (95% C.I.) |
|---|---|---|
| H. annuus vs. H. argophyllus | 0.226 (0.196–0.257) | 0.720 (0.629–0.812) |
| H. annuus vs. H. petiolaris | 0.094 (0.079–0.109) | 0.505 (0.422–0.587) |
| H. argophyllus vs. H. petiolaris | 0.222 (0.196–0.250) | 0.806 (0.724–0.887) |
Figure 3.—
The distribution of interspecific G′ST values in sympatric and allopatric pairs of sunflower species.
Predicting patterns of genomic differentiation:
With the exception of tight linkage to chromosomal breakpoints, none of the factors that we thought might predict patterns of genomic differentiation had a significant impact on G′ST (or FST), whether analyzed individually or in concert with other factors (Table 4). Markers within 5 cM of chromosomal breakpoints did exhibit an increase in G′ST, but the effect was modest (0.68 vs. 0.46; one-way ANOVA, P = 0.032; multi-way ANOVA, P = 0.053). However, the power of the test was weak, with the percentage of differences in means detectable with power equaling 0.8 ranging from ∼37% for the factors of microsatellite class, linkage group type, and proximity to QTL for species differences to 60% for the factors of proximity to chromosomal breakpoints and proximity to pollen-sterility QTL. Note that power for the latter two factors is particularly low because the sample sizes are unbalanced (12 in one group and 88 in the other).
TABLE 4.
Multi-way ANOVA of factors that affect the genomic distribution of genetic distance (G′ST) values
| Factor | d.f. | SS | F | P |
|---|---|---|---|---|
| Class of microsatellite | 1 | 0.044 | 0.253 | 0.616 |
| Kind of linkage group | 1 | 0.480 | 2.794 | 0.097 |
| Chromosomal breakpoints | 1 | 0.660 | 3.849 | 0.053 |
| Pollen-sterility QTL | 1 | 0.002 | 0.001 | 0.925 |
| QTL for interspecific differences | 1 | 0.182 | 1.057 | 0.307 |
| Error | 96 | 0.172 |
Sequence data, while limited, also failed to detect a significant difference between collinear and rearranged chromosomes (supplemental Table S2 at http://www.genetics.org/supplemental/), whether measured in terms of Gamma′ST (0.34 ± 0.03 collinear vs. 0.28 ± 0.06 rearranged; P = 0.41; two-tailed t-test) or FST (0.45 ± 0.04 vs. 0.35 ± 0.08; P = 0.31; two-tailed t-test). Likewise, exactly half of the sequences from both collinear and rearranged chromosomes had fixed differences (supplemental Table S2 at http://www.genetics.org/supplemental/).
Genetic differentiation along linkage groups:
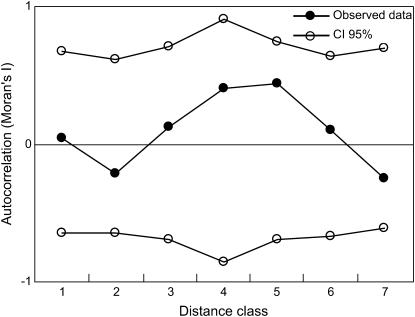
No correlations were observed in levels of genomic differentiation along linkage groups for any distance class, regardless of whether we used FST or G′ST as an estimator of differentiation or whether we employed a distance interval of 1, 2, 4, or 6 cM (Figure 4)
Figure 4.—
Autocorrelation of interspecific G′ST values as a function of genetic distance along linkage groups (in centimorgans).
Detection of loci under selection:
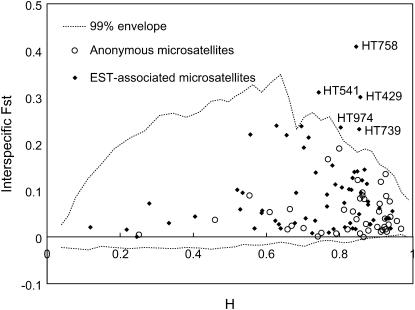
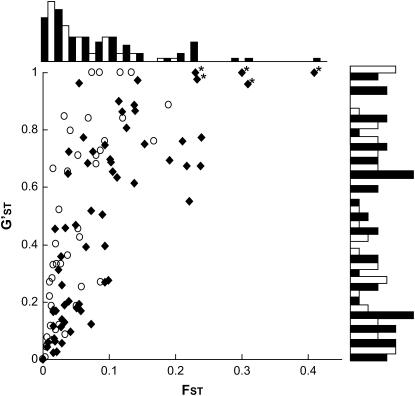
Using the coalescent simulations of Beaumont and Nichols (1996), five loci that behave differently between H. annuus and H. petiolaris at P < 0.01 were identified (Figure 5; Table 5). Note that because 108 tests were employed, one false positive is expected due to chance alone. All five outlier loci are EST-associated microsatellites, which might indicate that the ESTs themselves are the targets of selection rather than linked factors. However, given the correlation reported above between FST and allelic variance, it might also mean that the distribution of FST is not robust to demographic history (Flint et al. 1999; Storz et al. 2004). We therefore compared the distribution of FST values with that of G′ST, which is unaffected by genetic diversity levels. The loci detected as outliers in the FST tests were also outliers with respect to G′ST (Figure 6), so the results appear consistent with positive selection. The steep drop in G′ST seen between candidate selected loci and tightly linked markers (mean G′ST for candidate selected loci: 0.92; mean G′ST for markers within 3 cM of candidate selected loci: 0.40; P = 0.001, two-tailed t-test; supplemental Table S1 at http://www.genetics.org/supplemental/) also implies that the size of differentiated islands is small. For example, there is a decline in G′ST of 0.88 between the candidate selected locus HT758 and a completely linked marker, ORS798 (supplemental Table S1 at http://www.genetics.org/supplemental/). However, BLAST searches revealed significant hits (e-value <0.01) for only two of the five candidate selected loci and none had functions that can be easily related to ecological or phenotypic differences between H. annuus and H. petiolaris (Table 5).
Figure 5.—
Distribution of interspecific FST values as a function of heterozygosity (H). The envelope of values corresponding to neutral expectations (with FST = 0.078) under a stepwise mutation model was constructed according to Beaumont and Nichols (1996) (see Table 5).
TABLE 5.
Candidate loci for adaptive genetic divergence between populations of H. annuus and H. petiolaris
| Marker | Linkage group | FST | P | Homology |
|---|---|---|---|---|
| HT429 | 14 (R) | 0.300 | *** | Unknown protein (Arabidopsis thaliana) |
| HT541 | 4 (C) | 0.310 | ** | No hit |
| HT739 | 13 (R) | 0.231 | ** | No hit |
| HT758 | 17 (R) | 0.409 | *** | Glycine-rich protein (Nicotiana sylvestris) |
| HT974 | 1 (C) | 0.234 | ** | No hit |
R and C in parentheses indicate rearranged and collinear linkage groups, respectively. FST, interspecific FST with significance values calculated according to Beaumont and Nichols (1996). *P<0.05, **P<0.01, ***P<0.001.
Figure 6.—
Distributions of FST and G′ST of anonymous microsatellites (open circles and open bars) and EST-associated microsatellites (solid diamonds and solid bars). An asterisk indicates loci identified as outliers in the test of Beaumont and Nichols (1996) (see Table 5).
DISCUSSION
Interspecific gene flow:
Several lines of evidence suggest that the patterns of genomic differentiation reported here for a pair of sympatric sunflower species have been shaped by interspecific gene flow. First, the studied species have been hybridizing for as long as 200,000 generations, the contact zone is large, the frequency of F1 production is high (0–17%; Rieseberg et al. 1998), and early generation backcrosses are the most frequent category of plants in the contact zone (Rieseberg et al. 1999; Buerkle and Rieseberg 2001). Second, mean FST and G′ST values between H. annuus and H. petiolaris are much lower than values for the historically allopatric species pairs H. annuus vs. H. argophyllus and H. annuus vs. H. petiolaris (Figure 3; Table 3). Third, genetic distances are remarkably variable across loci between the sympatric species and much more so than between the pairs of allopatric species (Figure 3). Finally, genetic distances are greatest near chromosomal breakpoints (Table 4).
Are there other explanations for these patterns? One concern is that even in the absence of interspecific gene flow, the stepwise mutation process of microsatellites might lead to allele size convergence, which could be misinterpreted as evidence of introgression. While this may sometimes occur, it cannot easily explain the patterns of genomic differentiation observed since allele size convergence should be no more frequent in sympatric than in allopatric species.
Another worry relates to our use of FST and G′ST for estimating genetic differentiation. FST is known to be negatively correlated with the number of alleles, and we see the same effect in our data set (r = −0.33, P < 0.001). However, we also found that G′ST is positively correlated with heterozygosity (r = 0.56, P < 0.0001), although not with allele number. Thus, neither metric is ideal. Fortunately, the two estimates of differentiation are strongly correlated (r = 0.68, P < 0.0001), and we see the same pattern with both (significantly less differentiation in sympatric than in allopatric pairs of species), so our results are robust. Nonetheless, we cannot rule out the possibility that the greater genetic differentiation observed in the allopatric comparisons is due to the smaller effective population size of the allopatric species, H. argophyllus, which harbors less genetic diversity (H = 0.45 ± 0.03) than either of the sympatric species (H. annuus, H = 0.65 ± 0.03; H. petiolaris, H = 0.66 ± 0.02).
Finally, why are patterns of genomic differentiation poorly predicted by previous studies of introgression across hybrid zones between the same species (Rieseberg et al. 1999; Gardner et al. 2000; Buerkle and Rieseberg 2001)? In these studies, the introgression of RAPD markers was reduced on rearranged vs. collinear chromosomes. The main reason for the seeming discordance is experimental design. In the earlier studies, the behavior of species-specific markers was analyzed in early generation backcross hybrids within 10 m of the hybrid zone center. As a consequence, linkage disequilibrium was high, and differential introgression was observed at greater centimorgan distances from chromosomal breakpoints than reported here, where we analyzed locally allopatric populations that lacked early generation hybrids.
Also, in contrast to this study, the earlier work analyzed species-specific markers that presumably are linked to selected sites, so the long-term effects of hybridization on differentiation at neutral loci could not be tested. Interestingly, there was no difference in the fraction of taxon-specific RAPD markers found on collinear vs. rearranged chromosomes, which in hindsight predicts the results reported here.
The unit of genetic isolation:
Even given the differences in experimental design, we still expected to see greater differentiation in rearranged chromosomes or near factors that contribute to reproductive isolation. The weak effect of sterility QTL or chromosomal breakpoints is likely due to the low power of the ANOVA (see results), as well as the imprecision in QTL locations; patterns of genetic differentiation trend in the expected direction for sterility QTL and are significant for chromosomal breakpoints. On the other hand, the lack of an effect of chromosome type (collinear vs. rearranged) does not appear to be an artifact of limited power since the difference in means (while very small) is opposite that of the predicted direction. Also, no difference in levels of differentiation was found between collinear and rearranged chromosomes for sequenced loci (supplemental Table S2 at http://www.genetics.org/supplemental/).
Thus, the most likely explanation for the patterns of differentiation reported here is that the unit of isolation between these two sunflower species is smaller than we had anticipated, perhaps at the level of the individual gene, except in regions of low recombination such as near chromosomal breakpoints (Noor et al. 2001; Wu 2001; Turner et al. 2005). This would result in patterns of genomic differentiation that are mostly unaffected by linkage, at least on the scale that we could detect in this study. It also would explain why no correlations were observed in levels of genomic differentiation along linkage groups (Figure 4).
This explanation, however, would require some rethinking about the effects of plant chromosomal rearrangements on recombination. One of us recently argued that while plant chromosomal rearrangements typically do not suppress recombination directly, effective recombination rates are likely to be reduced because of selection against recombinant gametes, particularly for inversions (Rieseberg 2001). This would serve to extend the effects of isolation genes, thereby protecting larger fractions of the genome from interspecific gene flow. The results presented here do not necessarily contradict this hypothesis, but they do suggest that recombination suppression, which is characteristic of inversions in many animals, is more effective at limiting gene flow than is selection against recombinant gametes. Also note that the majority of karyotypic differences between these sunflower species result from large translocations (Rieseberg et al. 1995; Burke et al. 2004), which are not predicted to greatly restrict recombination rates in regions distal from breakpoints (Grant 1975).
The hypothesis that the genetic unit of isolation is small between these two hybridizing sunflowers is fully consistent with hybrid zone theory (Barton and Hewitt 1985). While gene dispersal across species barriers will initially be affected by linkage, if there is sufficient opportunity for recombination, then neutral or advantageous alleles may recombine into the genetic background of the other species. Once incorporated into the new genetic background, the movement of these “alien” alleles will depend largely on their own fitness effects rather than on those of linked alleles. This is a realistic scenario for these sunflower species since, as alluded to above, both are extremely abundant, the contact zone is long (stretching from Texas to Canada) and broad (most of the central United States), hybridization frequencies are high (0–17%; Rieseberg et al. 1998), and recombinant genotypes are common (Rieseberg et al. 1999).
Little is known about the lengths of chromosomal segments that are protected from interspecific gene flow in other taxa, although Ting et al. (2001) showed that regions just 2 kb from a sterility gene in Drosophila acted independently with respect to the retention of shared polymorphisms and/or history of introgression. In Anopheles, closely related forms exhibit differentiation in three genomic regions that range in size from 5 to 50 genes, all in areas of low recombination (Turner et al. 2005). However, a more detailed analysis of the largest of these three regions, which occurs near the centromere on the X chromosome, implies that the sizes of the differentiated regions may be underestimated (Stump et al. 2005). A recent study of two oak species (Scotti-Saintagne et al. 2004a) detected strong correlations among markers that were within 2 cM, with the sizes of differentiated regions ranging from ∼0.5 to 4 cM.
Why does the unit of isolation seem to be smaller in sunflowers than in Anopheles and oaks? One possibility is that marker density was not high enough in this study to detect hitchhiking effects and that, if a larger number of markers had been employed, we might have detected islands of differentiation. However, we did have 31 pairs of markers that were <1 cM apart, with no correlations observed between them, so this cannot be a full explanation. Another possibility is that hybridization rates in H. annuus and H. petiolaris (Rieseberg et al. 1999; Lai et al. 2005b) are much higher than in oaks, which would lead to greater opportunities for recombination.
Selection:
Selection may contribute to genetic differentiation in two ways. First, positive selection (and hitchhiking) may increase the frequency of intraspecific variants within one or the other species, thereby “creating” differentiation. Second, negative selection may eliminate foreign alleles, thereby “protecting” differences that have accumulated through either selection (see above) or genetic drift. While both kinds of selection are necessary to create and maintain divergence between populations that are connected by gene flow, the evolutionary dynamics of hybrid zones are usually dominated by negative selection (Barton and Hewitt 1985), while positive selection may be more important in allopatric populations (Morjan and Rieseberg 2004).
Given these considerations, the outlier analyses conducted in this article may have detected the footprints of recent positive selection. This is in contrast with previous studies of contemporary hybrid zones between this same pair of species that identified negatively selected genomic regions (Rieseberg et al. 1999; Buerkle and Rieseberg 2001). Like sunflower, outlier markers were mostly associated with genes rather than anonymous genomic regions in oaks (Scotti-Saintagne et al. 2005), but this pattern was not observed in salmon (Vasemagi et al. 2005). It is less clear whether ESTs associated with outlier markers (Table 5) are the direct targets of selection or whether they contribute to larger genomic islands of differentiation. However, the former hypothesis is consistent with the observations that only EST-associated markers were detected by the test for selection and that tightly linked markers exhibited a steep decline in interspecific genetic distances. Also, it should be remembered that these outliers are candidate loci only and that at least one false positive is expected by chance alone. Sequence and expression analyses are currently underway to strengthen the candidate status of outlier loci.
Conclusions and future directions:
Although reproductive isolation is traditionally viewed as a genomewide phenomenon (reviewed in Wu 2001), the extent of introgression between these two sunflower species appears to be greater and the unit of isolation smaller than implied by speciation texts (Mayr 1963; Coyne and Orr 2004). Indeed, our data imply that as long as a hybrid zone is ancient, the interface is long, and hybridization is frequent, even an extremely strong and genetically complex barrier may be porous to gene flow. This does not mean that reproductive barriers are of little importance to speciation. To the contrary, the barrier between H. annuus and H. petiolaris not only has served to maintain the differences between these species for >200,000 generations, but also has reduced gene migration rates to a sufficiently low level such that differentiation may occur at even very weakly selected loci.
Our results also imply that studies of contemporary hybrid zones may be misleading in two ways. First, the focus of many studies on taxon-specific markers (e.g., Rieseberg et al. 1999; Coart et al. 2002; Kelleher et al. 2005) may result in greater attention given to the differences between species rather than to the similarities. Second, because linkage disequilibrium is high in hybrid zones, the importance of linkage to divergently selected alleles maybe overemphasized relative to the power of recombination for breaking up these associations.
Future work will determine the size of differentiated regions between H. annuus and H. petiolaris by sequencing BACs that contain genes known to contribute to assortative mating and/or ecological divergence in Helianthus. Our plan is to assay SNPs from the sequenced regions in replicate transects of natural populations of each species that are located at different distances from the contact zone. This will lead to a better estimate of the “unit of isolation” between these species and may further our understanding of the escape and spread of alien genes from areas of contact into allopatric populations of the hybridizing species.
Acknowledgments
We thank Alex Buerkle, Mark Beaumont, and Ken Whitney for assistance with the data analyses and Daniel Ortiz-Barrientos, Troy Wood, and Yuval Sapir for helpful comments on an earlier version of the manuscript. This work was supported by grants from the National Science Foundation (DEB-0314654 and DBI0421630 to L.H.R.), the National Institutes of Health (GM059065 to L.H.R.), and the Japan Society for the Promotion of Science Postdoctoral Fellowships for Research Abroad to Y.Y.
References
- Arnold, M. L., 1997. Natural Hybridization and Evolution. Oxford University Press, New York.
- Barton, N. H., 1979. Dynamics of hybrid zones. Heredity 43: 341–359. [Google Scholar]
- Barton, N. H., and G. M. Hewitt, 1985. Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 16: 113–148. [Google Scholar]
- Beaumont, M. A., and R. A. Nichols, 1996. Evaluating loci for use in the genetic analysis of population structure. Proc. R. Soc. Lond. B Biol. Sci. 263: 1619–1626. [Google Scholar]
- Buerkle, C. A., and L. H. Rieseberg, 2001. Low intraspecific variation for genomic isolation between hybridizing sunflower species. Evolution 55: 684–691. [DOI] [PubMed] [Google Scholar]
- Burke, J. M., S. Tang, S. J. Knapp and L. H. Rieseberg, 2002. Genetic analysis of sunflower domestication. Genetics 161: 1257–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, J. M., Z. Lai, M. Salmaso, T. Nakazato, S. X. Tang et al., 2004. Comparative mapping and rapid karyotypic evolution in the genus Helianthus. Genetics 167: 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, J. M., C. C. Jan and B. H. Beard, 1986. Chromosomal differentiation among the annual Helianthus species. Syst. Bot. 11: 354–371. [Google Scholar]
- Cherdsak, L., C. E. Ritland, Y. A. El-Kassaby and K. Ritland, 2004. Single-copy, species-transferable microsatellite markers developed from loblolly pine ESTs. Theor. Appl. Genet. 109: 361–369. [DOI] [PubMed] [Google Scholar]
- Cho, Y. G., T. Ishii, S. Temnykh, X. Chen, L. Lipovich et al., 2000. Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.). Theor. Appl. Genet. 100: 713–722. [Google Scholar]
- Coart, E., V. Lamote, M. De Loose, E. Van Bodkstaele, P. Lootens et al., 2002. AFLP markers demonstrate local genetic differentiation between two indigenous oak species [Quercus robur L. and Quercus petraea (Matt.) Liebl] in Flemish populations. Theor. Appl. Genet. 105: 431–439. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 2004. Speciation. Sinauer Associates, Sunderland, MA.
- Doebley, J., 1989. Molecular evidence for a missing wild relative of maize and the introgression of its chloroplast genome into Zea perennis. Evolution 43: 1555–1559. [DOI] [PubMed] [Google Scholar]
- Don, R., P. Cox, B. Wainwright, K. Baker and J. Mattick, 1991. Touchdown PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19: 4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier, L., P. E. Smouse and J. M. Quattro, 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint, J., J. Bond, D. C. Rees, A. J. Boyce, J. M. Roberts-Thomson et al., 1999. Minisatellite mutational processes reduce F(st) estimates. Hum. Genet. 105: 567–576. [DOI] [PubMed] [Google Scholar]
- Gardner, K., A. Buerkle, J. Whitton and L. H. Rieseberg, 2000. Inferring epistasis in wild sunflower hybrid zones, pp. 264–279 in Epistasis and the Evolutionary Process, edited by J. Price, B. Brodie and M. J. Wade. Oxford University Press, Oxford.
- Gedil, M. A., C. Wye, S. T. Berry, B. Seger, J. Peleman et al., 2001. An integrated RFLP-AFLP linkage map for cultivated sunflower. Genome 44: 213–221. [PubMed] [Google Scholar]
- Gottlieb, L. D., 1972. Levels of confidence in analysis of hybridization in plants. Ann. Mo. Bot. Gard. 59: 435–446. [Google Scholar]
- Grant, V., 1975. Genetics of Flowering Plants. Columbia University Press, New York.
- Gross, B. L., A. E. Schwarzbach and L. H. Rieseberg, 2003. Origin(s) of the diploid hybrid species Helianthus deserticola (Asteraceae). Am. J. Bot. 90: 1708–1719. [DOI] [PubMed] [Google Scholar]
- Gross, B. L., N. C. Kane, C. Lexer, F. Ludwig, D. M. Rosenthal et al., 2004. Reconstructing the origin of Helianthus deserticola: survival and selection on the desert floor. Am. Nat. 164: 145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, R. G., 1990. Hybrid zones: windows on evolutionary processes. Oxf. Surv. Evol. Biol. 7: 69–128. [Google Scholar]
- Hedrick, P. W., 2005. A standardized genetic differentiation measure. Evolution 59: 1633–1638. [PubMed] [Google Scholar]
- Heiser, C. B., 1951. Hybridization in the annual sunflowers: Helianthus annuus × H. argophyllus. Am. Nat. 85: 64–72. [Google Scholar]
- Heiser, C. B., 1973. Introgression re-examined. Bot. Rev. 39: 347–366. [Google Scholar]
- Hey, J., Y. J. Won, A. Sivasundar, R. Nielsen and J. A. Markert, 2004. Using nuclear haplotypes with microsatellites to study gene flow between recently separated Cichlid species. Mol. Ecol. 13: 909–919. [DOI] [PubMed] [Google Scholar]
- Hudson, R. R., M. Slatkin and W. P. Maddison, 1992. Estimation of levels of gene flow from DNA sequence data. Genetics 132: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher, C. T., T. R. Hodkinson, G. C. Douglas and D. L. Kelly, 2005. Species distinction in Irish populations of Quercus petraea and Q. robur: morphological versus molecular analyses. Ann. Bot. 96: 1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, Z., K. Livingstone, Y. Zou, S. A. Church, S. J. Knapp et al., 2005. a Identification and mapping of SNPs from ESTs in sunflower. Theor. Appl. Genet. 111: 1532–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, Z., T. Nakazato, M. Salmaso, J. M. Burke, S. X. Tang et al., 2005. b Extensive chromosomal repatterning and the evolution of sterility barriers in hybrid sunflower species. Genetics 171: 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexer, C., D. M. Rosenthal, O. Raymond, L. A. Donovan and L. H. Rieseberg, 2005. Genetics of species differences in the wild annual sunflowers, Helianthus annuus and H. petiolaris. Genetics 169: 2225–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexer, C., A. Kremer and R. J. Petit, 2006. Shared alleles in sympatric oaks: recurrent gene flow is a more parsimonious explanation than ancestral polymorphism. Mol. Ecol. 15: 2007–2012. [DOI] [PubMed] [Google Scholar]
- Mayr, E., 1963. Animal Species and Evolution. Belknap Press of Harvard University Press, Cambridge, MA.
- Metzgar, D., W. J. Bytof and C. Wills, 2000. Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res. 10: 72–80. [PMC free article] [PubMed] [Google Scholar]
- Morjan, C. L., and L. H. Rieseberg, 2004. How species evolve collectively: implications of gene flow and selection for the spread of advantageous alleles. Mol. Ecol. 13: 1341–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir, G., and C. Schlötterer, 2005. Evidence for shared ancestral polymorphism rather than recurrent gene flow at microsatellite loci differentiating two hybridizing oaks (Quercus spp.). Mol. Ecol. 14: 549–561. [DOI] [PubMed] [Google Scholar]
- Navarro, A., and N. Barton, 2003. Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evolution 57: 447–459. [DOI] [PubMed] [Google Scholar]
- Nei, M., 1982. Evolution of human races at the gene level, pp. 167–181 in Human Genetics, Part A: The Unfolding Genome, edited by B. Bonne-Tamir, T. Cohen and R. M. Goodman. Alan R. Liss, New York.
- Nei, M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York.
- Noor, M. A. F., K. L. Grams, L. A. Bertucci and J. Reiland, 2001. Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. USA 98: 12084–12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashley, C. H., J. R. Ellis, D. E. McCauley and J. M. Burke, 2006. EST databases as a source for molecular markers: lessons from Helianthus. J. Hered. 97: 381–388. [DOI] [PubMed] [Google Scholar]
- Patterson, N., D. J. Richter, S. Gnerre, E. S. Lander and D. Reich, 2006. Genetic evidence for complex speciation of humans and chimpanzees. Nature 441: 1103–1108. [DOI] [PubMed] [Google Scholar]
- Petit, R. J., M. F. Deguilloux, J. Chat, D. Grivet, P. Garnier-Gere et al., 2005. Standardizing for microsatellite length in comparisons of genetic diversity. Mol. Ecol. 14: 885–890. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H., 1991. Homoploid reticulate evolution in Helianthus (Asteraceae): evidence from ribosomal genes. Am. J. Bot. 78: 1218–1237. [Google Scholar]
- Rieseberg, L. H., 2001. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16: 351–358. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H., and J. Wendel, 1993. Introgression and its consequences in plants, pp. 70–114 in Hybrid Zones and the Evolutionary Process, edited by R. Harrison. Oxford University Press, Oxford.
- Rieseberg, L. H., S. Beckstrom-Sternberg, A. Liston and D. Arias, 1991. Phylogenetic and systematic inferences from chloroplast DNA and isozyme variation in Helianthus sect. Helianthus. Syst. Bot. 16: 50–76. [Google Scholar]
- Rieseberg, L. H., C. Van Fossen and A. Desrochers, 1995. Hybrid speciation accompanied by genomic reorganization in wild sunflowers. Nature 375: 313–316. [Google Scholar]
- Rieseberg, L. H., S. Baird and A. Desrochers, 1998. Patterns of mating in wild sunflower hybrid zones. Evolution 52: 713–726. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H., J. Whitton and K. Gardner, 1999. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics 152: 713–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg, L. H., O. Raymond, D. M. Rosenthal, Z. Lai, K. Livingstone et al., 2003. Major ecological transitions in annual sunflowers facilitated by hybridization. Science 301: 1211–1216. [DOI] [PubMed] [Google Scholar]
- Rogers, S. M., and L. Bernatchez, 2005. Integrating QTL mapping and genome scans towards the characterization of candidate loci under parallel selection in the lake whitefish (Coregonus clupeaformis). Mol. Ecol. 14: 351–361. [DOI] [PubMed] [Google Scholar]
- Rosenthal, D. M., A. E. Schwarzbach, L. A. Donovan, O. Raymond and L. H. Rieseberg, 2002. Phenotypic differentiation between three ancient hybrid taxa and their parental species. Int. J. Plant Sci. 163: 387–398. [Google Scholar]
- Rosenthal, D. M., L. H. Rieseberg and L. A. Donovan, 2005. Re-creating ancient hybrid species' complex phenotypes from early-generation synthetic hybrids: three examples using wild sunflowers. Am. Nat. 166: 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., J.C. Sánchez-DelBarrio, X. Messeguer and R. Rozas, 2003. DnaSP: DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- Schilling, E. E., and C. B. Heiser, 1981 Infrageneric classification of Helianthus (Compositae). Taxon 30: 393–403.
- Schilling, E. E., C. R. Linder, R. D. Noyes and L. H. Rieseberg, 1987. Phylogenetic relationships in Helianthus (Asteraceae) based on nuclear ribosomal DNA internal transcribed spacer region sequence data. Syst. Bot. 23: 177–187. [Google Scholar]
- Schuelke, M., 2000. A poor man's approach to genotyping for research and high-throughput diagnostics. Nat. Biotechnol. 18: 233–234.10657137 [Google Scholar]
- Schwarzbach, A. E., and L. H. Rieseberg, 2002. Likely multiple origins of a diploid hybrid sunflower species. Mol. Ecol. 11: 1703–1715. [DOI] [PubMed] [Google Scholar]
- Scotti-Saintagne, C., S. Mariette, I. Porth, P. G. Goicoechea, T. Barreneche et al., 2004. a Genome scanning for interspecific differentiation between two closely related oak species [Quercus robur L. and Q. petraea (Matt.) Liebl.]. Genetics 168: 1615–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti-Saintagne, C., C. Bodenes, T. Barreneche, E. Bertocchi, C. Plomion et al., 2004. b Detection of quantitative trait loci controlling bud burst and height growth in Quercus robur L. Theor. Appl. Genet. 109: 1648–1659. [DOI] [PubMed] [Google Scholar]
- Scotti-Saintagne, C., E. Bertocchi, T. Barreneche, A. Kremer and C. Plomion, 2005. Quantitative trait loci mapping for vegetative propagation in pedunculate oak. Ann. For. Sci. 62: 369–374. [Google Scholar]
- Sokal, R., and N. L. Oden, 1978. Spatial autocorrelation in biology. 1. Methodology. Biol. J. Linn. Soc. 10: 199–228. [Google Scholar]
- Storz, J. F., B. A. Payseur and M. W. Nachman, 2004. Genome scans of DNA variability in humans reveal evidence for selective sweeps outside of Africa. Mol. Biol. Evol. 21: 1800–1811. [DOI] [PubMed] [Google Scholar]
- Stump, A. D., M. C. Fitzpatrick, N. F. Lobo, S. Traore, N. F. Sagnon et al., 2005. Centromere-proximal differentiation and speciation in Anopheles gambiae. Proc. Natl. Acad. Sci. USA 102: 15930–15935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, S., J. K. Yu, M. B. Slabaugh, D. K. Shintani and S. J. Knapp, 2002. Simple sequence repeat map of the sunflower genome. Theor. Appl. Genet. 105: 1124–1136. [DOI] [PubMed] [Google Scholar]
- Ting, C. T., S. C. Tsaur and C. I. Wu, 2001. The phylogeny of closely related species as revealed by the genealogy of a speciation gene, Odysseus. Proc. Natl. Acad. Sci. USA 97: 5313–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, T. L., M. W. Hahn and S. V. Nuzhdin, 2005. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 3: 1572–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer, M. C., S. J. E. Baird, J. Pan and L. H. Rieseberg, 1998. Rapid hybrid speciation in wild sunflowers. Proc. Natl. Acad. Sci. USA 95: 11757–11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasemagi, A., J. Nilsson and C. R. Primmer, 2005. Expressed sequence tag-linked microsatellitesas a source of gene-associated polymorphisms for detecting signatures of divergent selection in Atlantic salmon (Salmo salar L.). Mol. Biol. Evol. 22: 1067–1076. [DOI] [PubMed] [Google Scholar]
- Weir, B. S., and C. C. Cockerham, 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Welch, M. E., and L. H. Rieseberg, 2002. Patterns of genetic variation suggest a single, ancient origin for the diploid hybrid species Helianthus paradoxus. Evolution 56: 2126–2137. [DOI] [PubMed] [Google Scholar]
- Woodhead, M., J. Russell, J. Squirrell, P. M. Hollingsworth, K. Mackenzie et al., 2005. Comparative analysis of population genetic structure in Athyrium distentifolium (Pteridophyta) using AFLPs and SSRs from anonymous and transcribed gene regions. Mol. Ecol. 14: 1681–1695. [DOI] [PubMed] [Google Scholar]
- Wu, C. I., 2001. The genic view of the process of speciation. J. Evol. Biol. 14: 851–865. [Google Scholar]
- Yu, J. K., S. Tang, M. B. Slabaugh, A. Heesacker, G. Cole et al., 2003. Towards a saturated molecular genetic linkage map for cultivated sunflower. Crop Sci. 43: 367–387. [Google Scholar]