Abstract
Osmotic stress induces activation of an adaptive mitogen-activated protein kinase pathway in concert with disassembly of the actin cytoskeleton by a mechanism that is not understood. We have previously shown that the conserved actin-interacting MAP kinase kinase kinase Ssk2p/MEKK4, a member of the high-osmolarity glycerol (HOG) MAPK pathway of Saccharomyces cerevisiae, mediates recovery of the actin cytoskeleton following osmotic stress. In this study, we have employed in vitro kinase assays to show that Ssk2p kinase activity is activated for the actin recovery pathway via a noncanonical, Ssk1p-independent mechanism. Our work also shows that Ssk2p requires the polarisome proteins Bud6p and Pea2p to promote efficient, polarized actin reassembly but that this requirement can be bypassed by overexpression of Ssk2p. Formin (BNI1 or BNR1) and tropomyosin functions are also required for actin recovery but, unlike for Bud6p and Pea2p, these requirements cannot be bypassed by overexpression of Ssk2p. These results suggest that Ssk2p acts downstream of Bud6p and Pea2p and upstream of tropomyosin to drive actin recovery, possibly by upregulating the actin nucleation activity of the formins.
EUKARYOTIC cells have evolved to quickly detect and adapt to the highly variable osmotic conditions that they encounter in their natural environment. In mammalian systems, the p38 MAP kinase signaling pathway is the primary mediator of the osmotic stress response (reviewed by Sheikh-Hamad and Gustin 2004). Exposure of Saccharomyces cerevisiae to changes in external osmolarity activates the p38-homologous high-osmolarity glycerol response (HOG) MAPK pathway, resulting in global transcriptional changes and the accumulation of the osmolyte glycerol (reviewed by Hohmann 2002).
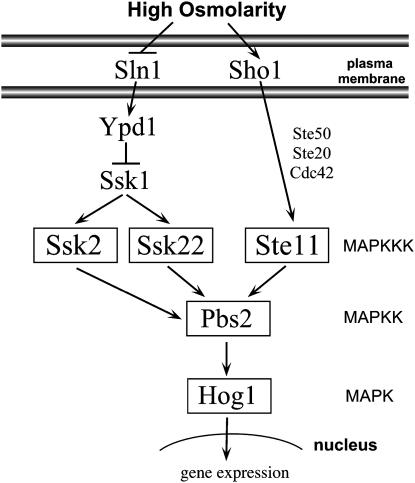
Activation of the HOG pathway is regulated by two membrane-bound osmosensors, Sln1p and Sho1p (Maeda et al. 1994, 1995). These proteins transmit signals through two independent branches of the pathway, which converge on the MEK Pbs2p (Figure 1). Osmotic stress induces the Sho1p branch to phosphorylate Pbs2p through the recruitment of an osmo-signaling complex composed of Cdc42p, Ste20p, Ste50p, and the MAPKKK Ste11p (Maeda et al. 1995; Posas and Saito 1997; O'Rourke and Herskowitz 1998; Posas et al. 1998; Wu et al. 1999; Reiser et al. 2000). Changes in turgor pressure activate the Sln1p branch of the HOG pathway (Reiser et al. 2003). Sln1p is a transmembrane histidine kinase that transmits signal through a three-component phosphorelay (Maeda et al. 1994; Posas et al. 1996). Under normal osmotic conditions the phosphorelay is active and a phosphate is transferred from Sln1p to Ssk1p. When the cell is exposed to stress, the phosphorelay is inhibited and Ssk1p is subsequently dephosphorylated. Unphosphorylated Ssk1p binds and activates the MAPKKKs Ssk2p and Ssk22p (Maeda et al. 1995). Binding of Ssk1p to the N terminus of Ssk2p causes the MAPKKK to autophosphorylate, and activated Ssk2p is then able to bind and phosphorylate Pbs2p (Maeda et al. 1995; Posas and Saito 1998). Pbs2p activates the MAPK Hog1p that accumulates in the nucleus and induces the expression of osmoregulatory and stress response genes.
Figure 1.—
The S. cerevisiae HOG MAP kinase pathway. Ssk2p functions in both the HOG MAP kinase pathway and the newly characterized actin recovery pathway following osmotic shock.
Osmotic stress also induces a transient and reversible disassembly of the actin cytoskeleton (Chowdhury et al. 1992). The actin cytoskeleton plays a vital role in the response to osmotic stress since many actin mutants exhibit osmosensitivity (Wertman et al. 1992). In nonstressed yeast cells, actin filaments form cables and cortical patches. Cables are typically oriented along the axis of growth and direct polarized growth and organelle segregation (reviewed by Pruyne and Bretscher 2000). Actin patches are highly dynamic and motile structures that are believed to play a role in endocytosis (reviewed by Engqvist-Goldstein and Drubin 2003). Patches initially localize to sites of apical growth before clustering at the mother-bud neck just before cytokinesis (Amberg 1998). Within 1 min after exposure to osmotic stress, actin cables begin to disassemble, followed by the slower depolarization of actin patches from sites of growth. Upon restoration of the internal hyperosmotic environment, after ∼1 hr, the actin cytoskeleton reassembles and polarized growth resumes (Chowdhury et al. 1992; Brewster and Gustin 1994).
Previous work from our laboratory has shown that the MEKK kinase Ssk2p mediates efficient recovery of the actin cytoskeleton after osmotic stress (Yuzyuk et al. 2002; Yuzyuk and Amberg 2003). Although Ssk1p is the only known upstream regulator of Ssk2p in the HOG pathway, previous work from our laboratory has suggested that Ssk1p is not required for Ssk2p-mediated actin recovery since an ssk1Δ strain is able to promote recovery of the actin cytoskeleton following osmotic stress (Yuzyuk et al. 2002). Ssk2p senses disassembly of the cytoskeleton as induced by osmotic stress by forming a 1:1 complex with actin. Actin binding stimulates the relocalization of Ssk2p from the cytoplasm to the mother-bud neck in a septin-dependent manner where the kinase promotes cytokinesis. In budding cells, Ssk2p concentrates at the bud tip via the polarisome protein Spa2p (Yuzyuk and Amberg 2003). Once at the bud tip, Ssk2p promotes the assembly of polarized actin cables. The polarisome complex, which includes Bni1p, Bud6p/Aip3p, Pea2p, and the scaffolding protein Spa2p, normally promotes polarized assembly of the actin cytoskeleton primarily through activation of the yeast formin Bni1p (Sheu et al. 1998; Evangelista et al. 2002; Sagot et al. 2002). Deletion of Ssk2p or the scaffold Spa2p, as well as a mutant of Ssk2p (ssk2ΔLD) that is unable to bind actin or properly localize to growth sites, results in a significant delay in actin recovery and bud emergence following osmotic stress. This role in cytoskeletal recovery is likely to be conserved in most eukaryotes as the mammalian homolog of Ssk2p, MEKK4, is able to functionally replace Ssk2p in the yeast actin recovery pathway (Yuzyuk and Amberg 2003). When expressed in yeast, MEKK4/MTK1 binds yeast actin in response to osmotic stress-induced disruption of the actin cytoskeleton, translocates to growth sites in a Spa2p-dependent manner, and can drive actin reassembly in the absence of the endogenous kinase Ssk2p.
In this report we have investigated the requirements for Ssk2p activation and the potential effectors of Ssk2p-mediated actin recovery. In support of previous studies, we show that Ssk2p functions in the actin recovery pathway independently of the HOG pathway. We further show that, in addition to the scaffold protein Spa2p and the actin-binding protein tropomyosin, Ssk2p-mediated actin recovery requires the polarisome proteins Bni1p, Bud6p, and Pea2p to promote new actin filament assembly via formin function.
MATERIALS AND METHODS
Yeast strains:
Strains used in this study are listed in Table 1. To confirm integration of the kanMX4 module at the proper open reading frame, the Research Genetics (Huntsville, AL) deletion strains (Invitrogen, San Diego) were analyzed by PCR using the following primer sets (see Table 2 for all primer sequences): DAo-AIP3-1 and DAo-AIP3-4 for ylr319cΔ, DAo-BNI1-1 and DAo-BNI1-4 for ynl271cΔ, BBo-BNR1-1 and BBo-BNR1-2 for yil159wΔ, and BBo-TPM2-3 and BBo-TPM2-4 for yil138cΔ. The ssk2Δ0∷NATr cassette was created with two-step fusion PCR (Amberg et al. 1995) using primers DAo-SSK2-8, -9, -10, and -11. The nourseothricin resistance (NATr) gene was amplified from p4339 (Tong et al. 2001) using primers m13 forward and reverse. The ssk22Δ0∷HGHr cassette was created with primers TYo-SSK1-1, -2, -3, and -4. The hygromycin gene (HGHr) was amplified from pAG32 (Goldstein and McCusker 1999) using primers F1 and R1. The sho1Δ0∷TRP1cg (Candida glabrata) cassette was amplified from SO1008 (O'Rourke and Herskowitz 2002) using primers TYo-SHO1-1 and TYo-SHO1-4. The ssk22Δ0∷KANr cassette was amplified from YCR073CΔ using primers TYo-SSK22-6 and TYo-SSK22-7. The ssk2ΔLD(aa 426–466)∷NATr cassette was created using primers BBo-SSK2/NAT-1 and DAo-SSK2-6 to amplify the NATr cassette from p4339, which was transformed into the unmarked TYY47 strain. The bni1-11∷NATr cassette was created by fusion PCR using primers BBo-BNI1-10 and BBo-BNI1/NAT-3 to amplify bni1-11 from ABY1827 (Evangelista et al. 2002) and primers BBo-BNI1/NAT-1 and BNI1/NAT-2 to amplify the NATr cassette from p4339. During construction of the bni1-11∷NATr cassette it was discovered that the bni1-11 allele has a single amino acid substitution, D1511G, and is missing the reported substitution K1601R (data not shown). The bnr1Δ0∷KANr cassette was amplified from ABY1285 (Evangelista et al. 2002) using primers BBo-BNR1-1 and BBo-BNR1-2.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Relevant genotype | Reference/source |
|---|---|---|
| FY23 | MATatrp1Δ63 ura3-52 leu2Δ1 | F. Winston |
| JTY143 | MATatrp1Δ63 ura3-52 leu2Δ1 his3-Δ200 pep4∷HIS3 prb1Δ1.6R can1r | This lab |
| TYYD6B | MATaura3-52 leu2Δ1 his3-Δ200 ssk2Δ∷URA3 | Yuzyuk et al. (2002) |
| TYY47 | MATatrp1Δ63 ura3-52 leu2Δ1 ssk2ΔLD (aa 426–466) | Yuzyuk and Amberg (2003) |
| SO1008 | MATassk1Δ∷HIS3cg sho1Δ∷TRP1cg | O'Rourke and Herskowitz (2002) |
| ABY1827 | MATahis3Δ1 ura3Δ0 bnr1Δ∷KANr bni1-11∷URA3 | Evangelista et al. (2002) |
| ABY1825 | MATahis3Δ1 ura3Δ0 bnr1Δ∷KANr | Evangelista et al. (2002) |
| BY4741 | MATaura3Δ0 leu2Δ0 his3Δ1 met15Δ0 | Research Genetics |
| ylr319cΔ | MATaura3Δ0 leu2Δ0 his3Δ1 met15Δ0 bud6Δ0∷KANr | Research Genetics |
| ynl271cΔ | MATaura3Δ0 leu2Δ0 his3Δ1 met15Δ0 bni1Δ0∷KANr | Research Genetics |
| yil159w Δ | MATaura3Δ0 leu2Δ0 his3Δ1 met15Δ0 bnr1Δ0∷KANr | Research Genetics |
| yer149cΔ | MATaura3Δ0 leu2Δ0 his3Δ1 met15Δ0 pea2Δ0∷KANr | Research Genetics |
| ynl079cΔ | MATaura3Δ0 leu2Δ0 his3Δ1 met15Δ0 tpm1Δ0∷KANr | Research Genetics |
| yil138cΔ | MATaura3Δ0 leu2Δ0 his3Δ1 met15Δ0 tpm2Δ0∷KANr | Research Genetics |
| ynr031cΔ | MATaura3Δ0 leu2Δ0 his3Δ1 met15Δ0 ssk2Δ0∷KANr | Research Genetics |
| ycr073cΔ | MATaura3Δ0 leu2Δ0 his3Δ1 met15Δ0 ssk22Δ0∷KANr | Research Genetics |
| BBY109 | MATatrp1Δ63 ura3-52 leu2Δ1 his3-Δ200 ssk2Δ0∷NAT | This study |
| BBY181 | MATatrp1Δ63 ura3-52 leu2Δ1 his3-Δ200 ssk1Δ0∷HGH | This study |
| BBY202 | MATatrp1Δ63 ura3-52 leu2Δ1 sho1∷TRP1cg ssk2Δ0∷NATr ssk22Δ0∷KANr | This study |
| BBY205 | MATatrp1Δ63 ura3-52 leu2Δ1 sho1∷TRP1cg ssk1Δ0∷HGHr ssk22Δ0∷KANr | This study |
| BBY208 | MATatrp1Δ63 ura3-52 leu2Δ1 his3-Δ200 sho1∷TRP1cg ssk1∷HGHr ssk2Δ0∷NAT ssk22Δ0∷KAN | This study |
| BBY240 | MATatrp1Δ63 ura3-52 leu2Δ1 ssk2ΔLD(a.a. 426-466)∷NATr sho1∷TRP1cg ssk1Δ0∷HGHr ssk22Δ0∷KANr | This study |
| BBY289 | MATatrp1Δ63 ura3-52 leu2Δ1 his3-Δ200 bni1-11∷NATr | This study |
| BBY293 | MATatrp1Δ63 ura3-52 leu2Δ1 his3-Δ200 bnr1Δ0∷KANr | This study |
| BBY297 | MATatrp1Δ63 ura3-52 leu2Δ1 his3-Δ200 bni1-11∷NATr bnr1∷KANr | This study |
| BBY358 | MATatrp1Δ63 ura3-52 leu2Δ1 sho1Δ∷TRP1cg ssk1Δ0∷HGHr ssk2Δ0∷NATr | This study |
TABLE 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| F1 | CGGATCCCCGGGTTAATTAA |
| R1 | GAATTCGAGCTCGTTTAAAC |
| DAo-AIP3-1 | CTGTGATCTGCTTGCTTC |
| DAo-AIP3-4 | CACCGTTCTCATGCAAATTGA |
| DAo-BNI1-1 | GAGAAGCAAGAAAGGAAGAG |
| DAo-BNI1-2 | GGAAACAATGGCACATAATA |
| DAo-SSK2-6 | TCCGTGGAATCGCCGTAAGGCTTATAATTGAAGTAGTCTGAATGCGACATACGCGGAACCAGATCCGATT |
| DAo-SSK2-8 | TGAAAAGAGTGATAAAGGTGGG |
| DAo-SSK2-9 | GTCGTGACTGGGAAAACCCTGGCGATGACTGCCTCATCTTGGAG |
| DAo-SSK2-10 | TCCTGTGTGAAATTGTTATCCGCTGGTTGAGCTGTTGATGGATC |
| DAo-SSK2-11 | GCGAGCGTTTTACATTAATCC |
| TYo-SSK1-1 | GCCCCTGGATTGAATCTTTGAC |
| TYo-SSK1-2 | GTCGTFACTGGGAAAACCCTGGCGGTCTATTCGTAGCCAAACCT |
| TYo-SSK1-3 | TCCTGTGTGAAATTGTTATCCGCTCAAATAGAATTGTGAGTTTGG |
| TYo-SSK1-4 | GACTGCGATGTGTTCATCCATG |
| TYo-SHO1-1 | GCTTTCGGAGTCGGGCTTTTG |
| TYo-SHO1-4 | GGGTTCTGCCATTACTGGTCCA |
| TYo-SSK2-NheI | CGGGCTAGCCTACTCTCTTTCCTCAG |
| TYo-SSK22-6 | GAGATAGCACCGGGAACTCTCAGG |
| TYo-SSK22-7 | GCCTCTCTCGTCATATCCAG |
| TYO-EPbs2-BamHI | CGCGGGGATCCCCATGGAAGACAAGTTTGCTAAC |
| TYO-EPbs2-XhoI | CCGCGCTCGAGCTATAAACCACCCATATGTAA |
| TYO-Pbs2-K/M-2 | TAGCTCCAAACGGACTTCCATCGTCGCCATAATAACATT |
| TYO-Pbs2-K/M-3 | AATGTTATTATGGCGACGATG GAAGTCCGTTTGGAGCTA |
| BBo-BNI1-10 | ATCCAGGGATATTTCTCAACAGTT |
| BBo-BNI1/NAT-1 | ATAGATCACTTATGATACACTTTACATTCATCACATGGAGGC CCAGAATACCC |
| BBo-BNI1/NAT-2 | AATGCAATAGTAGAGATCATTTGAGTAACAGTATAGCGACC AGCATTCAC |
| BBo-BNI1/NAT-3 | GATGAATGTAAAGTGTATCATAAGTGATCTAT |
| BBo-BNR1-1 | CACATCATTACCCTCCTCATGTTG |
| BBo-BNR1-2 | CTGTCCATCTCCAAATCTTAGTTC |
| BBo-pBB5-1 | CAACGCAATGGTTTCATCCAAAGC |
| BBo-pBB5-2 | ACATTATATCAATGAAAACGTACA |
| BBo-pBB5-4 | TCTTCTACTACATCAGCTTTTAGA |
| BBo-TAP-1 | CGCAGATCTATGTCTCAAGCTCCA |
| BBo-TAP-2 | GCTGCAGCTGAAAATTTGTATTTTCAA |
| BBo-TAP-3 | GCTGCAGCTGAAAATTTGTATTTTCAA |
| BBo-TAP-4 | GCTCTAGAGCGCGCATCGATGAGCTCGCGGCCGCAGCTGCAGCCAAAGCACCAGA |
| BBo-TPM2-3 | TCAACACACTATCGTACGTAGATT |
| BBo-TPM2-4 | TCATCACCTCCGTTGCCTTTAGCG |
| TAP-SSK2-NotI | AGGACTGCAGCGGCCGCGATGTCGCATTCAGACTACTTC |
| BBo-SSK2/NAT-1 | AGAAGATTAATGATAGCATTTTTTTATAATTGTTTTGATTTTGAGAACAGACATGGAGGCCCAGAATACCC |
| DAo-TAP-1 | GCGAGATCTGCTGCAGCTGAAAATTTGTATTTTCAAGGTGCTGCAGCTGAAAAAAGAAGATGGAAAAAAAATTTTATTGCTGTTTCTGCTGCAAATAGATTTAAAAAGATTTCTTCATCTGGTGCTTTGGCTGCAGCTGGATCCGCG |
| DAo-TAP-2 | CGCGGATCCAGCTGCAGCCAAAGCACCAGATGAAGAAATCTTTTTAAATCTATTTGCAGCAGAAACAGCAATAAAATTTTTTTTCCATCTTCTTTTTTCAGCTGCAGCACCTTGAAAATACAAATTTTCAGCTGCAGCAGATCTCGC |
Plasmid constructions:
Plasmids used in this study are listed in Table 3. Plasmid pBB34, containing a reengineered tandem affinity purification (TAP) tag, was created by amplifying the protein A repeat cassette from pProtA (Aitchison et al. 1995) using primers BBo-TAP-1 and BBo-TAP-2. The tobacco etch virus (TEV) cleavage site, calmodulin-binding peptide (CBP) sequence, and the cloning site were created by annealing oligonucleotides DAo-TAP-1 and DAo-TAP-2. The annealed product was amplified and a multiple cloning site was created with primers BBo-TAP-3 and BBo-TAP-4. The protein A cassette and the annealed oligonucleotides were combined using fusion PCR with primers BBo-TAP-1 and BBo-TAP-4. The resulting PCR product was digested with HindIII and NotI and ligated into pTS422 (Tim Stearns, personal gift). TAP-tagged SSK2 (pDA316) was created by amplifying the SSK2 locus with primers TAP-SSK2-NotI and TYo-SSK2-NheI. The PCR product was digested with NotI and NheI and ligated into pBB34 digested with XbaI and NotI. The construct was confirmed by sequencing with the primers BBo-pBB5-1, -2, and -4. To make GST-tagged inactive pbs2K389Mp (pTYPBS2-1), genomic DNA was amplified with two sets of primers (TYo-EPbs2-BamHI and TYo-Pbs2K/M-2 and TYo-EPbs2-XhoI and TYo-Pbs2K/M-3) that introduced an inactivating K-to-M substitution at position 389. The fusion product was digested with BamHI and XhoI before ligation into pGEX-5X-3 (GE Healthcare).
TABLE 3.
Plasmids used in this study
| Plasmid | Description | Reference/source |
|---|---|---|
| pBB34 | URA3 CEN ACT1p-TAP tag | This study |
| pDA316 | URA3 CEN ACT1p-TAP tag-SSK2 | This study |
| pBB111 | URA3 CEN ACT1p-TAP tag-ssk2ΔLD (aa 426–466) | This study |
| p366 | LEU2 CEN | This lab |
| pTY111U | GFP-SSK2 URA3 CEN | Yuzyuk et al. (2002) |
| pTY113U | GFP-ssk2K1295N URA3 | Yuzyuk et al. (2002) |
| pTY111L | GFP-SSK2 LEU2 CEN | Yuzyuk et al. (2002) |
| pTY113L | GFP-ssk2 K1295N LEU2 | Yuzyuk et al. (2002) |
| pTYPBS2 | GST-PBS2K389M | This study |
| p4339 | PAgTEF-natMX4 | Tong et al. (2001) |
| pAG32 | PAgTEF-hphMX4 | Goldstein and McCusker (1999) |
| pDA261 | URA3 CEN Act1p protein A repeats | This lab |
| Pts422 | URA3 CEN ACT1p-ACT1t | T. C. Doyle |
Purification of epitope-tagged proteins:
GST-tagged pbs2K389Mp (pTYPBS2-1) was expressed in Escherichia coli K12 UT5600 (New England Biolabs, Beverly, MA) using 1 mm IPTG. Cells were resuspended in buffer [50 mm Tris–HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, 0.1% 2-ME, 1% Triton X-100, 1 mm PMSF, 1% protease inhibitor cocktail set IV (Calbiochem)] and lysed by sonication. The cell extract was incubated with 500 μl of preequilibrated glutathione–sepharose beads for 30 min at 4°. After washing five times with buffer (50 mm Tris–HCl, pH 8.0, 2 mm DTT), proteins were eluted (20 mm glutathione, 50 mm Tris–HCl, pH 8.0, 2 mm DTT) and dialyzed overnight (PBS, 1 mm MgCl2).
TAP-tagged SSK2 was expressed in the protease-deficient yeast strain JTY143. Cells were grown in selective synthetic medium to a density of 2 × 107 cells/ml and were shocked with 0.9 m NaCl for 15 min before harvesting. Cells were resuspended in purification buffer [50 mm Tris–HCl, pH 7.5, 15 mm EDTA, 0.1% Triton X-100, 100 mm NaCl, 1 mm DTT, 1 mm PMSF, 0.5% protease inhibitor cocktail set IV (Calbiochem), and 0.5% phosphatase inhibitor cocktail set II (Calbiochem)] and lysed by one pass through a French press at 1100 psi. Proteins were bound to 200 μl of rabbit IgG beads (Sigma, St. Louis) for 2 hr at 4°. After extensive washing, beads were resuspended in 1 ml TEV cleavage buffer (10 mm Tris–HCl, pH 8, 0.5 mm EDTA, 0.1% Triton X-100, 50 mm NaCl, 1 mm DTT, 1 mm PMSF, 0.05% protease and phosphatase inhibitors) and 20 units of TEV protease (Invitrogen) and incubated overnight at 4°. The supernatant was collected and the beads were washed three times with 1 ml of TEV cleavage buffer to maximize recovery. The ∼4 ml of supernatant was concentrated to 200 μl using a Centricon YM-10 (Millipore, Bedford, MA).
In vitro kinase assay:
Sample protein (40 μl, 20% of total recovery from the TAP purification) was added to 160 μl of kinase buffer (40 mm Tris–HCl, pH 7.5, 1.5 mm DTT, 10 mm MgCl2) and mixed with 10 μl (1 μg) of substrate (GST or Pbs2-GST). Two microliters of [γ-32P]ATP (3000 Ci/mmol) was added for a final concentration of 30 nm and the reaction was incubated at 30° for 10 min. Reactions were quenched by the addition of 2 μl of 100 mm ATP for a final concentration of ∼1 mm. Reactions were terminated by the addition of 50 μl of 2× SDS loading buffer and proteins were analyzed by SDS–PAGE.
Cell synchronization and recovery:
For G1 synchronization, cultures at a density of 1 × 107 cells/ml were arrested in selective synthetic medium in the presence of 1 μg/ml α-factor (Sigma) for 2 hr at 25°. Cells were pelleted and released into selective medium ±0.9 m NaCl and allowed to recover at 25°. For each time point, >100 cells were counted; reported numbers are the averages calculated from three experiments. For experiments using temperature-sensitive strains, cells were released from α-factor arrest into prewarmed selective medium and allowed to recover at 37°.
RESULTS
Ssk1p is not required for Ssk2p activation:
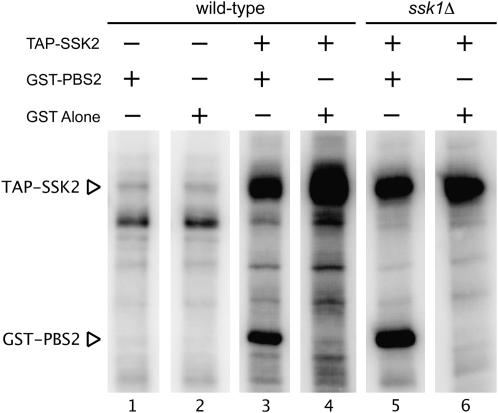
Previous work from this laboratory has shown that Ssk1p is not involved in regulating the localization of Ssk2p to growth sites upon osmotic shock (Yuzyuk et al. 2002; Yuzyuk and Amberg 2003). Additionally, cell cycle arrest experiments suggested that Ssk1p is not required for activation of Ssk2p in the actin recovery pathway. We therefore wanted to examine the in vitro activity of Ssk2p in response to osmotic stress in SSK1 and ssk1Δ backgrounds. To address this question, we modified the traditional TAP tag (Rigaut et al. 1999) to express four IgG-binding units of protein A from Staphylococcus aureus followed by a TEV cleavage site and a CBP at the N terminus of SSK2. We have found that increasing the number of IgG-binding repeats significantly increases retention in the first affinity purification step. Western blotting confirmed expression of a fusion protein of the correct size, and TAP-tagged Ssk2p was able to complement the HOG pathway defects of an ssk2Δ ssk22Δ sho1Δ strain and the actin recovery defects of an ssk2Δ strain (data not shown). We purified the fusion protein from an asynchronous population that had been osmotically shocked for 15 min with 0.9 m NaCl and measured its in vitro kinase activity using recombinant GST-Pbs2K389Mp as an exogenous substrate. TAP-Ssk2p from wild-type shocked cells was able to bind and phosphorylate the substrate (Figure 2, lane 3). The TAP tag alone, however, was not able to phosphorylate GST-Pbs2K389Mp (Figure 2, lane 1). Unfortunately, we were unable to measure the specific activity of the kinase due to the low concentration of the kinase in the cell. TAP-Ssk2p was then purified from an osmotically stressed ssk1Δ strain to examine the activation state of the protein. The kinase was fully active and able to phosphorylate GST-Pbs2K389Mp (Figure 2, lane 5). Therefore, even in the absence of its established HOG pathway activator, Ssk1p (Maeda et al. 1995), the kinase activity of Ssk2p can be activated.
Figure 2.—
Ssk2p kinase activity can be regulated independently of Ssk1p in response to osmotic shock. The protease-deficient strain JTY143 was transformed with a plasmid expressing the TAP tag (pBB34) or TAP-Ssk2p (pDA316). Cells were stressed with 0.9 m NaCl for 15 min, cell extracts were prepared, and the TAP tag was affinity precipitated with IgG beads. The protein was cleaved from the beads using TEV protease and used in an in vitro kinase assay with GST or GST-PBS2 as a substrate.
Activated Ssk2p can cross talk between the actin recovery pathway and the HOG pathway:
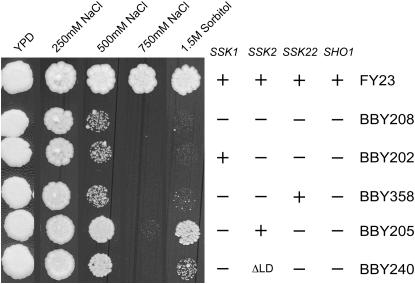
In the previous in vitro experiments, Ssk2p activated by the actin recovery pathway was able to phosphorylate Pbs2p, its HOG pathway substrate. We next asked if this level of activation is sufficient to activate the HOG pathway in vivo. Wild-type and mutant strains were plated on increasing concentrations of NaCl and sorbitol. The wild-type strain (FY23) was healthy in all concentrations of solute (Figure 3). The HOG pathway-deficient strain, with both branches of the pathway inactivated (ssk1Δ ssk2Δ ssk22Δ sho1Δ; BBY208), was sick in 500 mm NaCl and completely inviable in 750 mm NaCl and 1.5 m sorbitol. The presence of Ssk2p in the absence of its known activator, Ssk1p (ssk1Δ ssk22Δ sho1Δ; BBY205), raised viability to nearly wild-type levels in 500 mm NaCl and 1.5 m sorbitol. In contrast, adding back Ssk1p in the absence of Ssk2p (strain BBY202) did not increase viability. We were concerned that yeast MAP kinase kinase kinases might have a low level of basal activity that could explain the above results. However, the presence of Ste11p (another MEKK kinase of the HOG pathway) in strain BBY208 (ssk1Δ ssk2Δ ssk22Δ sho1Δ) did not enhance viability on higher concentrations of NaCl and sorbitol when compared to the presence of Ssk2p in strain BBY205 (ssk1Δ ssk22Δ sho1Δ). Further, in the absence of the Ssk1p activator, addition of Ssk22p (strain BBY358), which is highly homologous to Ssk2p but has been shown not to play a role in actin recovery after osmotic stress (Yuzyuk et al. 2002), did not increase viability over the quadruple delete strain BBY208. Together, these results suggest that the kinase activity of Ssk2p in these experiments is stimulated by an Ssk1p-independent mechanism, presumably by the actin recovery pathway. In agreement with this assertion was our observation that deletion of the ΔLD domain (aa 426–466; BBY240) of Ssk2p, which fails to polarize in response to osmotic stress and has actin recovery defects, resulted in minimally decreased viability on 500 mm NaCl but showed a more serious defect on 1.5 m sorbitol as compared to the strain expressing wild-type Ssk2p (strain BBY205). This suggests that actin binding can contribute to Ssk1p-independent activation of Ssk2p but that an additional factor(s) is also involved. Failure to bind actin could also lower the specific activity of the kinase or could hinder the ability of the kinase to find the proper substrate.
Figure 3.—
Ssk2p activated by an Ssk1p-independent mechanism can cross talk to the HOG pathway. Wild-type (FY23), ssk1Δ ssk2Δ ssk22Δ sho1Δ (BBY208), ssk2Δ ssk22Δ sho1Δ (BBY202), ssk1Δ ssk2Δ sho1Δ (BBY358), ssk1Δ ssk22Δ sho1Δ (BBY205), and ssk1Δ ssk2ΔLD ssk22Δ sho1Δ (BBY240) strains were diluted and plated on YPD and YPD + 250 mm, 500 mm or 750 mm NaCl, and YPD + 1.5 m sorbitol.
Ssk2p-mediated actin recovery employs the polarisome proteins Bni1p, Bud6p, and Pea2p:
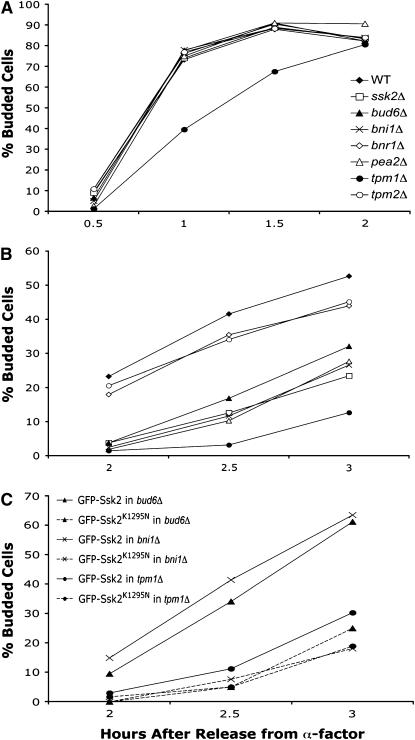
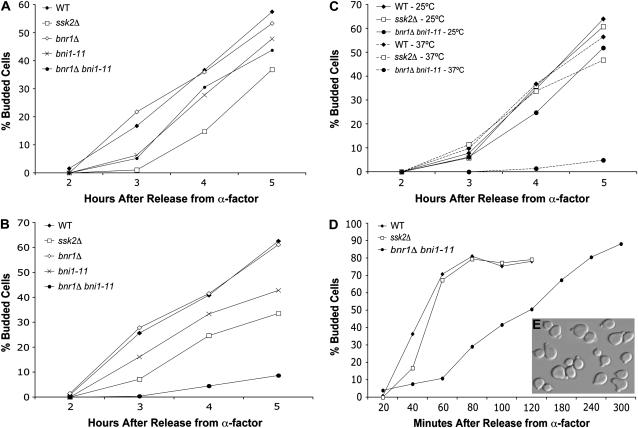
We previously showed that the polarisome scaffolding protein Spa2p is required for osmotic stress-induced polarization of Ssk2p as well as Ssk2p-mediated actin recovery (Yuzyuk and Amberg 2003). Recruitment of Ssk2p by Spa2p would allow the kinase to interact with other members of the polarisome, a multi-subunit protein complex that mediates polarized actin polymerization (Sheu et al. 1998; Pruyne and Bretscher 2000; Evangelista et al. 2002; Sagot et al. 2002). Members of the complex include the Spa2-interacting protein Pea2p and the actin-interacting proteins Bud6p and Bni1p. To examine the role of the polarisome proteins in actin recovery after osmotic stress, we synchronized deletion strains with α-factor and released them into medium with or without 0.9 m NaCl. When bni1Δ, bud6Δ, pea2Δ, and wild-type strains were released into normal-osmolarity medium, the cells were able to reestablish actin structures and within 1 hr were almost 80% budded (Figure 4A). Release into high-osmolarity medium, however, caused a significant delay in actin recovery and bud emergence in the bni1Δ, bud6Δ, and pea2Δ strains compared to wild-type cells (Figure 4B). The kinetics of recovery of the bni1Δ, bud6Δ, and pea2Δ strains were almost identical to the ssk2Δ strain. At 2.5 hr after release from α-factor into high-osmolarity medium, only 13% of ssk2Δ cells had begun to bud compared to 42% of wild-type cells, while only 10% of pea2Δ cells, 12% of bni1Δ cells, and 17% of bud6Δ cells were budded. Double mutants were constructed between ssk2Δ and bni1Δ, bud6Δ, and pea2Δ, and these double mutants were as comparably defective for bud emergence as the single ssk2Δ mutant (data not shown), consistent with these genes functioning in the same pathway of actin recovery.
Figure 4.—
The polarisome complex and tropomyosin are required for Ssk2p-mediated recovery of the actin cytoskeleton. Wild-type (BY4741), ssk2Δ (ynr031cΔ), bud6Δ (ylr319cΔ), bni1Δ (ynl271cΔ), bnr1Δ (yil159wΔ), pea2Δ (yer149cΔ), tpm1Δ (ynl079cΔ), and tpm2Δ (yil138cΔ) strains were synchronized with α-factor, released into (A) normal osmotic medium or (B) medium containing 0.9 m NaCl, and monitored for the percentage of budded cells. N ≥ 100. (C) α-Factor synchronized bud6Δ (ylr319cΔ), bni1Δ (ynl271cΔ), and tpm1Δ (ynl079cΔ) cells expressing GFP-Ssk2p (pTY111L) or GFP-Ssk2pK1295N (pTY113L) were released into selective medium containing 0.9 m NaCl.
Overexpression of wild-type Ssk2p is able to suppress the actin recovery defects of a spa2Δ strain, presumably by providing sufficient concentrations of the kinase at the appropriate sites of action in the absence of proper scaffolding (Yuzyuk et al. 2002). We next sought to determine whether overexpression of Ssk2p would suppress the actin recovery defects of the bni1Δ or bud6Δ strain after osmotic stress. bni1Δ and bud6Δ cells expressing wild-type GFP-Ssk2p and catalytically inactive GFP-Ssk2K1295N were synchronized with α-factor and released into high-osmolarity medium (Figure 4C). This construct has previously been shown to result in an approximately sevenfold overexpression of Ssk2p (Yuzyuk et al. 2002). The delay in actin recovery of bni1Δ and bud6Δ cells expressing Ssk2pK1295N was similar to that seen in bni1Δ and bud6Δ strains. Expression of wild-type GFP-Ssk2p, however, suppressed the actin recovery defects of bni1Δ and bud6Δ cells to approximately wild-type levels (compare B and C in Figure 4 2.5 hr after release).
Ssk2p-mediated actin recovery requires tropomyosin-1:
Another potential target for Ssk2p-mediated actin recovery are the tropomyosins Tpm1p and Tpm2p. Tropomyosin is an actin-binding protein that stabilizes growing actin cables nucleated by the formins (Liu and Bretscher 1989, 1992; Drees et al. 1995; Evangelista et al. 2002; Sagot et al. 2002). S. cerevisiae has two isoforms of tropomyosin called Tpm1p and Tpm2p, and deletion of both isoforms is lethal. Loss of the major form of tropomyosin, Tpm1p, causes slow growth and a lack of detectable actin cables (Liu and Bretscher 1989). Tpm2p is present at only about one-sixth the concentration of Tpm1p, and deletion of TPM2 does not cause any obvious cytoskeletal or morphological defects (Drees et al. 1995). To examine the role of tropomyosin in actin recovery after osmotic stress, tpm1Δ and tpm2Δ cells were synchronized with α-factor and released into normal medium (Figure 4A). Loss of Tpm2p had no effect on actin recovery and bud emergence, while deletion of TPM1 resulted in a minor defect in recovery and bud emergence. The tpm1Δ strain lagged behind the wild-type strain, eventually catching up at 2 hr after release. When the same strains were released into high-osmolarity medium, the tpm2Δ strain had a slight defect in bud emergence, showing 45% budded cells 3 hr after release compared to 53% for wild-type cells (Figure 4B). The tpm1Δ strain, however, had a serious delay with only 13% budded cells at 3 hr after release. This was even worse than the ssk2Δ strain that exhibited 24% budded cells. Expression of catalytically active GFP-Ssk2p in the tpm1Δ strain was only slightly effective in suppressing the actin recovery defects, raising the number of budded cells to 30% compared to 19% for the strain expressing catalytically inactive GFP-Ssk2K1295N (Figure 4B). These results suggest that Tpm1p plays an important role in Ssk2p-mediated actin recovery after osmotic shock and that Tpm2p is unable to perform that function. If one views the process of actin recovery as a distinct pathway, the inability to suppress the tpm1Δ deletion suggests that Tpm1p could be a potential substrate of the kinase or that under conditions of osmotic stress Tpm1p is an essential component of actin cytoskeleton recovery.
Ssk2p requires formin function to promote actin recovery and bud emergence after osmotic shock:
The formins are a family of actin-nucleating proteins that drive the polymerization of actin filaments (reviewed by Faix and Grosse 2006). S. cerevisiae has two formin isoforms called Bni1p and Bnr1p. Since Bni1p and Bnr1p have overlapping functions and each is capable of compensating for the loss of the other, neither formin is essential but deletion of both is lethal (Vallen et al. 2000; Ozaki-Kuroda et al. 2001). Deletion of either Bni1p or Bnr1p alone did not result in a notable delay in actin recovery when α-factor synchronized cells were released into normal medium (Figure 4). When released into high-osmolarity medium, the bnr1Δ strain showed only a very slight delay in actin recovery: 36% budded at 2.5 hr after release compared to 42% for the wild-type strain (Figure 4B). The bni1Δ strain, however, showed a delay in bud emergence that was similar to that of the ssk2Δ strain. Taken together, these results suggest that Bnr1p is able to promote actin polarization in a bni1Δ strain after release from α-factor synchronization but is not able to suppress the actin recovery defects of the same strain after release from α-factor into high-osmolarity medium. To examine this phenomenon further, we employed a temperature-sensitive allele of BNI1, bni1-11, which does not support cable formation or viability at a nonpermissive temperature (>35°) in a bnr1Δ background (Pruyne et al. 2002). It is possible, however, that this strain does have short disorganized filaments that are not detectable via rhodamine–phalloidin or immunofluorescent analysis. At permissive temperature (25°), the bnr1Δ bni1-11 strain exhibited only a slight delay in bud emergence after osmotic stress [48% budded cells at 5 hr compared to 58% for wild-type cells (Figure 5A)]. However, when the bnr1Δ bni1-11 strain was released into high-osmolarity medium from α-factor arrest, there was a severe defect in bud emergence (Figure 5B). At 5 hr after release, only 9% of bnr1Δ bni1-11 cells were budded compared to 63% of wild-type cells, 61% of bnr1Δ cells, and 43% of bni1-11 cells. Expression of GFP-Ssk2p was unable to suppress the actin recovery defects of the bnr1Δ bni1-11 strain, with only 5% of cells budded at 5 hr after release into high-osmolarity medium at nonpermissive temperatures compared to 57% of wild-type cells (Figure 5C). These results suggest that formin function is required for efficient actin recovery and bud emergence after osmotic stress of α-factor synchronized cells.
Figure 5.—
Formin-directed actin assembly is required for efficient recovery of the actin cytoskeleton after osmotic stress. Wild-type (FY23), ssk2Δ (BBY109), bni1-11 (BBY289), bnr1Δ (BBY293), and bni1-11 bnr1Δ (BBY297) strains transformed with the control vector p366 (to match the conditions used in the overexpression experiments) were synchronized with α-factor and released into (A) selective medium at the permissive temperature of 25° or (B) selective medium plus 0.9 m NaCl at the nonpermissive temperature of 37°. In C, wild-type (FY23), ssk2Δ (BBY109), and bni1-11 bnr1Δ (BBY297) strains overexpressing GFP-Ssk2p from plasmid pTY111L were synchronized with α-factor and released into selective medium at 25° and 37°. In D, wild-type (FY23), ssk2Δ (BBY109), bni1-11 (BBY289), bnr1Δ (BBY293), and bni1-11 bnr1Δ (BBY297) strains transformed with the control vector p366 were synchronized with α-factor and released into normal-osmolarity medium at 37°. In insert E, the bni1-11 bnr1Δ cells from D were visualized 5 hr after release from α-factor arrest into low-osmolarity medium.
Surprisingly, our results also suggest that formins are not essential for bud emergence under conditions of normal/low osmolarity. The bnr1Δ bni1-11 strain was able to initiate formation of a bud when released from α-factor synchronization at nonpermissive temperatures (Figure 5D). The recovery was much slower than what was observed in wild-type cells, and the strain arrested at the small-budded stage (Figure 5E). The ability of formin-deficient cells to initiate bud emergence under normal osmotic conditions is a novel observation that indicates that actin cables are not required for bud emergence. It is possible that the morphogenetic conditions of α-factor treatment create a unique environment that bypasses the need for formins in bud emergence, but we favor the idea that yeast normally employ turgor pressure, not actin nucleation, to initiate bud emergence. More will be said about this model in the discussion.
DISCUSSION
How is Ssk2p activated?
Previous work from our laboratory has suggested that Ssk1p is not required for Ssk2p-mediated actin recovery after osmotic shock (Yuzyuk et al. 2002). Here we show that the Ssk2p kinase is indeed active after osmotic shock, even in the absence of Ssk1p, and that the activated kinase is able to phosphorylate a known substrate (Pbs2p) (Figure 2). It is important to note, however, that although Ssk2p is able to phosphorylate the HOG pathway target Pbs2p in the ssk1Δ mutant, it is possible that the activity of Ssk2p toward the cytoskeletal substrate could be affected. Ssk1p-independent activation is also evident in vivo in the cross-talk experiment (Figure 3), where in the absence of Ssk1p, on intermediate concentrations of NaCl or sorbitol, Ssk2p is able to phosphorylate Pbs2p and promote growth. The other MEKK kinases of the HOG pathway, Ssk22p and Ste11p, are unable to promote viability at these concentrations, suggesting that their response to osmotic stress is contained within the HOG pathway and that they do not have the capacity to interact with this alternative activator. There is other evidence to support Ssk1p-independent activation of Ssk2p. Whole-genome transcript profiling of yeast cells exposed to osmotic stress reveals a number of genes whose induction is totally dependent on Pbs2p (O'Rourke and Herskowitz 2004). When the transcript profiles of a pbs2Δ strain and a ssk1Δ ste11Δ strain were analyzed in response to 0.5 m KCl, it was determined that a number of genes whose induction is totally dependent on Pbs2p were still significantly affected by osmotic stress in the ssk1Δ ste11Δ strain. This suggests that there is an Ssk1p/Ste11p-independent method of activation of Pbs2p in response to 0.5 m KCl. Ssk2p, activated by the actin recovery pathway and cross talking with the HOG pathway, could provide the moderate activation of Pbs2p observed in these experiments. If Ssk2p is activated in the absence of Ssk1p, why is the ssk1Δ ssk22Δ sho1Δ strain dead in high concentrations of NaCl? Under conditions of low osmotic stress, a small amount of Ssk2p that has been activated in the absence of Ssk1p might be able to interact with Pbs2p, which localizes to growth sites in a Sho1p-dependent manner in response to osmotic stress (Reiser et al. 2000). However, since Pbs2p and Ssk2p are ultimately held in alternative scaffolds (Ssk2p binding to the polarisome component Spa2p and Pbs2p binding to the membrane-bound Sho1p), this may provide enough insulation to prevent the level of cross talk needed to support viability at higher concentrations of NaCl. Interestingly, since Ssk2p is able to cross talk between the two pathways, it seems unlikely that there is an inherent difference between Ssk2p that has been activated by Ssk1p and Ssk2p that has been activated by the actin recovery pathway.
The pool of Ssk2p that is functioning within the actin recovery pathway is potentially regulated by actin itself, especially given the fact that the localization of Ssk2p changes in response to disassembly of the actin cytoskeleton as induced by latrunculin A. Actin binding could disrupt the autoinhibition of the kinase and/or allow the kinase to interact with Spa2p. Our results suggest, however, that actin is not the only mechanism of activation in the absence of Ssk1p (Figure 3), since the Ssk2ΔLD mutant, which is unable to interact with actin, is able to promote viability in moderate concentrations of NaCl. The observation that this mutant is less viable in 1.5 m sorbitol may reflect the possibility that salt stress invokes a slightly different adaptive response than osmotic stress alone and that this could induce additional, Ssk2p-independent mechanisms of cellular protection that boost the viability of the Ssk2ΔLD strain.
How does Ssk2p promote actin recovery after osmotic stress?
The polarisome protein complex, consisting of Bud6p, Pea2p, the formin Bni1p, and the scaffold Spa2p, is involved in establishing and maintaining polarized assembly of the actin cytoskeleton (Sheu et al. 1998). Ssk2p localizes to growth sites in a Spa2p-dependent manner in response to osmotic stress and also immunoprecipitates Spa2p (Yuzyuk and Amberg 2003). Recruitment of Ssk2p to the vicinity of the polarisome suggests that its components are potential targets for phosphorylation by the kinase. Indeed, both Bni1p and Bud6p have been shown to be phosphorylated in vivo (Goehring et al. 2003; Matheos et al. 2004; Moseley and Goode 2005). Deletion of BUD6 did not affect bud emergence after α-factor synchronization but had a significant effect on Ssk2p-mediated actin recovery after osmotic stress. Overexpression of the kinase was able to suppress these recovery defects, however, suggesting that although Bud6p plays an important role in Ssk2p-mediated actin recovery after osmotic stress, it is not an essential function. Recent work has shown that Bud6p and the actin monomer-binding protein profilin promote nucleotide exchange on actin monomers and work with Bni1p to add actin onto growing filaments (Moseley et al. 2004). In addition, Bud6p also increases the nucleation activity of Bni1p (Moseley and Goode 2005). It is possible that Ssk2p functions to stimulate the nucleation activity of Bni1p, perhaps by phosphorylating the formin. Increasing the amount of Ssk2p could thus suppress the loss of Bud6p's stimulating activity. Alternatively, Ssk2p could function to regulate localization of polarisome components, but in previous experiments we have observed no defects in Bud6p or Bni1p localization in osmotically stressed ssk2Δ cells (our unpublished results).
Deletion of Bni1p could also be suppressed by overexpression of Ssk2p, suggesting that the kinase either acts downstream of the formin or can employ the Bni1p homolog Bnr1p to promote recovery. In support of the latter hypothesis, the bud emergence defect resulting from the loss of both formin proteins could not be suppressed by higher concentrations of the kinase. Bni1p localizes to the bud tip and nucleates actin cables that direct polarized growth to the bud, while Bnr1p localizes to the bud neck and assembles cables that anchor at the neck and enter the mother cell (Pruyne et al. 2004). The loss of Bnr1p has only a minimal growth defect (Imamura et al. 1997), presumably because Bni1p functions at the neck during cytokinesis and is able to provide formin function in the absence of Bnr1p. The growth defects of a bni1Δ strain (Evangelista et al. 1997), however, suggest that, while Bnr1p is able to nucleate actin in the bud, it is not able to replace all of the functions of Bni1p. Our findings—that Bni1p is the primary player for actin nucleation in the bud during actin recovery from osmotic stress—support the previous observations. However, the ability to suppress actin recovery defects in the bni1Δ strain does suggest that Ssk2p can also upregulate the activity of Bnr1p, possibly through direct phosphorylation.
The tropomyosin Tpm1p is also involved in actin recovery from osmotic stress. The tpm1Δ strain was slightly defective for bud emergence even when released into normal-osmolarity medium, likely reflecting the actin cable stabilization defects of this strain. This deficiency was significantly pronounced when the strain was released into high-osmolarity medium. In fact, the delay was worse than with the ssk2Δ strain, confirming the importance of actin cable stabilization for actin recovery from osmotic stress. Overexpression of GFP-Ssk2p had only a minor effect on bud emergence in the tpm1Δ strain after osmotic stress. Thus Ssk2p is expected to exert its mechanism of action upstream of tropomyosin, most likely to promote or enhance actin polymerization by the formins rather than to stabilize growing filaments.
Our observation that bni1-11 bnr1Δ cells are able to form a bud at nonpermissive temperatures after α-factor synchronization suggests that actin cables are not required to initiate bud emergence under conditions of low osmolarity. In fact, the ability of bni1Δ bnr1Δ cells to initiate bud emergence has previously been reported (Ozaki-Kuroda et al. 2001): a bni1Δ bnr1Δ spore from the tetrad dissection of a double-heterozygous diploid strain arrested as a large cell with at least one bud upon germination. Additionally, a temperature-sensitive allele of the small Rho-GTPase Rho1p (rho1-104 D72N, C164Y) also causes cells to arrest as primarily small budded (Yamochi et al. 1994). GTP-bound Rho1p is known to directly interact with Bni1p (Kohno et al. 1996) and plays a role in the localization of Bni1p to growth sites (Fujiwara et al. 1998). More importantly, Rho1p is required to activate the formins Bni1p and Bnr1p at elevated temperatures (Dong et al. 2003). Consequently, a temperature-sensitive allele of Rho1p in cells at the nonpermissive temperature would have no formin function. Two other small Rho GTPases that bind the formins (Evangelista et al. 1997), Rho3p and Rho4p, are required for formin function at all temperatures (Dong et al. 2003). A rho3Δ rho4Δ strain is deficient for all formin function, resulting in lethality. Interestingly, depletion of Rho4p in a rho3Δ strain causes the cells to lose polarity and arrest in the small-budded stage (Matsui and Toh-e 1992).
Like all eukaryotic cells, S. cerevisiae maintains a higher internal osmolarity, causing water to flow into the cell. The rigid cell wall prevents expansion, causing a force that is termed turgor pressure. Cells can harness turgor pressure through carefully directed morphogenetic pathways to create localized growth (Harold 2002; Levin 2005; Slaughter and Li 2006). We hypothesize that, in the absence of the yeast formins, the cell is able to create localized weakening of the cell wall to exploit turgor pressure to initiate bud emergence. After osmotic shock, this turgor-pressure-directed process is greatly delayed as the cells slowly rebuild a higher internal osmolarity, and it is under these circumstances that bud emergence becomes dependent on localized actin assembly as directed by the formins.
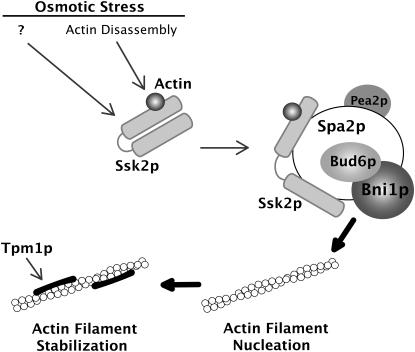
Our results support the following model (Figure 6). Osmotic shock induces the rapid depolymerization of the actin cytoskeleton. Ssk2p senses this osmotic shock, most likely through the formation of a 1:1 complex with actin, and translocates to growth sites via the polarisome scaffold protein Spa2p. In this process, the kinase activity of Ssk2p is activated (possibly by actin and additional factors) by relieving kinase auto-inhibition, resulting in auto-phosphorylation and activation. The kinase then stimulates the activity of the formins Bni1p and Bnr1p, perhaps by phosphorylation, which then nucleate actin cables that are subsequently stabilized by the tropomyosins. The actin cables are then used to reinitiate polarized secretion and polarized cell growth.
Figure 6.—
A model for Ssk2p-facilitated actin reassembly following osmotic stress. Ssk2p senses osmotic shock by forming a 1:1 complex with actin. The complex translocates to growth sites via the polarisome protein Spa2p and promotes actin recovery through the formin Bni1p at the bud tip. Bni1p nucleates actin cables that are stabilized by the tropomyosins and are used to direct polarized cell growth.
In summary, we have furthered our understanding of the activation of Ssk2p in response to osmotic stress, showing that the kinase is regulated by a mechanism distinct from the traditional Ssk1p-mediated pathway. Although the pool of Ssk2p that acts in the actin recovery pathway is spatially regulated by the Spa2p scaffold, under certain circumstances it is able to cross talk to the HOG pathway to assist in maintaining viability. Our suppression experiments suggest that Bni1p and Bnr1p are likely substrates of Ssk2p in facilitating the actin recovery function of the kinase. In addition to the two formin proteins, we have shown that other proteins involved in regulation of formin activity, such as Spa2p, Bud6p, and Pea2p, are important for Ssk2p-mediated actin recovery from osmotic stress. In addition, tropomyosin is required to maintain the actin recovery activity of this pathway.
Acknowledgments
We thank Brian Haarer for a critical reading of the manuscript, Anthony Bretscher and David Pruyne for strains ABY1825 and ABY1827, the laboratory of Ira Herskowitz for SO1008, Charles Boone for plasmid p4339, and all members of the Amberg laboratory for useful discussions and support. This work was supported by National Institutes of Health grant GM056189.
References
- Aitchison, J. D., M. P. Rout, M. Marelli, G. Blobel and R. W. Wozniak, 1995. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 131: 1133–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg, D. C., 1998. Three-dimensional imaging of the yeast actin cytoskeleton through the budding cell cycle. Mol. Biol. Cell 9: 3259–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg, D. C., D. Botstein and E. M. Beasley, 1995. Precise gene disruption in Saccharomyces cerevisiae by double fusion polymerase chain reaction. Yeast 11: 1275–1280. [DOI] [PubMed] [Google Scholar]
- Brewster, J. L., and M. C. Gustin, 1994. Positioning of cell growth and division after osmotic stress requires a MAP kinase pathway. Yeast 10: 425–439. [DOI] [PubMed] [Google Scholar]
- Chowdhury, S., K. W. Smith and M. C. Gustin, 1992. Osmotic stress and the yeast cytoskeleton: phenotype-specific suppression of an actin mutation. J. Cell Biol. 118: 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Y., D. Pruyne and A. Bretscher, 2003. Formin-dependent actin assembly is regulated by distinct modes of Rho signaling in yeast. J. Cell Biol. 161: 1081–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees, B., C. Brown, B. G. Barrell and A. Bretscher, 1995. Tropomyosin is essential in yeast, yet the TPM1 and TPM2 products perform distinct functions. J. Cell Biol. 128: 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist-Goldstein, A. E., and D. G. Drubin, 2003. Actin assembly and endocytosis: from yeast to mammals. Annu. Rev. Cell Dev. Biol. 19: 287–332. [DOI] [PubMed] [Google Scholar]
- Evangelista, M., K. Blundell, M. S. Longtine, C. J. Chow, N. Adames et al., 1997. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276: 118–122. [DOI] [PubMed] [Google Scholar]
- Evangelista, M., D. Pruyne, D. C. Amberg, C. Boone and A. Bretscher, 2002. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 4: 32–41. [DOI] [PubMed] [Google Scholar]
- Faix, J., and R. Grosse, 2006. Staying in shape with formins. Dev. Cell 10: 693–706. [DOI] [PubMed] [Google Scholar]
- Fujiwara, T., K. Tanaka, A. Mino, M. Kikyo, K. Takahashi et al., 1998. Rho1p-Bni1p-Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Biol. Cell 9: 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring, A. S., D. A. Mitchell, A. H. Tong, M. E. Keniry, C. Boone et al., 2003. Synthetic lethal analysis implicates Ste20p, a p21-activated potein kinase, in polarisome activation. Mol. Biol. Cell 14: 1501–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, A. L., and J. H. McCusker, 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Harold, F. M., 2002. Force and compliance: rethinking morphogenesis in walled cells. Fungal Genet. Biol. 37: 271–282. [DOI] [PubMed] [Google Scholar]
- Hohmann, S., 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66: 300–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura, H., K. Tanaka, T. Hihara, M. Umikawa, T. Kamei et al., 1997. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 16: 2745–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno, H., K. Tanaka, A. Mino, M. Umikawa, H. Imamura et al., 1996. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP-binding protein in Saccharomyces cerevisiae. EMBO J. 15: 6060–6068. [PMC free article] [PubMed] [Google Scholar]
- Levin, D. E., 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69: 262–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. P., and A. Bretscher, 1989. Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell 57: 233–242. [DOI] [PubMed] [Google Scholar]
- Liu, H., and A. Bretscher, 1992. Characterization of TPM1 disrupted yeast cells indicates an involvement of tropomyosin in directed vesicular transport. J. Cell Biol. 118: 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, T., S. M. Wurgler-Murphy and H. Saito, 1994. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369: 242–245. [DOI] [PubMed] [Google Scholar]
- Maeda, T., M. Takekawa and H. Saito, 1995. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269: 554–558. [DOI] [PubMed] [Google Scholar]
- Matheos, D., M. Metodiev, E. Muller, D. Stone and M. D. Rose, 2004. Pheromone-induced polarization is dependent on the Fus3p MAPK acting through the formin Bni1p. J. Cell Biol. 165: 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui, Y., and A. Toh-e, 1992. Yeast RHO3 and RHO4 ras superfamily genes are necessary for bud growth, and their defect is suppressed by a high dose of bud formation genes CDC42 and BEM1. Mol. Cell. Biol. 12: 5690–5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley, J. B., and B. L. Goode, 2005. Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. J. Biol. Chem. 280: 28023–28033. [DOI] [PubMed] [Google Scholar]
- Moseley, J. B., I. Sagot, A. L. Manning, Y. Xu, M. J. Eck et al., 2004. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol. Biol. Cell 15: 896–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke, S. M., and I. Herskowitz, 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12: 2874–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke, S. M., and I. Herskowitz, 2002. A third osmosensing branch in Saccharomyces cerevisiae requires the Msb2 protein and functions in parallel with the Sho1 branch. Mol. Cell. Biol. 22: 4739–4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke, S. M., and I. Herskowitz, 2004. Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol. Biol. Cell 15: 532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki-Kuroda, K., Y. Yamamoto, H. Nohara, M. Kinoshita, T. Fujiwara et al., 2001. Dynamic localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas, F., and H. Saito, 1997. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 276: 1702–1705. [DOI] [PubMed] [Google Scholar]
- Posas, F., and H. Saito, 1998. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 17: 1385–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas, F., S. M. Wurgler-Murphy, T. Maeda, E. A. Witten, T. C. Thai et al., 1996. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86: 865–875. [DOI] [PubMed] [Google Scholar]
- Posas, F., E. A. Witten and H. Saito, 1998. Requirement of STE50 for osmostress-induced activation of the STE11 mitogen-activated protein kinase kinase kinase in the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 18: 5788–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne, D., and A. Bretscher, 2000. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 113(Pt. 3): 365–375. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., M. Evangelista, C. Yang, E. Bi, S. Zigmond et al., 2002. Role of formins in actin assembly: nucleation and barbed-end association. Science 297: 612–615. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., L. Gao, E. Bi and A. Bretscher, 2004. Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol. Biol. Cell 15: 4971–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser, V., S. M. Salah and G. Ammerer, 2000. Polarized localization of yeast Pbs2 depends on osmostress, the membrane protein Sho1 and Cdc42. Nat. Cell Biol. 2: 620–627. [DOI] [PubMed] [Google Scholar]
- Reiser, V., D. C. Raitt and H. Saito, 2003. Yeast osmosensor Sln1 and plant cytokinin receptor Cre1 respond to changes in turgor pressure. J. Cell Biol. 161: 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann et al., 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17: 1030–1032. [DOI] [PubMed] [Google Scholar]
- Sagot, I., S. K. Klee and D. Pellman, 2002. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 4: 42–50. [DOI] [PubMed] [Google Scholar]
- Sheikh-Hamad, D., and M. C. Gustin, 2004. MAP kinases and the adaptive response to hypertonicity: functional preservation from yeast to mammals. Am. J. Physiol. Renal Physiol. 287: F1102–F1110. [DOI] [PubMed] [Google Scholar]
- Sheu, Y. J., B. Santos, N. Fortin, C. Costigan and M. Snyder, 1998. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol. Cell. Biol. 18: 4053–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter, B., and R. Li, 2006. Toward a molecular interpretation of the surface stress theory for yeast morphogenesis. Curr. Opin. Cell Biol. 18: 47–53. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader et al., 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. [DOI] [PubMed] [Google Scholar]
- Vallen, E. A., J. Caviston and E. Bi, 2000. Roles of Hof1p, Bni1p, Bnr1p, and myo1p in cytokinesis in Saccharomyces cerevisiae. Mol. Biol. Cell 11: 593–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertman, K. F., D. G. Drubin and D. Botstein, 1992. Systematic mutational analysis of the yeast ACT1 gene. Genetics 132: 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C., E. Leberer, D. Y. Thomas and M. Whiteway, 1999. Functional characterization of the interaction of Ste50p with Ste11p MAPKKK in Saccharomyces cerevisiae. Mol. Biol. Cell 10: 2425–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamochi, W., K. Tanaka, H. Nonaka, A. Maeda, T. Musha et al., 1994. Growth site localization of Rho1 small GTP-binding protein and its involvement in bud formation in Saccharomyces cerevisiae. J. Cell Biol. 125: 1077–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzyuk, T., and D. C. Amberg, 2003. Actin recovery and bud emergence in osmotically stressed cells requires the conserved actin interacting mitogen-activated protein kinase kinase kinase Ssk2p/MTK1 and the scaffold protein Spa2p. Mol. Biol. Cell 14: 3013–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzyuk, T., M. Foehr and D. C. Amberg, 2002. The MEK kinase Ssk2p promotes actin cytoskeleton recovery after osmotic stress. Mol. Biol. Cell 13: 2869–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]